Abstract
Precise reconstruction of heterotypic cell–cell interactions in vitro requires the coculture of different cell types in a highly controlled manner. In this article, we report a standing surface acoustic wave (SSAW)-based cell coculture platform. In our approach, different types of cells are patterned sequentially in the SSAW field to form an organized cell coculture. To validate our platform, we demonstrate a coculture of epithelial cancer cells and endothelial cells. Real-time monitoring of cell migration dynamics reveals increased cancer cell mobility when cancer cells are cocultured with endothelial cells. Our SSAW-based cell coculture platform has the advantages of contactless cell manipulation, high biocompatibility, high controllability, simplicity, and minimal interference of the cellular microenvironment. The SSAW technique demonstrated here can be a valuable analytical tool for various biological studies involving heterotypic cell–cell interactions.
Cell culture techniques have greatly benefited various studies in biology, biochemistry, and biomedical engineering, as well as providing in vitro platforms for drug screening in the pharmaceutical industry.1−4 However, cells residing in their in vivo niches experience a far more complex microenvironment than those maintained in conventional cell culture; they interact with neighboring cells, the extracellular matrix (ECM), and with soluble factors present in the microenvironment.5 In particular, interactions among different types of cells are crucial for the maintenance of normal cell function. For example, the interaction with nonparenchymal cells helps preserve the liver-specific functions of primary hepatocytes within coculture.6 To study heterotypic cell–cell interactions, cocultures of different cell types are needed. Traditionally, different types of cells are randomly mixed and grown together on cell culture plates to form cell cocultures. According to differential adhesion hypothesis (DAH), the differences in intercellular adhesiveness between different cell types can drive the movement and assortment of cells.7,8 However, the organization of cell coculture arising from DAH is governed by spontaneous rearrangement of cells with little control on the final cell pattern. Therefore, engineering approaches that offer on-demand control of cell arrangement are desirable for the reconstruction of physiologically relevant in vivo multicellular microenvironment.9−11
To address the unmet needs of cell coculture techniques with better controllability, researchers have developed many cell coculture platforms using micropatterned surfaces,12−26 cell printing,27−29 detachable substrates,30,31 physical barriers,32−35 microfluidic traps,36−38 and dielectrophoresis (DEP).39−41 Among these, micropatterning techniques are the most widely used and generate organized cell cocultures using an adhesion molecule- or microstructure-patterned substrate surfaces.12−26 However, cell adhesion to the micropatterned surfaces is a passive process with low controllability and cell behaviors within such cocultures might be influenced by the artificially introduced substrate heterogeneity. Cell-printing methods can actively deposit cells onto specific positions, but they have relatively low patterning resolution.27−29 A recently developed detachable substrate method enables reconfiguration of the formed cell coculture but is not suitable for multicellular construction.30,31 Using solid microstructures as physical barriers or microfluidic traps, researchers can control the positions of a group of cells or single cells within coculture in microfluidic channels.32−38 These platforms have fine resolution and good control of local cellular microenvironment,9 yet cells confined by these microstructures may experience unwanted mechanical stimuli potentially interfering with the study of heterotypic cell–cell interactions. The introduction of contactless DEP forces for cell patterning can overcome this limitation.39−41 However, for cell manipulation, DEP methods require a specially prepared culture medium with low ionic strength that is different from the cells’ in vivo microenvironment. Thus far, a cell coculture technique with high biocompatibility, high controllability, and minimal interference of the cellular microenvironment has yet to be realized.
Previously, our group has used standing surface acoustic waves (SSAWs) to precisely manipulate various micro/nano-objects (e.g., beads, cells, droplets, nanowires, and microorganisms).42−46 In particular, in our early work we demonstrated that our SSAW platform, the so-called acoustic tweezers technology, could create a pattern of suspended cells from a single cell type.42 In this work, we introduce the phase-shift approach and microfluidic cell culture technique to our SSAW cell-manipulation platform and demonstrate SSAW-based cell coculture (i.e., patterning of cultured cells from multiple cell types with desired arrangement). Our SSAW-based cell coculture technique can position multiple cell types using noninvasive, contactless acoustic forces with high precision and high tunability. It also has the flexibility to operate in literally any medium and with different types of adherent cells. For the validation of our platform, we demonstrate a coculture of epithelial cancer cells and endothelial cells and monitor the cell migration dynamics within the coculture. In our platform, cell behaviors (e.g., migration) are restricted by neither heterogeneous surface modifications nor physical barriers, which is advantageous over existing techniques (e.g., micropatterning, detachable substrates) in studying cell–cell interactions. We expect that the SSAW-based cell coculture platform demonstrated here will be a valuable tool for studying cell–cell interactions, tissue engineering, and drug screenings.47−49
Working Mechanism
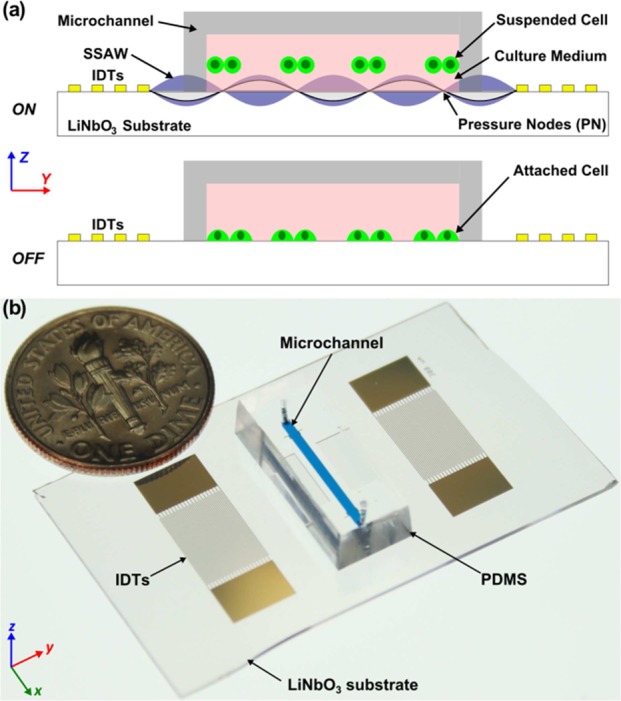
Figures 1 and 2 illustrate the working mechanism of our SSAW-based cell coculture platform, which is made by bonding a poly(dimethylsiloxane) (PDMS) microchannel between a pair of interdigital transducers (IDTs) fabricated on a lithium niobate (LiNbO3) piezoelectric substrate (Figure 1a). The pair of IDTs are aligned in parallel. When a radio frequency (RF) signal is applied to both IDTs, two series of identical surface acoustic waves (SAWs) propagating in opposite directions will be generated. The interference between these two series of SAWs forms a SSAW field, with a periodic distribution of pressure nodes (with minimum pressure amplitude) and pressure antinodes (with maximum pressure amplitude) on the piezoelectric substrate.50−54 When the resonating SSAW encounters the liquid medium inside the microchannel, it generates longitudinal-mode leakage waves, which cause pressure fluctuations in the medium.
Figure 1.
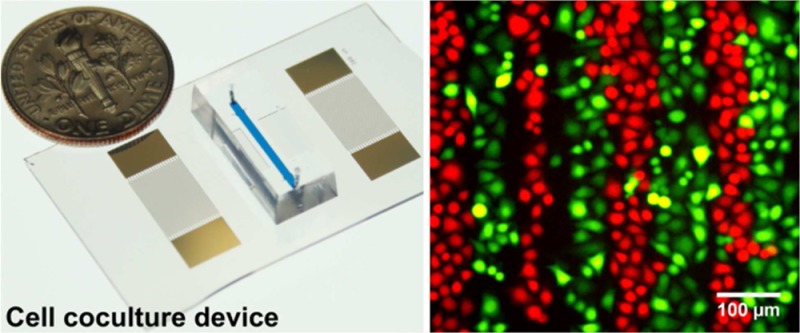
(a) Working mechanism of our SSAW-based cell coculture platform. (b) An optical image of our SSAW-based cell coculture device.
Figure 2.
Schematics of the SSAW-based cell coculture technique.
The primary acoustic radiation forces that act on any particle in the SSAW field can be expressed as55
| 1 |
| 2 |
where p0, λ, Vp, k, x, ρp, ρm, βp, and βm are the acoustic pressure, wavelength, volume of the particle, wave vector, distance from a pressure node, density of the particle, density of the medium, compressibility of the particle, and compressibility of the medium, respectively. Equation 2 describes the acoustic contrast factor, ϕ, which determines whether the particles move to pressure nodes or antinodes in the SSAW field: the particles will aggregate at pressure nodes if ϕ is positive and antinodes if ϕ is negative. The cells suspended in the medium will experience acoustic radiation forces caused by the pressure fluctuations and will be pushed toward pressure nodes in the SSAW field due to their positive acoustic contract factor.
As shown in Figure 1a, when the RF signal is on, cells suspended in the culture medium inside the microchannel will be aligned in parallel lines in the established SSAW field due to the acoustic radiation forces. When the RF signal is off, cells eventually settle down inside the microchannel and, in the absence of external flow, maintain their original pattern. Eventually, these cells will attach to the surface of the piezoelectric substrate forming a patterned cell culture. To form an organized cell coculture, different types of cells need to be patterned in different positions, which requires changing the distribution of pressure nodes and antinodes. In an established SSAW field, this can be achieved by tuning the relative phase between the RF signals applied to the pair of IDTs.56,57 With this phase-shift approach, different types of cells can be patterned sequentially in different positions to form an organized cell coculture.
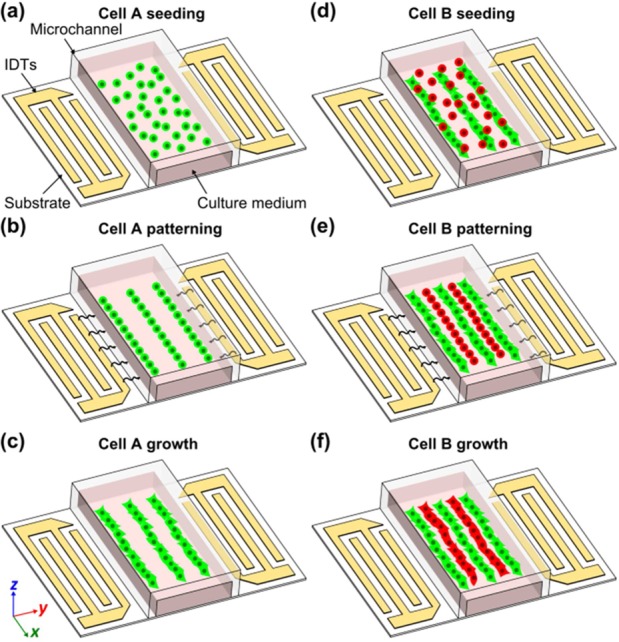
Figure 2 is a schematic of this phase-shift approach to forming cocultures of two cell types. First, cells of type A are seeded into the microchannel and patterned by SSAW (Figure 2, parts a and b). After type A cells attach to the bottom surface (Figure 2c), cells of type B are introduced into the microchannel (Figure 2d). To pattern type B cells in different positions from type A cells, we tune the relative phase between the RF signals applied to the pair of IDTs by 180°. Thus, type B cells are patterned in between type A cells with the same patterning period (Figure 2e). As a result, two types of cells will grow in alternate lines after type B cells attach to form an organized cell coculture (Figure 2f).
Materials and Methods
Device Fabrication
Figure 1b shows the optical image of our SSAW-based cell coculture device. To fabricate the device, a double layer of chrome and gold (Cr/Au, 50 Å/500 Å) was deposited on a photoresist-patterned 128° Y-cut LiNbO3 wafer (500 μm thick, double-side polished) using an e-beam evaporator (RC0021, Semicore, U.S.A.), followed by a lift-off process to form a pair of IDTs with a period of 300 μm. A single-layer PDMS microchannel (12 mm long, 1 mm wide, and 100 μm high) was fabricated by standard soft-lithography using SU-8 photoresist. After holes for an inlet and outlet were drilled into the PDMS microchannel with a Harris Uni-Core 1.00 mm punch, the PDMS microchannel was treated with oxygen plasma in a plasma cleaner (PDC001, Harrick Plasma, U.S.A.) for 3 min with the LiNbO3 substrate. The PDMS microchannel was then aligned and bonded to the LiNbO3 substrate between the IDTs. The entire device was then incubated overnight at 65 °C and sterilized under UV light for 30 min. The surface of the LiNbO3 substrate inside the microchannel was then coated overnight with 100 μg/mL collagen I rat tail (Gibco, Life Technologies, U.S.A.) in an ethanol solution before each experiment for better cell adhesion and growth.
Experimental Setup and Data Acquisition
All of the experiments were conducted on the stage of an inverted microscope (TE2000-U, Nikon, Japan) with an installed microscope incubation system (Chamlide TC, Live Cell Instrument, South Korea). In order to eliminate the virtual image introduced by using a double-side polished LiNbO3 substrate, we placed a polarizer in the light path adjusted at an angle. The SSAW is created by applying amplified RF signals to the IDTs using a function generator (AFG 3102C, Tektronix, U.S.A.) and power amplifier (25A250A, Amplifier Research, U.S.A.). A digital phosphor oscilloscope (load set at 1 MΩ) (DPO4104, Tektronix, U.S.A.) was used to determine the resonance frequency of the IDTs. A syringe pump (KDS210, KD Scientific, U.S.A.) was used to infuse fresh culture medium into the microchannel during long-term microfluidic cell culture. A charge-coupled device (CCD) camera (ORCA-Flash 2.8, Hamamatsu, Japan) was connected to the microscope for data acquisition. Cell movement trajectories were extracted from time-lapse images and analyzed using Nikon NIS-Elements Advanced Research (AR) software and plotted using R. Welch two-sample t test was conducted to compare cell movement data between two groups. All of the other image processes were conducted using ImageJ (NIH, Bethesda, MD, U.S.A.).
Cell Culture and Staining
HeLa cells were maintained in DMEM/F12 medium (Gibco, Life Technologies, U.S.A.), supplemented with 10% fetal bovine serum (Gibco, Life Technologies, U.S.A.) and 1% penicillin–streptomycin (Mediatech, U.S.A.). Human dermal microvascular endothelial (HMVEC-d) cells were purchased from ATCC and maintained in EndoGRO-LS complete media (Millipore, U.S.A.). For coculturing HeLa cells and HMVEC-d cells, EndoGRO-LS complete media was always used as the culture medium. A CO2 incubator (Nu-4750, NuAire, U.S.A.) was used to maintain a temperature of 37 °C and a 5% CO2 level during cell culture. CellTracker Green CMFDA and CellTracker Orange CMRA (Molecular Probes, Life Technologies, U.S.A.) were used to label cells with green and red fluorescence following the manufacturer’s standard protocols. Cells grown to 80–90% confluency were trypsinized (Trypsin-EDTA (0.05%), Gibco, Life Technologies, U.S.A.), washed with PBS, resuspended in fresh culture medium to desired cell concentrations, and seeded into the microchannel for the experiment. After the cells were treated and cultured in our SSAW-based devices, live/dead cell staining was conducted using Calcein AM and SYTOX orange (Molecular Probes, Life Technologies, U.S.A.) to assess cell viability in our device after being treated by SSAW and cultured in microfluidic devices.
Results and Discussion
Culture of Patterned Cells
We first validated the ability of our SSAW-based platform to create patterned cell culture. Micrographs of patterned HeLa cells cultured for up to 24 h are shown in Figure 3a–e. During this experiment, we applied a 12.78 MHz RF signal with a voltage of ∼20 Vpp to HeLa cells that were injected into the microchannel at a seeding density of 4 × 106 cells/mL. The resulting pattern of parallel lines (Figure 3a) had a period of approximately 150 μm, which matched well with the half-wavelength of the applied SAW (λ = ∼300 μm). After the cells were patterned in the SSAW field, we turned off the RF signal and stopped the fluid to allow the cells to settle down and adhere to the bottom surface of the microchannel. Although the cells were subject to movement of the residual flow before adhesion, this effect was minimal in the microfluidic environment. As shown in Figure 3b, after 1 h of culture in the incubation system, the cells maintained the original pattern, with only slight cell movement. The cell movement during the adhesion process was due to cell growth and expansion on the surface and did not affect cell patterns within 1 h of culture. We also found that all of the cells had attached within 1 h of culture; they were not flushed away by an external flow at this time point. For long-term culture of the patterned cells, after 1 h we infused fresh culture medium into the microchannel with a syringe pump at a flow rate of 0.5 μL/min. Patterned HeLa cells were then cultured in the SSAW-based microfluidic device with this continuous medium infusion until the cells grew to full confluency at 24 h, as shown in Figure 3c–e.
Figure 3.
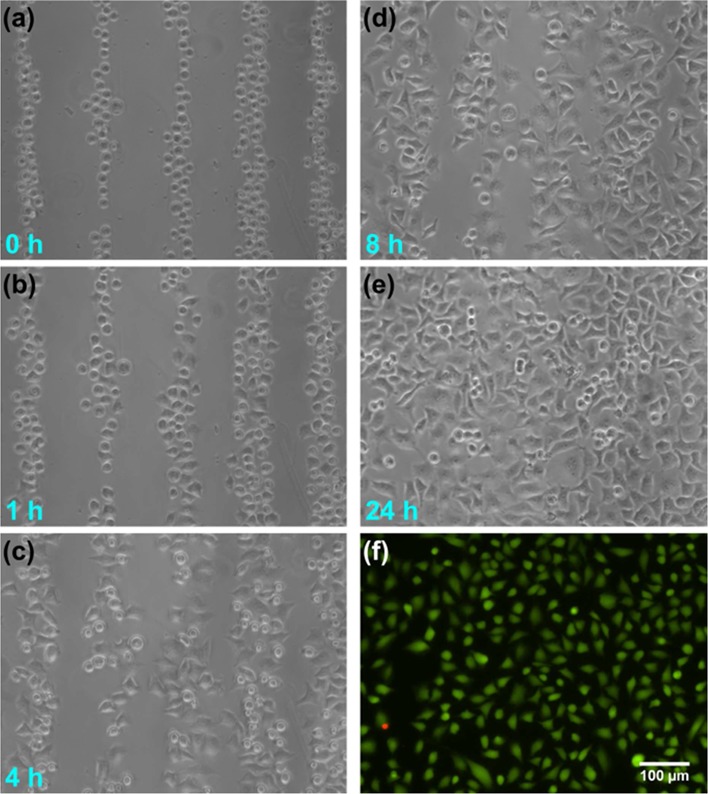
(a–e) Micrographs showing culture of patterned HeLa cells in our SSAW-based microfluidic device for up to 24 h. (f) Live/dead staining results indicate that most of the HeLa cells remain viable at the end of 24 h culture (green, live cells; red, dead cells).
During this phase of the experiment, cells were exposed to the SSAW field only during the patterning process and for less than 10 s. No obvious cell damage associated with acoustic exposure was observed during the 24 h microfluidic cell culture. In addition, we conducted live/dead cell staining using Calcein AM and SYTOX orange at the end of the 24 h cell culture, as shown in Figure 3f. A count of the stained cells in three different SSAW-exposed areas showed that 99.26% ± 0.38% of the cells remained viable at the end of the 24 h culture, proving the noninvasive nature of our SSAW technique.
The IDTs used in our experiment were designed to have a fixed period of 300 μm with a resonance frequency of 12.78 MHz, which was used in all subsequent experiments to generate the best cell patterning result. IDTs with different designs can be used to pattern cells with different periods. Furthermore, it is also feasible to change the period of cell patterning in a single device using tunable SSAW generated from slanted-finger IDTs or chirped IDTs,43,46 which can give our SSAW-based cell coculture platform an even greater degree of flexibility.
Coculture with Sequential Cell Patterning
After demonstrating a patterned culture of a single cell type, we examined the possibility of patterning and culturing multiple types of cells in one device. In our experiment, we employed the aforementioned phase-shift approach to form an organized coculture of two different cell types.
Figure 4 shows the experimental results validating this approach. In this experiment, green and red fluorescently labeled HeLa cells were used to represent two cell types. The green fluorescently labeled HeLa cells (type A cells) were first introduced into the microchannel at a seeding density of 4 × 106 cells/mL and patterned under an RF signal of 12.78 MHz, 20 Vpp, and 0° relative phase. After 2 h of culture with the SSAW off, the red fluorescently labeled HeLa cells (type B cells) were injected into the microchannel at the same seeding density. They were patterned under an RF signal of 12.78 MHz, 20 Vpp, but with a 180° relative phase. After another 2 h of culture with the SSAW off, fluorescent images were taken. As shown in Figure 4a, the change of the relative phase between the pair of IDTs from 0° to 180° will change the cell-patterning positions between the two rounds of cell seeding by switching between pressure nodes and antinodes. The fluorescent images in Figure 4, parts b and c, show the two groups of HeLa cells growing in patterned lines. From the merged image in Figure 4d, we can see that the patterned lines for the two groups of HeLa cells are in an alternate manner, with a separation of less than 75 μm (one-fourth of the applied SAW wavelength).
Figure 4.
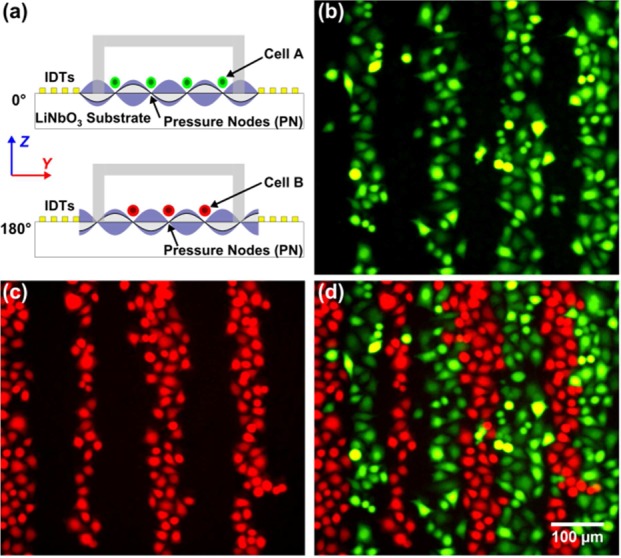
Coculture with SSAW-based sequential cell patterning. (a) Mechanism of patterning two types of cells in different positions with the phase-shift approach. (b) Green fluorescent image showing first-seeded HeLa cells. (c) Red fluorescent image showing second-seeded HeLa cells. (d) Merged image showing green and red HeLa cells grown in alternate lines.
In addition to its advantages in high controllability, our SSAW-based cell coculture platform requires only a small amount of cells. During each round of cell patterning, usually only 2 μL of cell suspension (approximately 4 × 106 cells/mL) was injected into the microchannel using a pipet, which means that <1 × 104 cells were required for coculture reconstruction for each cell type.
Investigation of Cancer Cell Movement during Coculture
To further examine the functionality of our SSAW-based cell coculture platform, we explored the interactions between epithelial cancer cells and endothelial cells in our system. For this study, we chose HeLa cells and HMVEC-d cells as our biological model. Prior to studying HeLa cell behaviors in coculture, we first explored patterned monoculture of HeLa cells. In the control experiment (Figure 5, parts a and b), two groups of HeLa cells were first labeled with green and red fluorescence, respectively. Then these two groups of HeLa cells were sequentially seeded and patterned in our device as described above. The seeding density for each round of patterning was 3 × 106 cells/mL, and the interval between two rounds of patterning was 1 h. Fluorescent images were then taken at 2 and 24 h time points. As shown in Figure 5a, green and red fluorescently labeled HeLa cells growing in alternate lines were formed at 2 h. When the HeLa cells grew to confluency at 24 h, this pattern of alternate lines was not disrupted, indicating that HeLa cell mobility in monoculture was low (Figure 5b). In our coculture experiment, we introduced green fluorescently labeled HMVEC-d cells at the seeding density of 3 × 106 cells/mL during the first round of cell patterning and red fluorescently labeled HeLa cells at the same seeding density after 1 h. At 2 h time point of coculture shown in Figure 5c, we can see that HMVEC-d cells and HeLa cells grew in alternate line regions. However, at 24 h time point, the organized HeLa cell pattern was clearly disrupted indicating high HeLa cell mobility when cocultured with HMVEC-d cells (Figure 5d).
Figure 5.
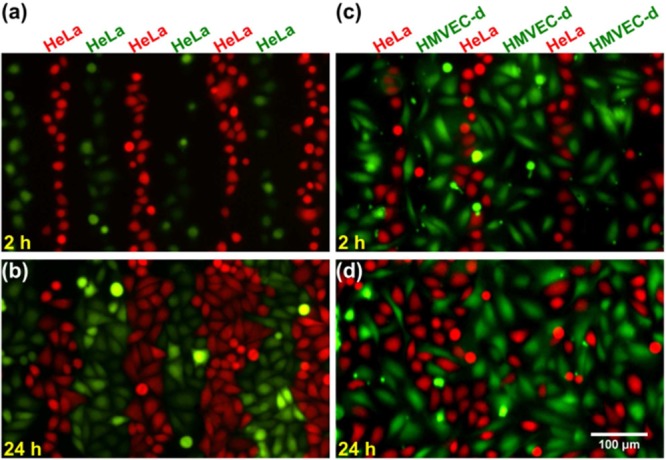
Fluorescent images of (a and b) HeLa cell monoculture at 2 and 24 h and (c and d) HeLa and HMVEC-d coculture at 2 and 24 h in our SSAW device.
In order to quantitatively investigate HeLa cell movement within monoculture or coculture, we cultured patterned HeLa cells when HMVEC-d cells (in EndoGRO-LS complete media) were present or absent in our SSAW device. As control experiments, off-chip monoculture and coculture of HeLa cells were also conducted in which cells were randomly seeded on a Petri dish. For coculture, HeLa cells were fluorescently labeled with Calcein AM for identification and seeded before the HMVEC-d cells. Time-lapse phase-contrast images were automatically taken every 20 min from 2 to 12 h for all the four groups (on-chip monoculture, on-chip coculture, off-chip monoculture, and off-chip coculture) to record cell positions. To analyze the cancer cell movements, we randomly picked 30 HeLa cells in each group and tracked their trajectories from 2 to 12 h. The time-lapse images with highlighted HeLa cell tracking for all the four groups are available in Supporting Information supplementary videos S1–S4.
Figure 6, parts a and b, shows three typical cell trajectories for on-chip monoculture and coculture, which illustrate that HeLa cells cocultured with HMVEC-d cells in our SSAW device had higher mobility than those cultured alone. This increased HeLa cell mobility was not caused by the higher cell density in on-chip coculture because HeLa cells within confluent off-chip monoculture also showed low level of mobility (Supporting Information supplementary video S3). The average movement path lengths of the 30 tracked HeLa cells at each time point in each group are plotted in Figure 6c. At 12 h, the average movement path lengths of the tracked HeLa cells in off-chip monoculture, on-chip monoculture, off-chip coculture, and on-chip coculture were 66.4 ± 11.2, 64.8 ± 19.5, 146.1 ± 21.3, and 174.0 ± 24.6 μm, respectively, as shown in Figure 6d. From the comparison, we can see that the mobility of HeLa cells in coculture is much higher than that in monoculture, either off-chip or on-chip. This increased cancer cell mobility in coculture can be attributed to the cross-talk initiated by endothelial cells, which enhances cancer cell survival and mobility through STAT3/Akt/ERK, α5β1 integrin, and GTPases signaling pathways.58,59 The average movement path length of the tracked HeLa cells in on-chip coculture was only slightly larger than in off-chip coculture, which indicates that the presence of endothelial cells increases the mobility of cancer cells either in a random coculture or in an organized coculture. However, when we compared HeLa cell trajectories between these two groups (Supporting Information supplementary videos S2 and S4), we found that the movements of HeLa cells in the two groups were different. In the organized on-chip coculture, HeLa cells tend to migrate away from their original positions, while in the random off-chip coculture, HeLa cells tend to wander around locally so that their final positions at 12 h were close to their original positions at 2 h. To illustrate this difference, we further plot the average distances from the original positions for the four groups at 12 h in Figure 6e. We can see that the average distance from the original positions for on-chip coculture (48.7 ± 24.9 μm) was twice that in off-chip coculture (23.1 ± 16.4 μm). One possible explanation is that, compared with the random cell arrangement in off-chip coculture, the organized cell arrangement in on-chip coculture better facilitates gradient formation for the signaling molecules secreted from HMVEC-d cells, which can guide HeLa cells to migrate away from their original positions. These results indicate that our SSAW-based cell coculture platform can be used for in vitro evaluation of the invasiveness of cancer cells and can be developed as an efficient tool for in vitro antitumor drug screenings.
Figure 6.
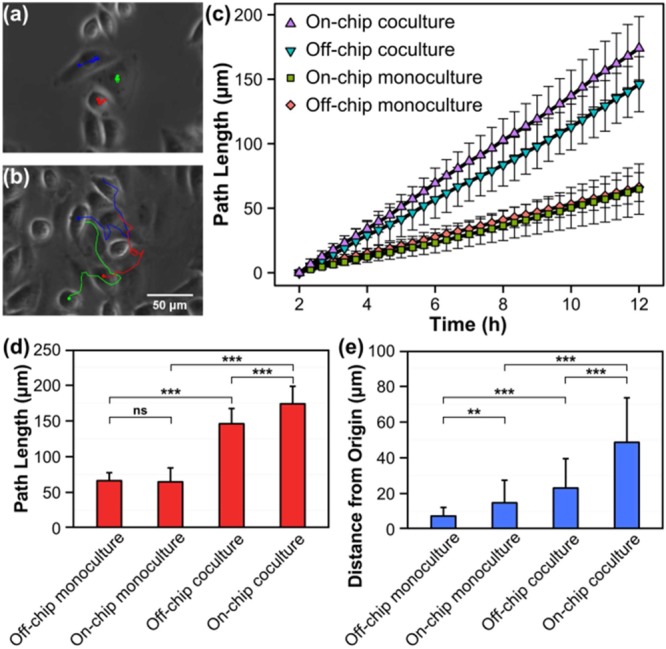
Quantitative analysis of HeLa cell movement in monoculture and coculture. (a and b) Three typical movement trajectories (in blue, green, and red lines) for HeLa cells in on-chip (a) monoculture and (b) coculture. (c) Average movement path lengths of the tracked HeLa cells plotted against culture time for the four groups. (d and e) Comparison of (d) average movement path length and (e) average distance from origin for the tracked HeLa cells at 12 h among four different groups. The error bars represent the standard deviation (n = 30 for each group; ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Conclusions
In conclusion, we have developed a SSAW-based cell coculture platform. Our platform takes advantage of the contactless, noninvasive nature of acoustic forces, thus exerting minimal interference on the cellular microenvironment while preserving high cell integrity. The use of contactless acoustic forces that pattern cells in a transient manner is valuable because cells are free of any major external stimulus (e.g., substrate heterogeneity, microstructure confinement, or mechanical stimulation) during culture. Thus, the interference of the cellular microenvironment by unwanted stimuli is minimized and the influence of heterotypic cell–cell interactions can be isolated for study.60,61 The SSAW-based cell coculture platform demonstrated here provides a novel analytical tool for real-time, dynamic observation of cell behaviors within coculture. The cellular-level resolution makes our SSAW-based cell coculture platform an excellent candidate for reconstructing heterotypic cell–cell interactions, which is important for probing cell communication and multicellular tissue construction.62−65
Acknowledgments
We thank Joey Rufo for manuscript revision and Dr. Suresh D. Sharma, Cheri Lee, and Sravani Banerjee from the Cameron Lab for the help in cell culture. This research was supported by National Institutes of Health (1 R01 GM112048-01A1 and 1DP2OD007209-01), Huck Innovative & Transformational Seed Fund, and the Penn State Center for Nanoscale Science (MRSEC) under Grant DMR-0820404. S.L. and C.E.C. were funded in part by Grant AI45818 from NIAID, NIH. Components of this work were conducted at the Penn State node of the NSF-funded National Nanotechnology Infrastructure Network (NNIN).
Supporting Information Available
Supplementary videos S1–S4 showing time-lapse images with highlighted HeLa cell tracking. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Barbulovic-Nad I.; Au S. H.; Wheeler A. R. Lab Chip 2010, 10, 1536–1542. [DOI] [PubMed] [Google Scholar]
- Astashkina A.; Mann B.; Grainger D. W. Pharmacol. Ther. 2012, 134, 82–106. [DOI] [PubMed] [Google Scholar]
- Clark A. M.; Sousa K. M.; Chisolm C. N.; MacDougald O. A.; Kennedy R. T. Anal. Bioanal. Chem. 2010, 397, 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. M.; Sousa K. M.; Jennings C.; MacDougald O. A.; Kennedy R. T. Anal. Chem. 2009, 81, 2350–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H.; Berk A.; Zipursky S. L.; Matsudaira P.; Baltimore D.; Darnell J.. Molecular Cell Biology, 4th ed.; W. H. Freeman: New York, 2000. [Google Scholar]
- Bhatia S. N.; Balis U. J.; Yarmush M. L.; Toner M. FASEB J. 1999, 13, 1883–1900. [DOI] [PubMed] [Google Scholar]
- Foty R. A.; Steinberg M. S. Dev. Biol. 2005, 278, 255–263. [DOI] [PubMed] [Google Scholar]
- Steinberg M. S. Science 1963, 141, 401–408. [DOI] [PubMed] [Google Scholar]
- Marimuthu M.; Kim S. Anal. Biochem. 2011, 413, 81–89. [DOI] [PubMed] [Google Scholar]
- Kaji H.; Camci-Unal G.; Langer R.; Khademhosseini A. Biochim. Biophys. Acta 2011, 1810, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salieb-Beugelaar G. B.; Simone G.; Arora A.; Philippi A.; Manz A. Anal. Chem. 2010, 82, 4848–4864. [DOI] [PubMed] [Google Scholar]
- Myers F. B.; Silver J. S.; Zhuge Y.; Beygui R. E.; Zarins C. K.; Lee L. P.; Abilez O. J. Integr. Biol. 2013, 5, 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.; Rajalingam B.; Selvarasah S.; Dokmeci M. R.; Khademhosseini A. Lab Chip 2007, 7, 1272–1279. [DOI] [PubMed] [Google Scholar]
- Chiu D. T.; Jeon N. L.; Huang S.; Kane R. S.; Wargo C. J.; Choi I. S.; Ingber D. E.; Whitesides G. M. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A.; Suh K. Y.; Yang J. M.; Eng G.; Yeh J.; Levenberg S.; Langer R. Biomaterials 2004, 25, 3583–3592. [DOI] [PubMed] [Google Scholar]
- Tien J.; Nelson C. M.; Chen C. S. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 1758–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe H.; Okuyama T.; Suzuki H.; Fukuda J. Acta Biomater. 2010, 6, 526–533. [DOI] [PubMed] [Google Scholar]
- Wang D.-Y.; Wu S.-C.; Lin S.-P.; Hsiao S.-H.; Chung T.-W.; Huang Y.-Y. Biomed. Microdevices 2011, 13, 517–526. [DOI] [PubMed] [Google Scholar]
- Tsuda Y.; Kikuchi A.; Yamato M.; Chen G.; Okano T. Biochem. Biophys. Res. Commun. 2006, 348, 937–944. [DOI] [PubMed] [Google Scholar]
- Bhatia S. N.; Yarmush M. L.; Toner M. J. Biomed. Mater. Res. 1997, 34, 189–199. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A.; Ferreira L.; Blumling J.; Yeh J.; Karp J. M.; Fukuda J.; Langer R. Biomaterials 2006, 27, 5968–5977. [DOI] [PubMed] [Google Scholar]
- Huang N. F.; Patlolla B.; Abilez O.; Sharma H.; Rajadas J.; Beygui R. E.; Zarins C. K.; Cooke J. P. Acta Biomater. 2010, 6, 4614–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc E.; El Kirat K.; Griscom L. Biomed. Microdevices 2008, 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Fukuda J.; Khademhosseini A.; Yeh J.; Eng G.; Cheng J.; Farokhzad O. C.; Langer R. Biomaterials 2006, 27, 1479–1486. [DOI] [PubMed] [Google Scholar]
- Yamato M.; Konno C.; Utsumi M.; Kikuchi A.; Okano T. Biomaterials 2002, 23, 561–567. [DOI] [PubMed] [Google Scholar]
- Liu N.; Liang W.; Liu L.; Wang Y.; Mai J. D.; Lee G.-B.; Li W. J. Lab Chip 2014, 14, 1367–1376. [DOI] [PubMed] [Google Scholar]
- Gaebel R.; Ma N.; Liu J.; Guan J.; Koch L.; Klopsch C.; Gruene M.; Toelk A.; Wang W.; Mark P.; Wang F.; Chichkov B.; Li W.; Steinhoff G. Biomaterials 2011, 32, 9218–9230. [DOI] [PubMed] [Google Scholar]
- Tavana H.; Mosadegh B.; Takayama S. Adv. Mater. 2010, 22, 2628–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.; Zhao W.; Zhu J.-M.; Albanna M. Z.; Yoo J. J.; Atala A. Biomaterials 2013, 34, 130–139. [DOI] [PubMed] [Google Scholar]
- Hui E. E.; Bhatia S. N. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 5722–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H.; Yokoi T.; Kawashima T.; Nishizawa M. Lab Chip 2009, 9, 427–432. [DOI] [PubMed] [Google Scholar]
- Zheng C.; Zhao L.; Chen G.; Zhou Y.; Pang Y.; Huang Y. Anal. Chem. 2012, 84, 2088–2093. [DOI] [PubMed] [Google Scholar]
- Torisawa Y.; Mosadegh B.; Luker G. D.; Morell M.; O’Shea K. S.; Takayama S. Integr. Biol. 2009, 1, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao A. Y.; Torisawa Y.; Tung Y.-C.; Sud S.; Taichman R. S.; Pienta K. J.; Takayama S. Biomaterials 2009, 30, 3020–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Majumdar D.; Jovanovic B.; Shaifer C.; Lin P. C.; Zijlstra A.; Webb D. J.; Li D. Biomed. Microdevices 2011, 13, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelley A. M.; Kirak O.; Suh H.; Jaenisch R.; Voldman J. Nat. Methods 2009, 6, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura B.; Liu Y.; Voldman J. Lab Chip 2014, 14, 2783–2790. [DOI] [PubMed] [Google Scholar]
- Hong S.; Pan Q.; Lee L. P. Integr. Biol. 2012, 4, 374–380. [DOI] [PubMed] [Google Scholar]
- Albrecht D. R.; Underhill G. H.; Wassermann T. B.; Sah R. L.; Bhatia S. N. Nat. Methods 2006, 3, 369–375. [DOI] [PubMed] [Google Scholar]
- Ho C.-T.; Lin R.-Z.; Chen R.-J.; Chin C.-K.; Gong S.-E.; Chang H.-Y.; Peng H.-L.; Hsu L.; Yew T.-R.; Chang S.-F.; Liu C.-H. Lab Chip 2013, 13, 3578–3587. [DOI] [PubMed] [Google Scholar]
- Yin Z.; Noren D.; Wang C. J.; Hang R.; Levchenko A. Mol. Syst. Biol. 2008, 4, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Ahmed D.; Mao X.; Lin S.-C. S.; Lawit A.; Huang T. J. Lab Chip 2009, 9, 2890–2895. [DOI] [PubMed] [Google Scholar]
- Ding X.; Lin S.-C. S.; Kiraly B.; Yue H.; Li S.; Chiang I.-K.; Shi J.; Benkovic S. J.; Huang T. J. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 11105–11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Ding X.; Lin S.-C. S.; Yang S.; Huang P.-H.; Nama N.; Zhao Y.; Nawaz A. A.; Guo F.; Wang W.; Gu Y.; Mallouk T. E.; Huang T. J. ACS Nano 2013, 7, 3306–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Ding X.; Guo F.; Chen Y.; Lapsley M. I.; Lin S.-C. S.; Wang L.; McCoy J. P.; Cameron C. E.; Huang T. J. Anal. Chem. 2013, 85, 5468–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Shi J.; Lin S.-C. S.; Yazdi S.; Kiraly B.; Huang T. J. Lab Chip 2012, 12, 2491–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. B.; Srigunapalan S.; Wheeler A. R.; Simmons C. A. Lab Chip 2013, 13, 2591–2598. [DOI] [PubMed] [Google Scholar]
- Neužil P.; Giselbrecht S.; Länge K.; Huang T. J.; Manz A. Nat. Rev. Drug Discovery 2012, 11, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-H.; Huang S.-B.; Lee G.-B. Lab Chip 2010, 10, 939–956. [DOI] [PubMed] [Google Scholar]
- Shi J.; Mao X.; Ahmed D.; Colletti A.; Huang T. J. Lab Chip 2008, 8, 221–223. [DOI] [PubMed] [Google Scholar]
- Shi J.; Huang H.; Stratton Z.; Huang Y.; Huang T. J. Lab Chip 2009, 9, 3354–3359. [DOI] [PubMed] [Google Scholar]
- Shi J.; Yazdi S.; Lin S.-C. S.; Ding X.; Chiang I.-K.; Sharp K.; Huang T. J. Lab Chip 2011, 11, 2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-C. S.; Mao X.; Huang T. J. Lab Chip 2012, 12, 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Li P.; Lin S.-C. S.; Stratton Z. S.; Nama N.; Guo F.; Slotcavage D.; Mao X.; Shi J.; Costanzo F.; Huang T. J. Lab Chip 2013, 13, 3626–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosioka K.; Kawasima Y. Acustica 1955, 5, 167–173. [Google Scholar]
- Meng L.; Cai F.; Zhang Z.; Niu L.; Jin Q.; Yan F.; Wu J.; Wang Z.; Zheng H. Biomicrofluidics 2011, 5, 044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloff N. D.; Dennis J. R.; Cecchini M.; Schonbrun E.; Rocas E.; Wang Y.; Novotny D.; Simmonds R. W.; Moreland J.; Takeuchi I.; Booth J. C. Biomicrofluidics 2011, 5, 044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke C. T. J. Biol. Chem. 2011, 286, 40025–40037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiva K. G.; Zhang Z.; Miyazawa M.; Warner K. A.; Karl E.; E. Nör J. Neoplasia 2009, 11, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y.; Mann J. M.; Chen W.; Fu J. Integr. Biol. 2014, 6, 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Fu J. Integr. Biol. 2013, 5, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. Y.; Huang P.-H.; Guo F.; Ding X.; Kapur V.; Mai J. D.; Yuen P. K.; Huang T. J. Lab Chip 2013, 13, 4697–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.; French J. B.; Li P.; Zhao H.; Chan C. Y.; Fick J. R.; Benkovic S. J.; Huang T. J. Lab Chip 2013, 13, 3152–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.; Lieu D. K.; Freschauf L.; Lew V.; Sharma H.; Wang J.; Nguyen D.; Karakikes I.; Hajjar R. J.; Gopinathan A.; Botvinick E.; Fowlkes C. C.; Li R. A.; Khine M. Adv. Mater. 2011, 23, 5785–5791. [DOI] [PubMed] [Google Scholar]
- Chen A.; Lee E.; Tu R.; Santiago K.; Grosberg A.; Fowlkes C.; Khine M. Biomaterials 2014, 35, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


