Abstract
The neocortex contains an unparalleled diversity of neuronal subtypes, each defined by distinct traits that are developmentally acquired under the control of subtype-specific and pan-neuronal genes. The regulatory logic that orchestrates the expression of these unique combinations of genes is unknown for any class of cortical neuron. Here, we report that Fezf2 is a selector gene able to regulate the expression of gene sets that collectively define mouse corticospinal motor neurons (CSMN). We find that Fezf2 directly induces the glutamatergic identity of CSMN via activation of Vglut1 (Scl17a7) and inhibits a GABAergic fate by repressing transcription of Gad1. In addition, we identify the axon guidance receptor Ephb1 as a target of Fezf2 necessary to execute the ipsilateral extension of the corticospinal tract. Our data indicate that co-regulated expression of neuron subtype–specific and pan-neuronal gene batteries by a single transcription factor is one component of the regulatory logic responsible for the establishment of CSMN identity.
During development, neurons of the CNS acquire distinct molecular, anatomical and physiological properties. Development of these traits is dictated by pan-neuronal and lineage-specific molecular cues, which in combination define neuronal subtype–specific identity and set the basis for neuronal diversity.
Among other regions of the mammalian CNS, the cerebral cortex stands as a prime example of how expanded neuronal diversity served the evolution of complex behavior. Here an extraordinary number of neuronal subtypes integrate into local and long-distance circuitry subserving sensory, motor and high-level cognitive functions1. The majority of cortical neurons are excitatory projection neurons. Their classification is largely based on axonal projection targets, but it is clear that multiple traits—molecular identity, primary and collateral axonal connections, somatodendritic morphology and electrophysiological activity—must be taken into account to accurately distinguish individual projection neuron subtypes2. The molecular regulatory logic that orchestrates acquisition of multiple class-specific features is not understood for any neuronal subtype of the cerebral cortex.
CSMN are one specialized class of cortical excitatory neurons, which connect layer Vb of the cortex to the spinal cord. At the molecular level, CSMN are the most extensively characterized class of cortical projection neurons3,4. A master transcription factor—forebrain expressed zinc finger 2 (Fezf2)—has been identified that is necessary for the fate specification of CSMN5–7. In the absence of Fezf2, CSMN fail to generate. In agreement, CSMN-specific genes are not expressed in layer Vb of the Fezf2−/− null cortex, a deficiency accompanied by changes in dendritic morphology and a lack of axonal projections to the spinal cord5–7.
Conversely, Fezf2 alone can cell-autonomously instruct the acquisition of CSMN-specific features when expressed in diverse, permissive cellular contexts in vivo. Ectopic overexpression of Fezf2 in progenitors and postmitotic neurons of different lineages is sufficient to induce the generation of neurons that express multiple CSMN genes and extend axons to subcerebral targets5,8–10. Beyond acquisition of lineage-specific CSMN features, neurons expressing Fezf2 also acquire necessary cortical pan-projection neuron features. They become glutamatergic and develop distinctive pyramidal morphology8. These data indicate that Fezf2 is sufficient to instruct the acquisition of multiple aspects of CSMN identity.
Here we demonstrate that Fezf2 is a selector gene for an individual class of neurons of the neocortex: corticospinal motor neurons. We found that activation and repression of large batteries of genes necessary to orchestrate the acquisition of CSMN defining traits was directly co-regulated downstream of Fezf2. Fezf2 was sufficient to induce vesicular glutamate transporter 1 (Vglut1) and to repress glutamate decarboxylase 1 (Gad1) expression to instruct the acquisition of glutamatergic identity. In addition, we uncovered a new role for EphB1 as a functionally relevant effector of Fezf2, required to execute the ipsilateral extension of the corticospinal tract through the ventral forebrain. Our data indicate a direct role for Fezf2 as a selector gene that coordinates the expression of pan-neuronal and class-specific genes required for the developmental acquisition of CSMN identity.
RESULTS
Fezf2 induces signature genes of corticospinal neurons
To understand the molecular logic underlying the acquisition of CSMN traits upon Fezf2 expression, we compared the in vivo gene expression of FACS-purified cortical progenitors that ectopically expressed Fezf2 to control progenitors. We used in utero electroporation to deliver Fezf2GFP or CtrlGFP expression vectors to neocortical progenitors at embryonic day (E) 14.5, when they primarily generate callosal projection neurons (CPN) of the upper layers (Supplementary Fig. 1a). Overexpression of Fezf2 in these progenitors is sufficient to instruct a fate switch resulting in the generation of CSMN and other subtypes of corticofugal projection neurons5,7,11. Fezf2GFP- and CtrlGFP-electroporated progenitors were FACS-purified 24 and 48 h after surgery (Supplementary Fig. 1b,c) and acutely profiled by microarrays.
A large set of genes was induced by Fezf2 as early as 24 h (263 genes; P < 0.001, fold change > 1.5), while others were induced not earlier than 48 h (441 genes; P < 0.001, fold change > 1.5) after electroporation. In addition, we identified a smaller set of genes repressed by Fezf2 (90 genes at 24 h; 89 genes at 48 h; P < 0.001, fold change > 1.5) (Supplementary Table 1).
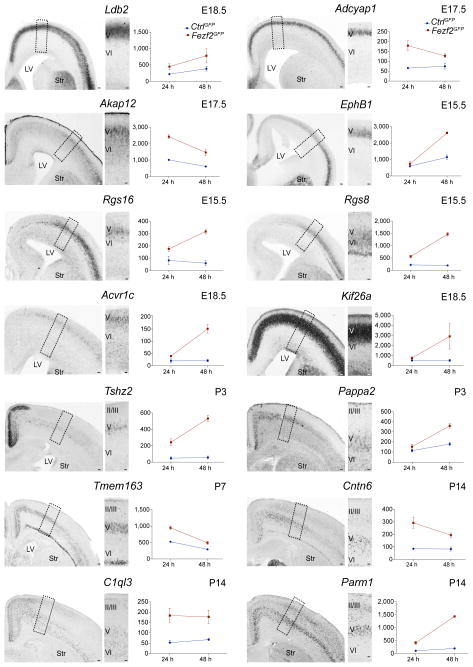
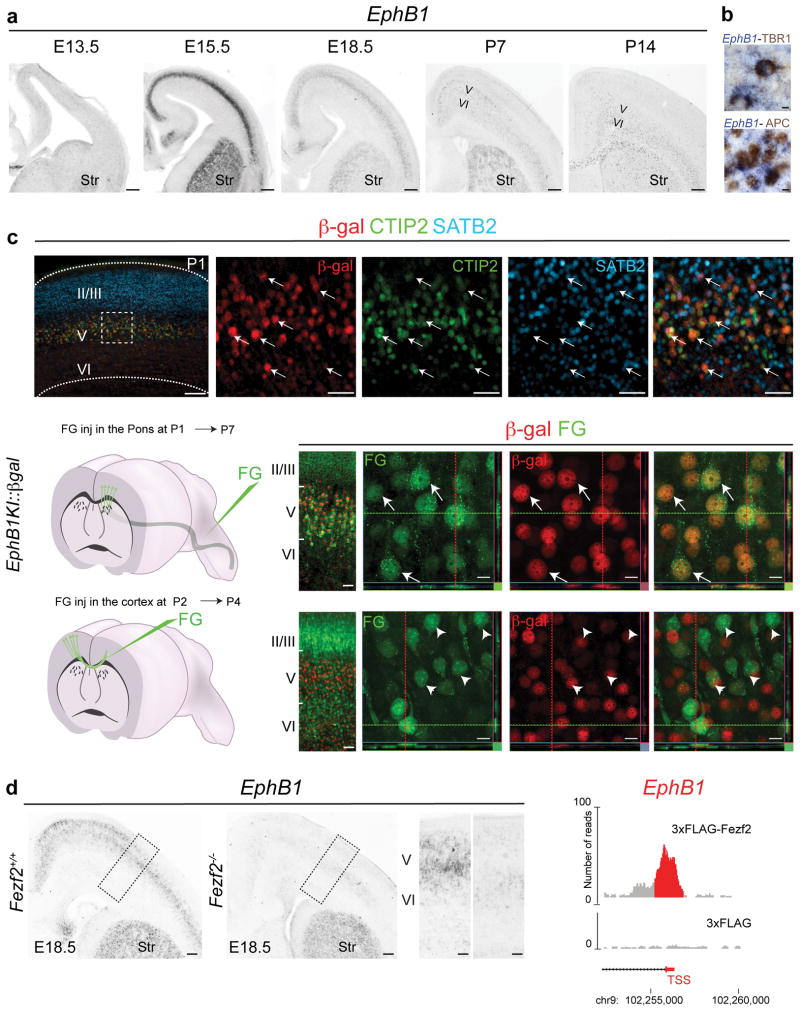
To investigate whether genes induced by Fezf2 mark developing CSMN, we performed in situ hybridization for selected candidates (Supplementary Table 1) at multiple stages of embryonic and postnatal development of the cerebral cortex (E15.5, E18.5, postnatal day (P) 3, P7 and P14; Fig. 1 and Supplementary Fig. 2). Remarkably, we identified a series of Fezf2 target genes (for example, Ldb2, Adcyap1, Akap12, Ephb1, Rgs16, Rgs8, Acvr1c, Pappa2, Kif26a) that labeled developing CSMN (and related subcerebral projection neurons, ScPN) in the early cortical plate (Fig. 1 and Supplementary Fig. 2), whereas other targets (for example, Kif26a, Tshz2, Tmem163, Cntn6, C1ql3, Parm1) were specific to later stages of CSMN development (Fig. 1 and Supplementary Fig. 2).
Figure 1.
Fezf2 overexpression in cortical progenitors induces genes that label corticospinal motor neurons. Left, in situ hybridizations on coronal sections of the cerebral cortex at different embryonic (E15.5, E17.5 and E18.5) and postnatal stages (P3, P7 and P14) (insets enlarged from boxed areas). Right, expression levels (normalized microarray intensity, see Methods) for each selected gene in CtrlGFP (blue) and Fezf2GFP (red) in utero electroporated cortical progenitors that were collected at 24 h (n = 3 litters per condition) and 48 h (n = 4 litters per condition). Vertical axes are normalized intensities (arbitrary units). Error bars indicate s.e.m. LV, lateral ventricle; Str, striatum. Scale bars, 100 μm; 50 μm in insets. Source data are shown in Supplementary Table 1.
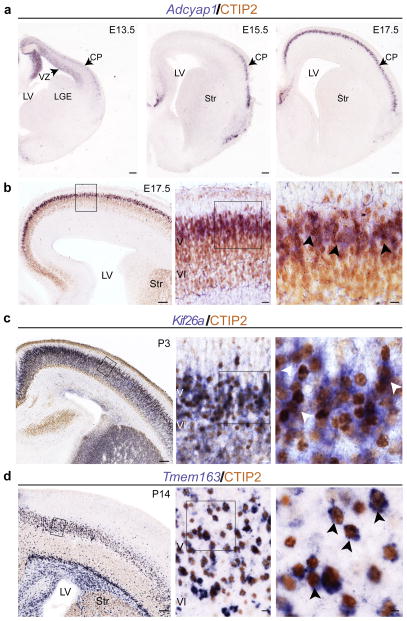
To precisely define the neuron subtype–specific expression of the identified transcripts, we performed colocalization analyses of all the selected Fezf2-induced genes with CTIP2, a well-established marker of CSMN and other ScPN3. As expected, all genes tested showed various degrees of colocalization with CTIP2 (Fig. 2 and Supplementary Fig. 3a). Among the embryonic CSMN marker genes, Adcyap1 showed restricted expression both in cortical progenitors at E13.5, the peak of CSMN neurogenesis, and subsequently in developing layer V (Fig. 2a). Adcyap1 expression molecularly defined a subpopulation within the broader group of CTIP2+ neurons (Fig. 2b). At postnatal stages, Kif26a expression similarly defined a subset of CTIP2+ neurons (Fig. 2c), whereas Tmem163 labeled most of the CTIP2+ population in the cortex (Fig. 2d).
Figure 2.
Fezf2-induced genes identify native CSMN and label subsets of the broad CTIP2-positive population in layer V. (a) In situ hybridization showing expression of Adcyap1 in E13.5 cortical progenitors (left) and young postmitotic subcerebral neurons in developing cortical plate (middle and right). (b–d) Immunocytochemistry for CTIP2 combined with in situ hybridization for Adcyap1, Kif26a and Tmem163 (boxed area enlarged in panel to the right). Examples of double-positive cells (arrowheads) are indicated in the right column. CP, cortical plate; LGE, lateral ganglionic eminence; LV, lateral ventricle; Str, striatum. Scale bars, 100 μm (a; b–d left panels), 20 μm (b–d, middle panels), 10 μm (b–d, right panels). The complete gene list is given in Supplementary Table 3.
To investigate whether Fezf2 is required for the expression of the identified target genes, we performed in situ hybridization for 13 genes on Fezf2−/− cortex. We found that, in the absence of Fezf2, the expression of all genes tested was abrogated or decreased in layer Vb. Notably, the effect of Fezf2 loss on expression was selective for layer Vb, as these genes were maintained in other layers (Supplementary Fig. 3b). These data indicate that Fezf2 induces numerous early and late CSMN genes and that Fezf2 is required for the expression of its targets in layer Vb.
Fezf2 induces a transcriptome shift toward nascent CSMN
To understand how early during development Fezf2 governs expression of CSMN genes, we interrogated our target gene list against available RNA sequencing data from E14.5 cortical plate, subventricular zone (SVZ) and ventricular zone (VZ)12. At E14.5 the cortical plate is mostly populated by early postmitotic corticofugal neurons (including nascent CSMN). In contrast, the SVZ and the intermediate zone (SVZ/IZ) primarily contain basal progenitors of upper layer CPN13–15 and early postmitotic upper layer CPN16. Finally, the VZ is mostly populated by apical progenitors generating upper layer CPN15. We found that Fezf2 overexpression upregulated 186 genes enriched in the cortical plate versus SVZ/IZ and VZ (Supplementary Fig. 4 and Supplementary Table 2).
We next compared the Fezf2-induced genes to the available transcriptome of purified CSMN3. Strikingly, within 48 h, Fezf2 overexpression had already induced 30 genes known to be restricted to early postnatal CSMN (Supplementary Table 3).
Next we investigated whether Fezf2 repressed genes of alternative neuronal fates. We found that 73 genes repressed by Fezf2 (at both 24 and 48 h) were preferentially expressed in the E14.5 VZ and SVZ/IZ, which mainly contain progenitors of upper layer projection neurons (Supplementary Fig. 5 and Supplementary Table 2). In agreement, comparative analysis with the available transcriptome of purified CPN17 showed that Fezf2 repressed 17 genes including Cux1, Zfhx4, Fzd1 and Btbd11. These included early markers of the CPN lineage, such as Cux2, shown to mark progenitors fated to an upper layer identity15. The data indicate that, when expressed in progenitors of upper layer CPN, Fezf2 induces a battery of transcripts that collectively define developing CSMN and represses genes of developing CPN.
Fezf2 associates with the proximal promoters of CSMN genes
To elucidate the strategy used by Fezf2 to instruct CSMN identity, we performed a genome-wide DNA binding analysis for Fezf2 in cortical progenitors, using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq)18 in combination with RNA-seq. Cortical neural progenitors were isolated at E14.5 and expanded in vitro for one passage as neurospheres. The neurospheres were infected with retroviruses encoding epitope-tagged Fezf2 (3xFlag-Fezf2) or control (3xFlag) and harvested 48 h after infection for anti-Flag ChIP (Supplementary Fig. 6a). The 3xFlag-tagged Fezf2 was confirmed to be functional by in utero electroporation (Supplementary Fig. 6b–e).
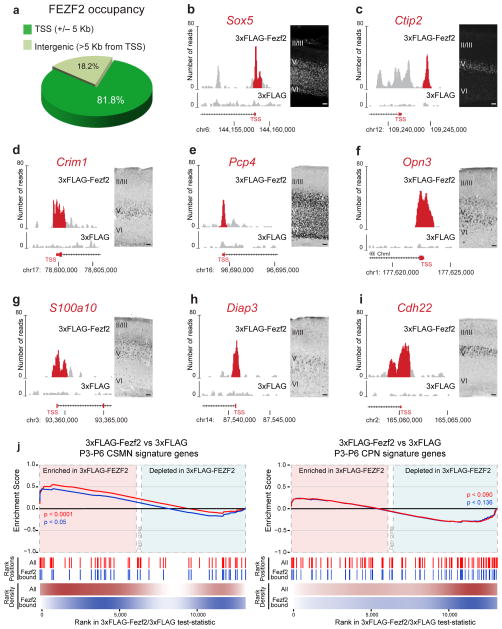
Two replicates of the ChIP-seq data sets were analyzed independently using the GEM (genome-wide event finding and motif discovery) integrative computational method19. In the two replicates, GEM analysis predicted 15,665 binding events with significant enrichment in 3xFlag-Fezf2 over control (P < 10−3; Supplementary Table 4). Of these, 81.8% fell within 5 kb of the transcription start site (TSS) of 12,860 annotated genes (Fig. 3a and Supplementary Table 4). Genes bound by Fezf2 contained over 63% of known CSMN genes3 (Supplementary Table 4), including those expressed at early (Fig. 3b–c), middle (Fig. 3d) and late stages of CSMN maturation (Fig. 3e–i).
Figure 3.
Genome-wide binding analysis for Fezf2 shows preferential association with proximal promoter regions of CSMN genes. (a) Fezf2 binding events preferentially occur in proximity to promoter regions, within 5 kb of the TSS of annotated genes. (b–i) Examples of 3xFlag-Fezf2 peaks at proximal promoters for early (Sox5 and Ctip2; b,c), middle (Crim1) (d) and late (Pcp4, Opn3, Diap3, S100a10 and Cdh22; e–i) CSMN genes. In situ hybridizations are shown for Crim1 (P21), Pcp4 (P21), Opn3 (P21), S100a10 (P21), Diap3 (P14) and Cdh22 (P21). Immunohistochemistry results are shown for SOX5 (E18.5) and CTIP2 (P1). Scale bars, 100 μm (b–i). (j) The “zero cross” area represents genes where the Cuffdiff2 test statistic is equal to 0 and no appropriate rank information is available. The color scales used for the rank density represent kernel density estimate of gene rank positions (white = 0, red/blue=max density). GSEA for CSMN and CPN signature gene sets. Signature gene sets (in red) and the corresponding subsets bound by Fezf2 (in blue) were assessed for enrichment in Fezf2-overexpressing neurospheres. Both CSMN signature genes and the subset bound by Fezf2 were significantly enriched in Fezf2-overexpressing neurospheres. Neither set of CPN signature genes showed significant enrichment. Source data are shown in Supplementary Table 4.
To assess the relationship between DNA binding and transcriptional regulation by Fezf2, we performed comparative RNA-seq analysis of 3xFlag-Fezf2 and 3xFlag neurospheres 48 h after infection (Supplementary Fig. 6a). We identified 1,295 transcripts differentially expressed upon Fezf2 induction, including 747 upregulated and 548 downregulated genes (false discovery rate = 10%; Supplementary Table 5). The fragments per kilobase RNA per million mapped reads (FPKM) values of Fezf2 indicate absence of transcription (FPKM = 0.42) in control progenitors and high levels upon Fezf2 overexpression (FPKM = 3,142, data not shown). Confirming Fezf2 overexpression in vivo, 224 of the genes induced by Fezf2 in neurospheres were preferentially expressed in the cortical plate and were enriched for postmitotic neuronal genes (Supplementary Fig. 7a and Supplementary Table 5). In contrast, 155 genes downregulated by Fezf2 were preferentially expressed in VZ and SVZ/IZ and were enriched for cell-cycle genes (Supplementary Fig. 7b and Supplementary Table 5). Genes upregulated by Fezf2 in neurospheres also showed enrichment for CSMN versus CPN signature genes by gene score enrichment analysis (GSEA) (Fig. 3j and Supplementary Table 4). Collectively, this indicates that Fezf2 induces similar transcriptional changes in isolated cortical progenitors to the ones observed in vivo.
We found that 64.1% of the genes induced and 77.4% of those downregulated by Fezf2 in cortical neurospheres were promoter-bound (Supplementary Fig. 8a,b). Similar percentages were observed when using transcripts regulated by Fezf2 in vivo (Supplementary Fig. 8c,d). These represent a significantly greater proportion of genes than expected by chance (Online Methods, Supplementary Fig. 8c–e and Supplementary Table 6). GSEA analysis of genes that were both transcriptionally regulated and promoter-bound by Fezf2, against sets of CSMN and CPN signature genes (Supplementary Table 4), showed that only CSMN signature genes were significantly enriched in the Fezf2-upregulated targets (Fig. 3j).
GEM motif-finding analysis identified a GC-rich sequence (CGCCGC) that is enriched at Fezf2-bound sites (Supplementary Fig. 9a). Since Fezf2-bound promoters are slightly more GC-rich than unbound ones (64.4% versus 49.6% GC content; Supplementary Fig. 9b), it is difficult to conclude whether Fezf2 directly binds to this motif. Nevertheless, this sequence was present in 42.9% of Fezf2-bound TSSs compared to 11.2% of unbound TSSs. In addition, electrophoretic mobility shift assay on selected target promoters containing this sequence (namely, Ascl1 and Ephb1) showed binding by Fezf2 (Supplementary Fig. 9c,d). Previously reported consensus sequences predicted either by SELEX (systematic evolution of ligands by exponential enrichment) in zebrafish20 or using protein-DNA arrays in the human protein-DNA interactome (hPDI)21 were present in 15.4% for hPDI and 3.9% for SELEX of all Fezf2-bound sites, respectively, while the GEM-defined motif was present in 48.6% of these sites (Supplementary Fig. 9b).
Together, these data indicate that Fezf2 activates and represses a broad program of neuron subtype–specific genes by binding to proximal promoters.
Fezf2 selects glutamatergic and represses GABAergic fate
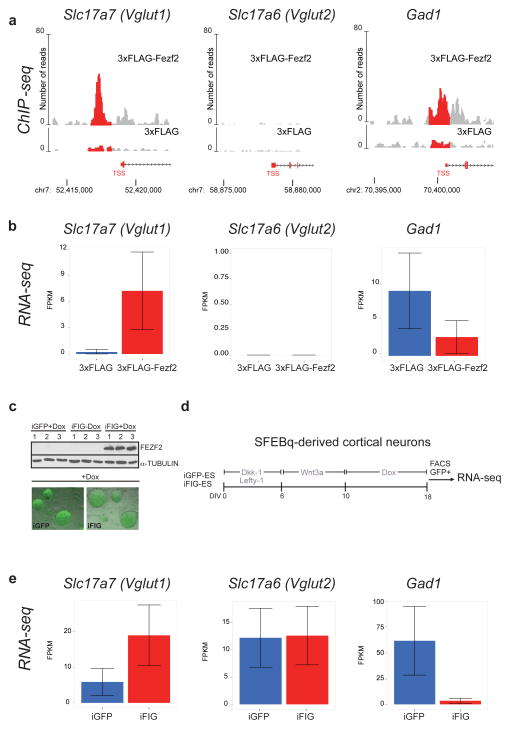
To functionally test whether genes regulated by Fezf2 control the acquisition of CSMN traits, we concentrated on two defining features of CSMN: glutamatergic identity and axonal extension to the spinal cord. Glutamate signaling is a terminal, necessary feature of all cortical projection neurons. It is unknown whether fate-specifying transcription factors for cortical neuron classes also instruct the establishment of this pan-neuronal trait. We found that Fezf2 bound to the promoter of Vglut1, the vesicular glutamate transporter used by most cortical projection neurons, including CSMN22, and was sufficient to induce its expression in cortical neurospheres (Fig. 4a,b). The binding appeared specific, as Vglut2 (Scl17a6), used only by glutamatergic neurons in cortical layer IV and outside of the cortex22, was neither bound nor transcriptionally regulated. Concomitantly, Fezf2 bound to the promoter of Gad1 (Gad67), necessary for establishment of GABAergic identity, but repressed its expression (Fig. 4a,b). To test the extent to which Fezf2 can instruct neurotransmitter choice, we established doxycycline-inducible Fezf2-IRES-GFP (iFIG) and control GFP (iGFP) mouse embryonic stem cell lines and induced neural differentiation using the Serum-free embryoid body quick (SFEBq) protocol23 (Fig. 4c,d and Supplementary Fig. 10). GFP+ cells were FACS-sorted and profiled by RNA-seq. Confirming our results in primary cortical progenitors, Fezf2 overexpression induced Vglut1, had no effect on Vglut2 and inhibited the expression of Gad1 (Fig. 4e). Together, these data indicate that Fezf2 is sufficient to select the appropriate effector genes to establish the neurotransmitter identity of CSMN, while repressing an alternative GABAergic fate.
Figure 4.
Fezf2 promotes glutamatergic and inhibits GABAergic neurotransmitter pathways. (a) ChIP-seq trace shows that 3xFlag-Fezf2 binds specifically to the promoters of Vglut1 (Slc17a7) and Gad1 but not Vglut2 (Slc17a6). (b) RNA-seq analysis shows the effect of 3xFlag-Fezf2 overexpression on these three genes in neural stem cells in vitro. (c) Inducible expression of single-copy Fezf2-IRES-GFP is show by immunoblotting and fluorescence microscopy. Uncropped original immunoblots are shown in Supplementary Figure 10. (d) Schematic representation of directed differentiation protocol used to instruct embryonic stem cell (ES) differentiation into mixed populations of cortical neurons23. (e) RNA-seq analysis showing the effect of Fezf2 expression on Vglut1, Vglut2 and Gad1 in ES-derived neurons. Clones used for RNA-seq were from the differentiation of n = 2 independently generated iGFP and n = 2 iFIG lines. the error bars represent 95% confidence intervals for the Cuffdiff2 model expression estimate as defined in Methods and described in Trapnell et al., 2013.
Fezf2 directly controls expression of Ephb1 in CSMN
Axonal connectivity to the spinal cord is a defining class-specific trait of CSMN. Directed axonal extension begins soon after CSMN fate specification and is strictly guided under the control of multiple axon guidance molecules (reviewed in ref. 24). Gene Ontology analysis of the Fezf2-induced genes showed a significant enrichment in axon guidance molecules. Fezf2 associated with the proximal promoters of 78% of these genes (Supplementary Table 4), including neuropilin 2 (Nrp2) [and Robo1, which are known to affect the extension and guidance of the corticospinal tract (Supplementary Fig. 11a). Among these, we selected for further analysis Ephb1 (Fig. 1), encoding a tyrosine kinase receptor that mediates axon guidance and is critical for midline repulsion decisions25–27 and whose function in CSMN remains unknown.
We first profiled Ephb1 expression in the developing cortex by in situ hybridization. Ephb1 mRNA peaked in the cortical plate at E15.5, when CSMN axonal extension begins. Ephb1 expression remained restricted to developing layer V, and at lower levels in layer VI, until approximately E18.5 (Fig. 5a). Postnatally, Ephb1 levels drastically decreased, and by P14 expression was weak and detectable only in layer VI (Fig. 5a) in both TBR1+ projection neurons28 and APC+ oligodendrocytes (Fig. 5b)29.
Figure 5.
Fezf2 controls Ephb1 selective expression in CSMN by direct association with the Ephb1 promoter. (a) In situ hybridization for Ephb1 in the forebrain at different stages of embryonic and postnatal development shows highest expression in developing cortical plate at E15.5, the time of initial CSMN axonal extension, and its restricted expression in developing layer V, until approximately E18.5. (b) In situ hybridization for Ephb1 and immunocytochemistry for TBR1 and APC show maintained expression of Ephb1 at postnatal stages in corticothalamic neurons (TBR1-positive in layer VI) and oligodendrocytes (APC-positive). (c) β-galactosidase immunocytochemistry in Ephb1 heterozygous mice at P1 shows that, within layer V, expression colocalizes with CTIP2, and not with SATB2 (arrows upper panel). The dotted rectangle area in the left panel is shown in high magnification in the four panels on the right. Retrograde labeling of ScPN from the pons of P1 Ephb1 heterozygous mice shows colocalization of FluoroGold with β-galactosidase in layer Vb (arrows). Retrograde labeling of CPN in P2 Ephb1 heterozygous mice shows no colocalization of FluoroGold with β-galactosidase in callosal neurons of layer Va (arrowheads). Confocal images of layer V were combined to produce three-dimensional reconstructions (Lower right panels). Sidebars represent projections along the x–z axes (right) and the y–z axes (below). (d) Left, in situ hybridization for Ephb1 on E18.5 wild-type (left inset) and Fezf2−/−(right inset) littermates shows that Ephb1 levels specifically decrease in layer V of the mutant cortices. Right, DNA regions spanning 5′ UTR and first exon of the Ephb1 gene show enrichment of 3xFlag-Fezf2 binding (q = 10−15) compared to control. Str, striatum. Scale bars, 100 μm (a; c, far left panels), 20 μm (b), 50 μm (c, top right panels; d), 20 μm (c, bottom right panels).
To define the neuron subtype–specific expression of Ephb1 within layer V, we used β-galactosidase (β-gal) as a proxy for endogenous Ephb1 expression in mice in which a lacZ cassette has been knocked into the Ephb1 locus (see Online Methods; Supplementary Fig. 11b). Virtually all of the β-gal+ neurons were CTIP2+ and negative for SATB2, a marker for CPN30,31 (Fig. 5c). In addition, we retrogradely labeled ScPN and CPN by injecting FluoroGold into the pons and the contralateral hemisphere, respectively. We found that β-gal was expressed in FluoroGold-labeled ScPN and excluded from CPN labeled from contralateral cortex (Fig. 5c). These data demonstrate that, in the developing cortex, Ephb1 is specifically expressed in neuronal subtypes that extend ipsilateral axonal projections and is excluded from neuronal subtypes that project through the cortical midline.
To investigate whether Fezf2 is required for Ephb1 expression, we performed in situ hybridization on cortical sections from Fezf2−/− mice at E18.5. In the absence of Fezf2, Ephb1 was no longer expressed in ScPN, and expression was maintained only at very reduced levels in layer VI (Fig. 5d). In agreement, our ChIP-seq data showed that Fezf2 associates with the proximal promoter of the Ephb1 gene, within a 310-base-pair region at the TSS (Fig. 5d and Supplementary Fig. 9d). Ephb1 expression was unchanged in the striatum of Fezf2−/− mice, a region that does not endogenously express Fezf2 (Fig. 5d). These data demonstrate that Fezf2 is required for the expression of Ephb1 in ScPN.
Abnormal anterior commissure axon crossing in Ephb1−/− mice
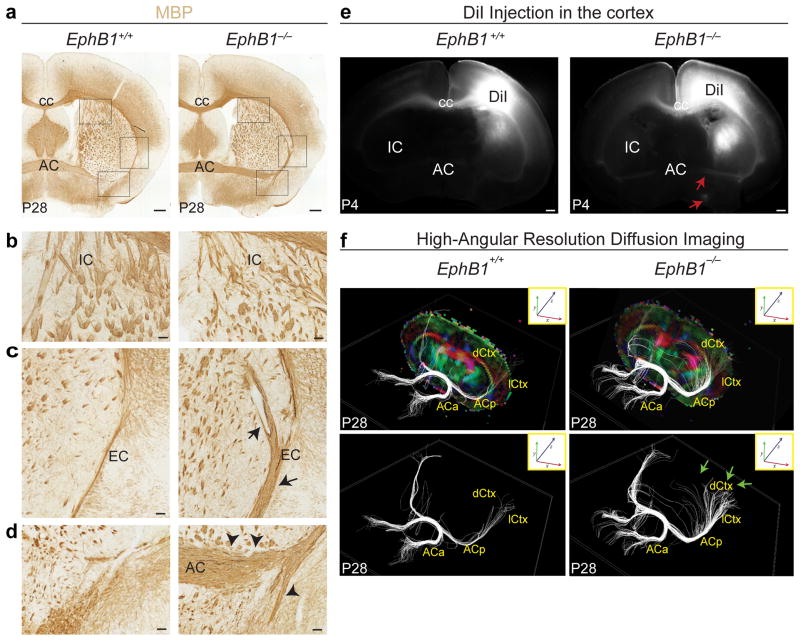
To decipher the role of EphB1 during CSMN development, we examined Ephb1−/− mice. Brains of Ephb1−/− and wild-type littermates (P28) were processed by immunohistochemistry for myelin basic protein to visualize myelinated axons. Mutant mice displayed clear abnormalities of major axon tracts (Fig. 6). The internal capsule comprised smaller than wild type and partly unfasciculated axon bundles (Fig. 6a,b). These abnormalities were accompanied by an expansion of the external capsule (Fig. 6a,c,d; arrows) and the anterior commissure (Fig. 6a,d; arrowheads). In addition, ectopic axon bundles extended within the internal capsule toward the external capsule and continued to grow ventrally (Fig. 6a,d).
Figure 6.
Cortical neurons aberrantly project through the anterior commissure in absence of EphB1. (a–d) Immunocytochemistry for myelin basic protein (MBP) on coronal sections of P28 wild-type and Ephb1−/− littermates shows internal capsule reduction and defasciculation, accompanied by an expansion of both the external capsule (arrows) and the anterior commissure (arrowheads). (e) DiI anterograde injections in deep layers of the somatosensory cortex of P2 wild-type (n = 3) and Ephb1−/− (n = 3) pups show axons ectopically crossing at the anterior commissure and extending ventrally and rostrally (red arrows) in the Ephb1 mutants but not in wild-type animals. (f) Color-coded three-dimensional reconstructions based on HARDI of P28 wild-type and Ephb1−/− littermates show distinct axon fibers originating in dorsal areas of the neocortex merging abnormally with the anterior commissure in the Ephb1−/− (top right) compared to wild-type (top left) brains. Red represents the anterior–posterior direction; green represents the medial–lateral direction; blue represents the dorsal–ventral direction. Green arrows indicate axon tracts penetrating into the cortex. AC, anterior commissure; ACa, anterior part of the anterior commissure; ACp, posterior part of the anterior commissure; cc, corpus callosum; dCtx, dorsal cortex; lCtx, lateral cortex; EC, external capsule; IC, internal capsule. Scale bars: 50 μm (a), 20 μm (b–d) 100 μm (e).
We hypothesized that, in the absence of EphB1, the trajectories of subcerebral axons are rerouted, abnormally crossing to contralateral targets via the anterior commissure. To test whether the ectopic axon tracts were of neocortical origin, we anterogradely traced neocortical output projections with the lipophilic tracer DiI injected in the sensorimotor cortex of Ephb1−/− and wild-type littermates at P2. DiI labeled axons were found in the corpus callosum and internal capsule in both wild-type and Ephb1−/− mice. However, only in the mutant mice did we observe distinct DiI-labeled axons crossing at the anterior commissure and extending ventrally and rostrally (Fig. 6e). In wild-type cortex, only neurons located in the most lateral cortical areas, the posterior perirhinal cortex, and the entorhinal cortex project contralaterally through the anterior commissure32. In the Ephb1−/− mice, however, neurons located in dorsal areas projected ectopically through this commissure.
To confirm and visualize the axonal projection routes in the absence of EphB1, we used high-angular-resolution diffusion imaging (HARDI) tractography to render a three-dimensional image of major axon tracts in mutant and wild-type brains33,34. We found that axon fibers in the dorsal areas of the neocortex merged with the anterior commissure in the Ephb1−/− but not in wild-type brains (Fig. 6f). To identify a potential source of EphB1 ligands, we performed in situ hybridization for the ephrin genes Efnb1, Efnb2 and Efnb3. We found that only Efnb3 was expressed at the ventral forebrain midline and thus might mediate EphB1-dependent repulsion of ipsilateral descending tracts (Supplementary Fig. 12).
Reduced corticospinal tract in Ephb1 null mutants
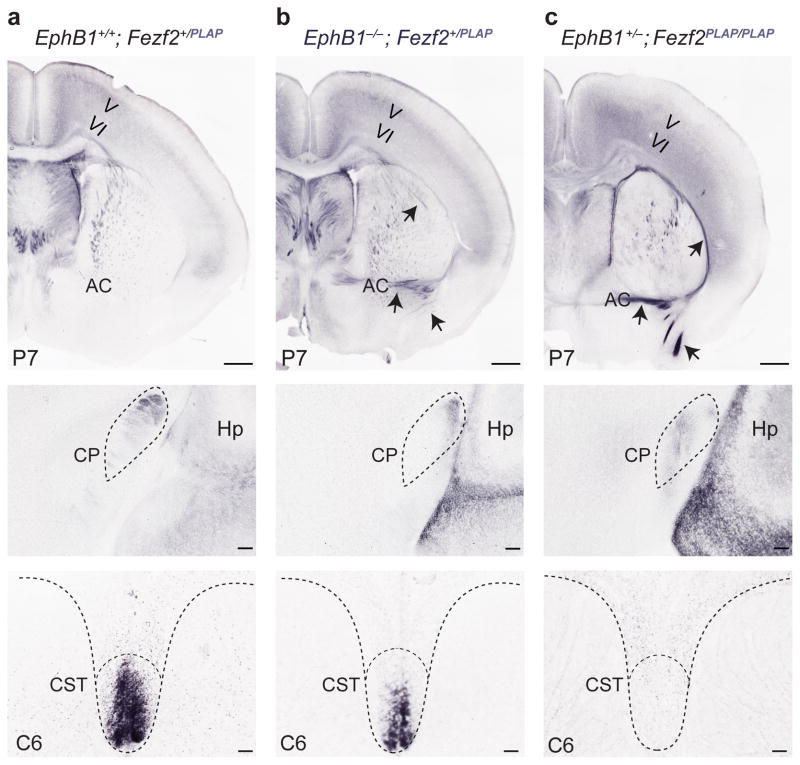
To determine whether the corticospinal tract is specifically affected by EphB1 loss, we used the Fezf2::PLAP (placental alkaline phosphatase) reporter line7 to genetically trace CSMN axons in control (Ephb1+/−;Fezf2PLAP/+) and EphB1-deficient (Ephb1−/−;Fezf2PLAP/+) mice at P7. In control mice, PLAP-positive axons were exclusively observed extending along ipsilateral, corticofugal trajectories (Fig. 7a). In contrast, in EphB1-deficient mice, a large proportion of PLAP-positive axons extended inappropriately to contralateral targets, crossing the midline via an enlarged anterior commissure (Fig. 7b). Most notably, this resulted in a drastic reduction of subcerebral axons reaching the cerebral peduncle (Fig. 7b) and of corticospinal axons in the dorsal funiculus of the spinal cord (Fig. 7b). These results demonstrate that, in the absence of EphB1, the ipsilateral axonal trajectories of the corticospinal tract are compromised and a distinct proportion of axons take contralateral, crossed routes through the anterior commissure.
Figure 7.
Ephb1−/− mice recapitulate the axon guidance phenotype observed in Fezf2 null mutants. (a–c) PLAP-positive axons inappropriately project through the anterior commissure in Ephb1−/− mutants (b, arrows), mimicking the Fezf2PLAP/PLAP axonal phenotype (c, arrows). Enlargement of the anterior commissure in Ephb1−/− mice is accompanied by a reduction of PLAP-positive axons found in the cerebral peduncle (b, middle panel) and the cervical spinal cord (b, bottom), canonical targets of the corticospinal tract in wild-type mice (a, middle and bottom). No axons are found in the cerebral peduncle or spinal cord of the Fezf2PLAP/PLAP mice (c, middle and bottom). AC, anterior commissure; Cp, cerebral peduncle; Hp, hippocampus; CST, corticospinal tract. C6, cervical vertebra. Scale bars: 100 μm (a–c, top panels), 50 μm (a–c, middle and bottom).
It is notable that Fezf2−/− mutants also show loss of axonal projections to the cerebral peduncle and the spinal cord (Fig. 7c and refs. 5–7), accompanied by an expansion of the anterior commissure (Fig. 7c and ref. 7). This phenotype is reminiscent of that observed in the EphB1-deficient mice (Fig. 7b and Supplementary Fig. 13). Beyond connectivity, in the absence of Fezf2 the global fate specification of CSMN (molecular identity, electrophysiological properties and connectivity to the spinal cord) is abolished5–7. In contrast, we found that loss of EphB1 did not affect the fate specification of projection neuron classes, which showed normal expression of the canonical projection neuron subtype-specific markers CTIP2, TBR1, SATB2 and CUX1 (Supplementary Fig. 13). The data indicate that EphB1 is an effector molecule directly downstream of Fezf2 that executes a specific modular aspect of the global Fezf2−/− phenotype.
DISCUSSION
Deciphering the molecular logic that orchestrates the acquisition of neuronal subtype–specific features in the mammalian cerebral cortex is fundamental to understanding how its neuronal diversity has evolved. These molecular rules are not yet defined for any subtype of cortical neuron. Here we report that acquisition of defining features of corticospinal motor neurons is at least partly achieved by co-regulated expression of batteries of CSMN effector molecules directly downstream of a single selector gene, Fezf2.
Precise control over activation and repression of transcription factors responsible for acquisition of class-specific traits in projection neurons is critical to resolve neuronal fates during cortical development. Tbr1 and Sox5 act as repressors of Fezf2 to negatively regulate the extension of the corticospinal tract and to promote connectivity to the thalamus35–38. Similarly, reciprocal repression of Fezf2, Satb2 and Ctip2 is employed to choose subtype-specific long distance connectivity39,40. These studies motivate a more global analysis of the molecular rules beyond single genes that govern the acquisition of lineage-specific traits.
We found that expression of Fezf2 in cortical progenitors fated to generate upper layer neurons resulted in both the induction of a large series of genes expressed in nascent CSMN and the repression of molecular programs of upper layer neurons. Given that this transcriptional regulation largely occurred via binding of Fezf2 to the promoters of both activated and repressed genes, the data indicate that lineage bifurcation decisions can be co-regulated by the same transcription factor from very early stages of differentiation. Our data also provide a molecular explanation for why, during development, cortical progenitors tune Fezf2 expression to the lower levels observed in late-stage cortical progenitors5,7,41. Maintenance of Fezf2 expression after the completion of neurogenesis of deep layer neurons may be molecularly incompatible with the initiation of upper layer neurogenesis.
Much work is still required to understand the nuances of how Fezf2 can act as an activator for some target genes and as a repressor for others. It is likely that factors such as post-translational modification of Fezf2, the choice of interacting cofactors and the presence of neighboring binding sites for different proteins at different loci critically contribute to these distinct transcriptional outcomes42.
Transcription factors endowed with properties of classic selector genes have been identified in different tissues as genes uniquely able to control ‘master’ molecular switches of distinct cellular fates. Elegant work in Drosophila has highlighted core properties of selector genes that clarify the logic guiding their activity. Multiple target genes are often regulated downstream of a single selector gene, and targets often include both activated and repressed genes43. Our evidence that Fezf2 acts as both an activator of CSMN genes and a repressor of genes for alternative fates further supports the idea that Fezf2 is a selector gene. To our knowledge, this is the first transcription factor endowed with these properties for any class of cortical neurons.
During development of CSMN, expression of lineage-specific and pan-neuronal genes alike display temporally dynamic regulation. It is intriguing that within 48 h of induction, Fezf2 is already associated with the promoters of CSMN genes whose expression is normally temporally spaced during CSMN differentiation. The data suggest that, in cortical progenitors, Fezf2 might be ‘priming’ the transcriptional territory of CSMN by also occupying genomic loci that are transcriptionally active in these neurons at later times. This implies that temporal resolution of gene expression over the several weeks of CSMN development likely relies on cofactors that operate by interaction with Fezf2 or work on the genomic loci that are primed by early Fezf2 binding. The fact that Fezf2 expression is maintained at all stages of CSMN differentiation in vivo5–7 supports the view that active binding by Fezf2 to CSMN loci may be required to poise them for transcriptional regulation at later stages of differentiation.
Precedents exist for the use of early priming of genomic loci in progenitor cells to enable the execution of specific differentiation programs at later stages of development. In Caenorhabditis elegans, asymmetric distribution of gene expression in bilaterally symmetrical ASE gustatory neurons relies on early priming of the Isy-6 microRNA locus in progenitors of only the left neurons44. MyoD has also been shown to associate with enhancers of both early and late myogenic genes45.
In his first definition of selectors, Antonio García-Bellido postulated that these genes would regulate effector molecules necessary for the acquisition of specific cellular features46. Here we identify EphB1 as an effector molecule of Fezf2. In turn, EphB1 executes CSMN axonal repulsion from the midline in the ventral telencephalon, enabling the ipsilateral extension of the corticospinal tract. Our data are in agreement with work demonstrating a key role for EphB1 in mediating midline repulsion decisions. In the spinal cord, EphB1 instructs the ventral trajectory of motor neuron axons by mediating their repulsion from the mesenchyme of the dorsal limb25. Similarly, in the optic chiasm, EphB1 mediates the repulsion of retinal ganglion cell (RGC) axons from the midline47. Notably, in the retina the transcription factor Zic2 is also upstream of genes critical for both axonal guidance decisions (EphB1) and neurotransmitter identity (Sert)48.
The regulation of axon guidance molecules in different classes of cortical projection neurons has just begun to be explored40. Whether expression of lineage-specific axon guidance molecules is directly controlled by the same transcription factors that govern neuron subtype fate specification remains unknown. It is striking that the phenotypic abnormalities observed in the absence of EphB1 mimic the axon guidance defects observed in the Fezf2−/− mutants, without affecting the overall fate specification of CSMN. While the possibility that expression of EphB1 in thalamus or striatum contributes to this phenotype49 cannot be ruled out, the fact that Fezf2 is not expressed in these regions and yet Fezf2−/− mice present similar axon guidance defects to Ephb1 mutants supports a cell-autonomous role of EphB1 in CSMN. Our data point to a direct regulation of key CSMN axon guidance receptors by the same selector gene that governs global programs of fate specification of this neuron subtype.
Not all selector genes have equal properties. Pioneering work in C. elegans has identified ones termed terminal selector genes sufficient to instruct specific neurotransmitter fates50. Our data indicate that Fezf2 controls the acquisition of glutamatergic identity by direct binding to the promoter of Vglut1 and other genes involved in the synthesis, transport and signaling of glutamate. This is in agreement with the fact that overexpression of Fezf2 in progenitors of GABAergic medium spiny neurons results in a switch to a glutamatergic fate in vivo8. In addition, like selector genes in C. elegans, Fezf2 expression is maintained in CSMN throughout life, suggesting a requirement for Fezf2 to both establish and maintain CSMN traits. In the future, conditional loss of Fezf2 in adult CSMN will test this possibility and define the extent to which Fezf2 fulfills the definition of a canonical terminal selector.
An understanding of the molecular regulatory architecture that shapes the identity of CSMN and other classes of cortical neurons is fundamental to gaining insights into how neuronal diversity develops in the cerebral cortex and will furthermore inspire tools to reprogram class-specific neuronal identity in the context of neurodegenerative disease.
ONLINE METHODS
Mice
Fezf2−/− mice were generated by Hirata et al.41 (Fezf2 GenBank accession code: AB042399). Ephb1−/− mice (Ephb1 GenBank accession code: NM_173447) were generated by Deltagen, Inc. (Menlo Park, CA). The line was generated by targeting the Ephb1 coding region with a cassette containing a 4.5-kb homology sequence upstream of exon 8, a Lac0-SA-IRES-lacZ-WT Neo/Kan cassette and a 2.2-kb homology sequence downstream of exon 8. The insertion of the Lac0-SA-IRES-lacZ-WT Neo/Kan cassette resulted in a 100-bp deletion of genomic DNA. Sequences of PCR primers employed to genotype the Ephb1−/− line are as follows: forward primer, ACGTGGGAGGACTCTAATCCTCTTC; reverse primer (wild-type allele), TCTAGGTTGCTGGCTACAGGACTTG; reverse primer (Neo), GGGCCAGCTCATTCCTCCCACTCAT. Fezf2::PLAP (Fezf2PLAP/+) mice were generated by Chen et al.7 and crossed with the Ephb1+/− mutant line to obtain Ephb1−/−;Fezf2PLAP/+ mutants. Timed-pregnant CD-1 mice were obtained from Charles River Laboratories (Wilmington, MA) for in utero electroporation. The day of the vaginal plug was designated embryonic day 0.5 (E0.5). The day of birth was designated postnatal day 0 (P0). All mice were maintained in standard housing conditions on a 12-h light/dark cycle with food and water ad libitum. No more than four adult animals were housed per cage. All mouse studies were approved by the Massachusetts General Hospital and Harvard University Institutional Animal Care and Use Committee and were performed in accordance with institutional and federal guidelines.
In utero electroporation
The Fezf2GFP and CtrlGFP construct and conditions for in vivo electroporation were described previously5,51. The 3xFlag-Fezf2 open reading frame was cloned into the CtrlGFP construct driven by the CAG promoter. Briefly, 800 nl of purified DNA (2 μg/μl) mixed with 0.005% fast green in sterile PBS was injected in utero into the lateral ventricle of CD1 embryos at E14.5 under ultrasound guidance (Vevo 770, VisualSonics). Five 40-V pulses of 50 ms were delivered at 1-s intervals in an appropriate orientation across the embryonic head using 1-cm-diameter platinum electrodes placed outside the uterus, using a CUY21EDIT square wave electroporator (Nepa Gene). Injected embryos were collected for FACS purification either 24 h or 48 h after electroporation.
FACS purification
Pregnant dams were deeply anesthetized 24 h or 48 h after surgery. Their embryos were removed and sensorimotor cortex was microdissected, as previously described52, from the electroporated cerebral hemisphere using a fluorescence dissecting microscope to precisely visualize the labeled regions. Fezf2GFP and CtrlGFP electroporated cells were purified by FACS directly into RNAlater53 and RNA was extracted as previously described54. To control for biological sample variability, we processed multiple independently collected samples for both Fezf2GFP and CtrlGFP electroporated cells at each time point of analysis (true biological replicates) for a total of fourteen samples derived from different litters, FACS purifications and microarray hybridizations (three each of Fezf2GFP and CtrlGFP at 24 h and four each at 48 h). Fezf2GFP- and CtrlGFP-labeled cortical tissues were enzymatically digested in dissociation medium (20 mM glucose, 0.8 mM kynurenic acid, 0.05 mM APV, penicillin-streptomycin (50 U/ml and 0.05 mg/ml, respectively), 0.09 M Na2SO4, 0.03 M K2SO4 and 0.014 M MgCl2) containing L-cysteine HCl (0.016 μg/μl) and papain (10 unit/ml; Worthington Biochemical Corp., NJ) at 37 °C for 30 min. Papain digestion was stopped with ovomucoid (10 mg/ml) and BSA (10 mg/ml) in dissociation medium at room temperature. Progenitors were mechanically dissociated to obtain a single-cell suspension by gentle trituration in iced OptiMEM containing glucose (20 mM), kynurenic acid (0.4 mM) and APV (0.025 mM). All chemicals were purchased from Sigma, and all media were purchased from GIBCO-BRL. GFP-positive cells were purified using a BD FACS Vantage SEM DiVa cell sorter. Cells were gated on the basis of on fluorescence, forward scatter and side scatter to select the appropriate population. FACS-purified progenitors and neurons from both time points were collected and stored in RNAlater before RNA extraction53.
Microarray and analysis
Approximately 10,000 to 30,000 FACS-sorted progenitors were used for each biological replicate. RNA was extracted using the StrataPrep Total RNA Micro Kit (Stratagene), and RNA quality was assessed using a Bioanalyzer (Agilent Technologies). RNA was amplified per Affymetrix small sample protocol using two consecutive rounds of linear in vitro transcription to obtain 15–20 μg of amplified and labeled cRNA for each hybridization. Microarray hybridization was performed with an Affymetrix Mouse Genome 430 2.0 Array according to standard Affymetrix protocol55. For microarray experiments, 3 or 4 replicates were used at each age, which allows the identification of differential gene expression with low false discovery rates3.
Using the Rosetta Resolver software (Rosetta Biosoftware), data from individual microarrays were normalized with the trend function and replicates were combined. Statistical significance of gene expression differences between Fezf2GFP and CtrlGFP experiments was determined by pairwise comparisons at each time point using the Rosetta Resolver error modeling method56 with P-value limit of <0.001 and fold change of >1.5. Similar results were obtained by normalizing with GCRMA in Bioconductor57 or with the Bioconductor implementation of MAS 5.0 and performing pairwise comparisons using significance analysis of microarrays (SAM)58.
For comparison with RNA-seq expression data from Ayoub et al.12, BAM files were downloaded from NCBI (accession code GSE30765), differential expression was determined with Cuffdiff, and analysis was performed with the Cummerbund package in the R language and environment for statistical computing and graphics59. From a list of genes with significant differential expression between VZ, SVZ and cortical plate (P < 0.001) in at least one sample, those with significant up- or downregulation induced by Fezf2 were identified and subsequently used for cluster analysis with Jensen-Shannon divergence as the distance metric in Cummerbund.
Western blot
Brains were dissected from P1 Ephb1 knockout, heterozygous and wild-type mice and lysed in RIPA buffer (150 mM NaCl, 1% NP40, 0.5% deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM Tris-HCl, pH 8) with proteinase inhibitors (S8830, Sigma-Aldrich). Mouse ES cells were treated with 2 μg/ml doxycycline for 48 h and harvested in RIPA buffer. After quantification by Bio-Rad protein assay (Bio-Rad), proteins were resolved in 4–12% SDS-PAGE gels and transferred to a nitrocellulose membrane. The western blot was performed using goat anti-EphB1 (M19, Santa Cruz, 1:250), rabbit anti-Fezf2/Fezf1 (F441, IBL, 1:500), rabbit anti-β-tubulin (9F3, Cell Signaling, 1:500) and mouse anti-α-tubulin (T9026, Sigma, 1:5,000). Anti-goat, anti-rabbit and anti-mouse HRP-conjugated secondary antibodies (HRP-linked anti-rabbit IgG, Cell Signaling Technology 7074, HRP-conjugated goat anti-mouse IgG, Cell Signaling Technology 7076, HRP-conjugated anti-goat IgG, Santa Cruz sc-2020, all at 1:5000), followed by ECL (Amersham), were used to visualize proteins of interest on the membrane.
Retrograde and anterograde labeling
Pups were anesthetized by hypothermia at P2 and injected with FluoroGold (for retrograde labeling) in the contralateral cortex or the pons, or with DiI (for anterograde labeling) in the motor cortex as previously described17,60. The site of DiI injection was identified by ultrasound guidance. Pups were returned to the care of their mother and deeply anesthetized at P4 and P7 in the retrograde tracer studies, and at P4 for the anterograde tracer study, before perfusion and collection of the brain for immunocytochemistry.
High-angular-resolution diffusion imaging (HARDI)
Four-week-old (P28) wild-type and Ephb1−/− littermates were perfused with saline followed by 4% paraformaldehyde, postfixed for 24 h, and subsequently fixed for an additional week in 4% paraformaldehyde solution containing 1 mM gadolinium (Gd-DTPA). For image acquisition, the brains were placed in Fomblin. Brains were scanned on a 4.7-T Bruker Biospec MR system. The pulse sequence used for image acquisition was a 3D diffusion-weighted spin-echo echo-planar imaging sequence, time repetition (TR)/time echo (TE) 1,000/45.47 ms, with an imaging matrix of 96 × 96 × 128 pixels. Spatial resolution was 125 × 125 × 125 μm. Sixty diffusion-weighted measurements (b = 4,000 s/mm2) and one non-diffusion-weighted measurement (b = 0) were acquired with δ = 12.0 ms, Δ = 24.2 ms. Total acquisition time was approximately 2 h for each imaging session. The HARDI method was used to reconstruct the orientation distribution function in each voxel and resulting tractography pathways were reconstructed using a streamline algorithm for diffusion tractography33,61 Diffusion Toolkit and TrackVis (http://trackvis.org/) were used to reconstruct and visualize axonal pathways. Trajectories were propagated by consistently pursuing the orientation vector of least curvature, and tracking was terminated when the angle between two consecutive orientation vectors was greater than the given threshold of 45° for each specimen. Brain mask volumes created by MRIcro (http://www.mccauslandcenter.sc.edu/mricro/mricro/mricro.html) were used to terminate tractography structures instead of the FA (Fractional Anisotropy) threshold as previously reported61,62. Commissural pathways were isolated as fibers passing through the manually drawn region of interest that included the entire anterior commissure at the midline, ensuring that other fiber pathways were not contributing, as previously described61.
Immunocytochemistry, PLAP staining and in situ hybridization
Brains for immunocytochemistry were processed as previously described55,63. Primary antibodies and dilutions used were as follows: rabbit anti-TBR1 antibody, 1:2,500 (gift from R. Hevner); rat anti-CTIP2 antibody, 1:1,000 (Abcam ab18465); rabbit anti-GFP, 1:500 (Invitrogen A11122); chicken anti–β-galactosidase, 1:500 (ICL lab CGAL-45A); mouse anti-SATB2, 1:50 (Abcam ab51502); rat anti-MBP, 1:500 (Millipore MAB386); and rabbit anti-CUX1, 1:100 (Santa Cruz CDP M-222). Goat anti-rabbit IgG Alexa Fluor 488, 546, and 647 (Life Technologies A11070, A-11071, A21246), Goat anti-chicken IgG Alexa Fluor 488, 546, 647 (Life Technologies A11039, A11040, A21449), Goat anti-rat 488, 546, 647 (Life Technologies A11006, A11081, A21247), Goat anti-mouse IgG Alexa Fluor 488, 546, 647 (Life Technologies A11017, A11018, A21237) secondary antibodies were diluted 1:750. PLAP activity was detected with alkaline phosphatase staining buffer (0.1 mg/ml 5-bromo-4-chloro-3-indolyl phosphate; 1 mg/ml nitro blue tetrazolium in 100 mM Tris-HCl, pH 9.5; and 100 mM NaCl) as previously described7. In situ hybridization and combined in situ hybridization with immunohistochemistry were performed following published methods and using 40-μm-thick sections cut on a vibrating microtome and mounted on Superfrost slides (Fisher)64. Riboprobes were generated as previously described54. The cDNA template clone for Ldb2 was provided by J. Macklis; all other riboprobes were generated from cDNA template clones using the primers listed below. Tissue sections were imaged using a Nikon 90i fluorescence microscope equipped with a Retiga Exi camera (Q-Imaging) and analyzed with Volocity image analysis software v4.0.1 (Improvision). All primary data from immunohistochemistry and in situ hybridization experiments were repeated at least three times and analyzed by one investigator, then confirmed by a second, independent investigator who was blinded to genotype and experimental conditions.
In situ hybridization riboprobe templates
cDNA templates for riboprobes were obtained by RT-PCR with the following primer pairs:
| Gene name | Forward primer | Reverse primer | Product length |
|---|---|---|---|
| Acvr1c | AGGACTTGCCTCGAAAGTGA | GAAAGCAGAAGCGGACATTC | 610 |
| Adcyap1 | AATGACTTGGGGAATTGCTG | GCATGAACAGCACTGGAGAA | 542 |
| Akap12 | GTTGGGACCTTGAGACCAAA | TGGCACACTCATCTGTCCAT | 691 |
| C1ql3 | AGTGTGGTCCTTCACCTGGA | CAAGAACCAAAGCTGACACG | 507 |
| Cntn6 | GTATCTGTCCGAGAAGGTCA | GTGGACCTTGGACACTTCTA | 238 |
| Ephb1 | CACATCCATCTCCCTTTGCT | TCCAGAAACCCTTTCCCTCT | 618 |
| Efnb1 | ACAAGCCACACCAGGAAATC | TGGGGGCAGTAGTTGTTCTC | 640 |
| Efnb2 | ACCACACAGCTATGCAGCAG | CGAACTCCACGTCTTCTGGT | 675 |
| Efnb3 | GCAGTGTGGACATGATGGAC | GCACACTAAAGAGCGGGAAG | 675 |
| Kif26a | TCCTCAGCTCCAGACTCCAT | GCGACAGTCTTTCCATCTCC | 851 |
| Pappa2 | CCATTGTTCCACAAATGCTG | CTCGCTCCACATGTTGCTTA | 900 |
| Parm1 | TGCACTGACCAAGCCAGATA | ACAAACAGCAAGGCAGTGAA | 700 |
| Rgs16 | AAAAGGCTGTGTGTGTGGAAC | CTATCACTTCTGAGTCTTACCG | 752 |
| Rgs8 | TGAGGTCATGTTTGGGTTCA | CAGGCTCTACGGACTTCTGG | 741 |
| Tmem163 | AGGGTCTCTGCTTGACAGGA | CCCTACATGTTGGCACACAC | 650 |
| Tshz2 | GGCGAAGAGGACACAGACTC | AAGGAGCGCTGTCGATAAAA | 710 |
| Tmem117 | AACTATGCCACAACGGTGCT | TTGTAGACAGTGGGCTGTGC | 754 |
Neurosphere isolation and viral infection
Primary cortical progenitors of the dorsal telencephalon were isolated from E14.5 brains and grown as neurospheres in the presence of growth factors as previously reported65. Neurospheres were expanded for a maximum of one passage and then plated as a monolayer on dishes coated with poly-D-lysine (VWR) and laminin (BD). E14.5 pregnant dams were deeply anesthetized and the embryos removed. The presumptive somatosensory area was microdissected in cold Hanks’ medium and mechanically triturated to obtain a single-cell suspension. Cells were spun at 5,000g for 5 min at room temperature, and the pellet was resuspended in neurosphere medium, containing 100 U/ml penicillin/streptomycin, 200 mM L-glutamine, N2 and B27 supplements (Gibco), as well as 20 ng/ml epidermal growth factor (EGF; Sigma) and 20 ng/ml β-fibroblast growth factor (FGF; Millipore) in order to promote the survival of neural stem cells. After quantification of cell density, neural stem cells were plated in 200-ml tissue culture flasks (untreated or low-adherence, Corning) at 200,000 cells/flask and incubated at 37 °C with 5% CO2. The medium was changed every 3–4 d and the cells were expanded as neurospheres for one passage. Subsequently, cells were plated as a monolayer on dishes previously coated with poly-D-lysine (VWR) and laminin (BD). 3xFlag-Fezf2 and 3xFlag were cloned into pRETROX-IRES-ZsGreen1 retroviral vectors (Clontech Laboratories, Inc.) to generate high-titer, replication-incompetent, VSVg-coated retroviral particles that were packaged in 293T cells (MOI = 5). 12–18 h after plating, neural stem cells were infected with retroviruses. 16–203h after infection, cells were switched into fresh medium. Cells were collected 48 h after infection for chromatin immunoprecipitation (ChIP, n = 2) and for RNA-seq (n = 2). Sample sizes for ChIP-seq and RNA-seq experiments on neural progenitors complied with the ChIP-seq ENCODE guidelines66.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed on 1.5 × 108 neural stem cells per condition (3xFlag-Fezf2ZsGreen1 and 3xFlagZsGreen1, 2 replicates each) as previously described67. Briefly, cells were dissociated with 0.25% trypsin (Invitrogen), washed with PBS and chemically cross-linked with formaldehyde solution (1%). Cells were disrupted in lysis buffer (50 mM Tris, pH 8.0; 10 mM EDTA; 1% SDS; and protease inhibitors) for 20 min on ice and sonicated with a Bioruptor to shear the DNA into 200–700 bp fragments. Dynal magnetic beads (sheep anti-mouse M-28, Invitrogen) were preblocked with 5 μg of anti-Flag M2 antibody per reaction (Sigma-Aldrich) rotating overnight at 4 °C. Beads were then washed and resuspended in fresh 0.5% BSA in PBS. Bead-antibody complexes were incubated at 4 °C overnight with rotation. Beads were resuspended twice in low-salt immune complex wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.1; and 150 mM NaCl), incubated for 5 min at 4 °C with rotation and resuspended in LiCl immune complex wash buffer (0.25 M LiCl; 1% NP40; 1% deoxycholate; 1 mM EDTA; and 10 mM Tris-HCl, pH 8.1) twice and incubated for 5 min at 4 °C with rotation. Beads were then rinsed with ice-cold 10mM Tris-HCl; 1mM EDTA, pH8.0 and DNA was eluted in 100 μl of elution buffer (50mM Tris-HCl; 10mM EDTA; 1%SDS, pH 8.0) at 65 °C for 15 min. Reverse crosslinking was performed by incubating the ChIP DNA overnight at 65 °C. DNA was purified on QIAquick PCR purification columns (Qiagen) for use as template for Solexa library construction. To prepare an input control, whole-cell-extract DNA (reserved from the sonication step) was also treated by reverse crosslinking and purified.
ChIP-seq data analysis
For both ChIP-seq replicates, high-throughput sequencing reads were aligned to the mouse genome (version mm9) using Bowtie version 0.12.5 with options “-q --best --strata -m 1 -p 4 --chunkmbs 1024,” and only uniquely mapping reads were retained for further analysis. GEM was used to detect binding events, using the options “--top 2000 --k_min 7 --k_max 12 --a 20 --q 3 --mrc 1 - constant_model_range --d_l 1000 --d_r 1000 --v 2 --nf --refine_pwm --gc0 0.42.” Reported peaks contain a ChIP-seq enrichment level that is significantly greater than 1.5 times the scaled read count from the corresponding region in the control experiment (P < 10−6, binomial test, adjusted for multiple testing using Benjamini & Hochberg’s method). GPS (genome positioning system) was also used to confirm binding locations, using the options “--top 2000 --a 20 --q 3 --mrc 1 --constant_model_range.” Regions of ChIP enrichment (1 Kbp) are highlighted in red or blue, centered on the GEM-predicted binding event locations. De novo motif discovery was also performed using GEM, which jointly estimates binding locations and primary DNA motif patterns. Motif frequencies at binding events and gene promoter regions were defined by scanning 100-bp windows around GPS binding locations or annotated TSSs for matches to each motif. Motif scoring thresholds were based on a false discovery rate of 10−3, defined using a third-order Markov model of the mouse genome. Comparison between peaks and gene features was performed using the Ensembl v62 mouse genome annotation. Genes were defined as bound if a Fezf2 binding event was located within 5 kb of a gene’s TSS. A chi-squared test was used to assess the significance of associations between Fezf2 binding and groups of differentially regulated genes in vivo.
RNA-Seq library preparation
Total RNA was isolated using QIAzol (Qiagen)/chloroform extraction followed by spin-column purification (RNeasy mini kit, Qiagen) according to the manufacturer’s instructions. RNA concentration and purity were measured with Nanodrop (Thermo Fisher), and RNA integrity was assessed on a Bioanalyzer (Agilent) using the RNA 6000 Nano Total RNA kit (Agilent). High-quality RNA samples (RNA integrity number ≥ 8) were used for library preparation. Poly(A)+ RNA-seq libraries were constructed using the TruSeq RNA Sample Preparation Kit (Illumina) following the manufacturer’s protocol. 200 ng of total RNA from neurospheres was used as input for the TruSeq libraries. Before sequencing, libraries were run on a Bioanalyzer DNA7500 chip to assess purity, fragment size and concentration. Libraries free of adaptor dimers and with a peak region (220–500 bp) area ≥80% of the total area were sequenced. Individually barcoded samples were pooled and sequenced on the Illumina HiSeq 2500 platform.
RNA-Seq analysis
Paired-end 101-bp reads were aligned to the mouse (mm9) reference genome assembly using Tophat2 (refs. 68,69) with default options. Aligned reads and the UCSC reference transcriptome .gtf file (UCSC genome browser; knownGene track) were used as input for Cuffdiff2 (ref. 70) for expression quantification in fragments per kilobase RNA per million mapped reads (FPKM) and differential testing between conditions using default options (P < 0.001). Cummerbund v2.1 (http://compbio.mit.edu/cummerbund/) was then used to process, index and visualize the output of the Cuffdiff2 analyses.
To test for enrichment for CSMN signature genes after Fezf2 overexpression in cortical progenitors, we performed a preranked gene set enrichment analysis (GSEA). Briefly, CSMN signature genes were selected from Arlotta et al.54 as the intersection of genes flagged as ‘present’ (Affymetrix; RMA) with a fold-change of CPN/CSMN ≤1/3 from the P3 and P6 time point comparisons. Inversely, CPN signature genes were created from the intersection of genes with a fold-change of CPN/CSMN ≥3 from the same two time points. Additional gene sets consisting of the subset of each gene list that was also selected as Fezf2-bound from our ChIP-Seq study were created.
We next created a ranked gene list of all expressed genes (FPKM ≥ 1) from our 3xFlag-Fezf2ZsGreen1 versus 3xFlagZsGreen1 cortical progenitor RNA-Seq assay. Expressed genes were rank-ordered using the value of the Cuffdiff2 test statistic. All derived gene sets were tested for enrichment or depletion after Fezf2 overexpression using the preranked GSEA approach71. CSMN signature and CSMN signature plus Fezf2-bound gene sets were determined to be significant with a nominal P value ≤ 0.01.
Gene ontology enrichment analysis was performed on Fezf2-induced genes with specific expression in the cortical plate (as determined by Ayoub et al.12) and Fezf2-repressed genes with specific expression in either the VZ or SVZ/IZ using DAVID72. Genes within each gene set were selected using a threshold specificity score S ≥ 0.65 (described below) for any given condition. Gene sets were tested for significant enrichment using the GO Biological Process FAT collection of gene sets.
We defined the specificity score Sgi for a gene (g) in a condition (i) as follows:
where JSD is the Jensen-Shannon divergence between pg, the expression profile of a gene g across all conditions expressed as a probability distribution, and qi, the unit vector of perfect expression in condition i. Where applicable, genes are specifically assigned to the condition with the highest S score above a threshold value. The threshold for specificity was selected empirically to be 0.65 through examination of the distribution of S scores across all genes.
Hypergeometric and bootstrap analyses
To determine enrichment for Fezf2-bound genes within the set of significantly differentially regulated genes in the E14.5 cortical neurospheres, we employed a hypergeometric test (phyper from the stats package in R) and a bootstrap method. For the bootstrap method, gene sets of comparable size were sampled (without replacement) from the universe of genes 10,000 times, and P values were estimated as the fraction of iterations where a greater than or equal number of Fezf2-bound genes were selected.
Derivation and differentiation of inducible Fezf2 and GFP mouse ES lines
Mouse Fezf2-IRES-GFP was PCR-cloned from the Fezf2GFP construct5 into P2lox-40 using the Gateway system (Life Technologies). The control GFP construct was a gift from E. Mazzoni73. The plasmid constructs were nucleofected into the A2loxCRE mouse ES line for inducible cassette exchange (ICE) as described in Iacovino et al.74 The cells were selected on DR4 mouse embryo fibroblasts (GlobalStem) with 300 μg/ml G418. Single inducible Fezf2-IRES-GFP (iFIG) clones and inducible GFP (iGFP) clones were manually picked and expanded in standard ES+LIF medium without doxycycline (15% FBS, 1× Glutamax, 1× non-essential amino acids, 1× penicillin/streptomycin and 1,000 U/ml mLIF (ESGROW, Millipore) in Knockout DMEM). All media were obtained from GIBCO BRL. The iFIG and iGFP clones were differentiated in vitro into cortical neurons using the SFEBq protocol as described in Eiraku et al.75 Expression of Fezf2 and GFP was induced at day 10 in vitro with 2 μg/ml doxycycline. The doxycycline was maintained in the culture until day 18, and the SFEBqs were dissociated and FACS-sorted for GFP+PI− populations, which were then analyzed by RNA-seq.
Electrophoretic mobility shift assay (EMSA)
GST-tagged truncated mouse Fezf2 (GST-tFezf2, AA291-455) protein was expressed in Rossetta DE3 E. coli (Millipore) and purified on GST-agarose with standard methods. Given that the full-length Fezf2 aggregates in solution, we chose a truncated version of Fezf2 (tFezf2) containing five of the six zinc finger motifs in the C-terminal domain of Fezf2, highly conserved across all vertebrates. IRDye end-labeled oligonucleotide probes (Integrated DNA Technologies) were designed against proximal promoter regions under ChIP-seq peaks of the Ascl1 and Ephb1 genes. Negative controls for each gene were chosen from neighboring sequences not containing ChIP-seq peaks. The probe sequences are as follows (with GEM motifs underlined):
Ephb1 positive probe:
GCCGAGCCCCAGCGGAGACGCGCCGCGTCCCAGGGCGCCGCTGCGCTCCCGGCGGGTGGCTAGCCACCCGCCGGGAGCGCAGCGGCGCCCTGGGACGCGGCGCGTCTCCGCTGGGGCTCGGC.
Ephb1 negative probe:
TGTCAGAAGACAGTCATAATGGGAAGACATGAAGAAACAGCCGAAAGTCTGACAACTTGTTAACAAGTTGTCAGACTTTCGGCTGTTTCTTCATGTCTTCCCATTATGACTGTCTTCTGACA.
Ascl1 positive probe:
GCTCTGAGCTGCCGCGGCCGCCGCCGCTGCCGCCGCCGCCGCGGTCGCAAAGAAGCAGGCGCCTGCTTCTTTGCGACCGCGGCGGCGGCGGCAGCGGCGGCGGCCGCGGCAGCTCAGAGC.
Ascl1 negative probe:
CCTAGTGGTTATTTTATTGCTGTAAAATAAATTTAACCCTTTCCTTACCAAGCTGCTTTTAAAAGCAGCTTGGTAAGGAAAGGGTTAAATTTATTTTACAGCAATAAAATAACCACTAGG.
EMSA was performed using the Odyssey EMSA Kit (LI-COR Biosciences, USA), following the manufacturer’s protocols. Briefly, 100 nM IRDye end-labeled probe was incubated with 0, 10 or 20 μg GST-tFezf2 at room temperature for 30 min in 20 μl reaction buffer (1× binding buffer, 0.2 mM DTT, 0.25% Tween 20, 0.05 μg/μl poly(dI.dC) and 5 mM MgCl2). The reactions were resolved on 4–20% Mini-PROTEAN TBE precast gel (Bio-Rad) and visualized by the Odyssey CLx infrared imaging system (LI-COR Bioscience, USA).
Statistics
Bar and line graphs represent mean values of all replicates and error bars represent s.e.m. except for RNA-seq results, in which the error bars represent 95% confidence intervals for the Cuffdiff2 model expression estimate. Animals were assigned to groups on the basis of genotype. Age-matched littermates were used as controls in all experiments. No randomization was used. For all animal experiments, data distribution was assumed to be normal, but this was not formally tested. For ISH and immunofluorescence studies, data collection and analysis were performed blinded to the conditions of the experiment. No animals were excluded from analyses. Except for RNA-seq and ChIP-seq sample sizes (explained above), no statistical methods were used to predetermine sample sizes. Our sample sizes are similar to those reported in previous publications3.
Supplementary Material
Acknowledgments
We would like to thank A. McMahon and J. Macklis for insightful advice in the early stages of the work; S. McConnell for her generous sharing of the Fezf2::PLAP line; J. Macklis (Harvard University), R. Hevner (Seattle Children’s Research Institute), A. Catic (Massachusetts General Hospital) and S. Sykes (Massachusetts General Hospital) for sharing of antibodies and expression vectors; C. O’Donnell and D. Cacchiarelli for discussions on ChIP-seq analysis.; Z. Trayes-Gibson, E. Stronge and A. Iannone for outstanding technical support and for reading the manuscript; and A. Goodwin for careful editing of the manuscript and comments. This work was supported by grants from the US National Institute of Health (NS062849, MH101268 to P.A.), (HD078561 and HD069001 to E.T.), the New York Stem Cell Foundation and the Harvard Stem Cell Institute to P.A. P.A. is a New York Stem Cell Foundation-Robertson Investigator.
Footnotes
AUTHOR CONTRIBUTIONS
S.L. and P.A. conceived the work, designed the experiments, analyzed the data and wrote the manuscript. S.L. performed the majority of the experiments. B.J.M. contributed to experimental design and performed microarray analysis. E.Z. performed the ChIP-seq experiments. L.A.G. analyzed the RNA-seq data. H.-H.C. performed the in vitro differentiation experiment and assisted in manuscript preparation. W.Y. performed the electrophoretic mobility shift assay experiment. A.M. assisted with FACS purification and the microarray experiments. E.T. performed the HARDI analysis. S.M. analyzed the ChIP-seq data. D.K.G. and J.L.R supervised the bioinformatics analyses. P.A. supervised all aspects of the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. GEO: raw and processed ChIP-seq data, GSE46707; RNA-seq data, GSE56446; microarray data, GSE56451. Mapped ChIP-seq data can be browsed at http://genome.ucsc.edu/cgibin/hgTracks?db=mm9&hgt.customText=http://nanog.csail.mit.edu/csmn/arlotta_fezf2.csmn.tracks&position=chr9:101716317-102365165.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 2.Migliore M, Shepherd GM. Opinion: an integrated approach to classifying neuronal phenotypes. Nat Rev Neurosci. 2005;6:810–818. doi: 10.1038/nrn1769. [DOI] [PubMed] [Google Scholar]
- 3.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 5.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat Neurosci. 2010;13:1345–1347. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat Cell Biol. 2013;15:214–221. doi: 10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De la Rossa A, et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat Neurosci. 2013;16:193–200. doi: 10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- 11.Lodato S, et al. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayoub AE, et al. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalczyk T, et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco SJ, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molyneaux BJ, et al. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLOS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Zheng J, Yang N, Li H, Guo S. Genomic selection identifies vertebrate transcription factor Fezf2 binding sites and target genes. J Biol Chem. 2011;286:18641–18649. doi: 10.1074/jbc.M111.236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Hu S, Blackshaw S, Zhu H, Qian J. hPDI: a database of experimental human protein-DNA interactions. Bioinformatics. 2010;26:287–289. doi: 10.1093/bioinformatics/btp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fremeau RT, et al. The expression of vesicular glutamate transporters defines two classes of excitatory synapse. Neuron. 2001;31:247–260. doi: 10.1016/s0896-6273(01)00344-0. [DOI] [PubMed] [Google Scholar]
- 23.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Canty AJ, Murphy M. Molecular mechanisms of axon guidance in the developing corticospinal tract. Prog Neurobiol. 2008;85:214–235. doi: 10.1016/j.pneurobio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron. 2008;60:1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Lee R, Petros TJ, Mason CA. Zic2 regulates retinal ganglion cell axon avoidance of ephrinB2 through inducing expression of the guidance receptor EphB1. J Neurosci. 2008;28:5910–5919. doi: 10.1523/JNEUROSCI.0632-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petros TJ, Shrestha BR, Mason C. Specificity and sufficiency of EphB1 in driving the ipsilateral retinal projection. J Neurosci. 2009;29:3463–3474. doi: 10.1523/JNEUROSCI.5655-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jouandet MLM, Lachat JJJ, Garey LJL. Distribution of the neurons of origin of the great cerebral commissures in the cat. Anat Embryol (Berl ) 1985;171:105–120. doi: 10.1007/BF00319060. [DOI] [PubMed] [Google Scholar]
- 33.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi E, et al. Developing neocortex organization and connectivity in cats revealed by direct correlation of diffusion tractography and histology. Cereb Cortex. 2011;21:200–211. doi: 10.1093/cercor/bhq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai T, et al. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci USA. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirata T, et al. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- 42.Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG. ‘Seq-ing’ insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623. doi: 10.1016/j.neuron.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 44.Cochella L, Hobert O. Diverse functions of microRNAs in nervous system development. Curr Top Dev Biol. 2012;99:115–143. doi: 10.1016/B978-0-12-387038-4.00005-7. [DOI] [PubMed] [Google Scholar]
- 45.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 46.García-Bellido AA. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;0:161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- 47.Williams SE, et al. Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron. 2003;39:919–935. doi: 10.1016/j.neuron.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 48.García-Frigola C, Herrera E. Zic2 regulates the expression of Sert to modulate eye-specific refinement at the visual targets. EMBO J. 2010;29:3170–3183. doi: 10.1038/emboj.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robichaux MA, et al. EphB receptor forward signaling regulates area-specific reciprocal thalamic and cortical axon pathfinding. Proc Natl Acad Sci U S A. 2014;111:2188–2193. doi: 10.1073/pnas.1324215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 52.Catapano LA, Arnold MW, Perez FA, Macklis JD. Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J Neurosci. 2001;21:8863–8872. doi: 10.1523/JNEUROSCI.21-22-08863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett MT, et al. High-quality RNA and DNA from flow cytometrically sorted human epithelial cells and tissues. Biotechniques. 2002;32:888–896. doi: 10.2144/02324rr06. [DOI] [PubMed] [Google Scholar]
- 54.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 55.Eberwine J, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng L, et al. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 57.Wu FX, Zhang WJ, Kusalik AJ. Modeling gene expression from microarray expression data with state-space equations. Pac Symp Biocomput. 2004;2004:581–592. [PubMed] [Google Scholar]
- 58.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosen GD, et al. Bilateral subcortical heterotopia with partial callosal agenesis in a mouse mutant. Cereb Cortex. 2013;23:859–872. doi: 10.1093/cercor/bhs080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi E, et al. Development of cerebral fiber pathways in cats revealed by diffusion spectrum imaging. Neuroimage. 2010;49:1231–1240. doi: 10.1016/j.neuroimage.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodato S, et al. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. J Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinrich C, et al. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat Protoc. 2011;6:214–228. doi: 10.1038/nprot.2010.188. [DOI] [PubMed] [Google Scholar]
- 66.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang DW, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 73.Mazzoni EO, et al. Embryonic stem cell–based mapping of developmental transcriptional programs. Nat Methods. 2011;8:1056–1058. doi: 10.1038/nmeth.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iacovino M, et al. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells. 2011;29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.