Abstract
To elucidate the effect of codon optimization and chaperone coexpression on the heterologous expression of mammalian cytochrome P450s in Escherichia coli, the expression of P450s 2B1, 2S1, 2U1, 2W1, and 27C1 were investigated. With codon optimization for N-terminus or the entire gene, the expression levels of P450 27C1, 2U1 and 2W1 increased 22-fold, 3.6-fold and 2.1-fold respectively, while those for P450s 2B1 and 2S1 remained unchanged. With coexpression of E. coli molecular chaperones GroEL/ES, the expression level increased up to 14-fold for P450 27C1, and 3- to 5-fold for P450s 2B1, 2S1, and 2W1. Simultaneous application of these two techniques resulted in synergetic effects.
Keywords: Chaperon, Codon, Cytochrome P450, Escherichia coli, Expression
Introduction
Cytochrome P450 (P450) superfamily is one of the most important groups of oxygenases (Ortiz de Montellano 2005). They are involved in various biological pathways, and the oxidation of numerous xenobiotics including a large fraction of drugs on the market (Guengerich 2005). Therefore, a substantial amount of active forms of mammalian cytochrome P450 enzymes are demanded for current drug metabolism and toxicity research. Bacterial expression systems have proven to be a valuable tool for that purpose because of their ease of use, low cost, and high protein production, and are chosen to produce a few commercially available P450s by main manufacturers such as New England Biolabs. Ever since the successful expression of active bovine P450 17A1 in E. coli in 1991 (Barnes et al. 1991), a variety of strategies have been established. However, functional expression in E. coli remains a challenge for some P450s, especially those of mammalian origins, which are all membrane-bound proteins.
It is well known that E. coli and mammalian codon preference display considerable divergence (Gustafsson et al. 2004). Codon bias has been identified as most highly associated with prokaryotic gene expression (Lithwick and Margalit 2003) and a very recent research links it to global translation efficiency and cellular fitness (Kudla et al. 2009). In our previous work on human orphan P450s, the utilization of codon optimization technique has been introduced, resulting in the successful heterologous expression of several P450s in E. coli (Wu et al. 2006a; Wu et al. 2006b; Stark et al. 2008a; Stark et al. 2008b). It is however unclear whether or to what extent this technique contributes to the expression process. We herein present a direct comparison of the expressions of several codon-optimized mammalian P450s with the same enzymes coded with original codons. The effects of coexpression of E. coli molecular chaperone GroEL and its co-chaperone GroES (GroEL/ES), introduced recently to mammalian P450 expressions (Yun et al. 2006), was also investigated. The chaperone is known to support the de novo protein folding by cycles of binding and release of the substrates after their release from ribosome (Mogk et al. 2002), therefore facilitates functional heterologous expression. Synergetic effects were observed with both techniques applied.
Materials and methods
Materials
The expression vector of P450s 2S1 with original codons (2S1org), and 2W1 & 27C1 with optimized codons (2W1opt & 27C1opt) were constructed in previous work with N-terminal modification (Wu et al. 2006a; Wu et al. 2006b). The cDNAs of P450s 2B1 (2B1org) and 2W1 with original codons (2W1org) were gifts from P. F. Hollenberg (Univ. Michigan, Ann Arbor) and M. Ingelman-Sundberg (Karolinska Institute, Stockholm) respectively. Oligonucleotides for cDNA synthesis and N-terminal modifications were purchased from Operon (Huntsville, AL) with salt-free quality and used directly without further purification.
PCR-based cDNA synthesis
The cDNAs of P450s 27C1 with original codons (27C1org) and 2B1 with codon optimized (2B1opt) to suit the codon preference bias of E. coli were synthesized using PCR-based oligonucleotide assembly as described previously (Wu et al. 2006a). The oligonucleotides were designed online using DNA Works program (http://helixweb.nih.gov/dnaworks/, see Supplementary Information) (Hoover and Lubkowski 2002). Correctly assembled cDNAs were constructed into a pCW vector for expression.
Construction of N-terminal codon-optimized vectors of P450s 2S1 and 2U1
Sequential site-directed mutagenesis was conducted using the QuickChange® mutagenesis kit and the supplier’s protocol (Stratagene). The primers were designed so that the mutated position was close to the middle with ~10–15 bases of template-complementary sequence on both sides (see Supplementary Information). For practical purpose, a pBluescript SK (+) vector of P450s 2S1 and 2U1 were first constructed and subjected to site-directed mutagenesis (Wu et al. 2005). The 1.5 kb mutated fragment was subcloned into the pCW vector using the NdeI and XbaI restriction sites. Two and three N-terminal regions (Fig. 1) with contiguous rare codons were mutated for P450s 2S1 and 2U1 respectively.
Fig. 1.

N-terminal regions of P450s 2S1 and 2U1 mutated to fit the codon bias of E. coli. The Frequency value represents the codon usage frequency fraction for the corresponding amino acid in E. coli K12 (http://openwetware.org/wiki/Escherichia_coli/Codon_usage).
N-Terminal modifications for P450s 2B1 and 2W1
N-Terminal mutations were introduced into the native constructs by PCR-based mutagenesis. The cDNAs of P450s 2B1org, 2W1org and 2B1opt were amplified between the NdeI and the XbaI sites using 5′-PCR primers containing the desired mutations (see Supplementary Information). PfuUltra™ High-Fidelity DNA polymerase was used for the PCR amplification at an annealing temperature of 55 °C. The products were purified, double-digested, and ligated with the monocistronic pCW vector. The modifications were confirmed by sequencing the ORF region of the new constructs.
Bacterial Expression of Mammalian P450s
Both plasmids pGro12 ES/EL encoding the chaperone protein GroEL/ES (Nishihara et al. 1998) and each of the tested constructs of mammalian P450s were transformed into E. coli DH5α competent cells. Single colonies were grown overnight in LB media fortified with 100 μg ampicillin/ml and 50 μg kanamycin/ml, at 37 °C with gyrorotary shaking at 250 rpm; 0.5 ml of each overnight culture was then inoculated into 50 ml of TB broth containing the same antibiotics in a 250 ml flask. The cultures were incubated at 37 °C with gyrorotary shaking at 220 rpm for 4 h on an INFORS Multitron shaker; induction of P450s and GroEL/ES transcription were then initiated with the addition of 1.0 mM IPTG and 1 mg arabinose/ml, respectively. The incubation continued at 27 °C with gyrorotary shaking at 200 rpm. Three sets of side by side experiments were performed for each sample.
Measurement of P450 concentrations
P450 concentrations were estimated using the reduced-CO versus reduced difference spectrum as described by Omura and Sato (Omura and Sato 1964). Cells harvested from 1.5 ml culture were resuspended in 3 ml potassium phosphate buffer (0.1 M, pH 7.4) containing 20% (v/v) glycerol and 0.2% (v/v) Emulgen 913. With the sample cuvette saturated with bubbles of CO, spectra from 400–500 nm were recorded until the 450-nm peak reached a maximum. P450 content is determined as follows: P450 (nmol/l) = [(A450-A490) baseline/0.091. Absorbance at 420 nm represents denatured forms of P450.
Results and Discussion
In the context of our series work on human P450s, bacterial expression system has been extensively used. This work was undertaken to shed light on the effect of codon optimization and chaperone coexpression. The active expression of mammalian P450s 2B1 (rat), 2S1 (human), 2U1 (human), 2W1 (human), and 27C1 (human) were investigated. In all cases, the N-terminal hydrophobic sequences were truncated or replaced before the well-conserved proline-rich region based on reported examples on successful expressions of P450s. The codon usage of P450s 2B1, 2W1 and 27C1 were optimized by resynthesizing the entire gene. For 2S1 and 2U1, only N-Terminal-contiguous rare codons were replaced (Fig. 1) since codon adaptation near the 5′ terminus is considered particular important (Gonzalez de Valdivia and Isaksson 2004).
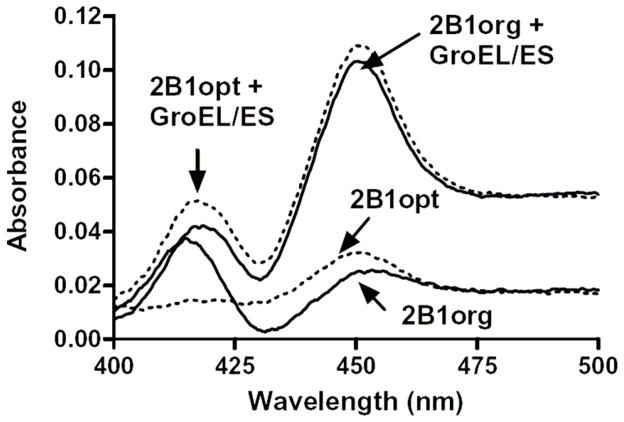
The results are given in Table 1 and typical CO differential spectra of P450 2B1 is shown in Figure 2. Both codon optimization and chaperone coexpression showed positive effects on the expression of active P450s in general, albeit to different extents. With codon optimization, the expression of active enzymes increased 22-fold for P450 27C1, 3.6-fold for P450 2U1, and 2.1-fold for P450 2W1, but remained practically unchanged for P450s 2B1 and 2S1. Coexpression of a molecular chaperone GroEL/ES brought the expression level up 14-fold for P450 27C1 and 3- to 5-fold for P450s 2B1, 2S1, and 2W1 (with or without codon optimization).
Table 1.
Comparison of functional expression of mammalian P450s in E. coli
| Entry | Mammalian P450s | Coexpression with GroEL/ES | P450 Yield (nmol/l)
|
||
|---|---|---|---|---|---|
| original | optimized | fold | |||
| 1 | 2B1 | Yes | 1180 | 1200 | 1.0 |
| 2 | 2B1 | No | 230 | 360 | 1.5 |
| 3 | 2W1 | Yes | 840 | 1800 | 2.0 |
| 4 | 2W1 | No | 230 | 350 | 1.5 |
| 5 | 2S1a | Yes | 600 | 670 | 1.1 |
| 6 | 2S1a | No | 120 | 120 | 1. |
| 7 | 2U1a | Yes | 50 | 180 | 3.6 |
| 8 | 27C1 | Yes | 60 | 1300 | 22 |
| 9 | 27C1 | No | 20 | 90 | 4.5 |
Partially optimized
Fig. 2.
Expression of P450s 2B1org and 2B1opt with or without chaperone coexpression.
During the process of constructing N-terminal optimized P450 2U1 vector, the expression of constructs with one or two regions optimized was tested. Construct with only the first region (PGPTP) optimized (Fig.1) showed 2-fold increase in active expression level, while those with the second (RRR) or third (RTR) region optimized (Fig.1) showed 3.6-fold increase, same as what was achieved later with all three regions modified. Although no synergetic effect was observed in this case, the replacement of N-terminal contiguous rare codons, especially those encoding arginine residues, showed a promising and convenient strategy to improve heterologous expression of mammalian proteins.
According to the nucleotide analysis from web server Optimizer (http://genomes.urv.es/optimizer/), the codon adaptation index (CAI) of 2B1opt, 2W1opt and 27C1opt increased similar amount compared to the original ones (see Supplementary Information), but their expression increments varied dramatically. It is well known that heterologous expression of proteins is effected by a variety of factors associated with both transcriptional and translational issues, and to produce functional proteins, more factors are involved. Codon bias adjustment and molecular chaperone coexpression only contribute to part of the process. Our results demonstrate the general enhancement effect of the two techniques on the bacterial expression of mammalian P450s. They are both easy to perform since both de novo gene synthesis service and some chaperone plasmids are commercially available, and in case of partial codon optimization, sequential site-directed mutagenesis would be a very straightforward approach. Taken together, the results suggest that these two techniques, plus the use of other available candidate chaperones (Yun et al. 2006), provide a feasible way to improve functional protein expression, and could be extended to the optimal expression of other mammalian P450s.
Supplementary Material
Acknowledgments
This work was supported in part by 100 Talents Program of the Chinese Academy of Sciences, the Provincial Sci & Tech Foundation for Young Scholars of Sichuan, China 08ZQ026-023, National Natural Science Foundation of China 20802073/B020104 (Z.-L.W.), and USPHS R37 CA90426 and P30 ES000267 (F.P.G.).
Footnotes
Electronic supplementary material The online version of this article (doi:) contains supplementary material, which is available to authorized users.
Contributor Information
Zhong-Liu Wu, Email: wuzhl@cib.ac.cn, Chengdu Institute of Biology, Chinese Academy of Sciences, P.O.Box 416, Chengdu 610041, China.
Jing Qiao, Chengdu Institute of Biology, Chinese Academy of Sciences, P.O.Box 416, Chengdu 610041, China. Graduate School of the Chinese Academy of Sciences, Beijing 100039, China.
Zhi-Gang Zhang, Chengdu Institute of Biology, Chinese Academy of Sciences, P.O.Box 416, Chengdu 610041, China. Graduate School of the Chinese Academy of Sciences, Beijing 100039, China.
F. Peter Guengerich, Department of Biochemistry, Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, TN, USA, 37232-0146.
Yan Liu, Chengdu Institute of Biology, Chinese Academy of Sciences, P.O.Box 416, Chengdu 610041, China.
Xiao-Qiong Pei, Chengdu Institute of Biology, Chinese Academy of Sciences, P.O.Box 416, Chengdu 610041, China.
References
- Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez de Valdivia EI, Isaksson LA. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, mechanism and biochemisty. 3. Kluwer Press; New York: 2005. pp. 377–531. [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. DNA Works: An automated method for designing oligonucleotides for PCR-based gene synthesis. Nucl Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithwick G, Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003;13:2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Mayer MP, Deuerling E. Mechanisms of protein folding: Molecular chaperones and their application in biotechnology. Chembiochem. 2002;3:807–814. doi: 10.1002/1439-7633(20020902)3:9<807::AID-CBIC807>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. Chaperone coexpression plasmids: Differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol. 1998;64:1694–1699. doi: 10.1128/aem.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- Ortiz de Montellano PR, editor. Cytochrome P450s: Structure, mechanism, and biochemistry. Kluwer Press; New York Stark: 2005. [Google Scholar]
- K, Dostalek M, Guengerich FP. Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J. 2008a;275:3706–3717. doi: 10.1111/j.1742-4658.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Wu ZL, Bartleson CJ, Guengerich FP. mRNA distribution and heterologous expression of orphan cytochrome P450 20A1. Drug Metab Dispos. 2008b;36:1930–1937. doi: 10.1124/dmd.108.022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-L, Bartleson CJ, Ham AJ, Guengerich FP. Heterologous expression, purification, and properties of human cytochrome P450 27C1. Arch Biochem Biophys. 2006a;445:138–146. doi: 10.1016/j.abb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Wu Z-L, Podust LM, Guengerich FP. Expansion of substrate specificity of cytochrome P450 2A6 by random and site-directed mutagenesis. J Biol Chem. 2005;280:41090–41100. doi: 10.1074/jbc.M508182200. [DOI] [PubMed] [Google Scholar]
- Wu Z-L, Sohl CD, Shimada T, Guengerich FP. Recombinant enzymes overexpressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol Pharmacol. 2006b;69:2007–2014. doi: 10.1124/mol.106.023648. [DOI] [PubMed] [Google Scholar]
- Yun CH, Yim SK, Kim DH, Ahn T. Functional expression of human cytochrome P450 enzymes in Escherichia coli. Curr Drug Metab. 2006;7:411–429. doi: 10.2174/138920006776873472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.