Abstract
Faithful repair of DNA double-strand breaks by homologous recombination is crucial to maintain functional genomes. The major Escherichia coli pathway of DNA break repair requires RecBCD enzyme, a complex protein machine with multiple activities. Upon encountering a Chi recombination hotspot (5′ GCTGGTGG 3′) during DNA unwinding, RecBCD’s unwinding, nuclease, and RecA-loading activities change dramatically, but the physical basis for these changes is unknown. Here, we identify, during RecBCD’s DNA unwinding, two Chi-stimulated conformational changes involving RecC. One produced a marked, long-lasting, Chi-dependent increase in protease sensitivity of a small patch, near the Chi recognition domain, on the solvent-exposed RecC surface. The other change was identified by crosslinking of an artificial amino acid inserted in this RecC patch to RecB. Small-angle X-ray scattering analysis confirmed a major conformational change upon binding of DNA to the enzyme and is consistent with two changes. We propose that, upon DNA binding, the RecB nuclease domain swings from one side of RecC to the other; when RecBCD encounters Chi, the nuclease domain returns to its initial position determined by crystallography, where it nicks DNA exiting from RecC and loads RecA onto the newly generated 3′-ended single-stranded DNA during continued unwinding; a crevice between RecB and RecC increasingly narrows during these steps. This model provides a physical basis for the intramolecular “signal transduction” from Chi to RecC to RecD to RecB inferred previously from genetic and enzymatic analyses, and it accounts for the enzymatic changes that accompany Chi’s stimulation of recombination.
Keywords: helicase–nuclease, recombination, limited proteolysis, SAXS, crosslinking
Introduction
Homologous genetic recombination catalyzes the reassortment of genetic markers that promote diversity and faithfully repairs DNA double-strand breaks in a chromosome by use of undamaged homologous DNA as template. Such reactions involve a series of complex interactions between duplex DNA substrates and the enzymes that act upon them. The major proteins involved in the early steps of bacterial recombination are the RecBCD multi-subunit nuclease–helicase enzyme and the RecA DNA strand-exchange protein. The RecBCD enzyme, or the functionally similar AddAB enzyme, is present in most bacterial species investigated [1].
The Escherichia coli RecBCD enzyme, whose structure in the DNA-bound form is shown in Fig. 1a, has ATP-dependent exonuclease and helicase activities, both of which are modulated by the specific DNA sequence 5′ GCTGGTGG 3′, also knownas Chi hotspots of recombination, encountered during un- winding of duplex DNA [2]. The enzyme has separate 3′ → 5′ and 5′ → 3′ helicase activities, in the RecB and RecD subunits, respectively, and a single nuclease domain in a separate domain of RecB, which can attack either 3′ or 5′ terminated strands during unwinding. The RecC subunit binds the RecB and RecD subunits, serves as a conduit for both DNA strands during unwinding, and recognizes Chi while the 3′ terminated unwound strand passes through RecC from the RecB helicase domain to the RecB nuclease domain.
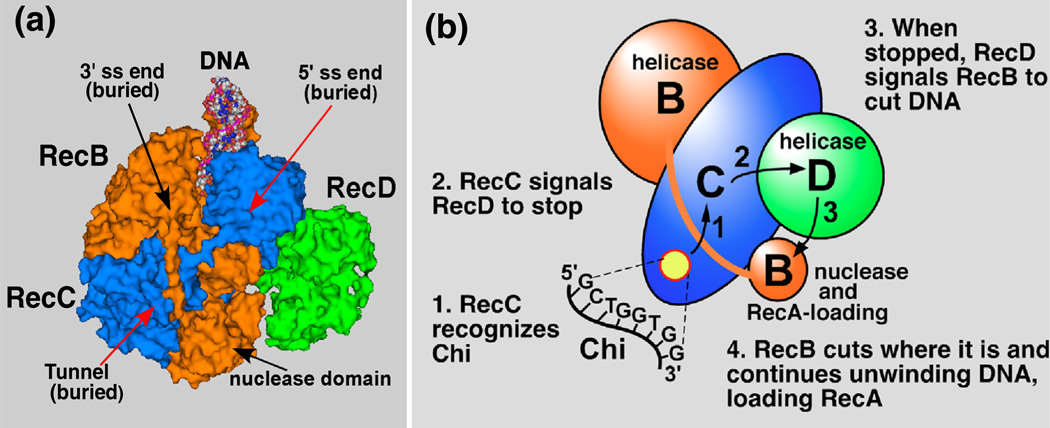
Fig. 1.
Structure of RecBCD enzyme bound to DNA and a “signal transduction” model for the Chi-dependent alteration of RecBCD enzyme. (a) The crystal structure of RecBCD bound to hairpin-shaped DNA (PDB entry 1W36) [18]. The RecB polypeptide is orange, RecC is blue, and RecD is green. In this structure, the 3′-ended strand would encounter the RecB nuclease domain upon exiting the RecC tunnel, in which Chi is likely recognized. (b) A “signal transduction” model for the Chi-dependent change of RecBCD [17]. When Chi is in the RecC tunnel (yellow disk), it prompts RecC to signal RecD to stop unwinding, which in turn signals RecB to nick the 3′-ended strand of DNA near Chi and to begin loading RecA.
The reaction of RecBCD enzyme on double-stranded (ds) DNA is initiated by the tight binding of an enzyme molecule to a DNA end, with the 3′-ended strand bound to the RecB helicase and the 5′-ended strand threaded through the RecC protein and bound to the RecD helicase (Fig. 1a) [3]. In the presence of ATP and Mg2+ ions, RecBCD rapidly unwinds the DNA. Because the RecD helicase is faster than the RecB helicase [4], a single-stranded (ss) loop accumulates on the 3′-ended strand and grows and moves along the DNA [5]. When RecBCD meets the Chi hotspot sequence 5′ GCTGGTGG 3′ on the 3′-ended strand [6,7], the activities of the enzyme are markedly changed. Under conditions with ATP in excess over Mg2+ ions, the RecB nuclease domain nicks the 3′-ended strand a few nucleotides to the 3′ side of Chi [8]. Under conditions with Mg2+ ions in excess over ATP, the nuclease switches from endonucleolytically nicking primarily the 3′-ended strand to nicking primarily the 5′-ended strand [9–11], and the enzyme begins loading the DNA strand-exchange protein RecA onto the 3′-ended ss DNA tail with Chi near its end [12]. At least under the former condition, the enzyme loses the ability to nick at a subsequently encountered Chi site [13], and later (probably at the end of the DNA), the three subunits disassemble and the enzyme remains inactive for an hour or more [14]. The RecA– ss DNA filament can pair with intact homologous DNA, and exchange of strands forms a D-loop [12], which can be further processed to form intact recombinant DNA [15,2].
Control by Chi of the RecBCD helicase, nuclease, and RecA-loading activities is critical for efficient recombination, as witnessed by the strong enhance- ment of recombination promoted by a single Chi site [16]. How these activities are regulated has remained a major unsolved problem in recombina- tion and DNA break repair. Understanding of the roles of the multiple subunits and activities of RecBCD enzyme of E. coli has been greatly aided by the in vivo and in vitro phenotypes of mutations altering the subunits of the enzyme. The properties of a special class of RecB mutants (in the helicase domain) allowed us to build on the enzymatic and physical properties of RecBCD enzyme and to propose a specific intramolecular signal transduction model for Chi’s regulation of the enzyme [17]. In that model (Fig. 1b), the 3′-ended strand passes from the RecB helicase into a tunnel in RecC, readily visible in the crystal structure of the enzyme bound to DNA, and emerges in the vicinity of the RecB nuclease domain [18,19]. When RecC engages the Chi sequence, RecC signals RecD to stop unwinding. This change in turn prompts RecD to signal RecB’s nuclease domain to nick the DNA near Chi and to begin loading RecA. Although this model accounts for many aspects of the change in RecBCD‧s activities, the physical change responsible for the enzymatic changes has been unknown. Because the Chi-dependent enzymatic changes occur with purified components (RecBCD, DNA, ATP, Mg2+, and buffer components; i.e., without additional enzymes), we hypothesized that the change is a conformational alteration in the RecBCD subunits, as opposed to regulation by other factors.
We report here multiple lines of evidence for conformational changes involving the RecC subunit of the enzyme and direct evidence that movement of part of RecB or RecC, relative to each other, is a Chi-mediated change. These results both provide evidence for our signal transduction model and demonstrate a conformational change in RecBCD enzyme upon meeting a Chi hotspot.
Results
Experimental design
We seek to characterize the conformational changes in RecBCD enzyme during the enzyme’s rapid (up to 1000 bp/s) unwinding of duplex DNA. We hypothesize these conformational changes to be the proximal cause of the profound changes in the activity of the enzyme that result from its interaction with a Chi sequence. The E. coli Chi sequence, 5′ GCTGGTGG 3′ [6], is recognized as a single strand [7] and recognized only if the enzyme approaches the abovementioned sequence from the 3′ side [8]. These observations, all consistent with the Chi sequence being “read” during the passage of a single strand through the tunnel in RecC [20–23], greatly constrain the design of physical assays of the hypothesized conformational change because the enzyme disassembles on exiting DNA after encountering a Chi site [14].
We used bacteriophage lambda DNA, naturally devoid of Chi sequences, as a control substrate, for comparison with lambda DNA bearing a X+D sequence 3.5 kb from the conventional right end of lambda (see Fig. 3e) [24]. RecBCD enzyme encounters that Chi, in an active orientation, a few seconds after entering the lambda DNA from the right end. RecBCD enzyme continues to unwind DNA after recognizing Chi [25] albeit at a reduced unwinding rate [26], with very high processivity [27], allowing us a minute or so to examine the change in the enzyme before it translocates off the distal end of the DNA. We have previously shown that, under the reaction conditions used here, Chi causes the enzyme to undergo a long-lasting disassembly as it exits the DNA [14]. We report here two types of experiments, using such lambda DNA substrates, which demonstrate conformational changes in the enzyme in response to binding DNA and to acting at Chi.
Fig. 3.
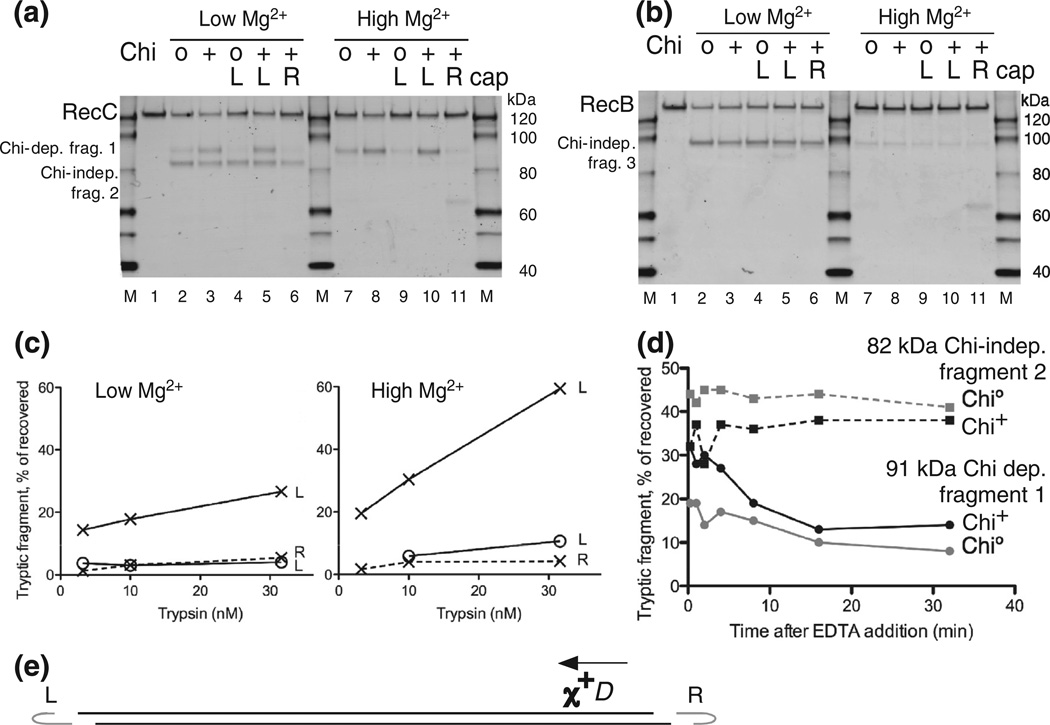
Further characterization of the Chi-dependent protease cleavage of RecC. (a) RecBCD enzyme was reacted with uncapped lambda Chi0 or Chi+ DNA, as in Fig. 2, or with lambda substrates capped at the left or right end of lambda, marked L and R, to block RecBCD entry at the capped end. Chi is active only if RecBCD enzyme enters from the right end [8]. Reactions were with 5 mM ATP and 2.5 mM Mg2+ (low Mg2+) or 8 mM Mg2+ (high Mg2+), as shown, and reacted with 10 nM trypsin for 1 min before Western blot analysis using RecC-specific polyclonal antibodies. (b) As in (a) but showing polypeptides detected with anti-RecB polyclonal antibodies. (c) Quantification of the Chi-dependent fragment in (a) and in parallel reactions that used 3.2 and 32 nM trypsin; x and o, Chi+ and Chi0 reactions; L and R, left- and right-end caps. (d) Persistence of RecC’s Chi-dependent sensitization to trypsin. RecBCD was reacted with Chi+ or Chi0 DNA for 1 min; the reactions were stopped by addition of EDTA to 23 mM, incubated for the further times indicated, and then treated for 1 min with 160 nM trypsin. RecC fragments were detected as in Fig. 2. Continuous and broken lines, ~91-kDa Chi-dependent fragment 1 and ~82-kDa Chi-independent fragment 2. (e) DNA substrate used in (a), (b), and (c). Phage lambda DNA (48.5 kb) with a Chi site (χ+D) 3.5 kb from the right end. Hairpin DNA (gray, data not drawn to scale) ligated in some cases to one strand at one or the other end of the lambda DNA substrate prevents RecBCD from unwinding from the capped end. Arrow, direction RecBCD must unwind DNA to be changed by χ+D [8].
RecC becomes protease sensitive during reaction in a Chi-dependent manner
We allowed RecBCD enzyme to bind to the end of lambda DNA (with or without x+D) in the absence of ATP, started the reaction by adding ATP, allowed 15 s for the enzyme to pass Chi, and then added trypsin to the ongoing reaction. Sixty seconds later, we terminated the trypsin reaction by addition of a high concentration of leupeptin, a rapid inhibitor of trypsin [28]. We then examined the proteolytic cleavage products after SDS-PAGE by Western blot analysis, using polyclonal antibodies raised against the individual RecB, RecC, or RecD poly-peptides, to allow separate visualization of digestion products from the three enzyme subunits. (For clarity, we use “cleavage” for proteases and “nicking” for nucleases.)
Figure 2 shows the results of such partial proteolysis during reaction of RecBCD enzyme with lambda DNA (with or without Chi) after 1 min of reaction with concentrations of trypsin varying from 3 to 1000 nM. RecC showed a prominent digestion product of ~82 kDa (Fig. 2a), and RecB showed a prominent product of ~98 kDa (Fig. 2b), neither of which was affected by the presence of Chi. As shown below, both of these products had the N-terminus of their respective polypeptides and correspond to previously identified protease-sensitive sites on exposed regions of the polypeptides [29,30]. Similar reactions using anti-RecD antibodies revealed no specific proteolytic fragments (unpublished data).
Fig. 2.
Reaction with Chi+ DNA sensitizes RecC to cleavage by trypsin. (a) RecBCD was treated for 1 min with trypsin (3.2, 10, 32, 100, 320,or 1000 nM) during reaction on phage lambda DNA (48.5 kb) lacking (Chi0) orbearinga Chi site 3.5 kb from the right end (Chi+). RecC polypeptides and their fragments were detected by Western blots using polyclonal antibodies. (b) As in (a) but showing polypeptides detected with anti-RecB polyclonal antibodies. (c) Quantification of Chi0 data in (a), with fragments identified by their molecular mass in kilodaltons (kDa). 91 + 55, sum of the 91-kDa and 55-kDa fragments, the latter likely resulting from trypsin cleavages at both sites 1 and 2. (d) Quantification of Chi+ data in (a). Diagrams at the bottoms of (a) and (b) indicate the positions of trypsin cleavages and the designations of the tryptic fragments.
A 91-kDa fragment of RecC, seen with low concentrations of trypsin, was much more prominent after reaction with Chi+ DNA than with Chi0 DNA (Fig. 2a). This result is clear evidence of a Chi-stimulated increase in the accessibility of trypsin to RecC, which we interpret as a change in the conformation of RecBCD at Chi. This fragment was readily visible at the lowest concentration of trypsin (3 nM) and rose to 22% of the recovered signal at 32 nM trypsin (Fig. 2d). The 91-kDa fragment, expressed as a fraction of total recovered signal in that lane, was 3- to 4-fold enhanced by Chi. We refer to this Chi-enhanced protease cleavage site as site 1, the Chi-independent site in RecC as site 2, and the site in RecB as site 3 (Fig. 2a) and the fragments produced by a single cleavage at these sites as fragments 1, 2, and 3, respectively. Because sites 2 and 3 reflect positions in RecBCD less specifically related to the change at Chi than site 1, we have focused our analysis on the Chi-dependent changes.
At higher trypsin concentrations, fragment 1 declined as a 55-kDa fragment increased: the size of that fragment is consistent with its resulting from RecC being cleaved at both sites 1 and 2 (Fig. 2a). We show below that fragment 1 had the native C-terminus and that fragment 2 had the native N-terminus, as expected from this interpretation, which is also supported by the decline in fragment 2 (Fig. 2c and d). The sum of fragment 1 and the 55-kDa fragment (Fig. 2c and d) is thus a measure of the Chi-stimulated cleavage and was 2- to 4-fold enhanced throughout the range of trypsin concentrations used, peaking at 41% of the recovered signal at the highest trypsin concentration used. The maximal reported efficiency of nicking of DNA at a Chi site after a single passage of RecBCD enzyme is ~40% [13]. The data in Fig. 2 are, to our knowledge, the first demonstration of a Chi-dependent change in the structure of RecBCD enzyme.
Chi-dependent protease cleavage of RecC at low and high [Mg2+]
The ratio of [Mg2+] to [ATP] profoundly affects the Chi-dependent nicking of DNA by RecBCD enzyme [10,11]. The nick of one strand of DNA seen at low [Mg2+] [8] is replaced at high [Mg2+] by a switch in the strand endonucleolytically nicked before and after Chi, and the frequency of nicking is increased [10,11]. The unifying feature of these reactions is the production of a single strand of DNA terminated at or near the Chi sequence [2].
To assess the effect of [Mg2+] on the protease sensitivity of RecBCD during reaction on DNA, we performed sets of reactions with or without oligonu-cleotide caps on the DNA to block RecBCD entry from the capped end (see the next section), in parallel with 2.5 or 8 mM Mg2+ and with 5 mM ATP. Chi-dependent cleavage at site 1 was modestly enhanced at 8 mM Mg2+, compared to that at 2.5 mM Mg2+ (Fig. 3a, lane 8 versus lane 3 and lane 10 versus lane 5). The Chi enhancement of cleavage previously seen at low Mg2+ (lanes 2 and 3) was maintained at high Mg2+ (lanes 7 and 8). Levels of fragment 1 were increased at high [Mg2+] with both Chi0 and Chi+ DNA, but the Chi-dependent enhancement seen at low [Mg2+] was maintained (lanes 2, 3, 7, and 8). Remarkably, in all cases, trypsin cleavage at Chi-independent sites 2 and 3 was almost eliminated at high [Mg2+]. The suppression of Chi-independent cleavages of RecB (Fig. 3b) and RecC (Fig. 3a) may reflect a more compact form of the enzyme at high [Mg2+]: both cleavage sites 2 and 3 are in exposed regions linking protein domains [29,30,18].
Unidirectional passage of RecBCD enzyme reduces Chi-independent cleavage at site 1 but leaves strong Chi-dependent cleavage
Oligonucleotide caps on duplex DNA force RecBCD enzyme to traverse DNA in only one direction [31,13] and hence to encounter a Chi site in only one orientation. With such capped substrates, we drew multiple important conclusions about the Chi-stimulated proteolytic cleavage of RecC.
In experiments presented above, RecBCD enzyme could enter either end of the lambda DNA substrate. RecBCD enzyme acts only on a Chi site that it approaches from the 3′ side of the sequence 5′ GCTGGTGG 3′ [8], and thus, it would be expected to react with Chi during only half of its passages along DNA, assuming that the two ends of the DNA bind the enzyme equally well. We therefore capped the left end of lambda DNA, to force RecBCD enzyme to encounter Chi in only the active orientation (Fig. 3). We expected that such caps would double the amount of site 1 cleavage seen, but we saw little or no increase at either high or low [Mg2+] (Fig. 3a, lane 5 versus lane 3 and lane 10 versus lane 8). We note that the constancy of cleavages at Chi-independent sites 2 and 3 (Fig. 3a and b), under the respective [Mg2+] conditions, serve as internal controls. We assume that differences between the nucleotide sequences at the ends of phage lambda DNA guide RecBCD enzyme to preferentially unwind lambda DNA from the right end and hence to encounter Chi in its active orientation even on uncapped DNA.We therefore omitted caps in experiments in subsequent sections designed solely to characterize the nature of the site cleavage. Here, we demonstrate that the Chi-enhanced protease cleavage depends on the orientation of Chi, as expected.
The addition of a cap to the left end of Chi0 lambda DNA reduced the amount of site 1 cleavage considerably but did not detectably affect site 2 or site 3 cleavage (Fig. 3, lane 4 versus lane 2 and lane 9 versus lane 7). We infer that RecBCD enzyme, during its occasional traverses of uncapped lambda DNA from left to right, reacts with Chi-like sequences that are active both in cells and with purified RecBCD enzyme [32,33]. Comparison of left-capped Chi0 and Chi+ DNA is thus a more sensitive measure of the magnitude of the Chi enhancement of cleavage of RecC at site 1, quantified below.
The addition of a cap to the left or right end of Chi+ DNA (lanes 6 and 11) permitted or prevented, respectively, RecBCD enzyme from encountering an active Chi and hence allowed us to compare trypsin cleavages on a Chi+ substrate (left capped) and an effectively Chi0 substrate (right capped) made from the same phage DNA preparation. The strong Chi dependence of site 1 cleavage, together with unchanged site 2 and site 3 cleavages (Fig. 3a and b, lane 5 versus lane 6 and lane 10 versus lane 11), validates the previous results with separate DNA preparations.
Quantification of the results in Fig. 3a, together with similar results at other trypsin concentrations (Fig. 3c and d), hence provides the best estimate of the magnitude of the Chi-enhanced cleavage of RecC. Left-capped Chi+ DNA reacted with RecBCD enzyme at 8 mM Mg2+ and treated with 32 nM trypsin yielded ~60% of the recovered RecC polypeptides cleaved at site 1 or 5.5 times the level seen with left-capped Chi0 DNA. Values at 2.5 mM Mg2+ showed a similar degree of enhancement but with lower absolute values.
We have thus confirmed that Chi-dependent cleavage at site 1 is a robust, reproducible feature of trypsin cleavage of RecBCD enzyme. This effect was seen under different reaction conditions, was barely present without Chi, and was augmented up to 5.5-fold by Chi. Cleavage at site 1 can be the major RecC reaction product after trypsin treatment during RecBCD’s reaction on Chi+ DNA.
Chi-dependent conformational change of RecBCD enzyme persists for several minutes
To estimate the lifetime of the conformational change, we allowed RecBCD to pass Chi and, by addition of ethylenediaminetetraacetic acid (EDTA), stopped it 60 s later when RecBCD was still unwinding the DNA. After further incubation for up to 32 min, trypsin was added and allowed to digest RecBCD for 60 s before being stopped as in the preceding experiments. The abundance of Chi-dependent fragment 1 was greatest when trypsin was added immediately after EDTA (Fig. 3d). The abundance of fragment 1 decreased over the next several minutes but remained above that observed in the absence of Chi. We estimate that the Chi-enhanced conformation-al change has a half-life of several minutes. This duration is compatible with the Chi-dependent change of RecBCD persisting to the end of the DNA, at which RecBCD likely disassembles [14], about 2 min after Chi is encountered on the substrate used here.
Proxy conditions that simulate Chi-dependent protease sensitivity of RecC
Further characterization of the Chi-enhanced conformational change required more RecBCD than could be conveniently obtained with reactions on lambda DNA, which has ~100 times the mass of RecBCD. We therefore sought proxy reaction conditions to produce larger quantities of reaction products for analysis. We had noted that, under some reaction conditions and trypsin concentrations, cleavage products apparently identical with that at site 1 could be produced in the absence of Chi reaction. For example, high concentrations of trypsin, acting on RecBCD enzyme in buffer without cofactors, can produce a great deal of such a fragment (Fig. S1). We found that, under several reaction conditions, RecC fragments were produced by cleavage apparently at site 1 (Fig. 4a and b). The activity of trypsin on an artificial substrate was essentially the same under all of the conditions used for these experiments (Table S1).
Fig. 4.
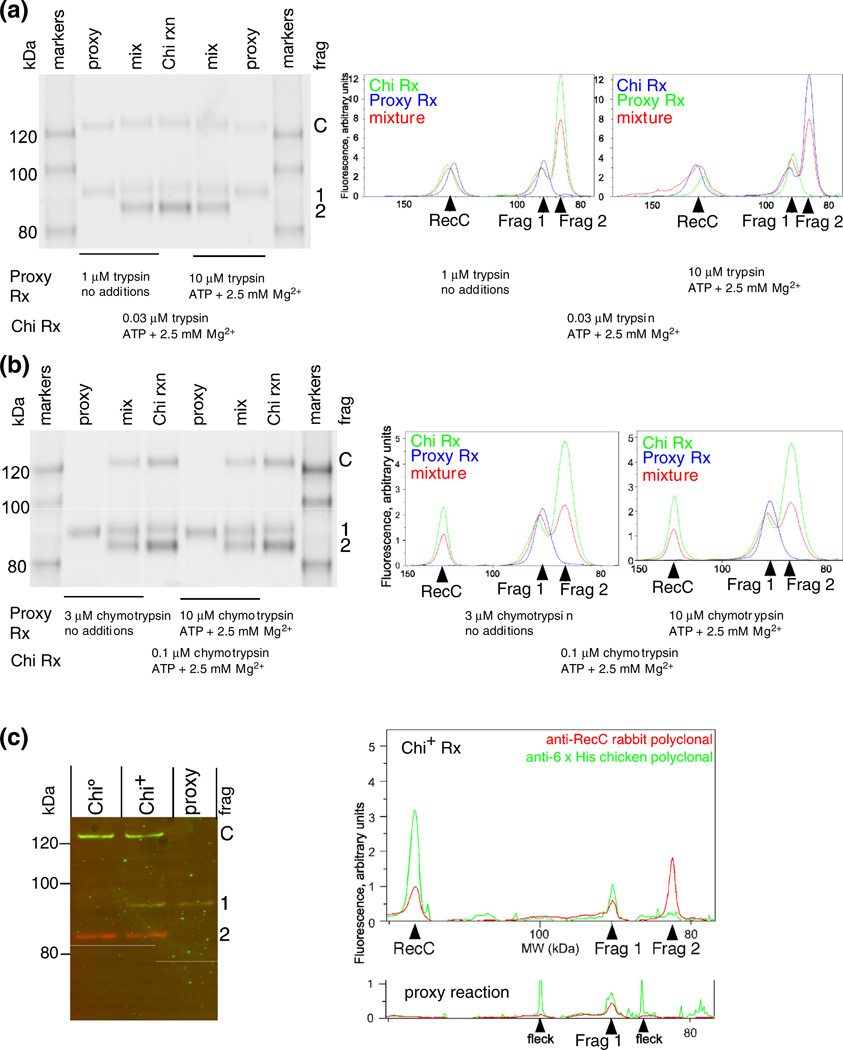
Chi-dependent and proxy fragments of RecC comigrate and retain the native carboxy-terminus. (a) RecBCD enzyme was treated with trypsin for 1 min under the conditions noted, and the reaction terminated with trichloroacetic acid and concentrated. Traces of lanes from the separate and mixed samples were aligned by the flanking size markers. The similarity of the width of the peak, without evidence of splitting into two peaks, for the ~91-kDa fragments (Frag 1) in the separate and mixed samples indicates that the tryptic cleavage site is at the same position (±3 amino acids, by interpolation from size markers) in the proxy and Chi reactions. (b) As above, but with chymotrypsin. (c) The Chi-dependent tryptic fragment and the fragment produced under proxy conditions extend from the same or similar position in RecC to the C-terminus of RecC. RecBCD tagged with six histidines at the C-terminus of RecC (RecBCHis6D) was digested under a proxy condition with 2.5 mM Mg2+ and 10 µM trypsin or during reaction on Chi0 or Chi+ DNA (digested with 32 nM trypsin). The products were analyzed using rabbit anti-RecC and chicken anti-His tag polyclonal antibodies, which were detected with fluorescent species-specific secondary antibodies (RecC, red; His tag, green). “Fleck” indicates a small, local imperfection in the scanned membrane.
We verified that the proxy and genuine site 1 cleavage products had indistinguishable lengths by showing their comigration in adjacent lanes or when mixed under gel conditions chosen for good separation in the range of interest. Trypsin (Fig. 4a) and chymotrypsin (Fig. 4b) were each tested under two different proxy conditions. Migration relative to markers of known sizes allowed us to estimate that the genuine and proxy site 1 fragments differed in length by less than three amino acids.
We verified that proxy and Chi-stimulated RecC fragments extended from the protease cleavage site to the C-terminus of RecC because both fragments were detected with rabbit anti-RecC and chicken anti-His polyclonal antibodies (Fig. 4c) (RecC was tagged at the C-terminus with six histidine residues). The comigration, within a lane, of the RecC fragment and C-terminus signals showed that the fragments from both Chi-stimulated and proxy reactions extend from an internal position in RecC to their C-terminus. The comigration of Chi-dependent and proxy reaction products in adjacent lanes (Fig. 4c) confirmed, for a third proxy condition, that the genuine fragment 1 and proxy reaction products were indistinguishable.
Chi-dependent protease cleavages occur on a small surface patch of RecC
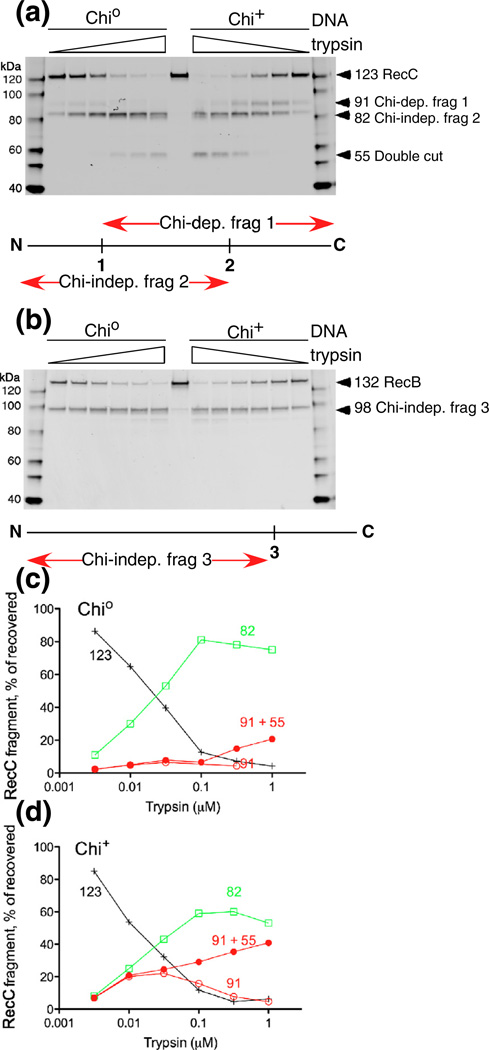
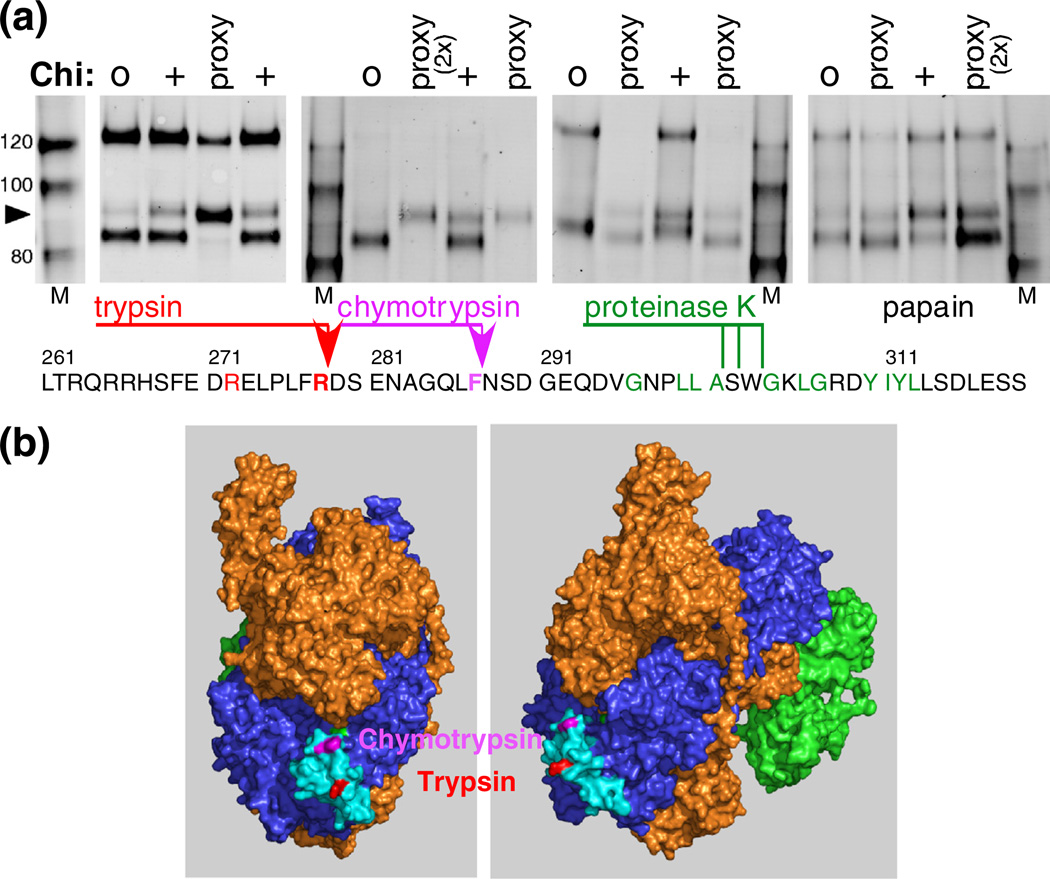
To determine more precisely the position of the proteolytic cleavages,weprepared sufficientRecBCD cleaved with trypsin or chymotrypsin under proxy conditions (i.e., in buffer with no additions, with ATP plus Mg2+, or with Mg2+ alone) to allow determination of the amino acid sequence of the novel RecC fragments’ N-termini by Edman degradation. These analyses indicated that, under each condition, trypsin cleaved RecC after Arg278 and that chymotrypsin cleaved after Phe287 (Fig. 5).
Fig. 5.
Location of Chi-dependent protease cleavages to a small exposed area of RecC. (a) RecBCD reacting with Chi0 or Chi+ DNA was treated with 32 nM trypsin, 100 nM chymotrypsin, 10 nM proteinase K, or 32 nM papain. RecBCD in buffer with DNA plus Mg2+ (proxy condition) was treated with the indicated protease (3 µM), and RecC was analyzed as in Fig. 2a. Sites of cleavage, determined by sequencing the trypsin- and chymotrypsin-derived Chi fragments (arrowheads), are shown on the RecC amino acid sequence. The location of the proteinase K cleavage was estimated by comigration with trypsin-cleaved substrate, using gels similar to those in Fig. 4. The substrate specificity of each protease includes the amino acids in color on the sequence. (b) The Chi-dependent sites of cleavage in the RecC polypeptide are in a disordered loop (cyan; residues 250–293) on the surface of the crystal structure [18]. The left view is rotated ~90° about the vertical axis relative to the right view.
A comparison of the mobilities of the fragment produced by proteinase K and tryptic fragment 1 indicated that proteinase K cleaved after amino acid 301, 302, or 303. These cleavage sites are after amino acids at which these enzymes are known to cleave (Fig. 5a). As expected, these residues are near each other on the surface of RecC in one of the two molecules in the asymmetric unit of the crystal structure [18] (Fig. 5b). This region of RecC (cyan in Fig. 5b) is disordered in the other RecBCD molecule [18] in the initial structure determination but is ordered in both molecules of the subsequent structure determination with a longer DNA substrate [19]. The disordered molecule suggests that this region is inherently disordered and may be ordered in the other molecules by crystal contacts, which are not expected to occur in solution. These considerations suggest that this patch of RecC is flexible and should be susceptible to proteases, unless a conformational change either stabilizes or sterically occludes this site.
Similar analysis also showed that the prominent Chi-independent fragment 3 of RecB and fragment 2 of RecC each had the native N-terminus of their polypeptides. The apparent molecular weights of the fragments thus confirm that they are identical with the previously reported proteolytic fragments of RecB and RecC [29,30], each of which resulted from cleavages in an exposed part of the protein.
DNA binding suppresses cleavage at the Chi-dependent site of RecC
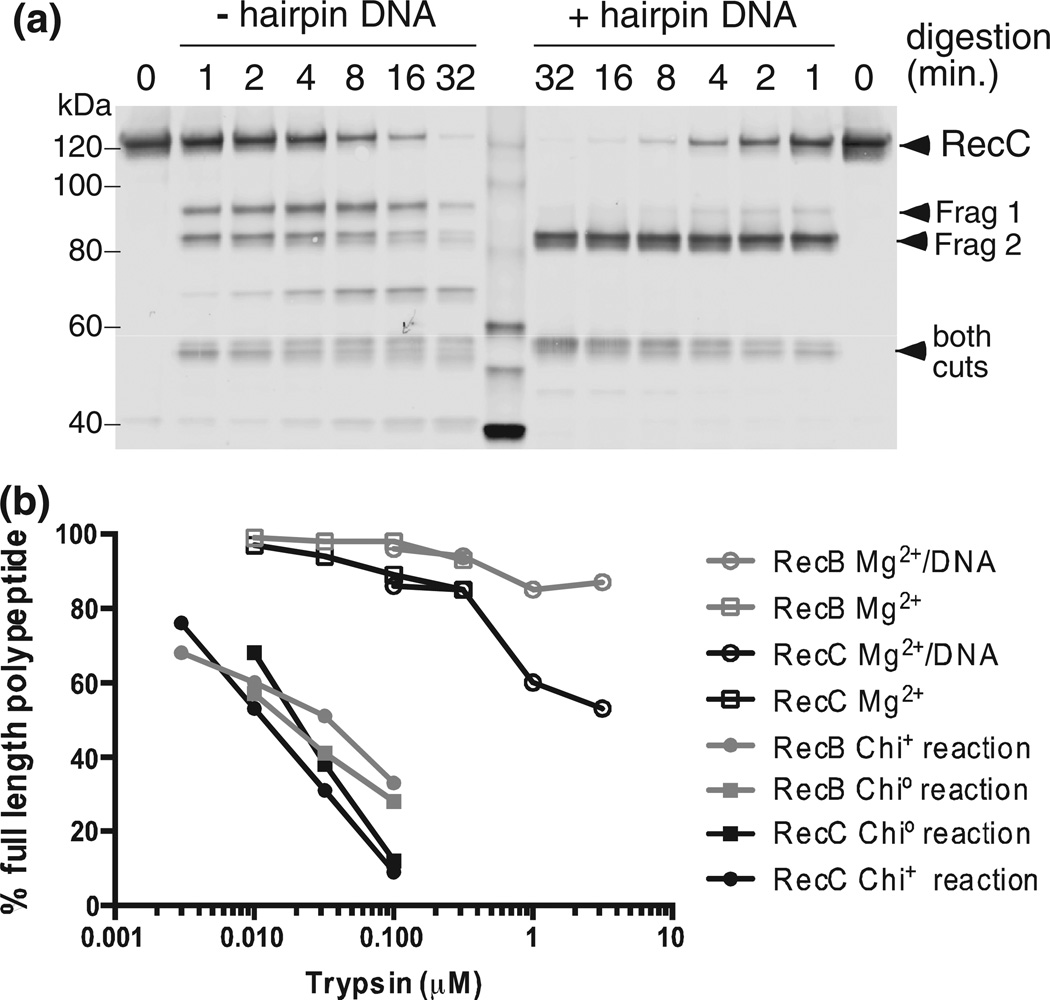
We noted above several proxy reaction conditions that stimulated cleavage at site 1 in the absence of RecBCD reaction. We show here that cleavage under one such condition (buffer without Mg2+) was strongly suppressed (Fig. 6a) if the enzyme was bound to a short hairpin-shaped duplex DNA similar to that in the crystal structure. Disappearance of fragment 1 upon DNA binding was not the result of cleavage at both sites 1 and 2: site 2 cleavage was not diminished during the time course of trypsin reaction with bound DNA. Binding of DNA to RecBCD, in the absence of its reaction, thus renders RecC resistant to cleavage at the site that becomes sensitive during reaction with Chi+ DNA.
Fig. 6.
Protease cleavage under proxy conditions is suppressed by DNA binding and requires much more trypsin. (a) RecBCD (35 nM) without or with 140 nM hairpin DNA similar to thatin the original crystal structure [18], with five additional T residues at the 5′ end, was treated for 1, 2, 4, 8, 16, or 32 min with 0.1 µM trypsin, and RecC was analyzed as in Fig. 2a. Similar results were obtained using K-PO4 buffer (Fig. S6). (b) Chi0, Chi+, and proxy reactions were performed and analyzed as in Fig. 2. The percentages of the visible bands that are full-length RecB or RecC are shown. Additional data are in Figs. S1 and S6.
While the changes in sensitivity are qualitatively similar for real and proxy reactions, cleavage of RecB and RecC under proxy conditions required ~100 times more protease than needed during enzyme unwinding (Fig. 6b). The inchworm movement of subunits, postulated to explain a helicase’s travel along DNA [34], may greatly sensitize the enzyme to protease cleavage. Hence, the similarities seen here between DNA binding (in the absence of reaction) and after reaction with Chi+ DNA may indeed reflect similar conformational changes in the enzyme but in the opposite direction.
A RecBCD mutant unable to nick DNA at Chi lacks Chi-dependent protease cleavage
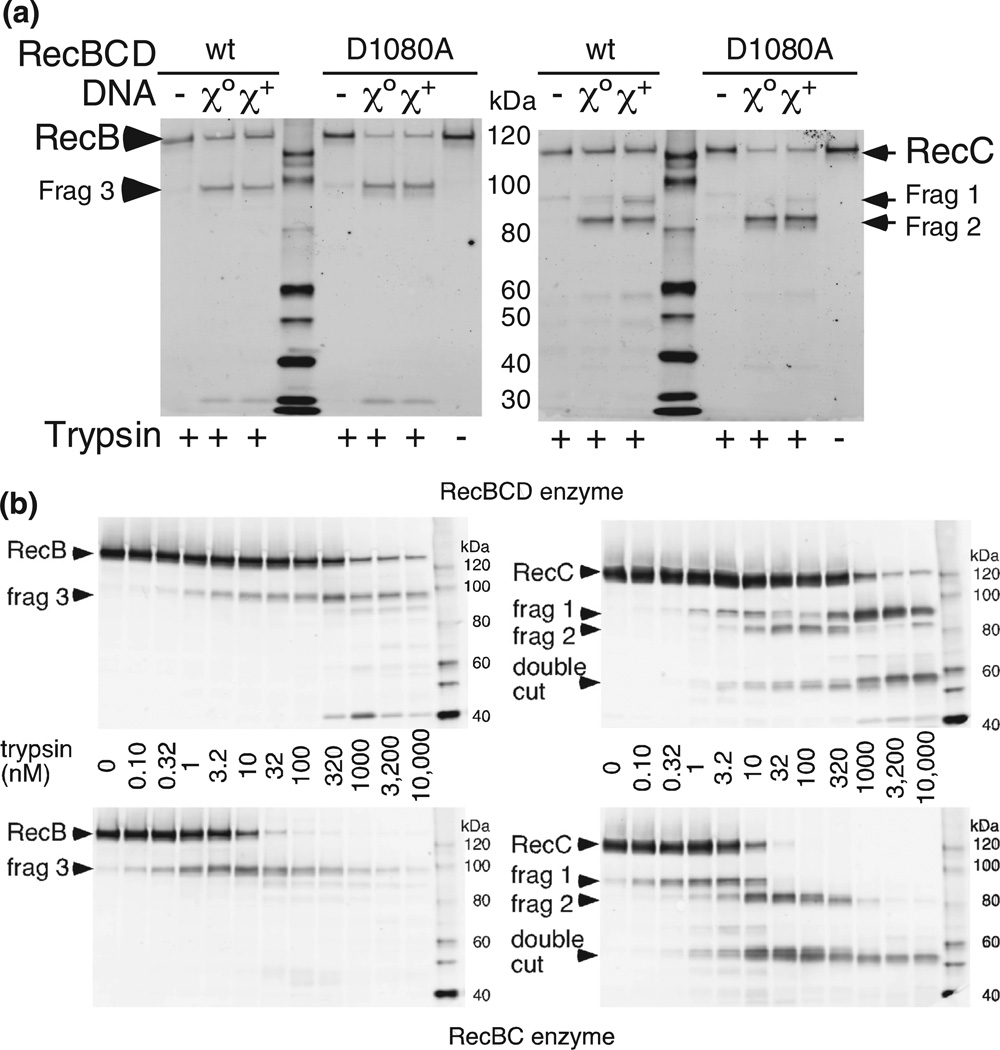
To test whether a step beyond mere recognition of Chi is required for the change in protease sensitivity, we tested a mutant RecBCD unable to nick DNA in response to Chi. The recB1080 (D1080A) mutation in the nuclease domain of RecB abolishes all of the enzyme’s nuclease activities [35]. We compared proteolysis of RecBCD and RecBD1080ACD enzyme during reaction with Chi0 or Chi+ DNA (Fig. 7a and Fig. S2). RecBD1080ACD enzyme yielded very little of the Chi-stimulated RecC fragment 1 but produced correspondingly more of fragment 2. However, both wild-type and mutant enzymes produced (at any trypsin concentration tested; Fig. S2) very similar yields of RecB fragment 3, indicating that the overall topologies of actively unwinding mutant and wild-type enzymes were similar. As noted before (Fig. 6), at the low trypsin concentrations used, very little proteolysis of enzyme that was not unwinding DNA was seen. These results indicate that a step after Chi recognition, that is, the nicking of DNA at Chi, is required for the long-lasting change in the conformation of RecBCD in response to Chi.
Fig. 7.
Chi-dependent protease cleavage is reduced in a nuclease-deficient mutant RecBCD enzyme but is enhanced in the absence of RecD. (a) RecBCD enzyme or RecBD1080ACD enzyme was reacted in the absence of DNA or during reaction with Chi0 or Chi+ DNA and treated with 32 nM trypsin. Additional data are in Fig. S2. (b) RecBCD enzyme or RecBC enzyme (50 nM in ME buffer) was digested for 1 min with trypsin at the indicated concentrations. Reaction and sample preparation were as in Fig. 2.
RecBC enzyme, lacking RecD, is hyper-sensitive to proteolysis of RecC
Like RecBD1080ACD, RecBC lacks nuclease activity and Chi hotspot activity [36,37], but unlike RecBD1080ACD, it also lacks the RecD subunit with its helicase domain [4]. We found that RecBC, in the absence of DNA, was more sensitive to trypsin cleavage at site 1 in RecC than was RecBCD (Fig. 7b, right panel). The concentration of trypsin that cleaves about half of the RecC polypeptide was about 100 times lower for RecBC (~3.2–10 nM) than for RecBCD (~320–1000 nM). (With RecBCD, a small amount of fragment 1 appeared at ~3.2– 10 nM, likely stemming from a small amount of RecBC present in RecBCD preparations.) RecB was also more sensitive, by a factor of ~30, in RecBCD than in RecBC (Fig. 7b, left panel). These data indicate that the RecD subunit provides partial protection of a site on RecC located far from RecD in the crystal structure [18] (Fig. 5b). Below, we discuss a mechanism for this allosteric effect.
Addition of one or more cofactors to RecBC, including complete reaction on Chi0 or Chi+ DNA, made the enzyme more resistant to trypsin, but no prominent fragments accumulated with increasing trypsin concentration (unpublished data). We infer that, after the first trypsin cleavage, RecBC becomes highly sensitive to trypsin and the polypeptides are degraded without leaving evidence for the most sensitive cleavage site.
Chi-stimulated crosslinking of RecB to RecC demonstrates a conformational change
To assess with another method Chi-dependent conformational changes in RecBCD, we prepared His-tagged RecBCD enzymes bearing p-benzoyl-L-phenylalanine (Bpa), an artificial UV-activated crosslinking amino acid [38], at four locations in the region of RecC identified above as sensitive to proteases in a Chi-enhanced manner. Two of them (at amino acid positions 277 and 287) yielded RecC– RecB crosslinked products enhanced by Chi and thus provided further evidence for novel Chi-stimulated conformational changes in the enzyme during unwinding and action at Chi.
The Bpa-containing RecBCF287BpaD enzyme was irradiated with UV during its reaction with lambda DNA, and reaction products were analyzed by Western blot. A slowly migrating polypeptide, most prominent after Chi+ reaction, was detected by both anti-RecB and anti-RecC antibodies and hence represents a UV- and Chi-stimulated crosslink of RecB to RecC (Fig. 8a, lanes 7 and 8). Polypeptides at this position were not detectable in the absence of UV (lanes 1 and 14) or in UV-irradiated His-tagged wild-type enzyme (i.e., RecBCD not containing Bpa; unpublished data); they were barely visible in the absence of ATP or DNA (lanes 2 and 13) and were less abundant in the presence of Chi0 or Chi+ DNA without RecBCD reaction (in the absence of ATP; lanes 3, 4, 11, and 12) than with reaction (lanes 6–9). The slowest migrating polypeptide, seen more clearly in the anti-RecB blot, was present without UV and was not stimulated by ATP or DNA (lanes 1–5): it may represent a pre-existing crosslink, perhaps induced by the UV monitor during purification.
Fig 8.
Chi stimulates UV-dependent crosslinking of RecC to RecB. (a) RecBCF287BpaD enzyme was reacted with Chi0 or Chi+ DNA. Reactions were started by addition of ATP and were irradiated at 259 nm from 15 s to 75 s. Samples were prepared and analyzed as in Fig 2. (b) Reactions as in (a), using the buffers (20 mM) shown. [Mg2+] was 5 mM except for samples marked with an asterisk, which contained 8 mM Mg2+. Reactions, in sets of three, were without DNA, with Chi0 DNA, and with Chi+ DNA. Additional data are in Figs. S3 and S4. (c) A surface view of a portion of RecBCD enzyme (PDB file 3K70); the left panel is an expansion of part of the right panel, which shows the entire RecBCD enzyme (Fig. 1). The RecB helicase domain is orange, and its nuclease domain is gray. RecC is blue, and RecD is green. RecC amino acid F287 (magenta) shows the position of the Bpa in RecBCF287BpaD and the adjacent chymotrypsin cleavage site (Fig. 5) [19]. RecC amino acid F277 (red) shows the position of Bpa RecBCF277BpaD and the nearby trypsin cleavage site (between amino acids 278 and 279). RecB amino acids A108 and R119, the most likely locations of the Bpa crosslink in RecBCF277BpaD, are cyan and green, respectively. The measured distances between the C alpha of F277 and A108 or R119 are indicated by red arrows.
Addition of ATP to allow unwinding of the DNA significantly increased the intensity of the cross-linked polypeptide (lanes 6 and 7 versus lanes 3 and 4; lanes 8 and 9 versus lanes 11 and 12). Chi stimulation of the yield of crosslinked product during unwinding reactions was modest (2- to 4-fold, lane 7 versus lane 6 and lane 8 versus lane 9) but was reproducible. This Chi stimulation of RecB–RecC crosslinks was insensitive to DNA substrate concentration (Fig. S3a), pH, or [Mg2+] (2.5, 5 or 8 mM Mg2+ with 5 mM ATP; Fig. 8b and Fig. S3b) but was less in K-PO4 buffer than in the other buffers tested. These observations, with those below, confirm the importance of the small region of RecC that is sensitive to proteolysis (Fig. 5) for Chi-mediated change in the enzyme and demonstrate the involvement of RecB in the changes.
Similar Chi- and UV-enhanced crosslinks were also seen with RecBCF277BpaD enzyme but not with RecBCW303BpaD enzyme (Fig. S4), perhaps because the latter enzyme produced a very high level of intra-RecC crosslinks. The latter enzyme was not studied further, but the high level of intra-RecC crosslinking seen (~50%) does confirm the high level of Bpa incorporation into the enzyme preparation. RecBCDenzyme withaRecCHis6 tag, butnoBpa substitutions, and RecBCF269BpaD showed no UV- or Chi-mediated crosslinks (Fig. S4; unpublished data).
Mass spectrometry analysis of UV-irradiated RecBCF277BpaD enzyme revealed a crosslink between the Bpa at position 277 of RecC and a tryptic peptide of RecB extending from amino acid 107 to amino acid 136, with the crosslink most likely to be at Ala108 or Arg119 (Fig. 8c; crosslink data in Fig. S4 and Fig. S5). The C alpha-C alpha distances between Phe277 of RecC and Ala108 and Arg119 of RecB in the RecBCD crystal structure are 27.5 Å and 31.5 Å, respectively. The maximum distance between C alpha in crosslinks between Bpa and its most likely targets is estimated as 17 Å or less [39,40]. This RecB–RecC crosslink was detected in two samples, one with and one without a bound hairpin oligonucleotide. While attempts to analyze, by mass spectrometry, similar crosslinks during RecBCD unwinding reactions were unsuccessful due to insufficient material, the observed crosslinks show that the enzyme can undergo at least a 10-Å movement in this region (which we dub “jaws”) relative to the crystal structure. The Chi-stimulated increase in the intensity of UV-induced crosslinks between RecB and RecC (Fig. 8) thus indicates a Chi-stimulated conformational change in this region of the enzyme, such as a closure of the jaws by ≥10 Å or a change in the flexibility of these regions of RecB and RecC that permits their crosslinking to one another.
Small-angle X-ray scattering reveals a major conformational change in RecBCD enzyme upon binding DNA
To test directly in another way for a conformational change of RecBCD under various conditions, we used small-angle X-ray scattering (SAXS) to examine RecBCD in free and DNA hairpin-bound states. The enzyme was unstable, over the time periods required for sample transport and analysis, in the Mops buffer typically used for our reactions. Data were therefore gathered in Mops buffer with 10% (v/v) glycerol to stabilize the enzyme or in K-PO4 buffer with or without glycerol. We confirmed that the effect of hairpin DNA on site 1 trypsin cleavage, seen in Fig. 6a with Mops buffer, was also observed in K-PO4 buffer, albeit only at longer trypsin incubation times (Fig. S6). Very similar SAXS results were obtained in each of these buffers (Fig. 9 and Fig. S7–Fig. S15; Tables S2 and S3).
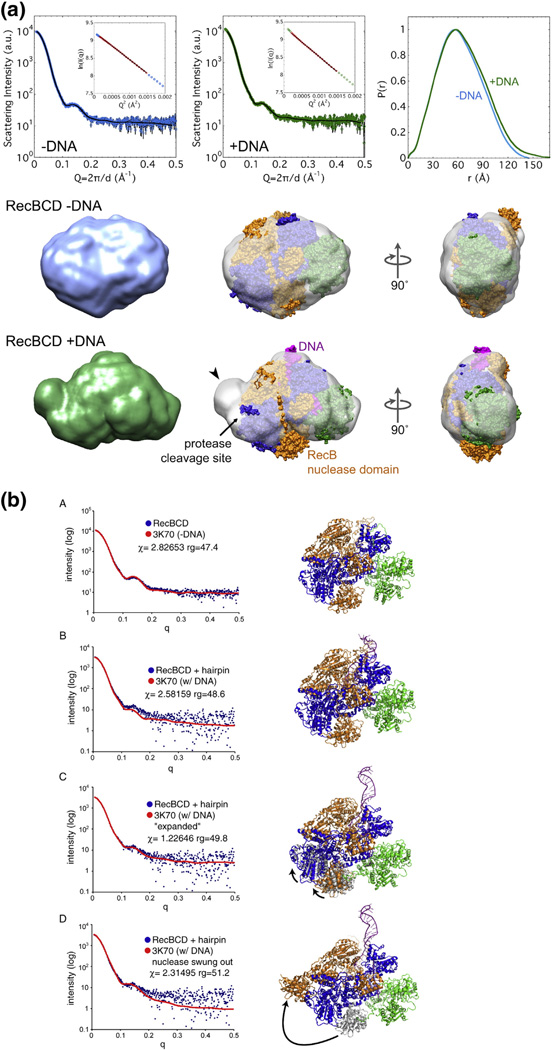
Fig. 9.
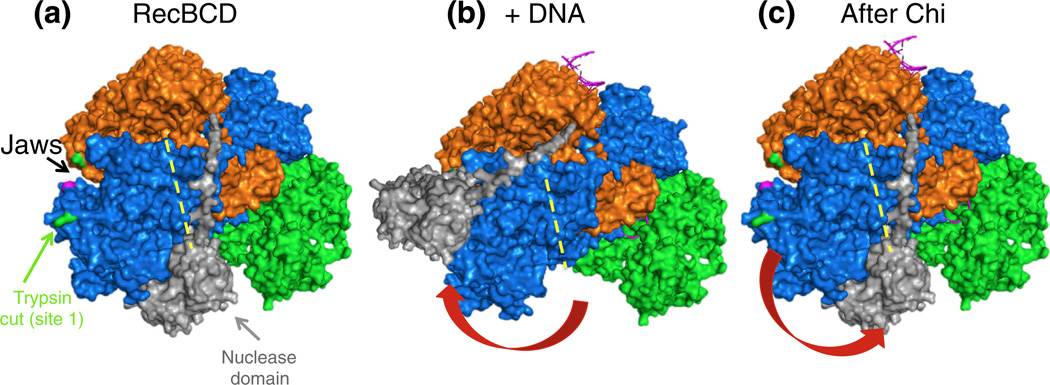
SAXS analysis of DNA-bound and unbound RecBCD enzymes. (a) Envelopes of RecBCD enzyme determined by SAXS (data in upper panels) of free enzyme and enzyme bound to hairpin DNA in K-PO4 buffer with glycerol (Figs. S7 and S8; Table S2). Ab initio shape reconstructions agree well with the data; Guinier analyses (insets) show linear fits. The crystal structure, with subunits colored as in Fig. 1a and oriented to be optimally docked within the SAXS envelope, is superimposed on the SAXS envelope. Note that swinging of the RecB nuclease domain (carat) improves the fit and accounts for the change of protease sensitivity (relatively sensitive −DNA and resistant + DNA). (b) SAXS all-atom modeling analysis of the above data. In (b–A), data from enzyme without DNA were tested against PDB structure 3K70 [19] with DNA computationally removed; in (b-B), data from enzyme with a bound DNA hairpin were tested against PDB structure 3K70; in (b–C) and (b–D), data from enzyme with a bound DNA hairpin were tested against two possible conformational changes in the enzyme, one (b–C) involving movement of the lower lobes of RecB and RecC moving toward the helicase domain of RecB, perhaps reflecting jaws closure, and the other (b–D) involving swinging of the nuclease domain to cover the protease-sensitive region. Additional data are in Figs. S7–S16.
Ab initio reconstructions of RecBCD conformation from the SAXS data indicate that unbound RecBCD in solution in fact resembled that seen in the crystal structure (Fig. 9a and Fig. S9) despite the presence of hairpin DNA only in the latter. Comparison of the experimental SAXS pattern and that calculated from the crystal structure (PDB ID 3K70) [18,19] with DNA computationally removed confirmed this conclusion. Upon binding to DNA, however, RecBCD in solution changed shape markedly, exhibiting a more elongated shape than free enzyme under a variety of conditions (Fig. 9a and Fig. S10). The ncreases in the radius of gyration (Rg) by ~2.0 Å and in the maximal dimension (Dmax) by ~30 Å were much larger than those expected solely from the addition of hairpin DNA, which would increase Rg by only ~0.4 Å and increase Dmax by only ~4 Å (Table S2). The shape of RecBCD allows a relatively unambiguous fit of the crystal structure into the SAXS-derived envelope, indicating changes in the region of the Chi-dependent protease cleavage site resulting from DNA binding. The increased volume at this position is consistentwith the RecB nuclease domain swinging ~50–100 Å to occupy this position. Alternatively, a conformational change involving the lower lobes of RecB and RecC swinging outward in tandem could account for the changes observed by SAXS; this latter possibility is equivalent to the jaw closure suggested by the crosslinking experiments reported above.
We also analyzed the SAXS data by all-atom ensemble modeling [41] (Fig. 9b). SAXS data from RecBCD enzyme without DNA in K-PO4 buffer without glycerol fit well with the structure of the RecBCD enzyme–DNA complex (PDB ID 3K70) with the DNA computationally removed, in accord with the ab initio analysis mentioned above. We tested the SAXS data from RecBCD enzyme bound to DNA against three models: RecBCD enzyme with DNA bound (PDB ID 3K70), that structure with a conformational change involving the lower lobes of RecB and RecC swinging outward in tandem, or that with the nuclease domain swung over to simulate the “nuclease swing” model discussed below. Both the fit of the measured SAXS data (blue dots) to the calculated SAXS pattern (red lines) in Fig. 9b and the Chi values (in the statistical sense) favor the model in which the lower lobes of RecB and RecC move toward the helicase domain of RecB in agreement with the RecB–RecC crosslinking data (Fig. S5).
Discussion
The RecBCD enzyme of E. coli nicks DNA at Chi recombination hotspot sites only during its rapid (up to 1000 nt/s) unwinding of duplex DNA, making it difficult to observe Chi-mediated changes in the enzyme. We have used three independent physical techniques to investigate the structural changes in RecBCD enzyme as it binds to duplex DNA, unwinds that DNA, and then encounters Chi on that DNA. Results from the multiple techniques cannot be easily interpreted in terms of a single conformational change, and we infer that the different assays detect multiple movements that occur as the recognition of Chi is telegraphed to the domain responsible for the DNA nicking and RecA protein-loading reactions that result from that recognition. Here, we propose that two major conformational changes occur during the multiple steps of RecBCD’s reaction—binding DNA, unwinding DNA, and nicking DNA at Chi—and account for the changes in its enzymatic activities and physical properties reported here.
A Chi-dependent protease-sensitive region of RecC
We found that a small region of the surface of the RecC subunit became sensitive to trypsin during unwinding of ds DNA but much more sensitive when that DNA contained an actively oriented Chi site. This specific protease sensitivity was also seen in RecBCD enzyme without DNA but disappeared on binding of DNA. Three other proteases also showed Chi-dependent increase in activity on that small stretch of RecC (amino acids 270–313), which appears in the crystal structure of the enzyme to be highly exposed to the solvent and hence readily accessible to proteases (Fig. 5b).
The Chi-dependent cleavage site in RecC, site 1 in Fig. 2, is one of the three most protease-sensitive cleavage sites on RecBCD enzyme (Fig. 2). Two other, but Chi-independent, sites are site 2, also on RecC [30], and site 3 on RecB [29], both of which are in exposed parts of the protein’s surface. After reaction with Chi+ DNA at higher [Mg2+], site 1 is by far the predominant protease-sensitive site on the enzyme (Fig. 3). The observation of very similar Chi-dependent RecC cleavage patterns at both low and high [Mg2+], coupled with similar crosslinking results, implies that the different RecBCD activities on DNA seen at low and high [Mg2+] [2] do not result from a radically different enzyme structure. The Chi-specific nature of cleavage at site 1 is consistent with the observation that RecBD1080ACD enzyme, mutant in the nuclease active site and lacking all nuclease activity [35], has a much reduced level of cleavage at site 1 but normal levels of cleavage at sites 2 and 3 (Fig. 7a) after reaction with Chi+ DNA.
The discovery and validation of proxy reactions that allowed protease cleavage at the Chi-dependent site allowed us to prepare enough material to determine the exact peptide bonds cleaved at site 1 by trypsin (between RecC amino acids 278 and 279) and by chymotrypsin (between RecC amino acids 287 and 288) (Fig. 5). Site 1 is highly exposed on the surface of the crystal structure [18], raising the questions of why it becomes protected from proteases when the enzyme binds DNA and why it becomes sensitive again during DNA unwinding in a Chi-enhanced manner. Crosslinking, however, becomes progressively more frequent as these steps proceed. Below, we reconcile these behaviors by proposing two separate conformational changes during RecBCD’s binding and reaction on DNA.
A nuclease domain swing model of Chi action
We propose the following model, inspired in part by our signal transduction model [17] (Fig. 1b), to bring together the proteolysis results and the known activities of RecBCD enzyme. Before binding DNA, the RecB nuclease domain is in the position shown in the crystal structure (Fig. 10a). Upon binding DNA, the nuclease domain swings on its ~30 amino acid tether to the left (Fig. 10b) and remains there, except for occasional transient returns to its initial position in the crystal structure near the exit of the RecC tunnel for DNA [18]. There, it is in position to nick, more frequently at high than at low [Mg2+], the 3′-ended strand of DNA but quickly returns to the left. When Chi is in the RecC tunnel (Fig. 1a and 10), the Chi-triggered signal transduction cascade [17] (Fig. 1b) causes the nuclease domain to swing back to the right (Fig. 10c), where it is in place to nick the 3′-ended strand nearby. We propose that, after swinging and nicking at Chi, the nuclease domain stays on the right but rotates to place its nuclease active site near the 5′-ended strand emerging from RecD and to place its RecA-loading surface [42] near the 3′-ended strand emerging from RecC. Before the determination of the crystal structure [18], Yu et al. proposed that the RecB nuclease domain swings from one position in the enzyme to another, to account for the change of DNA strand nicked before and after Chi under reaction conditions with excess Mg2+ [29]. While the position of the nuclease domain may be modulated by the concentration of Mg2+, as previously suggested [29], our results, in contrast, indicate that the nuclease domain resides principally on the left, even at high [Mg2+] (Fig. 3); the nuclease domain may be on the right more frequently and thereby nick the 3′-ended strand more frequently at high [Mg2+] [10,11]. We propose that change of the strand nicked at high [Mg2+], which is Chi dependent [9–11], reflects a rotation after swinging of the nuclease domain. Such a rotation would account for the lack of nicking at a subsequently encountered Chi site [13]. Furthermore, the nuclease domain may be in this rotated orientation in RecBC enzyme, which lacks RecD; this conformation could explain why RecBC is devoid of nuclease activity even though it contains the RecB nuclease domain [36,29] and why the Chi-dependent protease cleavage site 1 in RecC is more sensitive in RecBC than in RecBCD (Fig. 7b).
Fig. 10.
Model for the Chi-dependent conformational changes of RecBCD enzyme, based on the crystal structure [18] and the results reported here. Subunits are colored as in Fig. 8c. (a) In the absence of DNA, the nuclease domain (light gray) is on the right side of RecC, as in the crystal structure (Fig. 1a), next to the RecC tunnel (yellow broken line). A surface patch of RecC is exposed to cleavage at R278 (green) by trypsin or at F278 (magenta) by chymotrypsin (Figs. 2, 3, and 5). (b) Upon binding to DNA, the nuclease domain (positioned manually) swings to the left and protects the RecC surface patch from proteolysis (Fig. 6). In addition, the “jaws” between the RecB helicase domain and RecC close partially (hidden by the nuclease domain). (c) Upon addition of ATP plus Mg2+, RecBCD unwinds the DNA, and the jaws close more. When Chi moves through the RecC tunnel, the nuclease domain swings back to the right, again exposing the RecC surface patch to proteolysis (Figs. 2 and 3), and the jaws close even more (data not shown). The nuclease domain is then in position to nick the 3′-ended strand a few nucleotides to the 3′ side of Chi [8] and, upon rotation, to load RecA protein onto the 3′-ended strand [12,42] and, at high [Mg2+], to occasionally nick the 5′-ended strand [10,11].
The model predicts that, after the RecB nuclease domain has swung to the left and masked the observed protease-sensitive sites on RecC, we might be able to detect novel close contacts between RecB and RecC.
Chi-dependent RecC-to-RecB crosslinking
To locate novel contacts in RecBCD enzyme after Chi recognition, we prepared RecBCD enzyme bearing Bpa, an artificial, UV-activated crosslinking amino acid, at each of four locations in the region of RecC identified above by protease sensitivity. Two of the enzymes, with Bpa at amino acids 277 and 287, showed UV- and Chi-dependent crosslinking of RecC to RecB, and mass spectrometry analysis revealed a crosslink between the Bpa at position 277 of RecC and a tryptic peptide of RecB, with the crosslink most likely to be at Ala108 or Arg119 of RecB. We failed to detect crosslinks between the RecB nuclease domain and the protease-sensitive patch on RecC expected from the nuclease swing model (Fig. 10). There are, however, numerous reasons for such failure, including improper orientation of the Bpa in RecC and amino acids susceptible to crosslinking in RecB and incomplete coverage of peptides analyzed by mass spectrometry. Our analysis has focused on the observed crosslinks (Fig. 8 and Fig. S4 and Fig. S5).
Crosslinking between residues 277 and 287 in RecC with RecB requires considerable conforma-tional change in the crystal structure (Fig. 8c). Thus, our results suggest a Chi-stimulated conformational change in this part of the enzyme, which appears to result in a decrease of at least 10 Å in the gap between the protease-sensitive region of RecC and the adjacent helicase domain of RecB (Fig. 8) to allow UV-mediated Bpa crosslinking to occur. We term the significant gap between RecB and RecC in this part of the DNA-bound enzyme structure the “jaws” of the enzyme. Part of RecB or RecC may move, relative to the other, to allow crosslinking. Alternatively, the conformational change could be a change in the flexibility of these regions of RecB and RecC that allows their relative movement and hence crosslinking of these regions to one another. Recently, a conformational change of the related AddAB enzyme of Bacillus subtilis was reported upon lengthening of the bound DNA to place its Chi sequence in the AddB tunnel . The 1A and 2A helicase domains of AddA come closer to each other when an ATP analog is bound in the AddAB-DNA complex, and this conformation is reversed when Chi is in the AddB tunnel; whether this change is dependent upon the Chi sequence, rather than DNA length, was left unclear.
Two Chi-dependent conformational changes
We noted above that reaction with Chi-containing DNA stimulated protease cleavage at several sites and located two of them (shown in green and magenta on the blue surface of RecC in Fig. 10). It seems unlikely that the Chi-dependent increase in protease sensitivity could result from the closure of the jaws identified by Bpa crosslinking and suggested by all-atom analysis of the SAXS data discussed below (Fig. 9b). Proteolysis requires that a protease (e.g., trypsin, with a diameter of ~40 Å) approach closely enough for the target peptide linkage to enter the active site of the protease. If Chi-stimulated jaw closure excluded proteases from approaching RecC from the RecB side, Chi would inhibit protease cleavage, rather than stimulate it as observed (Fig. 2–Fig. 5 and Fig. 7).
The effects of DNA binding and enzyme reaction differ markedly between crosslinking and proteolysis experiments. RecB–RecC crosslinking is barely detectable without DNA, is stimulated by DNA binding, is stimulated further by reaction of RecBCD enzyme with ds DNA, and is yet more prominent during reaction with Chi-containing DNA (Fig. 8). The Chi-stimulated protease cleavage, by contrast, is seen with RecBCD enzyme without cofactors and is inhibited upon binding to DNA (Fig. 6). It is barely seen during reaction with ds DNA unless the DNA bears an actively oriented Chi site (Fig. 3). In other words, crosslinking becomes progressively more abundant as RecBCD binds DNA, unwinds it, and responds to Chi, but proteolytic sensitivity decreases and then increases again upon response to Chi. Thus, the dual conformational changes proposed in Fig. 10 account for the changes of enzymatic activities and proteolytic sensitivity, the crosslinking data, and the SAXS data better than either change alone. Because of the seeming disparity between protease and crosslinking results, we turned to direct physical measurements of protein conformation to seek confirmation of these changes.
Conformational changes seen by SAXS
We used SAXS analysis to investigate the structure of RecBCD enzyme with or without bound DNA. Data were interpreted using two independent methods. The ab initio method [44] produced molecular envelopes into which protein structures were automatically fit [45]. The structure of RecBCD enzyme without DNA (from PDB file 3K70 minus the DNA) was well contained in envelopes obtained, under all reaction conditions (Table S2), from RecBCD enzyme without DNA (Fig. 9a and Fig. S9). However, when data from RecBCD enzyme bound to hairpin DNA were fit to structure 3K70 (Fig. 9a and Fig. S10), there was clear evidence of a conformational change: the nuclease domain of RecB was outside the envelope, and there was an equivalent volume of empty space within the envelope, in the vicinity of the protease site 1 on RecC, in support of the nuclease swing model described above.
By a second method, data were interpreted by all-atom ensemble modeling [41], in which SAXS data were directly compared with an ensemble of molecular models. Again, data from RecBCD enzyme without DNA fit well with structure 3K70 with DNA computationally removed (Fig. 9b–A). Data from RecBCD enzyme bound to DNA were clearly not a good fit to structure 3K70 (Fig. 9b–B), confirming that the enzyme had undergone a conformational change upon DNA binding and that the structure of DNA-bound enzyme in our solution conditions was not that in the crystal. Comparison of simulations of our two inferred conformational changes (jaws closure, Fig. 9b–C; nuclease swing, Fig. 9b–D) showed that the solution structure of RecBCD enzyme bound to DNA was most consistent with the jaws closure conformational change.
While the two methods of analyzing the SAXS data generate results that can be interpreted in terms of two possible structural models for the DNA-induced conformational change, they do agree that the solution structure of DNA-free RecBCD reflects that of the crystal structure of DNA bound and, based upon the patterns of proteolytic susceptibility, RecBCD after it has encountered Chi (Fig. 2–Fig. 5). We infer that crystallization imparts a conformational constraint not present in solution, possibly due to crystal packing or the high levels of calcium, a RecBCD inhibitor [46], in the crystallization buffer. The trypsin sensitivity of RecC is concordant with swinging of the nuclease domain inferred from the SAXS structure under each condition tested when analyzed by ab initio methods (Fig. S7–Fig. 16; Tables S2 and S3). Further work is needed to determine unambiguously if one, the other, or both of these conformational changes accompany binding of DNA to RecBCD and its change at Chi.
We attempted to directly assess RecBCD subunit movement during its reaction on DNA by using Förster resonance energy transfer, but we were unable to synthesize sufficient RecBCD to allow incorporation of fluorescent moieties into enzyme containing two non-natural amino acids [47] (unpublished data). We wished to test a mutant RecBCD lacking the nuclease domain, but this enzyme proved to be too unstable for these biochemical experiments (unpublished data).
Multiple conformational changes result from interaction of RecBCD enzyme with Chi
We have examined physical changes to RecBCD enzyme during its binding to, and reaction with, duplex DNA bearing or lacking the Chi recombination hotspot, which strongly influences the enzyme’s multiple activities. Different physical techniques have provided evidence for two different conforma-tional changes.
Protease sensitivity experiments clearly demonstrate a profound change in RecBCD enzyme upon action on Chi-containing DNA. They inspired a nuclease swing model (Fig. 10, described above) that provides explanations both for the Chi-induced protease sensitivity of RecC and for how the enzyme integrates its multiple activities to catalyze homologous recombination and DNA break repair in E. coli. Our results do not exclude other conformational changes, however.
Site-specific UV-mediated crosslinks detected by SDS gels, characterized by mass spectrometry analysis, and confirmed by SAXS experiments indicate one change to be a Chi-dependent closing of the distance between the RecB helicase domain and the protease sensitive region of RecC (Fig. 8c) where the UV-crosslinking amino acid had been incorporated. The crystal structure of the enzyme [18] reveals a large cleft in that region that might facilitate the inter- and intra-subunit movements necessary for the inchworm-like movement of the enzyme as it travels along and unwinds duplex DNA [34].
This latter conformational change is clearly separate from the first one, but an enzyme as complex as RecBCD enzyme would be expected to exhibit multiple conformational changes during its unwinding and processing of duplex DNA.
Materials and Methods
RecBCD expression constructs
We constructed plasmids encoding wild-type RecBCD enzyme and derivatives of it containing (i)a C-terminal tag of six histidine residues (His6) on RecC to aid purification and identification of C-terminal proteolytic fragments, (ii) an inactivated RecB nuclease site in the D1080A substitution [35], and (iii) nonsense codons in recC to allow incorporation of Bpa into RecC at four independent sites [38]. Details of these constructions and the E. coli strains containing them for enzyme production are in Supplementary Information.
RecBCD enzyme purification
Wild-type RecBCD enzyme was purified from strain V2455 as previously described [4]. RecBD1080ACD enzyme was extracted from strain V3215 grown in Terrific Broth (Fisher Scientific) and purified by chromatography sequentially on columns containing GE Q FF, GE S300, GE Heparin agarose, and BioRad CHTII. Mutant enzymes containing Bpa were induced in strains listed in Table S3, which were grown in minimal medium containing 0.2 mM Bpa (Bachem) to an OD600 of 0.6, induced with 1 mM isopropylthiogalactoside, and then grown overnight [48]. Enzymes were purified by chromatography sequentially on columns GE HisTrapHP and GE HiTrap Heparin HP. All four enzyme preparations had ds exonuclease specific activities indistinguishable from that of wild-type enzyme (unpublished data).
DNA and oligonucleotides for RecBCD enzyme reactions
Lambda DNA (x0 and x+D123) was purified from induced lysogens [49]. Self-complementary 5′-OH terminated oligonucleotides (hairpin caps) were attached to the left or right ends of lambda DNA using E. coli DNA ligase. Excess oligonucleotides were removed on a Sepharose CL-2B column. The DNA was recovered by ethanol precipitation and dissolved in 20 mM Mops-KOH (pH 7.4) and 0.1 mM EDTA (ME buffer). Left cap (5′ AGGTCGCCGCCCGC GATCGCCCGAAACGGGCGATGCG 3′), right cap (5′ GGGCGGCGACCTGCGGCCGCGCCAAA GGCGCGGCCGC 3′), and hairpin (5′ TTTTTTCTAATGC GAGCACTGCTATTCCCTAGCAGTGCTCGCATTAGA 3′) oligonucleotides were from IDT (Coralville, IA). The sequence of the hairpin oligonucleotide is that in the initial crystal structure (PDB ID 1W36) with five extra T residues on the 5′-end.
Proteases
Trypsin (TPCK treated to inactivate chymotrypsin), α-chymotrypsin (TLCK treated to inactivate trypsin), and papain were from Sigma; proteinase K was from Invitro-gen; and leupeptin was from Roche. Protease solutions, in ME buffer or K-PO4 buffer [20 mM K-PO4 (pH 7.4) and 0.1 mM EDTA] containing 25 µg of bovine serum albumin per milliliter, were prepared fresh and stored on ice before use (less than 2 h); digestions of RecBCD contained 2.5 µg of bovine serum albumin per milliliter.
Antibodies
Polyclonal rabbit antibodies against RecB, RecC, or His-tagged RecD were raised using homogeneous polypeptides purified from overproducing E. coli strains, each of which lacked the genes for the other two polypeptides [50]. Polyclonal chicken anti-His tag antibody was from ProSci Inc. IRDye 800CW donkey anti-chicken IgG (Li-Cor) and Alexa Fluor 680 goat anti-rabbit IgG (Invitro-gen) secondary antibodies used in Western blots were detected using a Li-Cor Odyssey scanner and analyzed using GE ImageQuant software.
Reactions on DNA with and without Chi
Reactions (typically 10 µl per gel lane) were at room temperature and typically contained 5 nM (as molecules) lambda DNA, 2.5 nM RecBCD enzyme in 20 mM Mops-KOH (pH 7.4), and 2.5 or 8 mM magnesium acetate. RecBCD enzyme was allowed to bind DNA, and reactions were started by addition of ATP to 5 mM. After (typically) 15 s, trypsin was added, incubation continued for a further 60 s, and the trypsin reaction terminated by addition of leupeptin (Roche) to 16 mM, followed by chilling, addition of SDS-gel loading buffer (Invitrogen), and heating to 65 °C. Reactions with other proteases, or those requiring concentration before analysis, were terminated by addition of an equal volume of cold 20% trichloroacetic acid. Following addition of insulin to 0.25 mg/ml, we recovered precipitates by centrifugation, washed them with cold acetone, and dissolved them in loading buffer.
UV crosslinking
Light from a Sutter Instruments LS 300W Xenon Arc Lamp, controlled by a Sutter Instruments Lambda 10-3 filter changer and shutter, was passed through a 340- to 380-nm bandpass filter and led to the sample via a liquid lightguide (3 mm diameter lightpath). RecBCD reaction was started by addition of ATP, and the reaction mixtures (10–20 µl) were vortexed and centrifuged to the bottom of the 1.5-ml microfuge tube. The end of the arc lamp’s lightguide was supported above the sample by the walls of the tube, ~8 mm above the surface of the liquid. Light intensity, as measure by a Coherent LasermateQ meter, was 5.7 mW/mm2. The shutter was opened from 15 to 75 s after addition of ATP. Gel loading buffer was added and samples separated on gels. Unirradiated controls were handled in parallel.
Analysis of proteolytic fragments or crosslinked species
The experiment reported in Fig. 4c used a 4–12% NuPAGE Bis-Tris 1.5-mm gel in Mops buffer (Invitrogen); all other experiments used 3–8% NuPAGE Tris-acetate 1.5-mm gels in Tris-acetate running buffer (Invitrogen). Gels were typically run until tracking dye exited the gels, except for those used for comigration or crosslinking experiments, which were typically run about twice as long.
Following separation on SDS polyacrylamide gels, we transferred proteins to PVDF membranes (Millipore Immobilon-FL) and detected them by Western blotting, using appropriate antibodies. Membranes were blocked with Li-Cor Odyssey blocking buffer, and washes were carried out in PBS (phosphate-buffered saline) buffer with 0.1% (v/v) Tween-20. Primary and secondary incubations were in PBS-Tween with 10% (v/v) Odyssey blocking buffer, the latter also containing 0.01% SDS. Following a final wash in PBS, we scanned dried membranes on a Li-Cor Odyssey IR imager, and the images were analyzed using GE ImageQuant TL software. Antibodies were titrated and exposures set to ensure that all bands were in a linear response range. For amino acid sequence determination, proteolytic fragments were transferred to a PVDF membrane, excised and subjected to automated N-terminal protein sequencing at the Tufts University Core Facility.
Mass spectrometry identification of crosslinked species
RecBCD samples were analyzed by mass spectrom-etry as described in Supplementary Information. Briefly, the samples were denatured in trifluoroethanol, reduced with TCEP [tris(2-carboxyethyl)phosphine], and alky-lated with iodoacetamide prior to digestion with trypsin. Tryptic peptides were purified on C18 spin columns and analyzed using a Thermo Scientific Orbitrap Elite mass spectrometer with HCD fragmentation and serial mass spectrometry events. The RAW files were converted to mzXML files and analyzed by an in-house designed search algorithm RunBPAsearch to identify Bpa-cros-slinked peptides. All potential crosslinked peptide pairs were manually evaluated for the quality of the spectral match.
SAXS analyses
SAXS measurements were carried out at Stanford Synchrotron Radiation Laboratory Beam Line 4–2 [51] as detailed in Supplementary Methods. Measurements were gathered at RecBCD concentrations ranging from 0.4 to 1.8 mg/ml. Sample monodispersity was reflected by linearity of the Guinier regions (QRg < 1) and verified by dynamic light scattering. Analysis of SAXS data is detailed in Supplementary Methods.
Supplementary Material
Acknowledgements
We are grateful to Peter Schultz (The Scripps Research Institute) for plasmids directing incorporation of Bpa; Michael Berne (Tufts University Medical School) for amino acid sequence determinations; and Roland Strong, Adrian Ferré-d’Amaré, Steve Hahn, Ahmet Karabulut, Kyle Fowler, Randy Hyppa, and Naina Phadnis for helpful discussions and comments on the manuscript. This research was supported by research grants R01 GM031693 (G.R.S.), P50 GM076547 (J.R.), and R01 GM099989 (K.K.L.) from the National Institutes of Health of the United States of America. Data collection at Stanford Synchrotron Radiation Light-source was supported by grant P41-RR001209 from the National Center for Research Resources, a component of the National Institutes of Health.
Abbreviations used
- ds
double-stranded
- ss
single-stranded
- Bpa
p-benzoyl-l-phenylalanine
- SAXS
small-angle X-ray scattering
- EDTA
ethylenediaminetetraacetic acid
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2014.07.017.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Cromie GA. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J Bacteriol. 2009;191:5076–5084. doi: 10.1128/JB.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith GR. How RecBCD and Chi promote DNA break repair and recombination—a molecular biologist’s view. Microbiol Mol Biol Rev. 2012;76:217–228. doi: 10.1128/MMBR.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of Escherichia coli RecBCD enzyme. J Mol Biol. 1993;229:67–78. doi: 10.1006/jmbi.1993.1008. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 5.Taylor A, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- 6.Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL. Structure of Chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 7.Bianco PR, Kowalczykowski SC. The recombination hotspot X is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′- GCTGGTGG-3′. Proc Natl Acad Sci USA. 1997;94:6706–6711. doi: 10.1073/pnas.94.13.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation dependence of the cutting. Cell. 1985;41:153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- 9.Dixon DA, Kowalczykowski SC. The recombination hotspot x is a regulatory sequence that acts by attenuating the nuclease activity of the E coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 10.Taylor AF, Smith GR. Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme: modulation by ATP and magnesium levels. J Biol Chem. 1995;270:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- 11.Anderson DG, Kowalczykowski SC. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 12.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 13.Taylor AF, Smith GR. RecBCD enzyme is altered upon cutting DNA at a Chi recombination hotspot. Proc Natl Acad Sci USA. 1992;89:5226–5230. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AF, Smith GR. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 1999;13:890–900. doi: 10.1101/gad.13.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith GR. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 16.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amundsen SK, Taylor AF, Reddy M, Smith GR. Intersubunit signaling in RecBCD enzyme, a complex protein machine regulated by Chi hot spots. Genes Dev. 2007;21:3296–3307. doi: 10.1101/gad.1605807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 19.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domainin SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz DW, Taylor AF, Smith GR. Escherichia coli RecBC pseudorevertants lacking Chi recombinational hotspot activity. J Bacteriol. 1983;155:664–680. doi: 10.1128/jb.155.2.664-680.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundblad V, Taylor AF, Smith GR, Kleckner N. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5 . Proc Natl Acad Sci USA. 1984;81:824–828. doi: 10.1073/pnas.81.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handa N, Yang L, Dillingham MS, Kobayashi I, Wigley DB, Kowalczykowski SC. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, chi, by RecBCD enzyme. Proc Natl Acad Sci USA. 2012;109:8901–8906. doi: 10.1073/pnas.1206076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Handa N, Liu B, Dillingham MS, Wigley DB, Kowalczykowski SC. Alteration of chi recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc Natl Acad Sci USA. 2012;109:8907–8912. doi: 10.1073/pnas.1206081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith GR, Comb M, Schultz DW, Daniels DL, Blattner FW. Nucleotide sequence of the Chi recombinational hot spot χ+D in bacteriophage lambda. J Virol. 1981;37:336–342. doi: 10.1128/jvi.37.1.336-342.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- 26.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot Chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 27.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 28.Umezawa H. Low-molecular-weight enzyme inhibitors of microbial origin. Annu Rev Microbiol. 1982;36:75–99. doi: 10.1146/annurev.mi.36.100182.000451. [DOI] [PubMed] [Google Scholar]
- 29.Yu M, Souaya J, Julin DA. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli . Proc Natl Acad Sci USA. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amundsen SK, Taylor AF, Smith GR. A domain of RecC required for assembly of the regulatory RecD subunit into the Escherichia coli RecBCD holoenzyme. Genetics. 2002;161:483–492. doi: 10.1093/genetics/161.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor AF, Smith GR. Action of RecBCD enzyme on cruciform DNA. J Mol Biol. 1990;211:117–134. doi: 10.1016/0022-2836(90)90015-E. [DOI] [PubMed] [Google Scholar]
- 32.Cheng KC, Smith GR. Recombinational hotspot activity of Chi-like sequences. J Mol Biol. 1984;180:371–377. doi: 10.1016/s0022-2836(84)80009-1. [DOI] [PubMed] [Google Scholar]
- 33.Cheng KC, Smith GR. Cutting of Chi-like sequences by the RecBCD enzyme of Escherichia coli . J Mol Biol. 1987;194:747–750. doi: 10.1016/0022-2836(87)90252-x. [DOI] [PubMed] [Google Scholar]
- 34.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 35.Yu M, Souaya J, Julin DA. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- 36.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amundsen SK, Taylor AF, Smith GR. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination and DNA repair. Proc Natl Acad Sci USA. 2000;97:7399–7404. doi: 10.1073/pnas.130192397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis inE. coli . J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 39.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 40.Forne I, Ludwigsen J, Imhof A, Becker PB, Mueller-Planitz F. Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol Cell Proteomics. 2012:11. doi: 10.1074/mcp.M111.012088. http://dx.doi.org/10.1074/mcp.M111.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M, Weinkam P, Sali A, Lee KK. All-atom ensemble modeling to analyze small-angle x-ray scattering of glycosyl-ated proteins. Structure. 2013;21:321–331. doi: 10.1016/j.str.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Krajewski WW, Fu X, Wilkinson M, Cronin NB, Dillingham MS, Wigley DB. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature. 2014;508:416–419. doi: 10.1038/nature13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosamond J, Telander KM, Linn S. Modulation of the RecBC enzyme of Escherichia coli K12 by Ca2+ . J Biol Chem. 1979;254:8646–8652. [PubMed] [Google Scholar]
- 47.Kim J, Seo MH, Lee S, Cho K, Yang A, Woo K, et al. Simple and efficient strategy for site-specific dual labeling of proteins for single-molecule fluorescence resonance energy transfer analysis. Anal Chem. 2013;85:1468–1474. doi: 10.1021/ac303089v. [DOI] [PubMed] [Google Scholar]
- 48.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 49.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli . J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 50.Boehmer PE, Emmerson PT. Escherichia coli RecBCD enzyme: inducible overproduction and reconstitution of the ATP-dependent deoxyribonuclease from purified subunits. Gene. 1991;102:1–6. doi: 10.1016/0378-1119(91)90529-k. [DOI] [PubMed] [Google Scholar]
- 51.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J Appl Crystallogr. 2007;40:s453–s458. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.