Abstract
LNK (SH2B3) is an adaptor protein studied extensively in normal and malignant hematopoietic cells. In these cells, it down-regulates activated tyrosine kinases at the cell surface resulting in an antiproliferative effect. To date, no studies have examined activities of LNK in solid tumors. In this study, we found by in silico analysis and staining tissue arrays that the levels of LNK expression were elevated in high grade ovarian cancer. To test the functional importance of this observation, LNK was either overexpressed or silenced in several ovarian cancer cell lines. Remarkably, overexpression of LNK rendered the cells resistant to death induced by either serum starvation or nutrient deprivation, and generated larger tumors using a murine xenograft model. In contrast, silencing of LNK decreased ovarian cancer cell growth in vitro and in vivo. Western blot studies indicated that overexpression of LNK upregulated and extended the transduction of the mitogenic signal, whereas silencing of the LNK produced the opposite effects. Furthermore, forced expression of LNK reduced cell size, inhibited cell migration and markedly enhanced cell adhesion. LC-MS identified 14-3-3 as one of the LNK binding partners. Our results suggest that in contrast to the findings in hematologic malignancies, the adaptor protein LNK acts as a positive signal transduction modulator in ovarian cancers.
Keywords: LNK, SH2B3, ovarian cancer, mitogenic signaling
Introduction
The adaptor protein LNK (SH2B3) is a key negative regulator of the signaling pathway of hematopoietic receptors activated by growth factors 1, 2, thus playing a critical role in hematopoiesis 3. The protein contains a N-terminal proline-rich region which mediates dimerization, a pleckstrin homology (PH) domain and a Src homology 2 (SH2) domain which specifically binds to phosphorylated tyrosines and mediates signal transduction 1. Previous studies showed that LNK knockout mice had splenomegaly and abnormal lymphoid and myeloid homeostasis 4, 5. Subsequent studies revealed that LNK inhibited the activating signals mediated by thrombopoietin (TPO) 6–8 and erythropoietin (EPO) 9. On the other hand, overexpression of LNK inhibited proliferation of hematopoietic transformed cells through the suppression of the kinase activity of oncogenic JAK2 V617F 10, 11, MPL W515L 12, c-KIT 13, 14, c-FMS 15 , PDGFR/FIP1L1-PDGFRα and TEL-PDGFRβ 16, as well as FLT3-ITD 17. Recently, LNK mutations have also been found in patients with myeloproliferative neoplasms (MPN) 18–21, early T cell acute lymphoblastic leukemia (ALL) 22 , Ph-like ALL 23 , B-precursor ALL 24 and Down syndrome-related myeloid disorders 25. The mutational loss of function of LNK in hematopoietic cells releases its inhibitory activity against the activated tyrosine kinase receptors and its downstream JAK-STAT pathway, resulting in enhanced hematologic cell proliferation, thus fostering the development and progression of these hematopoietic neoplasms 10. The resultant phenotype is reminiscent of the MPN-like features of LNK−/− mice 26.
Most of the studies that have explored the function of LNK focused on either normal or transformed hematologic cells; recently, several studies have explored the effects of LNK on the function of normal endothelium cell 27–29. To our knowledge, the role of LNK in solid tumors has not been addressed. Previously, while studying hematopoiesis, control experiments noted that forced overexpression of LNK did not inhibit the growth of several nonhematopoietic cell lines (NIH3T3 and 293T) 6. In an overview survey, we found that selected solid cancers expressed LNK, and forced overexpression of LNK in several solid cancer cell lines did not have a significant effect on their proliferation, suggesting LNK may have a different role in solid tumor cells compared to hematologic dyscrasias 30. This observation prompted us to do an in depth study of the effect of LNK in ovarian cancer cells. We chose this cancer because we previously noted that some ovarian cancer cell lines had detectable levels of LNK mRNA 30.
Materials and methods
Antibodies and reagents
The following antibodies were used in this study: anti-human LNK antibody [AF5888, R&D system Inc, a sheep IgG against E.coli derived recombinant human LNK protein (amino acids 427–575)]; murine anti-β-actin monoclonal antibody (Sigma); V5 and pan 14-3-3 antibody (Abcam); antibodies against p-JAK2 (Tyr1007/1008), JAK2, p-p38 (Thr180/Tyr182), p-ERK1/2 (Thr202/Tyr204), p-JNK1/2 (Tyr183), p-PDK1 (Ser243), p-P70S6 (Thr421/Ser424), p-GSK3beta (Ser9), p-AKT (Ser473) and AKT, p-PI3K p110δ (Tyr485) (from either Cell Signaling Technology or Santa Cruz) and p-FAK Tyr861 (Epitomics).
Cell lines and cell culture
Ovarian cancer cell lines OVCA433, C13, A2008, CAOV-2 (provided by Ruby Huang, NUS) and melanoma cell lines M285, M368 (kindly provide by Antoni Ribas, UCLA) were maintained in RPMI 1640 containing 10% fetal bovine serum (FBS) with penicillin and streptomycin. The ovarian cancer cell line OVCAR5 was maintained in the same medium with 10 μg/ml insulin; OV7 and OV56 were maintained in DMEM Hi-Glucose/Ham’s F-12 [1:1] plus 10% FBS, 0.5 μg/mL hydrocortisone and 10 μg/mL insulin. HEK293T cells were cultured in DMEM medium with 10% FBS. Cells were grown at 37°C with 5% CO2 in humidified air.
Lentivirus and stable cell line generation
The pLKO.1-puro-CMV-TurboGFP lentivirus plasmid (SHC003) was obtained from Sigma. The entire coding region of huLNK, including the HIS tag and V5 tag, was amplified from pcDNA3 LNK using primers TGAGCTAGCATGAACGGGCCTGCCCTGCAGCC (Nhe I) and AAACACGTGCTCGAGCGGCCGCCACTGT (Pml I). The PCR product was ligated to pGEM-T vector and validated by Sanger sequencing. This construct was digested with Nhe I and Pml I, the LNK containing fragment was gel purified, and the GFP coding fragment of pLKO-CMV-GFP vector (SHC003) was replaced with the LNK open reading frame (pLKO-CMV-LNK). For gene silencing, shRNA plasmids targeted to LNK [TRCN0000265715 (shRNA15), TRCN0000265716 (shRNA16), TRCN0000256095 (shRNA95) and Scramble shRNA SHC002 were purchased from Sigma. shRNA plasmids targeted to AKT1 (TRCN0000010174), 14-3-3 Q (YWHAQ TRCN0000078169), 14-3-3 Z (YWHAZ TRCN0000029404) and 14-3-3 G (YWHAG TRCN0000078158) were generated according to the protocol described by Addgene (http://www.addgene.org/tools/protocols/plko/). The sequences of all the shRNA constructs were confirmed by Sanger sequencing using the U6 primer.
Ovarian cancer tissue array analysis
Human ovarian cancer tissue array (OVC1021) was purchased from Biomax US. Specifity of LNK antibody was validated by Immunohistochemical staining (IHC) of formalin fixed and paraffin embedded blocks either of silenced or overexpressed LNK OVCA433 cell. Detail process of IHC is described in the Supplemental Material.
Murine xenograft model
In vivo cell proliferative effects after either gain or lost of LNK was studied in a murine xenograft model. 5–6 weeks old Nod-SCID mice were used for the study. 2 million (CAOV2 and A2008) or 6 million (OVCAR5) cells were resuspened in 100 μl FBS and 100 μl Matrix gel (BD Biosciences), and subcutaneously injected into both flanks of immune deficient Nod-SCID mice. The mice were sacrificed, and the tumors were excised and weighted at the end of the experiments (days 18–28).
Microarray analysis
Microarray analyses were performed in triplicate using OVCA433 cells overexpressing LNK, compared to control cells containing GFP. The array hybridization was performed with Illumina Human HT-12 v4 Expression BeadChip, the pathway analysis was accomplished with KEGG and Biocarta database. Real time RT-PCR was performed to validate the significantly changed genes.
Co-Immunoprecipitation and LC-MS analysis
LNK overexpressing cell lines were place into the protein lysis buffer (0.5% Nonidet P40, 50 mM Tris/HCl pH 8.0, 150 mM NaCl with protease inhibitor cocktail and phosphatase inhibitors NaF and Na3VO4) at 4°C for 15 min, and centrifuged at 12000 rpm (15 min, 4°C) to remove cell debris. Protein lysates were shaken overnight with V5 antibody at 4°C and collected by precipitation with protein A/G beads. After washing with protein lysis buffer 3 times, the protein binding to the protein A/G beads were eluted with 5 x SDS loading dye; the eluted samples were subsequently used for LC-MS analysis.
Method in Supplemental Material
Detailed methods for Lentivirus packaging, western blot, immunofluorescence microscopy and the assays for cell proliferation, cell adhesion and detachment, cell motility and invasion, are provided in the Supplemental Material.
Result
Elevated expression of LNK in ovarian cancers
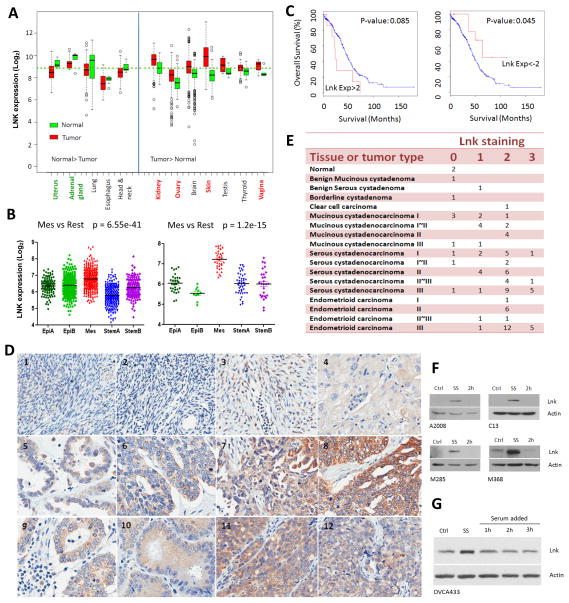
Previously, others 6 and our group 30 showed that overexpression of LNK inhibited cell growth and caused cell death in many leukemia cell lines, but similar experiments performed in several solid tumor cell lines had little effect on their proliferation 30, suggesting LNK might have a different role in solid tumor cells compared to those of the hematopoietic system. In silico analyses shown that rather than being down-regulated as anticipated for a tumor suppressor, LNK is upregulated in several types of tumors including skin (melanoma), kidney and ovarian cancers (Fig. 1A). Meanwhile, cancer copy number data from TCGA suggests the LNK locus is amplified in several solid tumors including sarcoma (5.8%, 3/52 cases in TCGA dataset or 2.4% 5/207 cases in MSKCC/Broad dataset), bladder cancer (3.8%, 1/26 cases) and ovarian cancer (2.3%, 7/311 cases) (Supplemental Fig. 1). Here, we focused on ovarian cancer. LNK expression was examined in the microarray data of 1,538 ovarian cancer patient samples and 142 ovarian cancer cell lines (molecular ovarian cancer subtypes were classified as described in 31). Among the five different ovarian subtypes (Epi-A, Epi-B, Mes, Stem-A and Stem-B), LNK mRNA expression is significantly elevated in the mesenchymal subtype in both patient samples and cell lines (Fig. 1B). Mesenchymal subtype includes more advanced staged including metastatic tumours; this subtype is associated with poorer outcomes 31. This was further analyzsed using cBio cancer genomics portal (http://www.cbioportal.org/public-portal/index.do) of the TCGA dataset. A total of 316 serous ovarian cancer sample were separated into LNK high and low expression groups, Patients with a higher LNK expression showed a worse outcome compared to those with a lower LNK expression (Fig.1C, left). In contrast, those patients having relative low LNK expression had a better survival (Fig.1C, right).
Fig. 1. Elevated expression of LNK in various cancers and serum-starved cells.
A. LNK expression in various human cancers compared to matched normal tissue. Left panel, LNK mRNA levels higher in the normal tissue v/s the cancer samples. Right panel, LNK mRNA levels higher in the cancer samples v/s normal tissues. Data are from GEO database. B. LNK expression in five different molecular subtypes 31 of ovarian cancer patients samples (left panel) and ovarian cancer cell lines (right panel). C. Kaplan-Meier plots of overall survival of 316 ovarian cancer patients: comparison of cases with high levels (EXP:>2, defined as: > mean + 2SD, left panel) versus low levels (EXP <-2, defined as: < mean - 2SD, right panel) of LNK mRNA. P values are calculated by log rank test and indicated on the graph. Data are obtained from TCGA (The Cancer Genome Atlas) ovarian cancer project using the cBio Cancer Genomics Portal. D. IHC staining of ovarian cancer tissue array stained with LNK antibody (Photo of the whole section core of each sample are provide in Supplemental Fig 4 and which appear in the same order): #1, Normal ovary 1; #2, Normal ovary 2; #3, Benign ovarian tumor; #4, Clear cell ovarian carcinoma; #5, Serous Cystadenocarcinoma of the ovary (Grade I); #6, Serous cystadenocarcinoma (Grade II); #7, Serous cystadenocarcinoma (Grade III); #8, Serous cystadenocarcinoma (Grade III); #9, Endometrioid carcinoma of the ovary (Grade I), #10, Endometrioid carcinoma (Grade II) #11, Endometrioid carcinoma (Grade III), #12, Endometrioid carcinoma (Grade III). E. IHC staining score of 84 ovarian samples (see Supplementary Material for scoring intensity). F. Western blots of endogenous LNK levels in ovarian cancer cell lines (A2008 and C13) and melanoma cell lines (M285 and M368) during serum starvation (18 hours, SS) and followed by addition of serum (2 hours). Confluent cultures were changed to serum-free medium for 18 hours, and then switched back to complete medium containing 10% FBS for 2 hours. Western blot was performed to examine the LNK protein at indicated time points. Ctrl (control cell grown in the complete medium containing 10% FBS). G Western blot of LNK in the ovarian cancer cell line OVCA433 with forced expressed LNK. Cells were serum-starved (18 hours) followed by serum stimulation (as described above) for 1–3 hours, and LNK protein was examined by western blot. β-actin was used as loading control.
In addition, moderate LNK immunohistochemical staining was observed in many serous cystadenocarcinoma and endometrioid carcinoma samples, expecially in high grade disease (Figs. 1D, E and Supplemental Figs. 3–5); only weak staining was observed in the benign serous cystadenoma and negative staining occurred in the normal ovary tissue.
Overexpression of LNK renders the cells resistant to cell death
Subsequently, we examined LNK protein expression in several ovarian cancer and melanoma cell lines by western blot. Unexpectedly, we found the LNK protein levels were significantly upregulated during serum-starvation which returned to baseline levels when the cells were restored to complete medium containing 10% FBS. This was noted in the ovarian cancer cell lines A2008 and C13, as well as the melanoma cell lines M285 and M368 (Fig. 1F). We also observed this phenomenon in cells which stably contained a LNK expression vector whose level of LNK transcription was constantly driven by the CMV promoter (Fig. 1G). Thus, accumulation of LNK protein with serum starvation is probably a post-translational event.
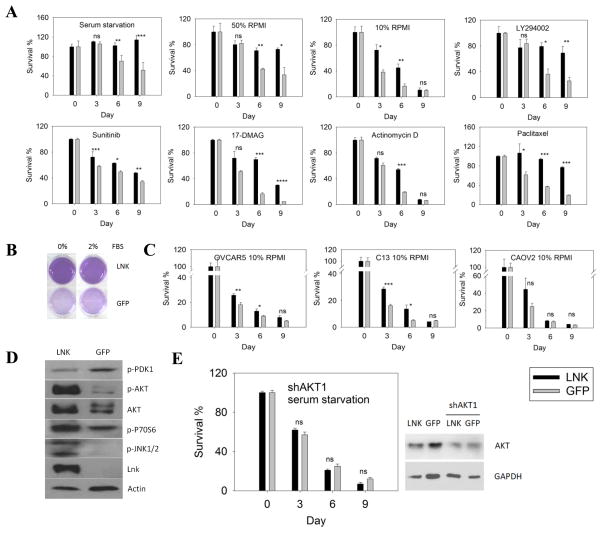
To test the functional importance of this finding, we studied both forced-expression and silencing of LNK in several ovarian cancer cell lines. Overexpression of LNK in the ovarian cancer cells OVCA433 significantly enhanced their cell survival during prolonged serum starvation. After 9 days culture in serum-free RPMI medium, over 60% of the control GFP cells were dead. In contrast, almost all the LNK overexpressing cells survived, suggesting that LNK promoted cell survival in serum-free adverse conditions [Fig. 2A (left), Fig. 2B]. In another set of complementary experiments, LNK protected ovarian cancer cells in even more adverse growth condition. LNK overexpressing cells displayed significantly higher survival rates than the GFP control cells after eliminating serum and growing in either 50% or 10% RPMI culture medium (replacing RPMI with PBS) (Fig. 2A, upper panel). Also, overexpression of LNK rendered the ovarian cancer cells resistant to death during exposure to either PI3 kinase inhibitor (LYS294002), multi-targeted receptor tyrosine kinase inhibitor (Sunitinib), inhibitor of transcription (Actinomycin-D), chemotherapy drug (Pacitaxel) or HSP90 inhibitor (17-DMAG), further confirming the pro-survival effect produced by the gain of LNK expression (Fig. 2A). This phenomenon was also observed to various degrees in three other ovarian cancer cell lines: CAOV2, OVCAR5 and C13 cultured in the absence of serum with the addition of only 10% RPMI and 90% PBS (Fig. 2C). Western blot revealed that the p-AKT pro-survival signal pathway in the LNK expressing cells was strongly activated during starvation (Fig. 2D). Phosphorylated P70S6 and JNK1/2 were also increased (Fig. 2D). When both cell lines (GFP or LNK) were infected with lentivirus containing shRNA targeted to AKT1, the pro-survival effect of LNK was diminished in the presence of serum starvation, suggesting that LNK activates the p-AKT pathway and protects the cells from cell death (Fig 2E).
Fig. 2. Forced expression of LNK renders the cells resistant to death.
A. OVCA433 cells with either forced express of LNK or GFP control, were cultured under conditions of either serum starvation or nutrient deprivation (50/50 % RPMI/PBS or 10/90 % RPMI/PBS), or treated with LY294002, Sunitinib, 17-DMAG, Actinomycin D or Paclitaxel. Cell survival was determined by MTT colorimetric assay. B. Cells overexpressing either LNK or GFP were cultured with RPMI medium with either 0% or 2% FBS for 9 days and the viability of the cells was examined by MTT assay. C. Cell resistance to death induced by nutrient deprivation (10/90% RPMI/PBS) of OVCAR5, C13 and CAOV2, ovarian cancer cells ± forced expression of LNK and MTT colorimetric assay was performed. D. Activation status of the AKT/MAPK signaling pathways in OVCA433 cells either with (left lane) or without (right lane) forced expression of LNK. Cells were serum-starved for 9 days and various total and phosphorylated (p-) proteins were examined by western blot. E. OVCA433 cells (forced expression of either LNK or GFP control) were infected with lentivirus containing shRNA targeting to AKT1 and then cultured under conditions of serum starvation. Cell survival was determined by MTT assay (left panel). Western blot was performed to examine the silencing of AKT (right panel). Significance was calculated by paired student t-test and was defined as: ns (not significant), p > 0.05; * p≤0.05; ** p≤0.01; *** p≤0.001; **** p≤0.0001.
LNK promotes tumor growth in an in vivo murine xenograft model
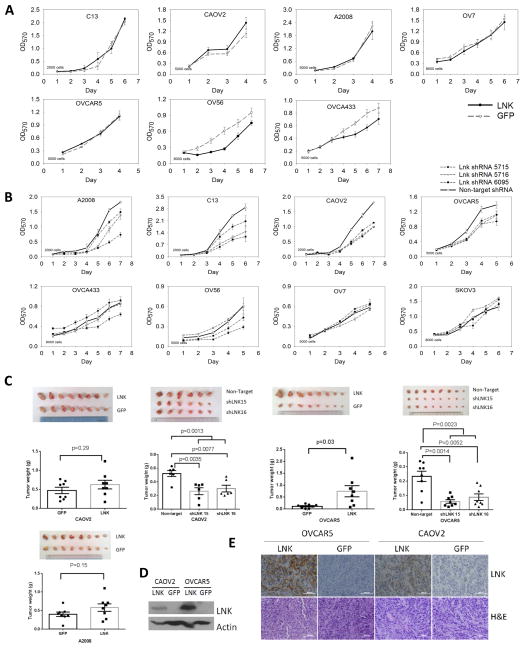
Under optimal growth conditions, forced expression of LNK in five out of 7 ovarian cancer cell lines had little effect on cell proliferation (Fig. 3A), while OV56 and OVCA433 had a slight inhibition of their cell growth. On the other hand, stable silencing of LNK using three different shRNAs slowed the proliferation of six of 8 ovarian cancer cell lines (Fig. 3B), including A2008, CAOV2, C13, OVCA433, OVCAR5 and OV56 (Fig. 3B).
Fig. 3. In vitro and In vivo effects of forced expression or silencing of LNK on cell growth.
A.B. In vitro effects of either forced expression (A) or silencing (B) of LNK on ovarian cancer cell growth. MTT assays were performed in 96 wells plates, and the number of cells seeded in each well at point zero is shown. (Mean ± SD, n=6). C. Effect of either forced expression or silencing of LNK on the growth of CAOV2, A2008 and OVCAR5 xenografts in Nod-SCID mice. 2 million (CAOV2 and A2008) or 6 million (OVCAR5) cells were resuspened in FBS, mix with matrigel and subcutaneously injected into both flanks of immune deficient Nod-SCID mice. Tumors size and weight were measured after the mice were sacrificed on either day 18 (CAOV2, A2008), day 21 (OVCAR5 cell force LNK) or day 28 (OVCAR5 silencing LNK). Tumor weights are shown as mean ± sem. D. Western blot of the protein extract from the xenograft tumor. E. IHC staining of the xenograft tumor (CAOV2 and OVCAR5) with either LNK antibody or H&E.
To observe the in vivo effect of LNK on cell growth, murine xenograft models were established by subcutaneous injection of either CAOV2, OVCAR5 or A2008 cells having either forced expression or silencing of LNK. Tumor sizes and weights were increased in all the cell lines with forced expression of LNK, expecially the OVCAR5 tumors (Fig. 3C, p value 0.03). In contrast, silencing of LNK in these cell lines reduced their tumor sizes and weights. Taken together, LNK promotes tumor growth in three murine xenograft models. Western blot and IHC staining confirmed the LNK overexpression in these tumors (Figs. 3D, E).
LNK regulates mitogenic signalling
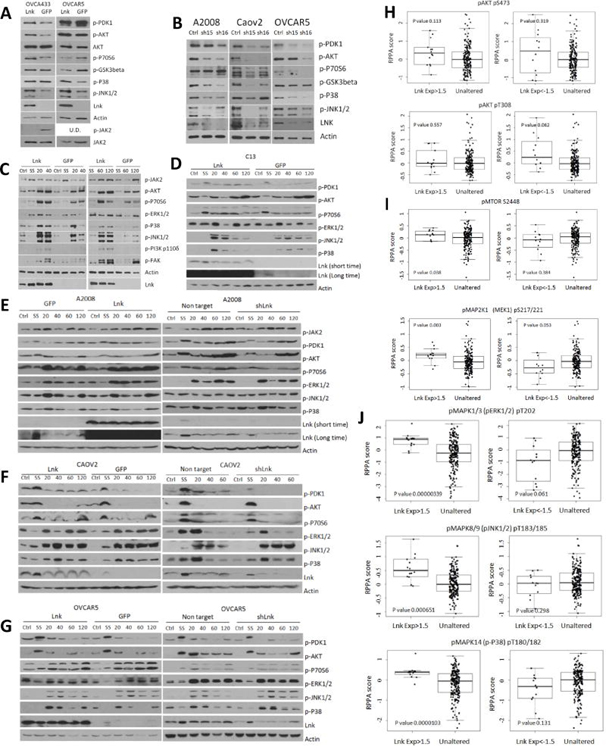
To investigate potential mechanisms by which LNK regulates cellular proliferation, major mitogenic signaling pathways were examined upon either gain or loss of LNK expression. In agreement with our notion that LNK might be a positive regulator in these cell lines, LNK expression enhanced several signaling pathways which are critical for cell proliferation and survival in two ovarian cancer cell lines, OVCA433 and OVCAR5 (Fig. 4A). Forced expression of LNK increased phosphorylation of AKT and JNK1/2 in both cell lines, as well as increasing phosphorylation levels of PDK1 and p70S6 in OVCA433, and phosphorylation level of p38 in OVCAR5. Of interest, similar to hematopoietic cells, LNK attenuated the modest basal levels of p-JAK2 in OVCA433 (Fig. 4A). In contrast, stably silenced LNK using two shRNA in three ovarian cancer cell lines (A2008, CAOV2 and OVCAR5) resulted in down-regulation of expression of p-PDK1, p-AKT, p-GSK3β and MAPK kinase (p-p38 and p-JNK1/2) (Fig. 4B). In further experiments, we examined in detail the alterations of mitogenic signaling upon serum stimulation in several ovarian cancer cell lines having either forced expression or silencing of LNK (Figs. 4C, D, E, F, G). These cells were serum-starved (SS) overnight followed by serum stimulation for different durations (20 to 120 mins). The serum stimulated LNK overexpressing cells showed higher levels of p-AKT, p-P70S6, p-ERK1/2 compared to similarly treated control cells, especially the OVCA433 and A2008 cells (Fig. 4C), while silencing of LNK generally generated the opposite effect (Figs. 4E, F, G, right panels).
Fig. 4. LNK upregulates mitogenic signaling pathways in ovarian cancer cells.
A. Phosphorylation status of a number of signaling pathways in OVCA433 and OVCAR5 cells with forced expression of LNK. Cells were grown in optimal conditions, lysed, total protein separated by SDS-PAGE and western bloted with indicated antibodies. B. Western blot analysis of phosphorylation status of signaling pathways in A2008, CAOV2 and OVCAR5 cells stably silenced for expression of LNK with shRNA. Ctrl (non-target shRNA), sh15 and sh16 (shRNA 5715 and 5716, both target LNK). Figs. C.D.E.F.G: Phosphorylation status of AKT and MAPK pathways in OVCA433 (C), C13 (D), A2008 (E), CAOV2 (F) and OVCAR5 (G) ovarian cancer cells with either forced expression or silencing of LNK after serum-starvation overnight, followed by serum- stimulation for indicated durations, and Western blot was performed with indicated antibodies. Ctrl cells in normal growth conditions, SS (serum-starved overnight), 20–120 min (serum-starved overnight followed by serum stimulation for 20–120 minutes). Figs. H, I, J: Correlation of expression levels of LNK mRNA and activated (phosphorylated) AKT (panel H), MTOR and MAP2K1 (MEK1) (panel I), MAPK (panels J, includes p-ERK1/2, p-JNK1/2 and p-p38) in 316 ovarian cancer samples [protein data from TCGA ovarian cancer, reverse phase protein array (RPPA) dataset].
In agreement with our observations, a significant correlation can be noted between the endogenous levels of LNK and the activation status of MAPK pathway in the TCGA 316 ovarian cancer samples [Reverse Phase Protein Array (RPPA)]. Ovarian cancer samples with high expression of LNK displayed high levels of p-AKT, p-mTOR, p-MAP2K1 and p-MAPK (p-ERK1/2, p-JNK1/2 and p-p38), whereas samples with lower LNK mRNA expression had lower phosphorylation levels of the above proteins (except p-AKT) (Figs. 4H, I and J).
LNK enhances cell attachment and inhibits cell migration
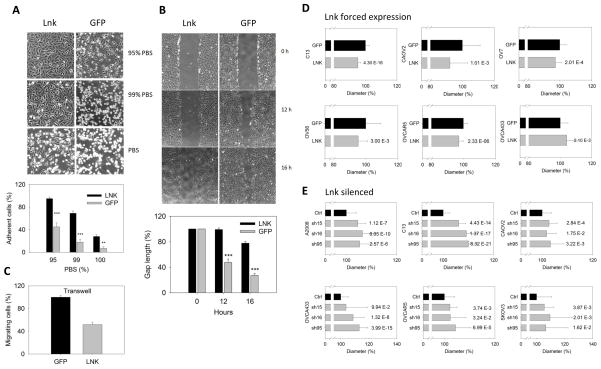
During cell passage, we noted that stable LNK overexpressing OVCA433 cells displayed a strong attachment to the tissue culture plate, requiring either a longer duration or higher concentration of trypsin to release them from the plates compared to control cells. In contrast, stable knockdown of LNK in these cell lines slightly reduced the length of exposure to trypsin in order to release them from the plates (data not shown). We, therefore, considered that LNK enhanced cell adhesion. To explore this hypothesis, OVCA433 cells were allowed to adhere to culture dishes in six well plates until they reached confluence. Plates were washed with PBS (not containing calcium) and cultured in different percentages of PBS (95–100%) and RPMI (5–0%) for 3 hours. This lead to disruption of calcium-dependent cell contact junction/adhesion and slowly release the cells. As anticipated, compared to GFP control cells, those with forced expression of LNK protein significantly retained their cell attachment to the plates as the culture media were replaced with increasing amounts of calcium-free PBS (Fig. 5A). In the presence of 99% PBS for 3 hours, a total of 69% of the LNK expressing cell were still attached to the plates, compared to 18% of control cells.
Fig. 5. LNK regulates cell adhesion, migration and cell size.
A. Overexpression of LNK renders cells more adhesive to tissue culture plate. Cell detachment was measured by replacing regular RPMI media with different ratios of PBS to RPMI, resulting in media with either 95 %, 99 % or 100 % PBS. Upper panel, photographs of cells taken 3 hours after media replacement; lower panel, graphic representation of data. B. Upper panel: representative images from wound healing experiments using confluent OVCA433 cells overexpressing either GFP or LNK. Pictures were collected at 0, 12 and 16 hours. Lower panel: graphic display of the size of the gap at 12 and 16 hours after ‘wounding’, expressed as a percentage of the gap of control at time zero. Results represent mean ± SD of 4 experiments. C. Transwell experiments of OVCA433 cells stably expressing either LNK or GFP. Cells migrating through the membrane were determined by MTT dye staining. Data are expressed as a percentage of migrating cells, expressed as a percent of control cells (48 hours), * p≤0.05; ** p≤0.01; *** p≤0.001. D. Overexpression of LNK reduces the cell diameter in a panel of ovarian cancer cell lines. Ovarian cancer cell lines with either forced expression of LNK or GFP controls were grown in similar, non-confluent conditions, trypsinized, centrifuged, resuspended in complete media and their cell diameters were measured using the Vi-CELL Cell Viability Analyzer (Beckman). E. Silencing of the LNK enlarges the diameter of ovarian cancer cell lines. Ovarian cancer cell lines which were stably expressing shRNA targeted either to LNK (5715, 5716 and 6095) or non-target shRNA as control (Ctrl), were collected and their cell diameters were measured. For each cell line, 500~2,000 cells were measured to generate the mean value of the cell diameter. Significance was calculated by paired student t-test; and the P values are indicated after each bar.
Cell migration was measured using the wound healing assay (Fig. 5B). At 16 hours after the wound scratch, LNK overexpressing OVCA433 cells maintained a nearly intact gap. In contrast, the GFP-control cells nearly closed the healing gap, indicating that LNK indeed plays a role in regulation of cell migration. In addition, transwell experiments found that LNK overexpressing ovarian cancer cells migrated significantly less than the GFP control cells (Fig. 5C).
LNK regulates cells size
Consistent with the notion that LNK may play a role in the cytoskeleton organization and regulate cellular shape, overexpression of LNK reduced the cell diameter of five of 6 ovarian cancer cell lines (Fig. 5D). In contrast, silencing of LNK generated the opposite effect (Fig. 5E). The cell diameter increased to various extents in the six ovarian cancer lines having one of three shRNA targeted to different regions of LNK. This was most prominent in A2008, C13, OVCA433 and SKOV3 ovarian cancer cell lines (Fig. 5E).
Pathway analysis of microarray data
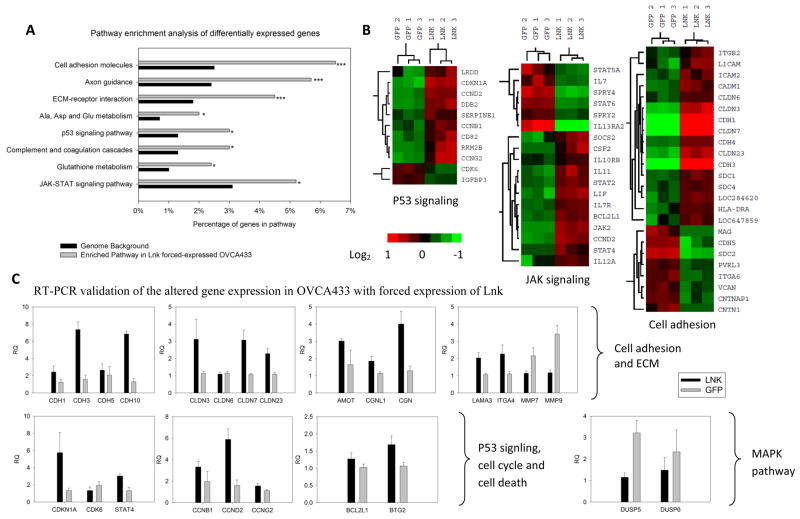
RNA microarray analysis was performed to examine the gene expression changes after forced expression of LNK in OVCA433. Compared to vector control cells, the experimental cells had a prominent upregulation/alteration of genes related to extracellular matrix (ECM) receptor interaction (p-value 7.91 E−4), cell adhesion/attachment (p-value 1.46 E−4), and molecules associated with cell-cell tight junctions (Fig. 6A). A heat map displays genes with altered expression related to cell adhesion, p53 signaling and the JAK-STAT pathway (Fig. 6B).
Fig. 6. Altered gene expression in LNK overexpressing OVCA433 cells.
A. Microarray analysis was performed in triplicate using OVCA433 cells either overexpressing LNK or GFP (control). Pathway enrichment analysis of genes differentially expressed between LNK and control cells was analyzed using the KEGG database. * p≤0.05; *** p≤0.001. B. Heat map of altered genes in p53, JAK-STAT and cell adhesion pathways. C. Real time PCR validation of prominently altered genes related to cell adhesion and ECM (upper panel), p53 signaling, cell cycle and cell death (lower left panel) and MAPK pathway (lower right panel) in LNK versus control cells. RQ (Relative Quantification of the gene expression after normalization to beta-actin). Results represent mean ± SD of quantitative measurments.
Quantitative real time RT-PCR was performed to confirm some of the interesting genes. Several members of the cadherin family (CDH1, 3, 5, 10) (contributes to cell attachment) and members of the claudin family (CLDN3, 6, 7, 23) (critical for the cell-cell tight junction formation) were indeed upregulated in the LNK overexpressing cells (Fig. 6C, top panel). Additional genes related to cell adhesion and cell-cell connection including AMOT, CGNL1, CGN, Laminin (LAMA3) and integrin (ITGA4) were also upregulated in the LNK overexpressing cells, as confirmed by real time PCR (Fig. 6C, upper panel). In contrast, two matrix metallopeptidases associated with extracelluar matrix degradation and enhanced cell migration (MMP9 and MMP7) were downregulated in the LNK overexpressing cells (Fig. 6C, upper panel). Interestingly, several cyclins (CCNB1, CCND2 and CCNG2), as well as BCL2L1 (a potent inhibitor of cell death) were upregulated in cells with overexpression of LNK, which is consistent with their resistance to cell death. In addition, two phosphatases (DUSP5 and DUSP6) were down-regulated in the LNK overexpressing cells (Fig. 6C, lower panel), which may contribute to the elevated MAPK signaling in the LNK overexpressing cells.
Liquid chromatography–mass spectroscopy (LC-MS)
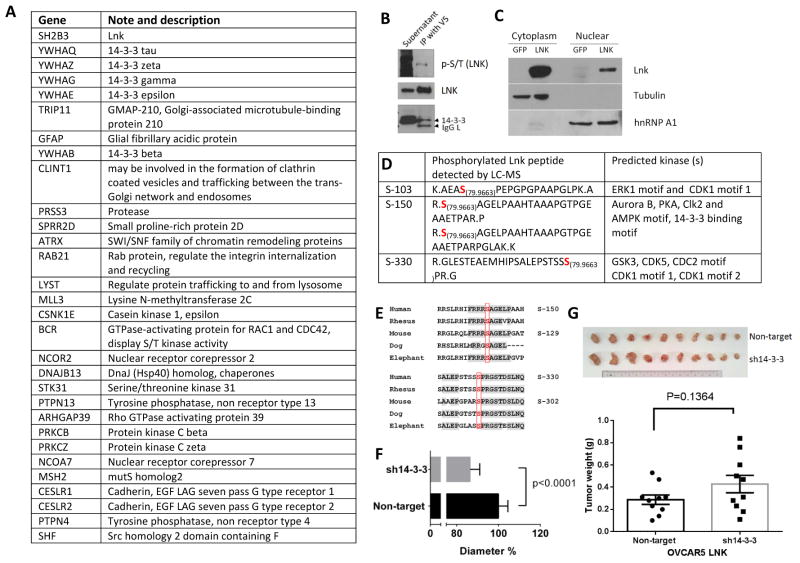
To investigate further the potential mechanism by which LNK regulates signal transduction, LC-MS was performed to identify possible LNK binding proteins. LNK and the binding proteins were initially co-immunoprecipitated with V5 antibody from OVCA433 cells stably expressing V5 tagged LNK; the immunoprecipitated protein complex was extensively washed and analyzed by LC-MS. Proteins also present in the control (OVCA433 cells stably expressing GFP immunoprecipitated with either V5 antibody or normal IgG) were removed from the results, and the candidate binding partners are summarized (Fig 7A). The protein 14-3-3, recently identified as a LNK binding partner in hematologic cells 32, appears at the top of the list (Fig. 7A). This binding was further confirmed by immunoprecipitation and western blot (Fig. 7B), suggesting that LNK also bound to this scaffold protein in ovarian cancer cells. Interestingly, several binding candidates (MLL3, ATRX, NCOR2, and NCOR7) are nuclear protein having important role in regulation of histone methylation, chromatin remodeling and control of transcription. Their presence on the list raises the possibility that LNK may also be active in the nucleus. SH2B1β, another member of the same family as LNK, shuttles between the plasma membrane, cytosol and nucleus; and its nuclear-cytosol cycling is critical to promote NGF-mediated preneuronal cell differentiation 33. Our protein fractionation and western blot experiments showed a small amount of LNK was indeed present in the nuclear protein fraction (Figs. 7C); the role of the nuclear LNK requires further study.
Fig. 7. LC-MS identification of binding partners and phosphorylation sites of LNK.
A. Potential LNK binding partners identified by mass spectrometry. Ranking based on the mass spectrometry Log (e) value. LNK protein expressed in OVCA433 was immunoprecipitated using V5 antibodies; potential binding proteins were detected by LC-MS as described in Materials and Methods. B. Western blot confirms the serine/threonine phosphorylation (p-S/T) of LNK and LNK binding to 14-3-3. LNK protein was pulled down from OVCA433 cells overexpressing V5 tagged LNK using V5 antibodies and examined by western blot with antibodies against total phosphorylated S/T (upper panel). The membrane was stripped and re-probed with antibodies to LNK (middle panel) and 14-3-3 (lower panel). IgG L indicated the IgG light chain non-specific band. C. Total proteins from OVCA433 cells overexpressing LNK were separated into cytoplasmic and nuclear fractions, and western blot was performed. Successful separation of cytoplasmic and nuclear proteins was confirmed by probing blots with antibody against α-tubulin (cytoplasmic protein) and hnRNP (nuclear protein). D. Phosphorylation of LNK identified by LC-MS. 79.9663 indicates the possible phosphorylation site (molecular weight of phosphoryl group). The motifs recognized by the possible binding kinases were predicted using Scansite (http://scansite.mit.edu/). E. Comparison of the LNK amino acid sequences surrounding the phosphorylation sites (S-150 and S-330) in humans and other mammals (phosphorylation sites are highlighted with red). F. Silencing of 14-3-3 further reduces the cell diameter of OVCAR5 with stable forced expression of LNK. Result are expressed as a percentage of the cell infected with non-target shRNA (but force expression of LNK). G. Silencing of 14-3-3 modestly enhanced in vivo tumor growth of OVCAR5 stably expressing LNK. A total 6 million cells were resuspened in FBS, mix with matrigel and subcutaneously injected into both flanks of immune deficient Nod-SCID mice. The mice were sacrificed on day 15, tumor are pictorially shown and their weights shown as mean ± sem of 10 tumors.
Three phosphorylation modification of LNK were identified by mass spectroscopy (Fig 7D): S-103, S-150 and S-330, and the phosphorylation modification was further confirmed by antibodies specifically targeting total phosphorylated S/T (Fig. 7B). Among these sites, Serine-150 is the human LNK homolog of murine LNK S-129 (Fig.7E), which after phosphorylation allows LNK to bind avidly to 14-3-3 32. Phosphorylation (S-150) of human LNK is also predicted to mediate binding of the 14-3-3 (http://scansite.mit.edu/). This is consistent with our immunoprecipitation studies showing that 14-3-3 is bound to LNK (Figs. 7A, B). Also, both sites (S-150 and S-330), and their surrounding amino acids are highly conserved across many species (Fig.7E), suggesting their biological importance. Our MS results covered the peptide fragments containing Y44, Y203, Y273, Y401, Y555 and Y572 of LNK, and no phosphorylation of tyrosine residues in these fragments was detected.
We further investigate the binding effect of the 14-3-3 to the LNK protein. OVCAR5 cells with forced expression of LNK was further infected with lentivirus containing either non-target shRNA or shRNA mixture targeted to three 14-3-3 members: tau, zeta and gamma. Reduction of the cell diameter (Fig 7F) and generating slightly larger tumors (Fig 7G) was observed in these cells, compared to tumors with forced expression of LNK alone (Fig 7G). Taken together, these result suggested release or loss of the 14-3-3 binding will further enhanced the effect of LNK. 14-3-3 may suppress the activity of LNK by modulating the amount of “freely available” LNK to participate in regulation of the signaling pathway. Base on these observation, an working model of LNK in ovarian cancer is proposed (Supplemental Fig 9).
Discussion
LNK is a well-studied tumor suppressor gene in hematopoiesis 6, 34, 35. It is mutated in hematopoietic malignancies including 3~5% of MPN samples, 10% of MPN evolved to acute myeloid leukemia, and 5% of early T cell leukemia (reviewed in 34). These mutations appear to be a loss of function, i.e. dampening an activated tyrosine kinase. Paradoxically, we show that LNK can exert tumor survival properties in ovarian cancer cells and promote tumor growth in in vivo models. Growth-factor/cytokine signaling often governs the uptake of nutrients, which in turn determines concentrations of intracellular metabolites. Nutrient depletion, as well as other stresses may trigger altered cellular pathway leading either cell survival or apoptosis 36. The AKT and MAPK pathways are among the major downstream signals activated by growth factors, which are often deregulated in cancer. Increased expression of LNK in ovarian cancer cells up-regulated and prolonged the activation of AKT and MAPK and rendered the cells resistant to death induced by either serum/nutrient starvation or drug treatments. In contrast, silencing of LNK in these cells decreased activated AKT and MAPK signaling and slowed their cell proliferation. A similar observation has been reported recently, showing that LNK protected normal endothelium cells from death and rendered them resistant to apoptosis induced by tumor necrosis factor (TNF) and actinomycin-D 29, suggesting this pro-survival effect of LNK is not only limited to ovarian cancer cells. Our RNA array analysis suggested that metabolic pathways are enhanced in cells overexpressing LNK. We speculate that LNK enhancement of signal transduction pathways (e.g. AKT and MAPK pathways), allows for sufficient nutrient uptake, even when cells are exposed to low nutrient/stress conditions, which in turn enables the cells to maintain their metabolic needs.
The mammalian Target of Rapamycin (mTOR) plays a central role in regulating cell growth (cell mass) and cell proliferation (cell numbers) in response to environmental cues such as nutrient availability and different types of stress 37. LNK overexpression in ovarian cancer cells enhanced their proliferation and reduced their cell size, while silencing of LNK had the opposite affects. The phosphorylation levels of AKT ( upstream of mTOR) and p70S6 (downstream of mTOR) were both increased upon LNK overexpression, suggesting that the mTOR pathway is up-regulated by LNK in ovarian cancer cells.
The in vivo xenograft experiments showed that LNK can promote cell growth and generate bigger tumor. Why does LNK appear to function differently in leukemia cells verses ovarian cancer/normal endothelial cells? LNK does not have enzymatic activity; its function is totally dependent on its binding partners. LNK may behave similar to a “buffer molecule” modulating direction, amplitude and duration of signal transduction of the binding partner. Notably, the other two members [SH2B1 and APS (SH2B2)] of this family of proteins, share similar sequence homologies 38–40, and their stimulatory and inhibitory roles also appear to be cell-type and pathway dependent. Many hematopoietic malignancies including MPN, act as classical “activated kinase diseases” driven by a mutant activated receptor tyrosine kinase (e.g., FLT3-ITD, mutant c-KIT) or the downstream kinase e.g. JAK2 to propel proliferation and survival. Typically, these activated mutant tyrosine kinases phosphorylate LNK at a tyrosine residue which becomes the scaffold for other SH2 domain containing proteins to attenuate the activated tyrosine kinase. This represents a feedback mechanism. In contrast, most of the ovarian cancers contain mutations associated with alterations of pathway signaling by RB, PI3K/RAS, Notch, BRCA1/2 and the FOXM1 transcription factor 41. Thus, we hypothesize that these ovarian cancer cells do not depend on either mutant, activated JAK2 and/or mutant, activated type III receptor tyrosine kinase pathways. Instead, oncogenic drivers of ovarian cancer cells are downstream of these receptor tyrosine kinases. Thus, LNK may not have a significant inhibitory effect on transformed epithelial cells. Meanwhile, a recent study in drosophila revealed that LNK regulates and enhances the insulin signaling by binding to the insulin receptor 42, 43. Female flies who are homozygously mutant for LNK have much smaller ovaries containing few oocytes and are sterile due to an arrest in oogenesis 42. These studies suggest that LNK is required for normal ovarian development in the Drosophila.
Our studies show that LNK affects cell adhesion, ECM interaction and possibly the integrin pathway of ovarian cancer cells. We observed that overexpression of LNK inhibited cell migration. Concordantly, bone marrow mast cell (BMMC) from LNK −/− mice were reported to display increased migration to a cytokine compare to the wild type mast cells 13. The authors attributed the phenotype of these BMMC to upregulation of the RAC1, p-JNK1/2 and p-P38 pathway in the LNK−/− mice cells 13. However, in ovarian cancer cells, our data suggest that the activated p-JNK1/2 and p-P38 pathway is associated with LNK expression and decreased migration. Consistent with our results, a recent study suggested that overexpression of LNK inhibits cell migration of normal endothelial cells; and the authors reached the conclusion that this phenomenon is associated with enhanced turnover of focal adhesion complexes and activation of the integrin pathway 27. Additionally, SH2B1 recently was also found to play a role in the regulation of focal adhesion complexes 44. Of particular interest, our microarray analysis showed that a number of genes related to cell adhesion and ECM interaction (e.g., E-cadherin and the cell-cell tight junction claudin family members), are up-regulated in ovarian cancer cells that have high expression of LNK. At the same time, our mass spectroscopy results suggest that LNK might bind to the small G protein RAB21, which is critical for integrin internalization, trafficking and redistribution 45. Together, our data suggest that LNK may enhance cell attachment and attenuate cell migration by multiple mechanisms.
Besides participating in cell adhesion and ECM-interaction, pathway analysis suggested a possible role of LNK in the JAK2-STAT signaling pathways; this is consistent with the function of LNK in hematopoietic cells. Interestingly, pathway analysis also suggests that LNK participates in the amino acid metabolism pathway (Fig. 6A). In line with this observation, a prior study indicated that LNK was dysregulated when cells were placed in a cysteine-free environment 46.
Our mass spectroscopy results suggest that some nuclear proteins interact with LNK, which prompted us to examine the subcellular localization of LNK. Our protein fractionation studies found a small amount of LNK localized in the nucleus, implying that LNK may have some biological function in the nucleus. Of note, the well known binding partner of LNK, JAK2, enters the nucleus and regulates transcription by phosphorylating the core histone H3 47. Although we did not identify JAK2 binding to LNK by our mass spectroscopy, this is worthy of further exploration. Also of interest, several proteins related to the endocytic pathway (CLINT1, RAB21 and LYST) appear to bind to LNK as detected by mass spectroscopy. Generally in hematopoietic cells upon ligand stimulation, their receptor tyrosine kinases enter the endocytosis pathway and in part undergo lysosomal degradation to prevent overactivation of these activated signaling pathways. LNK may aid in the attenuation of the tyrosine kinase activity through endocytic degradation.
In summary, our data identified for the first time several unique functions of LNK in ovarian cancer cells. LNK augmented the p-AKT and p-MAPK pathways, enhanced cell adhesion and slowed cell migration, and promoted the in vivo tumor growth in murine xenograft model. 14-3-3 was identified as one of the LNK binding partner which can suppress LNK activity. Our results suggest that in contrast to the findings in hematologic malignancies, the adaptor protein LNK acts as a positive signal transduction modulator in ovarian cancers. We believe that our observations are novel and open a new area of inquiry for this important adaptor protein.
Supplementary Material
Acknowledgments
This research is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under the Research Centres of Excellence initiative, and by the National Institutes of Health of the USA (R01CA026038-33) and the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research Investigator Award to H. Phillip Koeffler.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Devalliere J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem Pharmacol. 2011;82:1391–1402. doi: 10.1016/j.bcp.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, He X, Schembri-King J, Jakes S, Hayashi J. Cloning and characterization of human Lnk, an adaptor protein with pleckstrin homology and Src homology 2 domains that can inhibit T cell activation. J Immunol. 2000;164:5199–5206. doi: 10.4049/jimmunol.164.10.5199. [DOI] [PubMed] [Google Scholar]
- 3.Rudd CE. Lnk adaptor: novel negative regulator of B cell lymphopoiesis. Sci STKE. 2001:2001, pe1. doi: 10.1126/stke.2001.85.pe1. [DOI] [PubMed] [Google Scholar]
- 4.Velazquez L, Cheng AM, Fleming HE, Furlonger C, Vesely S, Bernstein A, et al. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med. 2002;195:1599–1611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takaki S, Morita H, Tezuka Y, Takatsu K. Enhanced hematopoiesis by hematopoietic progenitor cells lacking intracellular adaptor protein, Lnk. J Exp Med. 2002;195:151–160. doi: 10.1084/jem.20011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong W, Lodish HF. Lnk inhibits Tpo-mpl signaling and Tpo-mediated megakaryocytopoiesis. J Exp Med. 2004;200:569–580. doi: 10.1084/jem.20040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buza-Vidas N, Antonchuk J, Qian H, Mansson R, Luc S, Zandi S, et al. Cytokines regulate postnatal hematopoietic stem cell expansion: opposing roles of thrombopoietin and LNK. Genes Dev. 2006;20:2018–2023. doi: 10.1101/gad.385606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seita J, Ema H, Ooehara J, Yamazaki S, Tadokoro Y, Yamasaki A, et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci U S A. 2007;104:2349–2354. doi: 10.1073/pnas.0606238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong W, Zhang J, Lodish HF. Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood. 2005;105:4604–4612. doi: 10.1182/blood-2004-10-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baran-Marszak F, Magdoud H, Desterke C, Alvarado A, Roger C, Harel S, et al. Expression level and differential JAK2-V617F-binding of the adaptor protein Lnk regulates JAK2-mediated signals in myeloproliferative neoplasms. Blood. 2010;116:5961–5971. doi: 10.1182/blood-2009-12-256768. [DOI] [PubMed] [Google Scholar]
- 11.Gery S, Cao Q, Gueller S, Xing H, Tefferi A, Koeffler HP. Lnk inhibits myeloproliferative disorder-associated JAK2 mutant, JAK2V617F. J Leukoc Biol. 2009;85:957–965. doi: 10.1189/jlb.0908575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gery S, Gueller S, Chumakova K, Kawamata N, Liu L, Koeffler HP. Adaptor protein Lnk negatively regulates the mutant MPL, MPLW515L associated with myeloproliferative disorders. Blood. 2007;110:3360–3364. doi: 10.1182/blood-2007-05-089326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon C, Dondi E, Chaix A, de Sepulveda P, Kubiseski TJ, Varin-Blank N, et al. Lnk adaptor protein down-regulates specific Kit-induced signaling pathways in primary mast cells. Blood. 2008;112:4039–4047. doi: 10.1182/blood-2008-05-154849. [DOI] [PubMed] [Google Scholar]
- 14.Gueller S, Gery S, Nowak V, Liu L, Serve H, Koeffler HP. Adaptor protein Lnk associates with Tyr(568) in c-Kit. Biochem J. 2008;415:241–245. doi: 10.1042/BJ20080102. [DOI] [PubMed] [Google Scholar]
- 15.Gueller S, Goodridge HS, Niebuhr B, Xing H, Koren-Michowitz M, Serve H, et al. Adaptor protein Lnk inhibits c-Fms-mediated macrophage function. J Leukoc Biol. 2010;88:699–706. doi: 10.1189/jlb.0309185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueller S, Hehn S, Nowak V, Gery S, Serve H, Brandts CH, et al. Adaptor protein Lnk binds to PDGF receptor and inhibits PDGF-dependent signaling. Exp Hematol. 2011;39:591–600. doi: 10.1016/j.exphem.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Lin DC, Yin T, Koren-Michowitz M, Ding LW, Gueller S, Gery S, et al. Adaptor protein Lnk binds to and inhibits normal and leukemic FLT3. Blood. 2012;120:3310–3317. doi: 10.1182/blood-2011-10-388611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado C, Erquiaga I, Aranaz P, Migueliz I, Garcia-Delgado M, Novo FJ, et al. LNK can also be mutated outside PH and SH2 domains in myeloproliferative neoplasms with and without V617FJAK2 mutation. Leuk Res. 2011;35:1537–1539. doi: 10.1016/j.leukres.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Ha JS, Jeon DS. Possible new LNK mutations in myeloproliferative neoplasms. Am J Hematol. 2011;86 :866–868. doi: 10.1002/ajh.22107. [DOI] [PubMed] [Google Scholar]
- 20.Lasho TL, Pardanani A, Tefferi A. LNK mutations in JAK2 mutation-negative erythrocytosis. N Engl J Med. 2010;363:1189–1190. doi: 10.1056/NEJMc1006966. [DOI] [PubMed] [Google Scholar]
- 21.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, Jr, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Garcia A, Ambesi-Impiombato A, Hadler M, Rigo I, Leduc CA, Kelly K, et al. Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood. 2013;122:2425–2432. doi: 10.1182/blood-2013-05-500850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 26.Bersenev A, Wu C, Balcerek J, Jing J, Kundu M, Blobel GA, et al. Lnk constrains myeloproliferative diseases in mice. J Clin Invest. 2010;120:2058–2069. doi: 10.1172/JCI42032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devalliere J, Chatelais M, Fitau J, Gerard N, Hulin P, Velazquez L, et al. LNK (SH2B3) is a key regulator of integrin signaling in endothelial cells and targets alpha-parvin to control cell adhesion and migration. FASEB J. 2012;26:2592–2606. doi: 10.1096/fj.11-193383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitau J, Boulday G, Coulon F, Quillard T, Charreau B. The adaptor molecule Lnk negatively regulates tumor necrosis factor-alpha-dependent VCAM-1 expression in endothelial cells through inhibition of the ERK1 and -2 pathways. J Biol Chem. 2006;281:20148–20159. doi: 10.1074/jbc.M510997200. [DOI] [PubMed] [Google Scholar]
- 29.Chatelais M, Devalliere J, Galli C, Charreau B. Gene transfer of the adaptor Lnk (SH2B3) prevents porcine endothelial cell activation and apoptosis: implication for xenograft's cytoprotection. Xenotransplantation. 2011;18:108–120. doi: 10.1111/j.1399-3089.2011.00629.x. [DOI] [PubMed] [Google Scholar]
- 30.Gery S, Gueller S, Nowak V, Sohn J, Hofmann WK, Koeffler HP. Expression of the adaptor protein Lnk in leukemia cells. Exp Hematol. 2009;37:585–592. e582. doi: 10.1016/j.exphem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5:983–998. doi: 10.1002/emmm.201201823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Balcerek J, Rozenova K, Cheng Y, Bersenev A, Wu C, et al. 14-3-3 regulates the LNK/JAK2 pathway in mouse hematopoietic stem and progenitor cells. J Clin Invest. 2012;122:2079–2091. doi: 10.1172/JCI59719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maures TJ, Chen L, Carter-Su C. Nucleocytoplasmic Shuttling of the Adapter Protein SH2B1β (SH2-Bβ) Is Required for Nerve Growth Factor (NGF)-Dependent Neurite Outgrowth and Enhancement of Expression of a Subset of NGF-Responsive Genes. Mol Endocrinol. 2009;23:1077–1091. doi: 10.1210/me.2009-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gery S, Koeffler HP. Role of the adaptor protein LNK in normal and malignant hematopoiesis. Oncogene. 2012 doi: 10.1038/onc.2012.435. [DOI] [PubMed] [Google Scholar]
- 35.Oh ST. When the brakes are lost: LNK dysfunction in mice, men, and myeloproliferative neoplasms. Therapeutic Advances in Hematology. 2011;2:11–19. doi: 10.1177/2040620710393391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 37.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed Z, Pillay TS. Adapter protein with a pleckstrin homology (PH) and an Src homology 2 (SH2) domain (APS) and SH2-B enhance insulin-receptor autophosphorylation, extracellular-signal-regulated kinase and phosphoinositide 3-kinase-dependent signalling. Biochem J. 2003;371:405–412. doi: 10.1042/BJ20021589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishi M, Werner ED, Oh BC, Frantz JD, Dhe-Paganon S, Hansen L, et al. Kinase activation through dimerization by human SH2-B. Mol Cell Biol. 2005;25:2607–2621. doi: 10.1128/MCB.25.7.2607-2621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian X, Ginty DD. SH2-B and APS are multimeric adapters that augment TrkA signaling. Mol Cell Biol. 2001;21:1613–1620. doi: 10.1128/MCB.21.5.1613-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The-Cancer-Genome-Altas-Research-Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werz C, Kohler K, Hafen E, Stocker H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 2009;5:e1000596. doi: 10.1371/journal.pgen.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almudi I, Poernbacher I, Hafen E, Stocker H. The Lnk/SH2B adaptor provides a fail-safe mechanism to establish the Insulin receptor-Chico interaction. Cell Commun Signal. 2013;11:26. doi: 10.1186/1478-811X-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanning NJ, Su HW, Argetsinger LS, Carter-Su C. Identification of SH2B1beta as a focal adhesion protein that regulates focal adhesion size and number. J Cell Sci. 2011;124:3095–3105. doi: 10.1242/jcs.081547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellinen T, Tuomi S, Arjonen A, Wolf M, Edgren H, Meyer H, et al. Integrin trafficking regulated by Rab21 is necessary for cytokinesis. Dev Cell. 2008;15:371–385. doi: 10.1016/j.devcel.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Lee JI, Dominy JE, Jr, Sikalidis AK, Hirschberger LL, Wang W, Stipanuk MH. HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics. 2008;33:218–229. doi: 10.1152/physiolgenomics.00263.2007. [DOI] [PubMed] [Google Scholar]
- 47.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.