Abstract
Plants produce hundreds of glycosidases. Despite their importance in cell wall (re)modeling, protein and lipid modification, and metabolite conversion, very little is known of this large class of glycolytic enzymes, partly because of their post-translational regulation and their elusive substrates. Here, we applied activity-based glycosidase profiling using cell-permeable small molecular probes that react covalently with the active site nucleophile of retaining glycosidases in an activity-dependent manner. Using mass spectrometry we detected the active state of dozens of myrosinases, glucosidases, xylosidases, and galactosidases representing seven different retaining glycosidase families. The method is simple and applicable for different organs and different plant species, in living cells and in subproteomes. We display the active state of previously uncharacterized glycosidases, one of which was encoded by a previously declared pseudogene. Interestingly, glycosidase activity profiling also revealed the active state of a diverse range of putative xylosidases, galactosidases, glucanases, and heparanase in the cell wall of Nicotiana benthamiana. Our data illustrate that this powerful approach displays a new and important layer of functional proteomic information on the active state of glycosidases.
Carbohydrates are present in all kingdoms of life and are particularly prominent in plants (1). Plants produce carbohydrates as one of their major constituents through their photosynthetic activity. The simplest synthesized forms of carbohydrates are monosaccharide sugars such as glucose, which provides energy for various cellular activities. Carbohydrates also exist in very complex forms. Monosaccharide sugars are attached to one another through covalent glycosidic linkage, which generates di-, oligo-, and polysaccharides. Carbohydrates also attach to non-carbohydrate species (lipids, proteins, hormones) through a glycosidic linkage to form glycoconjugates (2).
Glycosidic bonds are hydrolyzed by a group of enzymes termed glycosyl hydrolases (GHs)1 or glycosidases (3). Because of the tremendous carbohydrate diversity, there are a vast variety of glycosidases, including glucosidases, xylosidases, and galactosidases, that preferentially hydrolyze their respective glycoside substrates. In general, the number of glycosidase-related genes in plants (for instance, Arabidopsis) is relatively high when compared with that in other sequenced organisms (for instance, human) (4). This signifies the unique importance of glycosidases in plants as opposed to other organisms. Based on protein sequence similarities, glycosidases are classified into different GH families. Members of the same GH family share a common mechanism of glycosidic bond cleavage (5).
Mechanistically, glycosidases are classified as retaining or inverting enzymes (6). To hydrolyze the glycosidic bond, both retaining and inverting enzymes carry two catalytic glutamate or aspartate residues (or both) (7). Of these two catalytic residues, one acts as a proton donor and the other as a nucleophile/base. The distance between these catalytic residues in the active site of the glycosidases determines the mechanism of hydrolysis. Retaining enzymes have two catalytic residues separated by a distance of ∼5.5 Å, and their hydrolysis mechanism retains the net anomeric configuration of the C1 atom in the sugar molecule. In contrast, inverting enzymes have catalytic residues that are ∼10 Å apart, and these enzymes invert the overall anomeric configuration of the C1 carbon atom in the released sugar (8).
Both retaining and inverting glycosidases are present abundantly in plants. The genome of Arabidopsis thaliana encodes for 400 glycosidases, of which 260 are retaining enzymes and 140 are inverting enzymes. Genetic, molecular, and biochemical approaches revealed that glycosidases are localized in different cellular compartments and are important for various biological processes. The majority of plant glycosidases reside in the cell wall, and these enzymes can play major roles in cell wall restructuring (9). Other characterized glycosidases reside in other compartments to regulate glycosylation of proteins and hormones. Despite the importance of GH enzymes, physiological and biochemical functions are assigned to only a few glycosidases (9).
Activity-based protein profiling (ABPP) is a powerful tool for monitoring the active state of multiple enzymes without knowledge of their natural substrates (10, 11). ABPP involves chemical probes that react with active site residues in an activity-dependent manner. Thus ABPP displays the availability and reactivity of active site residues in proteins, which are hallmarks for enzyme activity (12). ABPP is particularly attractive because the profiling can be done without purifying the enzymes and can be performed in cell extracts or in living cells. Another key advantage of ABPP is that the activities of large multigene enzyme families can be monitored using broad-range probes. ABPP has had a significant impact on plant science. After the introduction of probes for papain-like cysteine proteases (13, 14), these probes revealed increased protease activities in the tomato and maize apoplasts during immune responses (15, 16) and that these immune proteases are targeted by unrelated inhibitors secreted by fungi, oomycetes, and nematodes (17–24). Likewise, probes for the proteasome displayed unexpected increased proteasome activity during immune responses (25) and revealed that the bacterial effector molecule syringolin A targets the nuclear proteasome (26). We anticipate that more regulatory mechanisms will be discovered through the use of probes introduced for serine hydrolases, metalloproteases, vacuolar processing enzymes, ATP binding proteins, and glutathione transferases (27–32).
Cyclophellitol-aziridine-based probes were previously used in animal proteomes to target retaining glucosidases (33). Here we established and applied glycosidase profiling in plants. We discovered that cyclophellitol-aziridine-based probes targeted an unexpectedly broad range of glycosidases representing members of at least seven different GH families. We used these probes to study the active state of glycosidases present in living cells, in different organs and plant species, and in the apoplast of Nicotiana benthamiana.
EXPERIMENTAL PROCEDURES
Probe Synthesis
The synthesis of JJB70 and JJB111 is described in the supplemental material. The synthesis of KY371 and JJB111 has been described previously (33). Aliquots of these compounds are available upon request.
Biological Materials
A. thaliana plants (ecotype Columbia, Col-0) were grown at 24 °C (day)/20 °C (night) in a growth chamber under a 12-h light regime. Leaves from rosettes of 4-week-old Arabidopsis plants were used for protein extraction. Cell cultures of A. thaliana (ecotype Landsberg erecta) were grown and weekly subcultured in L9 liquid medium (4.41 g/l MS-Basal medium including nitsch vitamins (Duchefa M0256), 30 g/l sucrose, 5.4 μm α-naphthyleneacetic acid, 0.23 μm kinetin, adjusted to pH 5.6). N. benthamiana plants were grown at 22 °C and 60% relative humidity under a 12-h light regime. 4-week-old N. benthamiana plants were used for apoplastic fluid isolation. Commercially available xylosidase was purchased from PROZOMIX (Nothumberland, UK).
Sample Preparation for Labeling Assays
Extracts for Small-scale Labeling
Leaves of various plant species or different organs of Arabidopsis plants were homogenized with 500 μl of 50 mm MES buffer at pH 6.0 or 50 mm MOPS buffer at pH 7.5. For the pH course experiment, the leaves were homogenized in sterile water. Two leaf discs (1.5-cm diameter) were taken from the leaves of different plant species and homogenized with 500 μl of 50 mm MOPS buffer at pH 7.5. After the tissues had been ground in a 1.5-ml tube, the samples were centrifuged at 10,000g and 4 °C for 10 min to remove cell debris, and the supernatant containing the soluble proteins was used for labeling.
Extracts for Large-scale Labeling
2 g of Arabidopsis leaf rosettes were frozen with liquid nitrogen and ground with a mortar and pestle. 5 ml of extraction buffer (50 mm MES buffer at pH 6.0, 0.1% Triton (X-100), and 10% glycerol) were added to the leaf powder, vortexed, and centrifuged at 4000 rpm and 4 °C for 30 min to remove cell debris. The supernatant containing the soluble proteins was used for large-scale labeling.
Apoplastic Fluid Isolation
The apoplastic fluids were isolated by means of vacuum infiltration and centrifugation (34, 35). In brief, the leaves of N. benthamiana were submerged in ice-cold sterile water, and vacuum was applied. The water infiltrated the intercellular spaces of leaves as the vacuum was slowly released. The leaves were then dried briefly and centrifuged at 3000 rpm and 4 °C for 15 min. Protease inhibitor mixture solution (cOmplete Protease Inhibitor Tablets, Roche Applied Sciences) was added to the isolated apoplastic fluids at 4× final concentration.
Labeling Plant Extracts
Leaf or organ extracts of Arabidopsis containing ∼1.0 to 1.5 mg/ml total soluble proteins were pre-incubated with 50 μm KY371 or DMSO for 30 min at pH 6.0 or 7.5. These extracts were incubated with 2 μm JJB70 or JJB111 for 1h at room temperature in the dark at 50 μl total reaction volume. Equal volumes of DMSO were added for no probe control. For the pH course experiment, Arabidopsis leaf extracts were incubated with suitable pH buffer (50 mm Sodium acetate buffer for pH 4.0, 50 mm MES buffer for pH 5.0–6.5, 50 mm MOPS buffer for pH 7.0–7.5, Tris buffer for pH 8–10) and 2 μm JJB70 in the dark for 1h at room temperature at 50 μl total reaction volume.
Peptide-N-Glycosidase F (PNGaseF) Treatment of Labeled Proteins
10 μl of JJB70 or DMSO incubated leaf extracts of Arabidopsis were treated with 1.5 μl of 10X glycoprotein denaturing buffer (New England BioLabs) and heated at 100 °C for 10 min. The denatured proteins were treated with 3 μl of H2O, 2 μl of 10% Nonidet P-40, 2 μl of 10X G7 buffer (New England BioLabs), 1.5 μl PNGaseF (New England BioLabs) or H2O and incubated at 37 °C for 1h.
Labeling Apoplastic Fluids
Apoplastic fluids of N. benthamiana were pre-incubated with 50 μm KY371 or DMSO for 30 min at pH 5.0 with 50 mm MES buffer. These apoplastic fluids were incubated with 2 μm JJB70 or DMSO in the dark for 1h at room temperature for labeling at 50 μl total reaction volume. For pH course experiment, apoplastic fluids were incubated with suitable pH buffer (50 mm Sodium acetate buffer for pH 4.0, 50 mm MES buffer for pH 5.0–6.5, 50 mm MOPS buffer for pH 7.0–7.5, Tris buffer for pH 8–10) and 2 μm JJB70 in the dark for 1h at room temperature at 50 μl total reaction volume.
Analysis of Labeled Proteins
The labeling reactions were stopped by adding gel loading buffer containing β-mercaptoethanol at 1X final concentration and heating at 95 °C for 10 min, unless indicated otherwise. The labeled proteins were separated on 12% protein gels at 200 volts for 1h. The JJB70-labeled proteins were detected in the protein gels with the Typhoon FLA 9000 scanner (GE Healthcare Life Sciences) using excitation wavelength at 472 nm and BPB1 filter (GE Healthcare Life Sciences). The fluorescence of the labeled proteins was quantified using ImageQuant (GE Healthcare Life Sciences). JJB111-labeled proteomes were transferred from the protein gels onto PVDF membranes, incubated with streptavidin HRP (ultra sensitive, Sigma) and detected using chemiluminescent substrates (SuperSignal West Chemiluminescent substrates, Thermo scientific).
In Vivo Labeling of Arabidopsis Cell Cultures
500 μl of an Arabidopsis cell cultures were transferred to a 12-well tissue culture plate. The medium was removed from the cell culture, replaced by 500 μl fresh L9 medium and kept shaking gently for 30 min before labeling. The cell cultures were preincubated with 50 μm KY371 or DMSO for 30 min under gentle shaking. The cell culture was labeled by the addition of 2 μm JJB70 under gentle shaking for 1 h. After labeling, the medium from the cell culture was removed and precipitated with 1 ml of 100% ice-cold acetone. 20 μl of 1× gel loading buffer (containing β-mercaptoethanol) was added to the precipitated proteins, and the sample was heated at 95 °C for 10 min. The labeled cells were ground with 50 μl of 4× gel loading buffer (containing β-mercaptoethanol), and the sample was heated at 95 °C for 10 min. For Control 1, 500 μl of washed and unlabeled cells were homogenized with 1 μl of 1 mm JJB70 for 30 s and heated at 95 °C for 10 min in the presence of 50 μl of 4× gel loading buffer (containing β-mercaptoethanol). For Control 2, washed and unlabeled cells were homogenized with 50 μl of 4× gel loading buffer containing 2 μm JJB70 and β-mercaptoethanol. The labeled proteins were analyzed as explained above.
Large-scale Labeling and Affinity Purification
2 ml of Arabidopsis leaf extracts containing ∼2.0 mg/ml total leaf proteins were incubated with 5 μm JJB111 at room temperature for 1 h. To label apoplastic fluids of N. benthamiana, 10 ml of apoplastic fluids were incubated with 5 μm JJB111 at room temperature for 1 h. Labeling was stopped by precipitating the total proteins via the chloroform/methanol precipitation method (36). In brief, 4 volumes of ice-cold methanol, 1 volume of ice-cold chloroform, and 3 volumes of water were added. The samples were vortexed and centrifuged at 4000 rpm and 4 °C for 30 min. The precipitated proteins were dissolved in 2 ml of 1.2% SDS dissolved in 1× PBS solution (SDS/PBS) by sonication for 4 to 5 s and heating at 95 °C for 10 min. This solution was diluted to 0.2% SDS/PBS by the addition of 10 ml of 1× PBS solution. The resulting solution was incubated with 100 μl of pre-equilibrated streptavidin beads (high-capacity streptavidin beads, Thermo Scientific) for 1 h at room temperature. The beads containing the labeled proteins were isolated via centrifugation at 1400g for 3 min. These beads were washed successively three times with 5 ml of 1.2% SDS/PBS solution, thrice with 5 ml of 1.0% SDS/PBS solution, and thrice with 10 ml of 1× PBS buffer. The final wash was with 10 ml of water. The captured proteins were eluted by boiling the beads at 95 °C in 30 μl of 4× gel loading buffer. The eluted proteins were separated on 12% protein gels at 200 V for 1 h, and the protein gels were stained overnight with SYPRO Ruby (Invitrogen). To detect the proteins, we scanned the gels at an excitation wavelength of 473 nm with an LPG filter in a Typhoon FLA 9000 scanner (GE Healthcare Life Sciences).
On-bead Trypsin Digestion
The streptavidin beads from the no-probe control sample were treated with 500 μl of 6 m urea dissolved in 10 mm phosphate-buffered saline (PBS) and 25 μl of 200 mm Tris (2-carboxyethyl) phosphine solution, incubated at 65 °C for 15 min, and cooled to 35 °C. 25 μl of 400 mm iodoacetamide solution in water was added and incubated at 35 °C for 30 min. 950 μl of 10 mm PBS solution was added to dilute the reaction, and the supernatant was removed by centrifuging the beads at 1400g for 2 min. A trypsin digestion solution containing 200 μl of 2 m urea dissolved in 10 mm PBS, 2 μl of 100 mm calcium chloride, and 4 μl of trypsin (20 μg of trypsin in 40 μl of trypsin buffer) were added to the beads, and the beads were incubated at 37 °C overnight. The supernatant containing the trypsin-digested peptides was identified via mass spectrometry.
In-gel Digestion and MS
Bands were excised by hand and treated with trypsin as described elsewhere (37). Digests were separated on a Thermo/Proxeon Easy nLC II in a two-column configuration (precolumn 3 cm × 100 μm, 5 μm C18AQ medium, analytical column 10 cm × 75 μm, 3 μm C18AQ) coupled to an LTQ-Velos ion trap (Thermo Scientific). Peptides were separated over a 35-min gradient running from 5% to 32% acetonitrile/H2O with 0.1% formic acid. MS/MS spectra were acquired on multiply charged precursors with m/z between 400 and 1600 Da using a Top20 method with active exclusion for 30 s in a window from 0.2 Da below to 1.5 Da above the precursor mass. The resulting RAW files were de-noised and converted to MGF format using MS Convert from the Proteowizard 2.1 package, accepting only the six strongest peaks per 100-Da window.
Database Searching
The N. benthamiana database (v. 0.4.4, 76,379 sequences) was downloaded from the SOL genomics network. The Arabidopsis database was based on TAIR10 (35,384 sequences). 1095 common artifact sequences were added to both databases, and both were then reversed and concatenated to provide decoys for false discovery rate calculation. MS/MS spectra were searched against the described databases using MASCOT 2.4, permitting JJB111 (735.386 Da) as an additional variable modification of Asp and Glu residues in the searches. For both searches, a Mascot score of 38 represented a 95% certainty cutoff.
Other Mascot parameters were as follows: precursor mass tolerance, 0.4 Da; fragment mass tolerance, 0.5 Da; one C13 permitted; one tryptic miscleavage allowed; fixed modifications of cysteine (57.02 Da, carbamidomethyl); variable modifications of methionine (15.99 Da, oxidation)
Bioinformatics
Glycosyl Hydrolase Family Classification
GH family classification of identified glycosidases in the Arabidopsis leaf proteome was performed based on the annotations done for the genome of A. thaliana in the CAZY database (38). GH family classification of identified glycosidases in the secreted leaf proteome of N. benthamiana was done by submitting protein sequences to the Pfam database (39).
Phylogenetic Tree Construction
Protein sequences of 258 retaining glycosidases in the genome of Arabidopsis were retrieved from the TAIR database (40). The protein sequences of AT4G33850 and A IG002N01.5 were not available in the TAIR database, so they were retrieved from the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/). The sequences were aligned with ClustalX2 (41). The pairwise alignment gap opening penalty 30 and gap extension penalty 0.75 were used, whereas for multiple alignment gap opening penalty 15 and gap extension penalty 0.3 were used. The alignment files were saved in phylip format, and R script was implemented to construct the tree (42, 43). A neighbor-joining algorithm was used for tree construction from the calculated distance matrix in the program.
A. thaliana Leaf Transcriptome/Proteome Analysis
The transcript levels (average of three replicate experiments) of all 260 retaining glycosidases from the vegetative rosette (age 1.09) were retrieved from the AtGenExpress database (44). Spectral counts (Qty spectra) of all 260 retaining glycosidases detected in the juvenile leaves of Arabidopsis were retrieved from the pep2pro database (45).
RESULTS
Labeling of Leaf Proteomes with Cyclophellitol-Aziridine Activity-based Probes
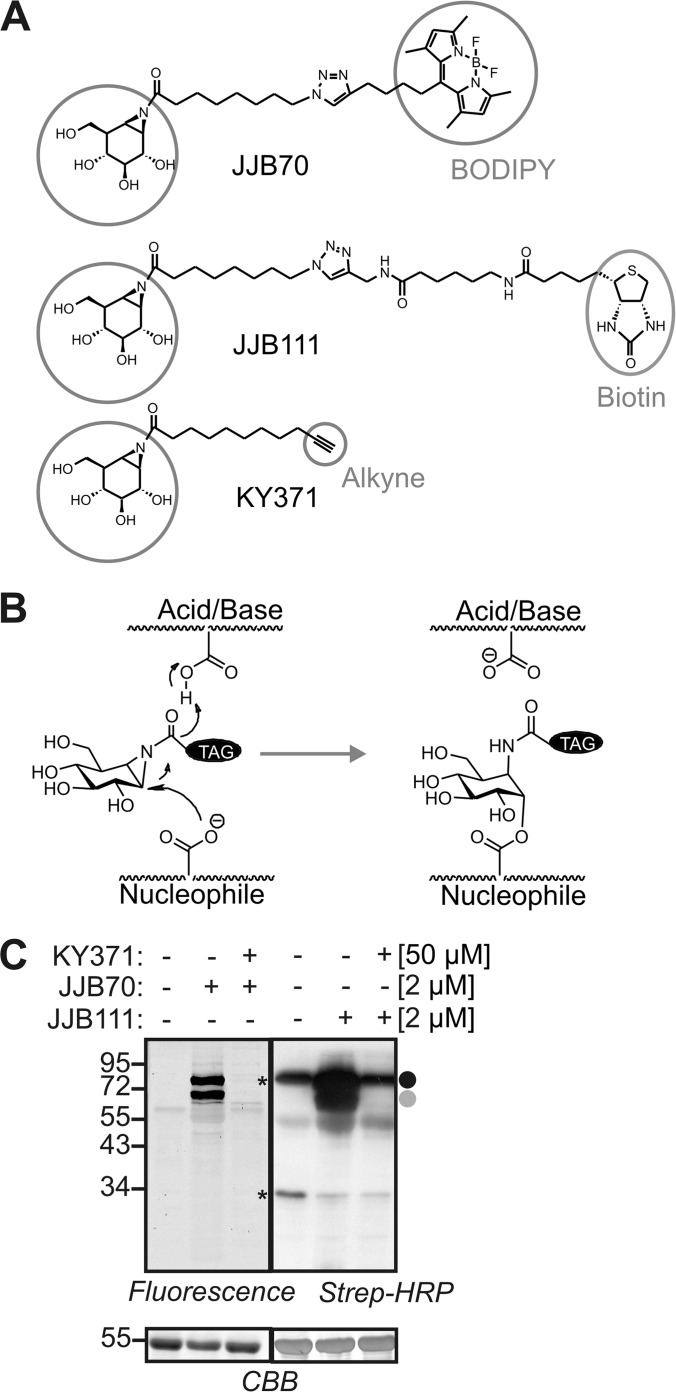
JJB70, JJB111, and KY371 are the three activity-based probes that carry an aziridine analog of cyclophellitol as their warhead. JJB70 carries a bodipy as its reporter tag and is slightly different from the previously described ABP3 (33) in that it has the triazole linker in reverse orientation (Fig. 1A). JJB111 (previously called ABP4) and KY371 carry biotin and alkyne minitags, respectively (Fig. 1A) (33). The electrophilic acyl-aziridine group in the probes reacts with the nucleophilic active site glutamate/aspartate residue of retaining glycosidases, resulting in a reversible covalent ester bond (Fig. 1B).
Fig. 1.
Glycosidase profiling with cyclophellitol aziridine activity-based probes. A, Structures of cyclophellitol aziridine activity-based probes. All the three probes have an aziridine analog of cyclophellitol as warhead followed by a linker. JJB70 has a BODIPY fluorescent reporter tag. JJB111 has an extended linker and a biotin reporter tag. KY371 carries an alkyne minitag. B, Mechanism of labeling retaining glycosidases (Kallemeijn et al., 2012). The nucleophilic oxygen of one of the two catalytic glutamic acids attacks the electrophilic carbon next to nitrogen in the aziridine ring to form a covalent, reversible ester bond. C, Labeling profiles of JJB70 and JJB111 on Arabidopsis leaf extracts. Arabidopsis thaliana leaf exacts containing ∼1.5 mg/ml total soluble proteins were pre-incubated with and without 50 μMKY371 for 30 min and labeled with 2 μM JJB70 or JJB111 for 1 h at pH 7.5. The labeled proteins were either analyzed by in-gel fluorescent scanning (left) or detection on protein blot using streptavidin conjugated to horseradish peroxidase (strep-HRP) (right). The two major signals are indicated by grey and black circles; *, endogenously biotinylated proteins; CBB, Coomassie-Brilliant Blue.
To test plant extracts for labeling, Arabidopsis leaf extracts were incubated with and without 2 μm JJB70 or JJB111. After 1 h of labeling, proteins were separated on protein gels, and labeled proteins were detected via fluorescent scanning (JJB70) or protein blot analysis using streptavidin-linked horseradish peroxidase (for JJB111). Two major signals were detected at 68 kDa (gray circle) and 75 kDa (black circle) (Fig. 1C). These signals were not detected in the no-probe control and were suppressed in the samples preincubated with KY371 (Fig. 1C). These data indicate that JJB70 and JJB111 target similar proteins in Arabidopsis leaf extracts and that labeling with these probes is specific. Further characterization of labeling parameters revealed that a one-hour labeling time is sufficient to reach maximum labeling, that labeling is saturated at 1 μm JJB70, and that labeling is optimal at slightly acidic pH (supplemental Fig. S1). These experiments show that labeling strongly depends on labeling conditions.
Active Site Labeling of Myrosinases TGG1 and TGG2
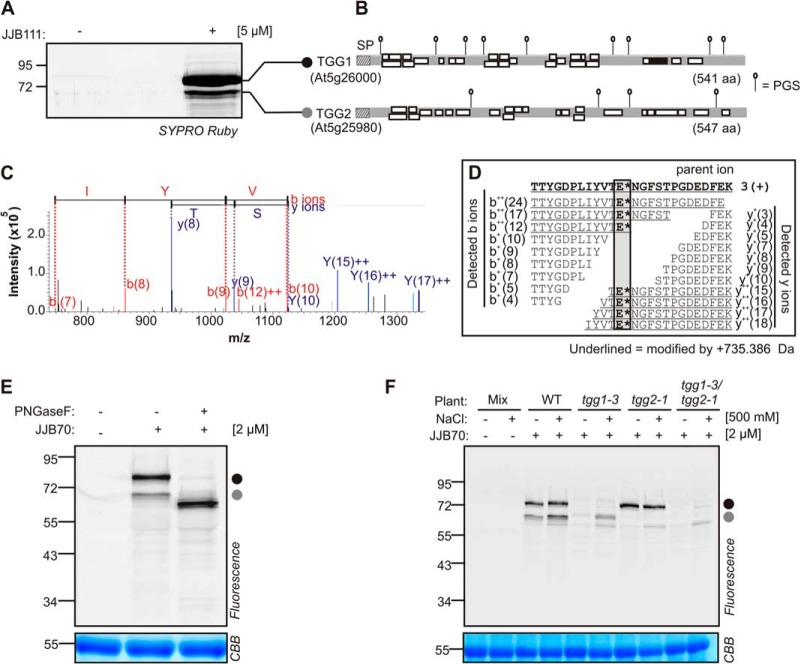
To identify the labeled proteins, large-scale labeling was performed from JJB111-labeled leaf extracts. The labeled proteins were enriched on streptavidin beads, separated on a protein gel, stained with SYPRO Ruby, and detected via fluorescent scanning. The detected protein bands were excised and treated with trypsin, and the peptides were identified via ion-trap mass spectrometry. Similar to small-scale labeling, two strong signals at 68 and 75 kDa were enriched, and these signals were absent in the no-probe control (Fig. 2A). MS analysis showed that the majority of the peptide spectra from the 68-kDa signal originated from β-thioglucoside glucohydrolase TGG2, whereas the 75-kDa signal contained peptides mostly from TGG1 (Fig. 2B). TGG1 and TGG2 are myrosinases that mediate the conversion of glucosinolates during herbivore attack (46). Myrosinases have one catalytic glutamate residue; the second catalytic residue has been replaced by glutamine residue (47).
Fig. 2.
Myrosinase activity profiling in Arabidopsis leaf extracts. A, Identification of proteins labeled by JJB111. Large scale labeling was performed by incubating Arabidopsis leaf extracts with and without 5 μM JJB111. Biotinylated proteins were purified with streptavidin beads and separated on protein gels. The proteins were visualized by SYPRO Ruby staining and fluorescent scanning. Bands were excised, treated with trypsin and the peptides were identified by ion trap mass spectrometry. B, Peptide coverage of myrosinases TGG1 and TGG2. The identified peptides of TGG1 and TGG2 are indicated by white boxes. The black box represents the labeled active site peptide of TGG1. The signal peptide (SP) and putative N-glycosylation sites (PGS) are indicated. C, Fragmentation of modified active site peptide of TGG1. Region of MS2 fragmentation spectrum showing ions that indicate the labeling site. Peaks from the annotated b- and y-ions are indicated. D, Summary of all detected ions of modified peptide in the MS2 fragmentation spectrum. E*, the active site residue. All underlined ions carry an extra modification by the probe (735.386 Da). E, TGG1 and TGG2 migrate at their predicted molecular weight upon deglycosylation. Leaf extracts of wild type (WT) plants were incubated at pH 6.0 with and without 2 μM JJB70. The labeled proteomes were denatured treated with and without PNGaseF, separated on protein gel and detected by in-gel fluorescence scanning. F, Labeling of mutant Arabidopsis lines confirms that signals are caused by TGG1 and TGG2. Leaf proteins of wild type (WT) and tgg1-3, tgg2-1 and tgg1-3/tgg2-1 mutant plants were extracted with and without 500 mMNaCl and labeled with 2 μM JJB70. Mixtures of these extracts (mix) were used as no-probe-control.
To identify the labeling site, the expected modification by JJB111 (735.386 Da) on potentially reactive residues (E and D) was included in the searches. One labeled peptide of TGG1 was identified (black in Fig. 2B). Further analysis of the MS2 fragmentation spectrum revealed that b-ions up to T419 and y-ions down to E420 of this peptide were unmodified. In contrast, b-ions beyond E420 or y-ions starting at T419 or before carried the modification (Fig. 2C). These data demonstrate that the modification was at either E420 or T419. This is consistent with the fact that E420 has been annotated as the nucleophilic active site residue (48). Notably, in addition to the labeled active site peptide, the spectra of the unmodified active site peptide were found among the peptide spectra. This can be explained by the fact that the ester bond between the probe and the active site (Fig. 1B) can hydrolyze in water. This is the first time that a modification of the active site with cyclophellitol-aziridine has been identified through MS.
TGG1 and TGG2 Cause the Major Signals in Arabidopsis Leaf Extracts
The TGG1 and TGG2 signals migrate at a higher apparent molecular weight (MW) than calculated from the plain amino acid sequence of the mature enzyme (59 and 60.4 kDa, respectively). The increased apparent MW could be due to N-glycosylation, as TGG1 and TGG2 carry nine and four putative N-glycosylation sites, respectively (Fig. 2E). To examine whether TGG1 and TGG2 are N-glycosylated, we incubated the labeled leaf proteins with and without the deglycosylation enzyme PNGaseF. After PNGaseF treatment, labeled TGG1 and TGG2 migrated in their expected MWs (Fig. 2E), demonstrating that TGG1 and TGG2 are N-glycosylated. The presence of N-glycosylation is also consistent with the absence of tryptic peptides carrying putative glycosylation sites from the MS data (Fig. 2B).
To confirm that the major signals were caused by myrosinases TGG1 and TGG2, leaf extracts of wild-type, tgg1-3, tgg2-1, and tgg1-3/tgg2-1 knockout plants (46) were incubated with and without JJB70. Surprisingly, both the major signals were absent in the tgg1-3 and tgg1-3/tgg2-1 mutants, and only the 68-kDa signal was absent in the tgg2-1 mutant (Fig. 2F). It has previously been noted that the solubility of TGG2 depends on the presence of TGG1, unless the proteins are extracted in the presence of 500 mm NaCl (48). Indeed, the 68-kDa signal was recovered by extraction in 500 mm NaCl only in the tgg1-3 mutant and remained absent in tgg2-1 and tgg1-3/tgg2-1 mutants. A third signal at 60 kDa was detected in all plants upon extraction in 500 mm NaCl. Taken together, these data demonstrate that TGG1 and TGG2 cause the two major signals in Arabidopsis leaf extracts labeled with JJB70 and JJB111.
JJB Probes Label a Second Layer of Glycosidases Representing New GH Families
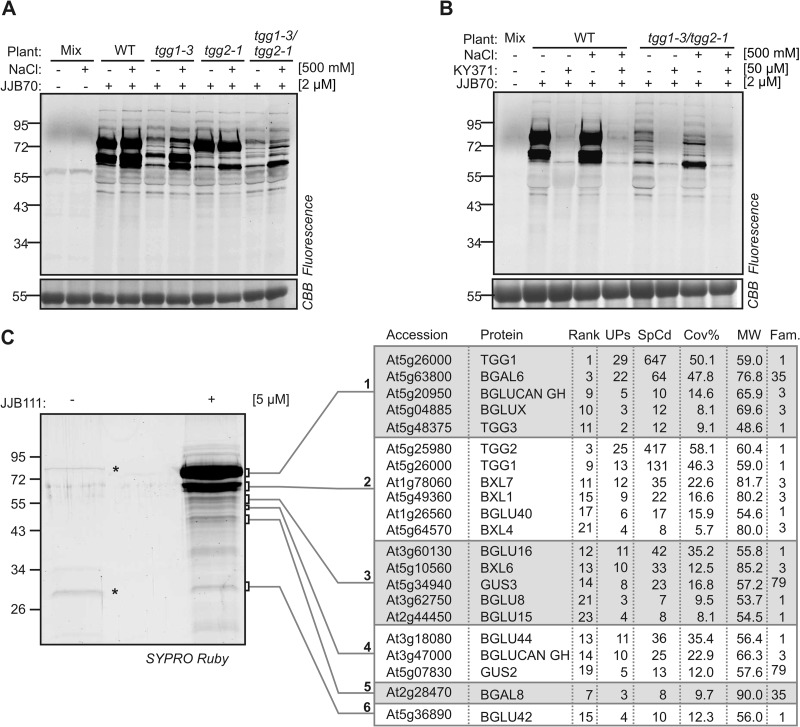
Further examination of the labeling profiles of the tgg1-3/tgg2-1 double mutant revealed another 13 weak signals at longer exposure times (Fig. 3A). These signals were also present in wild-type plants but were covered by the strong TGG1 and TGG2 signals (Fig. 3A). Importantly, pre-incubation with KY371 blocked the labeling of all these proteins in both wild-type and tgg1-3/tgg2-1 plants (Fig. 3B). These data demonstrate that JJB70 labeled proteins in addition to myrosinases TGG1 and TGG2.
Fig. 3.
JJB probes label a second layer of glycosidases. A, Prolonged exposure of protein gels revealed additional proteins labeled by JJB70. The labeled proteins in Figure 2E were detected by prolonged exposure. B, Competition assay confirms specific labeling of additional proteins. Leaf proteins of WT and tgg1-3/tgg2-1 plants were extracted with and without 500 mM NaCl, pre-incubated with and without KY371 for 30 min and labeled with JJB70 at pH 6.0. Mixtures of these proteins were used as no-probe-control (mix). C, The additional labeled proteins are glycosidases. Purified JJB111-labeled proteins (Figure 2A) were detected by prolonged exposure of the SYPRO Ruby-stained protein gel. Six bands were excised, treated with trypsin and analyzed by MS. Summarized are: rank of protein in the list of identified proteins; number of unique peptides (Ups); spectral counts of all peptides (SpCd); sequence coverage (Cov%); expected molecular weight (MW) and GH family (Fam).
To identify these additional labeled proteins, we extended the MS analyses of JJB111-labeled proteins by analyzing the weaker signals. This analysis revealed an additional 18 proteins that were annotated as glycosidases in the CAZY database (Fig. 3C and supplemental Table S1). This included myrosinase TGG3 with two unique peptides (Fig. 3C). TGG3 was previously classified as a pseudogene with no ascribed functions (49). Detection of TGG3 upon JJB111 labeling demonstrates that this enzyme is in an active state in Arabidopsis leaves. Nine additional proteins included the endogenously biotinylated proteins BCCP2 and MCCA and abundant proteins such as ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). These non-glycosidases are usually also detected in no-probe controls (see supplemental Table S3) and were not included in further analysis.
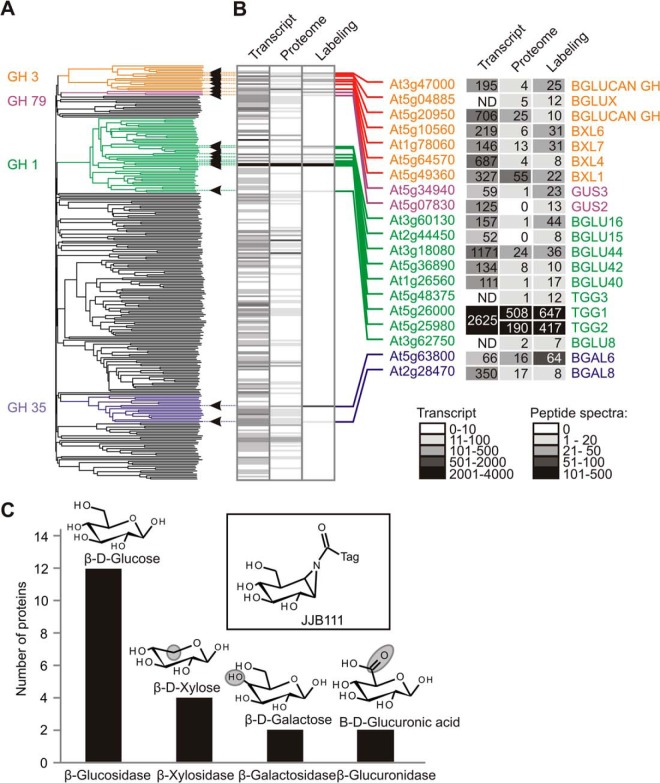
The genome of A. thaliana encodes for 260 retaining glycosidases subdivided into 24 GH families. Mapping the identified glycosidases onto a genome-wide phylogenetic tree of Arabidopsis retaining glycosidases showed that the detected glycosidases were not clustered in the tree and represented members from families GH1, GH3, GH35, and GH79 (Fig. 4A). This broad range of GH families extends the target list of these probes beyond the previously described GH1, GH3, and GH30 families (33). Of the 20 identified glycosidases, only eight enzymes had been previously characterized for their biochemical and molecular functions. This included three enzymes from the GH1 family (TGG1, TGG2, and BGLU44), three enzymes from the GH3 family (BXL1, BXL4, and BXL6), one enzyme from the GH35 family (BGAL6), and one enzyme from the GH79 family (GUS2) (48, 50–55). These data show that the probes displays a broad range of active glycosidases.
Fig. 4.
Identified glycosidases are diverse in phylogeny and have related putative substrates. A, Identified glycosidases represent four major families. The unrooted phylogenetic tree of Arabidopsis retaining glycosidases was constructed with protein sequences of 260 retaining glycosidases annotated in the CAZY database. The extended lines on the right indicate the proteins identified in this study. B, Comparison of transcript and protein abundance with activity of retaining glycosidases in leaf proteomes. The transcript levels (transcript: retrieved from the AtGenExpress database) and the protein levels (proteome: spectral counts retrieved from the pep2pro database) of each retaining glycosidases detected in leaves is compared with the spectral count of JJB111 labeled proteins (labeling). ND, not determined. C, Identified glycosidases have related but distinct putative substrates. The putative substrate for the glycosidases was identified based on UniProtKB and NCBI databases. The key differences with the probe (inset) are indicated with circles.
Selectivity of Glycosidase Labeling
To investigate possible preferential labeling of certain glycosidases (e.g. TGG1 and TGG2) in Arabidopsis leaves, we compared our data with the abundance of transcript and peptide spectra present in the AtGenExpress database and the Arabidopsis proteome database, respectively. We retrieved the transcript levels of the vegetative rosette from the Botany Array Resource (44) and raw spectral details of juvenile leaves from the pep2pro database (45) for all 260 retaining glycosidases. The majority of the glycosidases were detected at various transcript levels (Fig. 4B). Peptides of 90 retaining glycosidases were detected in the juvenile leaves of Arabidopsis with different frequencies (Fig. 4B). Next, we compared transcript levels and the spectral counts with the spectra of our MS data (Fig. 4B). We are aware that such a comparison is not fully meaningful because the used materials and methods are not identical. Nevertheless, this comparative analysis indicated that major signals caused by TGG1 and TGG2 in leaf extracts are caused not by a greater affinity of the probe for TGGs, but by the high abundance of these enzymes relative to other glycosidases. Additional glycosidases that we detected occur in relatively low abundance in leaf extracts. However, we did not detect all retaining glycosidases present in leaf proteomes, despite the fact that some occur at reasonable abundance. This might be because of a different affinity for the probe, or because these proteins are not active under the chosen extraction and labeling conditions.
To analyze the diversity of the 20 identified glycosidases, we annotated substrates for these enzymes based on the names in the UniProtKB and NCBI databases. In these databases, protein names are assigned based on either experimental evidence or homology with characterized enzymes. All nine identified enzymes from the GH1 family are classified as β-d-glucosidases. Three GH3 family members are assigned as β-d-glucan exohydrolase-like proteins (At3g47000 and At5g20950) or β-glucosidase related protein (At5g04885). We consider these enzymes as β-d-glucosidases because the β-d-glucan exohydrolases show β-d-glucosidase activity (56). The other four enzymes from the GH3 family were classified as β-d-xylosidases. Both enzymes from the GH35 family were assigned as β-galactosidases, and both enzymes from the GH79 family were assigned as β-glucuronidases. The structural differences in the glycoside substrates of these enzymes deviate only slightly from the reactive group of the JJB probes (Fig. 4C), explaining why these proteins are labeled despite having different substrates.
To confirm that glycosidases other than glucosidases can be labeled, we tested whether commercially available xylosidase can also be labeled with JJB70. This well-characterized xylosidase is from the soil bacterium Opitutus terrae PB90–1 (ACB77584) and belongs to the GH52 family. This experiment revealed that this classical xylosidase can be labeled with JJB70 and that labeling is competed with KY371 and is pH dependent (supplemental Fig. S2). These experiments demonstrate that JJB probes have a much broader range of target retaining glycosidases than originally thought.
Glycosidase Profiling Is Widely Applicable
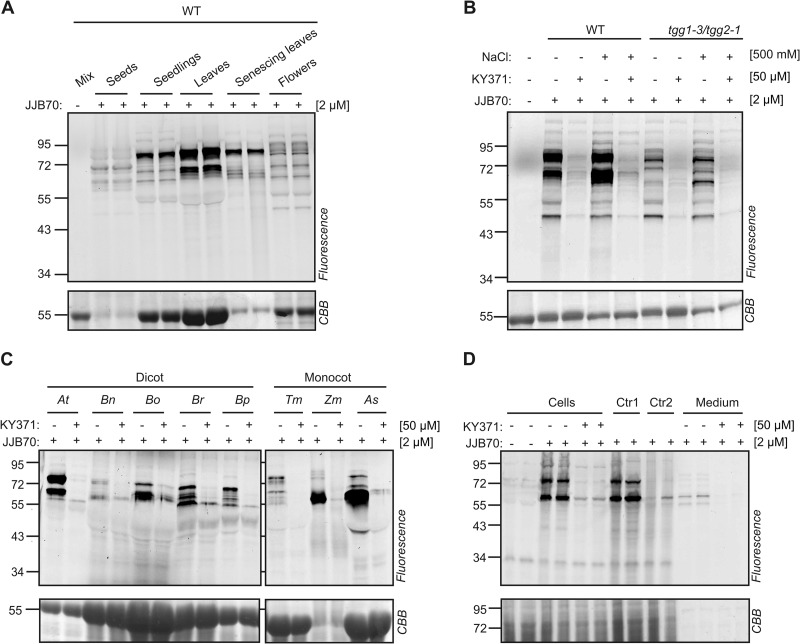
To investigate whether glycosidase profiling could be extended to different organs of Arabidopsis, we labeled extracts from seeds, seedlings, leaves, senescing leaves, and flowers with JJB70. Profiles were very different between organs (Fig. 5A). Notably, many signals were detected in flower extracts. Labeling of these proteins was blocked upon pre-incubation with KY371, and most signals remained in the tgg1-3/tgg2-1 double mutant extracted with and without NaCl (Fig. 5B).
Fig. 5.
Glycosidase profiling is broadly applicable. A, Glycosidase activity profiles differ in different organs of wild-type Arabidopsis thaliana. Organ extracts containing ∼1.0–1.5 mg/ml soluble proteins were labeled with and without 2 μM JJB70 for 1 h. The different organ proteomes were mixed and used as a no-probe-control (mix). B, Flowers contain a diversity of active glycosidase in addition to TGG1 and TGG2. Flower proteins of WT and tgg1–3/tgg2–1 plants were extracted with and without 500 mM NaCl, pre-incubated with and without KY371 for 30 min and labeled with JJB70 at pH 6.0 C, Glycosidase profiling in different plant species. Leaf extracts of various dicot and monocot plants were pre-incubated with and without 50 μM KY371 for 30 minutes and incubated with 2 μM JJB70 for 1 h. The labeled proteins were detected by in-gel fluorescent scanning. At, Arabidopsis thaliana; Bn, Brassica napus; Bo, Brassica oleracea; Br, Brassica rapa; Bp, Brassica pekinensis; Tm, Triticum monococcum; Zm, Zea mays; As, Avena sativa. D, JJB70 labels glycosidases in living cells. Arabidopsis cell cultures were pre-incubated with and without 50 μM KY371 for 30 minutes and incubated with 2 μM JJB70 for 1 h. The cell cultures were ground with SDS-containing gel-loading buffer (GLB) to stop the labeling reaction. For control-1 (Ctr1), untreated cell cultures were ground with JJB70 for 30s, followed by adding GLB. For control-2 (Ctr2), untreated cell cultures were ground in GLB containing 2 μM JJB70. Liquid medium of cell cultures incubated with the probes was separated and concentrated using acetone precipitation.
To extend glycosidase profiling to different plant species, leaf extracts of other dicot and monocot plants were incubated with and without JJB70. Labeling was observed in all tested plant extracts with different profiles and different intensities (Fig. 5C). Labeling of these proteins was suppressed by pre-incubation with KY371. Taken together, these data demonstrate that glycosidase activity profiling is broadly applicable in plant science.
To test whether JJB70 can label proteins in living cells, Arabidopsis cell cultures were pre-incubated with and without KY371 and labeled with JJB70 in duplicate. Proteins were extracted by grinding the cell cultures with SDS-containing gel loading buffer to stop the labeling reaction. JJB70 labeling caused two major signals at 56 and 72 kDa, and these signals were suppressed by KY371 pre-incubation (Fig. 5D). Protein extraction in the presence of JJB70 also caused labeling (Control 1), but this ex vivo labeling was completely abolished if the extraction was performed in SDS-containing gel loading buffer (Control 2; Fig. 5D). This shows that JJB70 does not label proteins under denaturing conditions and demonstrates that the probe can label proteins in living cells. Interestingly, the labeling profile of the cell culture medium was distinct from that of the cells, as it displayed two faint signals at 55 kDa that were suppressed upon pre-incubation with KY371, indicating that the probe also displayed activities of secreted glycosidases.
Glycosidase Profiling of Secreted Proteomes
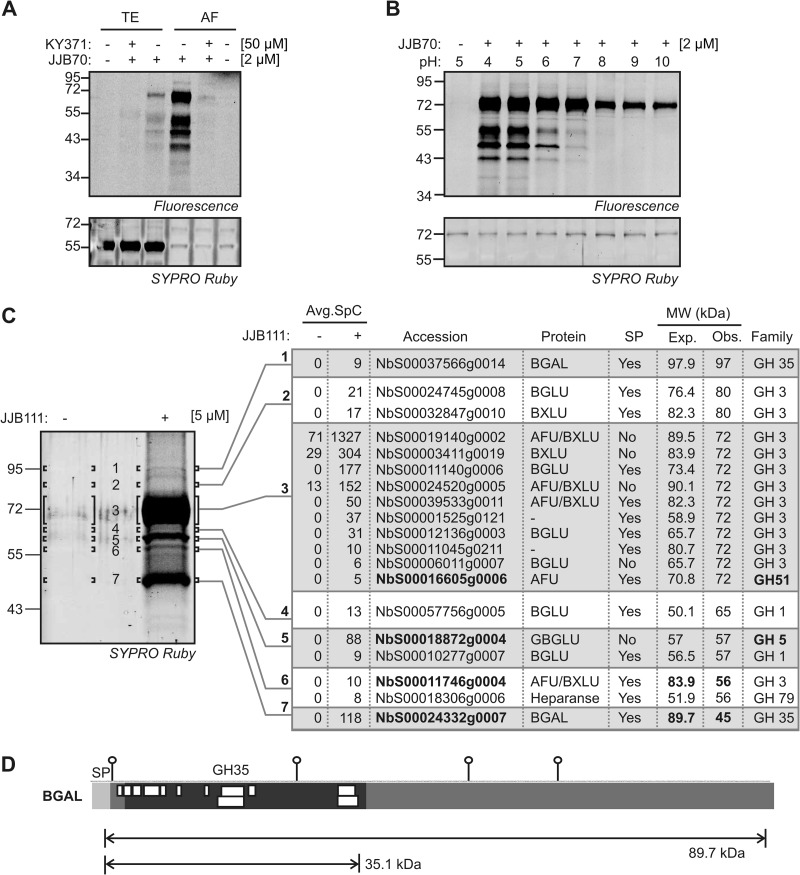
To investigate the secreted glycosidases further, we identified active glycosidases in the extracellular space (apoplast) of N. benthamiana, an important model plant having large leaves that are ideal for apoplast extraction. The apoplast of plants contains many glycosidases involved in cell wall remodeling and defense (57, 58). To monitor the active state of apoplastic glycosidases, we isolated apoplastic proteomes from N. benthamiana leaves and incubated them with and without 2 μm JJB70 for 1 h. The labeled proteins were separated on protein gel and detected via fluorescent scanning. Three major signals at 45, 55, and 72 kDa; a weak signal at 43 kDa; and a series of weaker signals below 40 kDa were detected in the probe-labeled sample (Fig. 6A). Similar but much weaker signals were detected in the total extracts (Fig. 6A). These signals were absent in the no-probe control and suppressed in the samples pre-incubated with KY371. Furthermore, labeling was optimal at acidic pH (Fig. 6B), consistent with the pH of the apoplast.
Fig. 6.
The plant apoplast contains a diversity of active glycosidases. A, Labeling of total extracts (TE) and apoplastic fluids (AF) isolated from N. benthamiana leaves. TE and AF were isolated from leaves of N. benthamiana, pre-incubated with and without 50 μM KY371 for 30 min and labeled with 2 μM JJB70 at pH 5. B, Labeling is optimal at acidic (apoplastic) pH. Apoplastic fluids were incubated with 2 μM JJB70 at pH 4–10 for 1 h. A–B, Labeled proteins were detected by in-gel fluorescent scanning. The gel was stained with coomassie to show the equal loading. C, Identification of the JJB111-labeled glycosidases. Large scale labeling was performed by incubating 10 ml of apoplastic fluids from N. benthamiana leaves with and without 5 μM JJB111. Biotinylated proteins were purified, separated on protein gels and visualized by fluorescent scanning of SYPRO Ruby-stained protein gels. Bands were excised, treated with trypsin and the peptides were identified by ion trap mass spectrometry by performing two independent MS runs. Proteins identified in both runs were selected and the average spectral counts are summarized. Band 3 was sub-sliced into 3a, 3b, and 3c and the peptides were identified from each sub-slice. Avg.SpC, average spectral counts; SP, predicted signal peptide; MW, molecular weight; Exp., expected MW; Obs, observed MW. D, Peptide coverage of a truncated beta-galactosidase (BGAL). The identified peptides of BGAL are represented by white boxes. Putative N-glycosylation sites (PGS) and the theoretical molecular weight of GH35 domain in BGAL and mature protein without signal peptide (SP) are indicated.
To identify the labeled proteins, we purified JJB111-labeled proteins, separated them on protein gels, and detected them through SYPRO Ruby staining. Labeled proteins were enriched in the JJB111-labeled sample, and these signals were not detected in the no-probe control (Fig. 6C). Seven protein bands from the JJB111-labeled sample and the corresponding regions from no-probe control were excised and treated with trypsin. MS analysis of peptides identified 19 different glycosidases from six different GH families: GH1, GH3, GH5, GH35, GH51, and GH79 (supplemental Table S2). Members of the GH5 and GH51 families had not been detected before in labeling experiments, which signifies the potential for JJB probes to label retaining glycosidases from additional GH families. Of the 19 identified glycosidases, BGAL (NbS00024332g0007) and α-l-arabinofuranosidase/β-d-xylosidase (NbS00011746g0004.1) migrated at apparent MWs of 45 and 56 kDa, respectively, which are much lower than their theoretical MWs (89.7 and 83.9, respectively; Fig. 6C). The MS analysis identified 12 unique peptides from BGAL and four unique peptides of α-l-arabinofuranosidase/β-d-xylosidase. All the identified peptides of BGAL originated from the GH35 catalytic domain, and not from the C-terminal half of this protein (Fig. 6D). This indicates that the active state of BGAL is a 45-kDa truncated protein consisting of only the catalytic domain.
DISCUSSION
We introduced broad-range activity profiling of glycosidases and demonstrated its potential to uncover unexpected post-translational regulation of these enzymes. Using cyclophellitol-aziridine probes, we detected the active state of nearly 40 different retaining glycosidases of Arabidopsis and N. benthamiana. These identified glycosidases belong to seven different GH families and are likely to have different substrate specificities. The majority of the detected glycosidases are uncharacterized enzymes. Glycosidase activity profiling revealed the active state of apoplastic glycosidases in N. benthamiana.
Activity-based Glycosidase Profiling
Several experiments have demonstrated that glycosidase labeling is activity dependent. First, glycosidase labeling depends on pH. For example, myrosinases TGG1 and TGG2 and apoplastic glycosidases were labeled intensively at slightly acidic or acidic pH, consistent with in vitro studies of these enzymes (48, 59) and the location of these enzymes in the acidic apoplast, lysosome, or vacuole (60). In general, biomolecules have evolved to perform their functions in the conditions of the compartments where they are located (61). Second, the identified glycosidases are all retaining enzymes. This selectivity is in agreement with the proposed mechanism of these probes to label the active site of retaining glycosidases with a covalent glycosyl-enzyme intermediate. Third, spectral analysis of the JJB111-labeled peptide of TGG1 indicated that the nucleophilic glutamate active site residue was indeed labeled by the probe. This confirms that the warhead of the probe enters the substrate-binding pocket of glycosidases to label the active site. Identification of both labeled and unlabeled versions of active site peptides from TGG1 is explained by the reversible nature of the ester bond formed between the probe and the enzymes, sensitive for hydrolysis during the chemical proteomics workflow. This also explains why we did not identify labeled peptides from the other detected glycosidases. Fourth, labeling of proteins in cell cultures was abolished under denaturing conditions, implying that a native protein structure is essential for labeling. Fifth, competition assays by pre-incubation with glycosidase inhibitor KY371 blocked labeling. Taken together, these results show that the probes labeled glycosidases based on their active state rather than their abundance.
Broad-range Glycosidase Profiling
We discovered the potential of JJB probes to label a greater number of glycosidases than originally thought. Cyclophellitol-aziridine-based probes were initially designed to label retaining glucosidases (33). In addition to glucosidases, we detected glycosidases with additional specificities including xylosidases, galactosidases, glucuronidase, and glucanases. Interestingly, we also identified an α-l-arabinofuranosidase and heparanase, whose substrates are α-l-arabinofuranoside and heparan sulfate, respectively. Furthermore, labeling of a well-characterized xylosidase confirmed that the JJB probes do not target only glucosidases. Detection of a large number of different glycosidases in plant proteomes is consistent with the fact that plant genomes have more glycosidase-related genes than animals (4). We detected the active state of 39 different glycosidases representing six different retaining GH families of Arabidopsis and N. benthamiana. The labeled purified xylosidase is a representative of the seventh GH family. In addition to labeling members of GH1, GH3, and GH30 (33), we have now detected labeling of members of the GH5, GH35, GH79, GH51, and GH52 families, which constitute 100 genes in Arabidopsis. Thus JJB probes offer great potential for studying the active state of a large number of glycosidases in plants.
Opportunities Offered by Glycosidase Profiling
Being a large superfamily, glycosidases are challenging to characterize. Glycosidase profiling can be used to study when and which of these enzymes are in their active state. Some glycosidases do not show activity when heterologously expressed and purified. For example, recombinant TGG2 did not have myrosinase activity upon heterologous expression and in in vitro enzyme assays (62). This highlights the importance of profiling activities of enzymes under native conditions. To our knowledge, activities contributed by TGG1 or TGG2 have not been studied in wild-type Arabidopsis plants before. In order to study myrosinase activities contributed by TGG2, a tedious extraction and purification procedure had to be followed using tgg1 mutant plants (48). We displayed the active states of TGG1 and TGG2 in wild-type plants without using knockout lines and without complex extraction procedures. Thus glycosidase activity profiling can significantly simplify studies of myrosinases and other glycosidases.
The synthesis of putative substrates via organic chemistry to characterize glycoside specificities is a bottleneck in glycobiology (58). Furthermore, some glycosidases, such as plant α-d-xylosidases, hydrolyze only their natural substrates and not commercially available synthetic substrates (58, 63). Glycosidase activity profiling bypasses these bottlenecks by displaying active site availability and reactivity, which are hallmarks of enzyme activity (12). Thus active glycosidases can be monitored with ABPP without knowledge of the natural substrates. We also detected active glycosidases that were assumed to be inactive. Detection of TGG3, for example, showed that this enzyme was active, even though it was originally classified as a pseudogene (49). Glycosidase profiling is not restricted to the proteomes from which we have identified labeled proteins. We also profiled for active glycosidases in extracts of different organs and leaves of different dicot and monocot plant species. Thus glycosidase profiling can be used to unravel the roles of glycosidases in other plant species. We have also demonstrated that both the fluorescent probe JJB70 and the inhibitor KY371 enter living cells and label glycosidases in vivo. These experiments demonstrate that KY371 is an excellent broad-range glycosidase inhibitor in living cells. KY371 can now be used in chemical knockout studies to display phenotypes associated with a loss of broad-range glycosidase function at a time point and dose of choice.
Active Glycosidases in the Apoplast
The extracellular space and the cell wall jointly constitute an important part of a plant called the apoplast. In this study, we chose N. benthamiana as a model system to study the active glycosidases in the leaf apoplast. Apoplastic glycosidases can selectively alter cell wall polysaccharides to remodel the cell wall architecture during plant growth and development. By labeling apoplastic proteomes, we detected the active state of four putative bi-functional α-l-arabinofuranosidase/β-d-xylosidases, two putative β-d-xylosidases, five putative β-d-glucosidases, two putative β-d-galactosidases, one putative heparanase, one putative β-d-glucanase, and one putative α-l-arabinofuranosidase. Glycosidases with these substrate-hydrolyzing activities process the major cell wall polysaccharides, including glucan, xylan, arabinoxylan, galactan, and arabinan, during cell wall assembly and reorganization (9). Thus glycosidase profiling reveals the active state of glycosidases that likely play a role in cell wall remodeling. Furthermore, we also detected two GH3 family enzymes with unknown glycosidic activities. Thus glycosidase profiling allows for the detection of the active state of these enzymes without knowledge of their substrates.
The activity of glycosidases is tightly regulated at the post-translational level. For example, cell wall proteome analysis suggests that glycosidases can be regulated by proteases (64). Consistent with this, we found that the labeled BGAL (NbS00024332g0007) was a truncated protein consisting of only the GH35 catalytic domain. Such truncation has also been observed in an animal lysosomal β-galactosidase where the C-terminal half is removed to release a mature enzyme in an acidic environment (65). Interestingly, a C-terminal truncation of a bacterial β-galactosidase resulted in an efficient transgalactosylase (66). The truncated BGAL in the plant apoplast might have acquired transglycosylase activity and might catalyze the transfer of galactose to acceptor molecules. In addition, the loss of the C-terminal half of the protein might also have altered the location of this enzyme in the apoplast. The role of proteolytic regulation of BGAL is therefore an interesting topic for further studies.
Supplementary Material
Acknowledgments
We thank Georg Jander (Boyce Thompson Institute for Plant Research, Ithaca, NY) and Pawel Bednarek (Institute of Bioorganic Chemistry, Polish Academy of Science, Poland) for kindly providing tgg mutants, and Dr. Johana Misas-Villamil for technical support. The mass spectrometry data have been deposited in the Peptide Atlas database (www.peptideatlas.org) under accession code PASS00525.
Footnotes
Author contributions: B.C. and R.v.d.H. designed research; B.C. performed research; J.J. and H.S.O. contributed new reagents or analytic tools; B.C., T.C., A.E.E., A.H., H.S.O., and R.v.d.H. analyzed data; B.C. and R.v.d.H. wrote the paper; T.N.H. performed apoplastic proteome isolation; J.G.V. supported the pull-down experiment.
* This work was supported by ERA-IB project “PRODuCE,” the Max Planck Society, Deutsche Forschungsgemeinschaft projects HO 3983/9-1 and HO 3983/13-1, the University of Oxford, and COST program Chemical Proteomics CM1004.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GH
- glycosyl hydrolase
- ABPP
- activity-based protein profiling
- DMSO
- dimethyl sulfoxide
- MW
- molecular weight
- BGAL
- β-d-galactosidase.
REFERENCES
- 1. Sturgeon R. J. (2003) Carbohydrates. In eLS, John Wiley & Sons, online article available at: 10.1038/npg.els.0000690 [DOI] [Google Scholar]
- 2. Cheeke P. R. (2001) Glycosides: naturally occurring. In eLS, John Wiley & Sons, online article available at: 10.1038/npg.els.0000692 [DOI] [Google Scholar]
- 3. Hancock S. M., Withers S. G. (2014) Glycosidases: functions, families and folds. In eLS, John Wiley & Sons, online article available at: 10.1002/9780470015902.a0020548.pub2 [DOI] [Google Scholar]
- 4. Coutinho P. M., Starn M., Blanc E., Henrissat B. (2003) Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 8, 563–565 [DOI] [PubMed] [Google Scholar]
- 5. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinnott M. L. (1990) Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 90, 1171–1202 [Google Scholar]
- 7. Mccarter J. D., Withers S. G. (1994) Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4, 885–892 [DOI] [PubMed] [Google Scholar]
- 8. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 9. Minic Z. (2008) Physiological roles of plant glycoside hydrolases. Planta 227, 723–740 [DOI] [PubMed] [Google Scholar]
- 10. Cravatt B. F., Wright A. T., Kozarich J. W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 11. Kolodziejek I., Van der Hoorn R. A. L. (2010) Mining the active proteome in plant science and biotechnology. Curr. Opin. Biotechnol. 21, 225–233 [DOI] [PubMed] [Google Scholar]
- 12. Kobe B., Kemp B. E. (1999) Active site-directed protein regulation. Nature 402, 373–376 [DOI] [PubMed] [Google Scholar]
- 13. Richau K. H., Kaschani F., Verdoes M., Pansuriya T. C., Niessen S., Stuber K., Colby T., Overkleeft H. S., Bogyo M., Van der Hoorn R. A. L. (2012) Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 158, 1583–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van der Hoorn R. A. L., Leeuwenburgh M. A., Bogyo M., Joosten M. H., Peck S. C. (2004) Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 135, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., Van der Hoorn R. A. L. (2008) Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 20, 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van der Linde K., Mueller A. N., Hemetsberger C., Kashani F., Van der Hoorn R. A. L., Doehlemann G. (2012) The maize cystatin CC9 interacts with apoplastic cysteine proteases. Plant Signal. Behav. 7, 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dong S. M., Stam R., Cano L. M., Song J., Sklenar J., Yoshida K., Bozkurt T. O., Oliva R., Liu Z. Y., Tian M. Y., Win J., Banfield M. J., Jones A. M. E., Van der Hoorn R. A. L., Kamoun S. (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555 [DOI] [PubMed] [Google Scholar]
- 18. Hörger A. C., Ilyas M., Stephan W., Tellier A., Van der Hoorn R. A. L., Rose L. E. (2012) Balancing selection at the tomato RCR3 guardee gene family maintains variation in strength of pathogen defense. PLoS Genet. 8, e1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., Ilyas M., Win J., Kamoun S., Van der Hoorn R. A. L. (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lozano-Torres J. L., Wilbers R. H. P., Gawronski P., Boshoven J. C., Finkers-Tomczak A., Cordewener J. H. G., America A. H. P., Overmars H. A., Van 't Klooster J. W., Baranowski L., Sobczak M., Ilyas M., Van der Hoorn R. A. L., Schots A., de Wit P. J. G. M., Bakker J., Goverse A., Smant G. (2012) Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109, 10119–10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mueller A. N., Ziemann S., Treitschke S., Assmann D., Doehlemann G. (2013) Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9, e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rooney H. C., Van't Klooster J. W., Van der Hoorn R. A. L., Joosten M. H., Jones J. D., de Wit P. J. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308, 1783–1786 [DOI] [PubMed] [Google Scholar]
- 23. Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., Van der Hoorn R. A. L., Kamoun S. (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 106, 1654–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. (2007) A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gu C., Kolodziejek I., Misas-Villamil J., Shindo T., Colby T., Verdoes M., Richau K. H., Schmidt J., Overkleeft H. S., Van der Hoorn R. A. L. (2010) Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J. 62, 160–170 [DOI] [PubMed] [Google Scholar]
- 26. Kolodziejek I., Misas-Villamil J. C., Kaschani F., Clerc J., Gu C., Krahn D., Niessen S., Verdoes M., Willems L. I., Overkleeft H. S., Kaiser M., Van der Hoorn R. A. L. (2011) Proteasome activity imaging and profiling characterizes bacterial effector syringolin A. Plant Physiol. 155, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu C., Shannon D. A., Colby T., Wang Z. M., Shabab M., Kumari S., Villamor J. G., McLaughlin C. J., Weerapana E., Kaiser M., Cravatt B. F., Van der Hoorn R. A. L. (2013) Chemical proteomics with sulfonyl fluoride probes reveals selective labeling of functional tyrosines in glutathione transferases. Chem. Biol. 20, 541–548 [DOI] [PubMed] [Google Scholar]
- 28. Kaschani F., Gu C., Niessen S., Hoover H., Cravatt B. F., Van der Hoorn R. A. L. (2009) Diversity of serine hydrolase activities of unchallenged and Botrytis-infected Arabidopsis thaliana. Mol. Cell. Proteomics 8, 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lenger J., Kaschani F., Lenz T., Dalhoff C., Villamor J. G., Koster H., Sewald N., Van der Hoorn R. A. L. (2012) Labeling and enrichment of Arabidopsis thaliana matrix metalloproteases using an active-site directed, marimastat-based photoreactive probe. Bioorg. Med. Chem. 20, 592–596 [DOI] [PubMed] [Google Scholar]
- 30. Misas-Villamil J. C., Toenges G., Kolodziejek I., Sadaghiani A. M., Kaschani F., Colby T., Bogyo M., Van der Hoorn R. A. L. (2013) Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 73, 689–700 [DOI] [PubMed] [Google Scholar]
- 31. Nickel S., Kaschani F., Colby T., Van der Hoorn R. A. L., Kaiser M. (2012) A para-nitrophenol phosphonate probe labels distinct serine hydrolases of Arabidopsis. Bioorg. Med. Chem. 20, 601–606 [DOI] [PubMed] [Google Scholar]
- 32. Villamor J. G., Kaschani F., Colby T., Oeljeklaus J., Zhao D., Kaiser M., Patricelli M. P., Van der Hoorn R. A. L. (2013) Profiling protein kinases and other ATP binding proteins in Arabidopsis using acyl-ATP probes. Mol. Cell. Proteomics 12, 2481–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kallemeijn W. W., Li K. Y., Witte M. D., Marques A. R. A., Aten J., Scheij S., Jiang J. B., Willems L. I., Voorn-Brouwer T. M., van Roomen C. P. A. A., Ottenhoff R., Boot R. G., van den Elst H., Walvoort M. T. C., Florea B. I., Codee J. D. C., van der Marel G. A., Aerts J. M. F. G., Overkleeft H. S. (2012) Novel activity-based probes for broad-spectrum profiling of retaining beta-exoglucosidases in situ and in vivo. Angew Chem. Int. 51, 12529–12533 [DOI] [PubMed] [Google Scholar]
- 34. Hong T. N., Van der Hoorn R. A. L. (2014) DIGE-ABPP by click chemistry: pairwise comparison of serine hydrolase activities from the apoplast of infected plants. Methods Mol. Biol. 1127, 183–194 [DOI] [PubMed] [Google Scholar]
- 35. Joosten M. H. A. J. (2012) Isolation of apoplastic fluid from leaf tissue by the vacuum infiltration-centrifugation technique. Plant Fungal Pathog. Methods Protoc. 835, 603–610 [DOI] [PubMed] [Google Scholar]
- 36. Wessel D., Flugge U. I. (1984) A method for the quantitative recovery of protein in dilute-solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 37. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 38. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The PFAM protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lamesch P., Berardini T. Z., Li D. H., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D. L., Garcia-Hernandez M., Karthikeyan A. S., Lee C. H., Nelson W. D., Ploetz L., Singh S., Wensel A., Huala E. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 42. Charif D., Lobry J. (2007) SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution (Bastolla U., Porto M., Roman H. E., Vendruscolo M., eds), pp. 207–232, Springer, Berlin [Google Scholar]
- 43. Paradis E., Claude J., Strimmer K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 [DOI] [PubMed] [Google Scholar]
- 44. Tougfighi K., Brady S. M., Austin R., Ly E., Provart N. J. (2005) The botany array resource: e-Northerns, Expression Angling, and promoter analysis. Plant J. 43, 153–163 [DOI] [PubMed] [Google Scholar]
- 45. Hirsch-Hoffmann M., Gruissem W., Baerenfaller K. (2012) pep2pro: the high-throughput proteomics data processing, analysis, and visualization tool. Front. Plant Sci. 3, 00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barth C., Jander G. (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46, 549–562 [DOI] [PubMed] [Google Scholar]
- 47. Burmeister W. P., Cottaz S., Driguez H., Iori R., Palmieri S., Henrissat B. (1997) The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 5, 663–675 [DOI] [PubMed] [Google Scholar]
- 48. Zhou C., Tokuhisa J. G., Bevan D. R., Esen A. (2012) Properties of beta-thioglucoside hydrolases (TGG1 and TGG2) from leaves of Arabidopsis thaliana. Plant Sci. 191–192, 82–92 [DOI] [PubMed] [Google Scholar]
- 49. Zhang J., Pontoppidan B., Xue J., Rask L., Meijer J. (2002) The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant 115, 25–34 [DOI] [PubMed] [Google Scholar]
- 50. Arsovski A. A., Popma T. M., Haughn G. W., Carpita N. C., McCann M. C., Western T. L. (2009) AtBXL1 encodes a bifunctional beta-D-xylosidase/alpha-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol. 150, 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dean G. H., Zheng H., Tewari J., Huang J., Young D. S., Hwang Y. T., Western T. L., Carpita N. C., McCann M. C., Mansfield S. D., Haughn G. W. (2007) The Arabidopsis MUM2 gene encodes a beta-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19, 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eudes A., Mouille G., Thevenin J., Goyallon A., Minic Z., Jouanin L. (2008) Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 49, 1331–1341 [DOI] [PubMed] [Google Scholar]
- 53. Hruba P., Honys D., Twell D., Capkova V., Tupy J. (2005) Expression of beta-galactosidase and beta-xylosidase genes during microspore and pollen development. Planta 220, 931–940 [DOI] [PubMed] [Google Scholar]
- 54. Minic Z., Rihouey C., Do C. T., Lerouge P., Jouanin L. (2004) Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol. 135, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu Z., Escamilla-Trevino L., Zeng L., Lalgondar M., Bevan D., Winkel B., Mohamed A., Cheng C. L., Shih M. C., Poulton J., Esen A. (2004) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367 [DOI] [PubMed] [Google Scholar]
- 56. Hrmova M., Harvey A. J., Wang J., Shirley N. J., Jones G. P., Stone B. A., Hoj P. B., Fincher G. B. (1996) Barley beta-D-glucan exohydrolases with beta-D-glucosidase activity. Purification, characterization, and determination of primary structure from a cDNA clone. J. Biol. Chem. 271, 5277–5286 [DOI] [PubMed] [Google Scholar]
- 57. Gunl M., Neumetzler L., Kraemer F., de Souza A., Schultink A., Pena M., York W. S., Pauly M. (2011) AXY8 encodes an alpha-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23, 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Minic Z., Jouanin L. (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 44, 435–449 [DOI] [PubMed] [Google Scholar]
- 59. Rather M., Mishra S. (2013) Beta-glycosidases: an alternative enzyme based method for synthesis of alkyl-glycosides. Sustain. Chem. Process. 1, 7 [Google Scholar]
- 60. Grignon C., Sentenac H. (1991) pH and ionic conditions in the apoplast. Annu. Rev. Plant Phys. 42, 103–128 [Google Scholar]
- 61. Talley K., Alexov E. (2010) On the pH-optimum of activity and stability of proteins. Proteins 78, 2699–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Andersson D., Chakrabarty R., Bejai S., Zhang J. M., Rask L., Meijer J. (2009) Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 70, 1345–1354 [DOI] [PubMed] [Google Scholar]
- 63. Fanutti C., Gidley M. J., Reid J. S. G. (1991) A xyloglucan-oligosaccharide-specific alpha-D-xylosidase or exo-oligoxyloglucan-alpha-xylohydrolase from germinated nasturtium (Tropaeolum majus L) seeds—purification, properties and its interaction with a xyloglucan-specific endo-(1-]4)-beta-D-glucanase and other hydrolases during storage-xyloglucan mobilization. Planta 184, 137–147 [DOI] [PubMed] [Google Scholar]
- 64. Minic Z., Jamet E., Negroni L., Arsene der Garabedian P., Zivy M., Jouanin L. (2007) A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 58, 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Van der Spoel A., Bonten E., d'Azzo A. (2000) Processing of lysosomal beta-galactosidase—the C-terminal precursor fragment is an essential domain of the mature enzyme. J. Biol. Chem. 275, 10035–10040 [DOI] [PubMed] [Google Scholar]
- 66. Jorgensen F., Hansen O. C., Stougaard P. (2001) High-efficiency synthesis of oligosaccharides with a truncated beta-galactosidase from Bifidobacterium bifidum. Appl. Microbiol. Biot. 57, 647–652 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.