Abstract
Molecular combing and flow-induced stretching are the most commonly used methods to immobilize and stretch DNA molecules. While both approaches require functionalization steps for the substrate surface and the molecules, conventionally the former does not take advantage of, as the latter, the versatility of microfluidics regarding robustness, buffer exchange capability, and molecule manipulation using external forces for single molecule studies. Here, we demonstrate a simple one-step combing process involving only low-pressure oxygen (O2) plasma modified polysilsesquioxane (PSQ) polymer layer to facilitate both room temperature microfluidic device bonding and immobilization of stretched single DNA molecules without molecular functionalization step. Atomic force microscopy and Kelvin probe force microscopy experiments revealed a significant increase in surface roughness and surface potential on low-pressure O2 plasma treated PSQ, in contrast to that with high-pressure O2 plasma treatment, which are proposed to be responsible for enabling effective DNA immobilization. We further demonstrate the use of our platform to observe DNA-RNA polymerase complexes and cancer drug cisplatin induced DNA condensation using wide-field fluorescence imaging.
INTRODUCTION
A wide variety of platforms has been developed over the past two decades to enable single molecule studies.1–5 Advantages of the single molecule approach lie in its ability to manipulate individual molecules and serve as a platform to observe and analyze their interaction with other biomolecules.6,7 Specifically, single DNA molecule studies have gained an immense interest and hence different methods, such as atomic force microscopy (AFM), optical and magnetic tweezers, are developed to study DNA and its interactions with proteins, membranes, drugs, etc.2,4,8 The semi-flexible nature of DNA molecules and their physiological random coil conformation in bulk environment make direct visualization of DNA and its interactions challenging. Immobilizing DNA molecules in its stretched form can overcome this problem. Among the commonly used methods,9–13 molecular combing and flow stretching techniques are widely practiced, as they are simple, inexpensive, and versatile.8 Bensimon's group was the first to report DNA stretching using molecular combing, where tens of DNA molecules were stretched on a functionalized hydrophobic surface by a receding water meniscus.14 Weiss et al. reported the use of poly-l-lysine coated coverslides for effective stretching and immobilization of DNA molecules and demonstrated high resolution mapping of T7 RNA polymerase (RNAP) binding sites on T7 DNA molecules.10,15 Schwartz and his coworkers used trimethylsilane and dimethyldiethoxysilane (DMDES) coated coverslides to render a hydrophobic surface for effective DNA combing.16,17 Larson's group reported a novel protein assisted DNA immobilization (PADI) on micro- or nanofluidic surfaces, in which restriction enzymes or RNAP was used to anchor DNA molecules with limited surface interactions.9 Other notable methods involve the use of biotin-streptavidin coupling, neutravidin coated surfaces, polystyrene coated coverslides, aminopropyltriethoxysilane (APTES) coated surface, receding meniscus on micro-structured polydimethylsiloxane (PDMS) substrates, etc., for DNA stretching and immobilization.8,18–20 Generally, such experiments are conducted open-top or have DNA molecules sandwiched between two coverslides. Using a microfluidic channel will provide better control in sample handling and imaging, making the platform more robust. But, the use of microfluidic channels makes surface functionalization (SF) difficult, requiring flow-in of buffer containing SF molecules, incubation, and washing steps to remove unbound SF molecules. For the ease of applications, it is imperative to have a platform that does not require multiple surface functionalization steps to facilitate DNA immobilization and stretching in a microfluidic channel. In this work, we demonstrated a thin spin-coated polysilsesquioxane (PSQ) layer on a coverslide, when modified by low-pressure (LP) O2 plasma, could be used for room-temperature microfluidic device bonding and immobilization of stretched single DNA molecules. We propose the working mechanism of this simple approach and show its applications for DNA-protein and DNA-drug studies.
PSQ is a silicon resin, synthesized using trifunctional organosilane compounds. It is an organic-inorganic hybrid polymer, the siloxane bond contributing to its inorganic characteristics, and the side-chain functional group contributing to its organic properties. PSQ thus shows excellent heat, electrical, chemical resistance, and mechanical properties.21 PSQ is also highly transparent, good for optical observation, and its high Young's modulus (∼800 MPa) makes it a superb material for micro-nanofluidic device bonding much less prone to cambering or collapsing. When PSQ-coated coverslides are treated with O2 plasma, open silanol (Si-O-H) groups are formed, which upon interaction with silanol groups of silicon or fused silica substrate forms Si-O-Si bonds through condensation reaction. Based on this reaction, Gu et al.22 first demonstrated a simple and effective room-temperature bonding method using PSQ, and this method was later extended to obtain micro-nanofluidic devices for wide variety of applications.12,13,23,24
While O2 plasma has been widely used for polymer-assisted bonding using polymers such as PDMS,25,26 polymethylmethacrylate (PMMA),27 and PSQ,22 here we demonstrate the use of low-pressure oxygen (O2) plasma treatment to fine tune surface properties of PSQ, such that it serves the purpose of both a polymer intermediate in room-temperature fluidic channel bonding and an activated surface to immobilize single DNA molecules afterwards. Further, we demonstrate immobilization of DNA molecules at one end or at multiple points under different plasma treatment conditions.
MATERIALS AND METHODS
Surface preparation and device fabrication
First, standard No. 1 coverslides (25 × 25 mm, Goldseal) were cleaned thoroughly using piranha solution (98% Con. H2SO4 and 30% H2O2 in 1:1 ratio) for 15 min, then rinsed by de-ionized (DI) water, and followed by blow-drying using N2 gun. Dried coverslides were briefly baked for dehydration at 200 °C for 2 min, to ensure complete removal of water molecules from the coverslide surface. PSQ was freshly prepared before experiments by mixing two parts of xylene with one part of Hardsil (AP grade, Gelest, Inc.), followed by filtration using 0.45 μm polytetrafluoroethylene (PTFE) membrane (Basic Life, Inc.) to remove any large debris. Cleaned coverslides were spin-coated with PSQ on one side, at 3000 rpm for 30 s. PSQ-coated coverslides were then baked at 240 °C for 30 min and then exposed to O2 plasma using a reactive ion etching (RIE) machine (Anela DEM-451T). First, we fixed the radio frequency (RF) power and the O2 flow rate and used two different process pressures (high pressure (HP) and low pressure). HP corresponds to 0.2 mbar (150 mTorr) and LP corresponds to 0.02 mbar (15 mTorr). Second, we varied exposure time for plasma treatment (1.5, 3, and 5 min) while keeping other parameters (power, pressure, and flow rate) constant.
For experiments involving microfluidic devices, a simple microchannel (100 μm in width and 1 μm in depth) with reservoirs at its ends was used. Microchannels were obtained by photolithography (EVG620 mask aligner) on fused silica substrates (Semiconductor Wafers, Inc.), followed by dry etching using an inductively coupled plasma (ICP) machine (RIE-10ip, Samco). Through holes on the substrate were obtained by sand blasting. Plasma treated PSQ/coverslide was brought into contact with the microfluidic chip and pressed gently with tweezers to enable bonding. Finally, glass reservoirs were glued on top of the loading holes using UV-curable adhesive (No. 108, Norland Optical Adhesives), to assist sample loading. PSQ offers a very robust bonding, but we can debond chips using commercially available removers. For this, bonded devices were soaked in Dynasolve 210 (Dynaloy, Inc.) with ultra-sonication for 2–3 h or overnight without sonication. After this process, PSQ-coated coverslides can be separated easily from the fused silica chips and further cleaned with isopropyl alcohol (IPA), rinsed thoroughly with DI water, and dried using a N2 gun. The same chip can then be reused effectively for 2–3 times with re-coated fresh PSQ, without compromising the bonding quality.28
AFM and Kelvin probe force microscopy (KPFM) measurements
AFM experiments (Bruker Innova) were conducted to measure the surface roughness of O2 plasma treated PSQ coverslides. Additionally, surface potential measurements were conducted using KPFM (Digital Instruments/Veeco Bioscope SZ) combined with an inverted optical microscope (Nikon TE2000-U) on both HP and LP O2 plasma treated PSQ coverslides. For this, PSQ was coated on one half of the coverslide, keeping the other half masked using a 3M tape. The tape was removed before hard baking PSQ at 240 °C. The 3M tape used here was to protect the coverslide instead of materials like photoresists, to avoid use of solvents for removal of protecting layer, which could possibly alter the PSQ surface. No cleaning agents were used after stripping the 3M tape and AFM result showed no marks/contamination from the 3M tape (see supplementary Figure S1).29 All other steps in PSQ coating process remain the same, as mentioned in the surface preparation and device fabrication section. This way, potential difference between bare coverslide surface and PSQ-coated surface was obtained.
For KPFM experiments, conductive tips (PPP-EFM, Nanosensors) were used for scans in lift mode, with two-pass scan system (trace and retrace). In first pass scan, standard topographic images were obtained in tapping mode with no applied tip bias. During the second pass scan, the topography was retraced with AFM tip lifted to 30 nm above sample surface (hlift) with an applied tip bias voltage (Vtip), to measure the surface potential. The coverslide was grounded, by placing it on a metal substrate connected to the sample bias control of the scanning probe microscope (SPM) controller.
DNA combing experiments
Microfluidic devices bonded with HP and LP treated PSQ coverslides were used for this purpose. DNA combing (immobilization and stretching) experiments were carried out using T4 DNA (166 Kb, Wako Chemicals), λ-DNA (48 Kb, New England Biolabs), and calf thymus DNA (22 Kb, Sigma). Briefly, sample containing 0.1 μg/ml DNA in observation solution composed of 0.5× tris-borate-EDTA (TBE) buffer with 2.5% (w/w) polyvinylpyrrolidone (PVP, Sigma), 10% (w/v) glucose (Sigma), and 1% (v/v) Tween 20 (Sigma) was prepared. An oxygen scavenging system comprising of 50 μg/ml glucose oxidase (Sigma), 10 μg/ml catalase (Roche), and 0.5% (v/v) β-mercaptoethanol (BME, Sigma) was added to the observation solution, to suppress photobleaching and nicking of DNA by YOYO-1 intercalating dye (Invitrogen, 1:5 dye to base pair ratio). In all the experiments, post-staining was exercised to avoid interference of YOYO-1 dye molecules during DNA combing, and DNA interaction with drug or proteins.
DNA-protein complex experiments
Bacteriophage λ-DNA complexed with multi-subunit E. coli RNAP using formaldehyde cross-linking scheme was used for this purpose. Observation solution, oxygen scavenging system, and dye labeling scheme remain the same as in DNA combing experiments. E. coli RNAP was labeled with quantum dots (QDs) using a primary antibody-secondary antibody coupling scheme.28
Cisplatin-complexed DNA experiments
Cisplatin [Pt(NH3)2Cl2] is a widely used anti-cancer drug, known to form DNA adducts leading to inter- and intra-strand crosslinks, causing DNA condensation and programmed cell death.30,31 Disadvantage of cisplatin lies in its high cytotoxicity which includes mitochondrial damage, decreased ATPase activity, and altered cellular transport mechanisms, leading to numerous undesired side effects.32,33 Higher cisplatin concentrations induce long-range intra- and interstrand crosslinks, leading to complete DNA condensation.31
Stock solution of cisplatin (Sigma-Aldrich, 1 mg/ml) was prepared using distilled water and stored at room temperature. After several days of incubation, the stock solution contains 35% undissociated and 65% mono-aquated cisplatin. Calf thymus DNA and λ-DNA were used to observe cisplatin induced DNA condensation. Ultrapure calf thymus DNA (20 Kb) or λ-DNA (48 Kb) was complexed with cisplatin at 0.05, 0.2, and 0.5 Pt/nucleotide ratios. Cisplatin-DNA incubation was carried out in 10 mM NaClO4 (pH 6) for various reaction times at 37 °C in a dark place. Reactions were stopped at desired times by adding NaCl and EDTA to a final concentration of 100 mM and 50 μM, respectively. Observation buffer, oxygen scavenging system, and dye labeling details are the same as in DNA combing experiments. DNA concentration in observation solution was ∼0.1 μg/ml.
RESULTS AND DISCUSSION
Surface properties of O2 plasma treated PSQ
RIE is a widely used dry etching technique in which plasma is generated by ionizing gases using a RF electromagnetic field in an evacuated chamber. Plasma thus generated is a mixture of ions, electrons, and free radicals that interacts with the sample, resulting in etching, and altered surface properties. RF power, pressure, and gas flow rate are among the major factors that influence RIE process.34
O2 is the most common gas among the multitude of gases used in RIE. O2 plasma treatment plays a crucial role in our sample preparation step. The plasma source can induce physical reaction on the substrate surface, due to the bombardment of energetic ions, as well as chemical reaction due to the chemically active ionized oxygen or free radicals.34 Both physical and chemical reactions can occur, based on the reaction parameters (power, pressure, and gas flow rate), and one factor may be dominant than the other. Earlier studies have shown an increase in physical interaction with decreasing pressures.35 Many works have reported the use of O2 plasma to modify polymer surfaces, which introduces or removes certain functional groups, thus altering the surface chemistry.35–37 Adsorption of oxygen by the topmost layers of the polymer surface leads to increase in functional polar groups, increasing the surface energy, and yielding a hydrophilic surface. O2 plasma treatment is thus advantageous for micro- and nanofluidic device bonding using polymer intermediates.
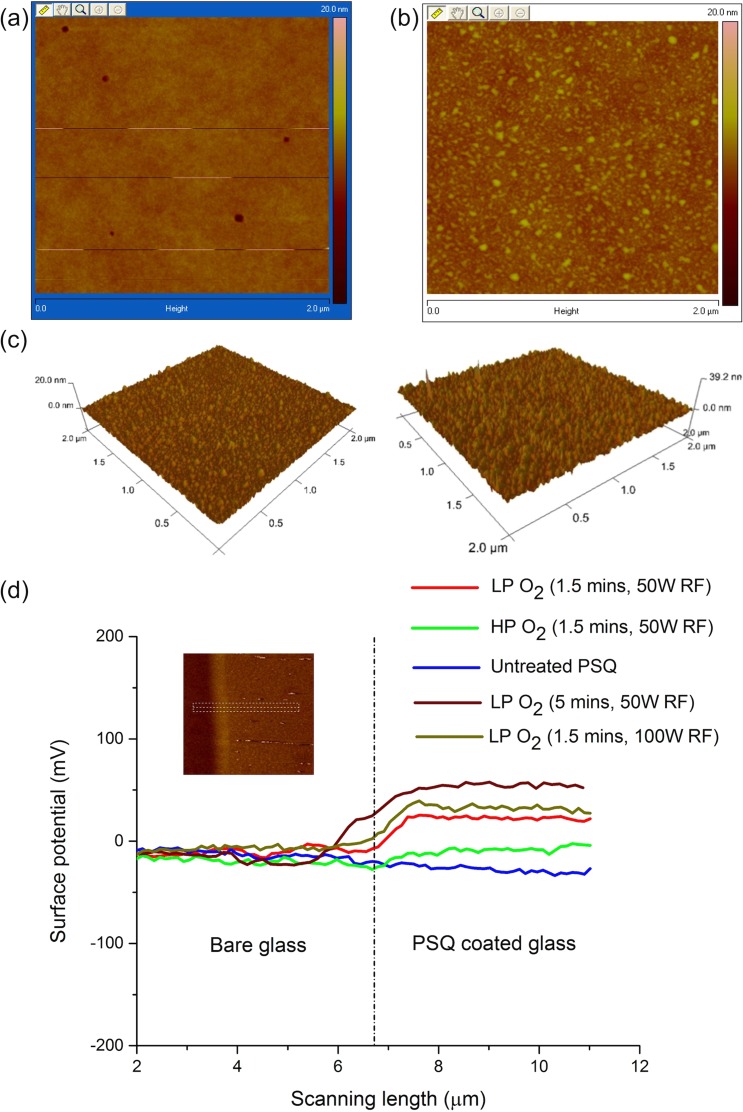
PSQ bonding opens up the possibility of achieving sub-10 nm deep nanochannels and has advantages over other commonly used bonding methods.22,23 Although successful device bonding could be achieved using wide range of plasma conditions, surface adsorption of biomolecules was pronounced for LP O2 plasma exposed PSQ. AFM results showed less surface roughness for HP O2 plasma (Figure 1(a)) treated PSQ compared to the LP case (Figure 1(b)). Many small structures of heights 3–5 nm were observed in LP O2 plasma treated PSQ surface (Figure 1(b)).
FIG. 1.
AFM profile of (a) HP (0.2 mbar pressure) and (b) LP (0.02 mbar pressure) O2 plasma treated PSQ surface. Both were processed with 20 sccm O2, 50 W RF power, and 1.5 min exposure. (c) Comparison of LP O2 plasma treated PSQ surfaces for different RF powers: 50 W (left) and 100 W (right), showing roughness of 0.7 ± 0.1 nm (with ∼5 nm crests) and 1.8 ± 0.2 nm (with ∼10 nm crests), respectively (see supplementary Fig. S2).29 (d) KPFM measurements of LP and HP O2 plasma treated and untreated PSQ surfaces. PSQ was coated on one half of a coverslide for measurements. Inset shows representative image obtained during surface potential measurements.
During the plasma treatment, applied RF field ionizes O2 gas molecules, thus generating plasma.34 Active species such as free radicals and ions present in the plasma react with the coated PSQ surface. Earlier studies have shown that, at pressures above 100 mTorr, the mean free path of the gas molecules decreases, rendering an isotropic plasma.38 Ions formed near the substrate surface have higher probability to reach and react with the surface. The ions in such conditions impart smaller momentum transfer to the substrate surface. Thus, at high-pressure conditions, surface modification is carried out without inflicting much damage. On the other hand, low-pressure conditions (10 mTorr in our experiments) with greater mean free paths mean ions impart larger momentum transfer to the substrate surface, possibly damaging the backbone of PSQ polymer. This explains the increased roughness at low-pressure conditions. Although small crests created by LP O2 plasma exposure explain hooking and stretching of DNA molecules observed, we anticipated an increase in surface potential during O2 plasma treatment, rendering the surface positive for effective DNA immobilization.
KPFM measurements were performed for PSQ surface with respect to bare coverslide surface to verify this speculation, and the results did show an increase in surface potential for both LP and HP O2 plasma treated PSQ surfaces (20 sccm O2, 50 W RF power, and 1.5 min exposure). LP case had the most positive surface potential (∼20 mV) compared to HP (∼0 mV) and untreated PSQ (∼−10 mV) cases. Additionally, surface potential for 5-min exposure using LP O2 plasma (20 sccm O2 and 50 W RF power) was measured to be around 60 mV. Surface potential changes respective to varying RF power were also tested, for which PSQ surface was treated using LP O2 plasma for 1.5 min at 100 W RF power, resulting a potential of ∼35 mV, while surface roughness doubly increased to 1.8 ± 0.2 nm (with ∼10 nm crests) from 0.7 ± 0.1 nm (with ∼5 nm crests) of LP O2 plasma treatment (1.5 min) carried out at 50 W RF power (Figures 1(c) and 1(d) and supplementary Figure S2).29 Bare coverslide showed negative surface potential (∼ −10 mV) for all four cases due to the silanol dangling bond on the surface.
Surface potential measurements using KPFM were carried out on a dry surface, but one could use zeta potential measurements on plasma treated PSQ surfaces to avail more information on the contribution of surface charges in the presence of buffer solution from experiments.39,40 But, zeta potential measurements in aqueous solutions based on electrophoretic mobility or streaming potential are very sensitive to small changes in solution conditions, temperature, etc., and might require experiments under controlled environments to obtain useful information.41 Adsorption of non-ionic species such as Tween-20 used in our DNA experiments could alter the zeta potential and it is quite possible that the surface can have an inherent surface potential but no measurable zeta potential.41,42 Specific adsorption of ions present in the buffer solution could also contribute to the charge in the surface-sample interface.42 We also understand that zeta potential measurements on hydrophobic surfaces (PSQ is hydrophobic without plasma treatment) are not well characterized due to the uncertainties in the chemistry of water-hydrophobic surfaces.42 To our knowledge, no previous study has reported zeta potential measurements for PSQ surface, both in dry and wet conditions, and can be a separate work on its own to compare the two conditions.
Surface potential change measured using KPFM is hence attributed to the prolonged DNA immobilization on PSQ surface in LP plasma conditions, with some initial assistance from crests and valleys on the surface (surface roughness). Though DNA immobilization and stretching have been demonstrated earlier using polymers like SU-8 and PDMS,43,44 after subjecting to O2 plasma treatment, none of these works come up with a clear mechanism to explain the phenomena as we are attempting here. Zeta potential measurements of O2 plasma treated PSQ surfaces using buffer conditions in our experiments could further reveal information on the contribution of surface charges but would be a separate work on its own to compare the two (dry and wet) conditions.
Combing of DNA molecules
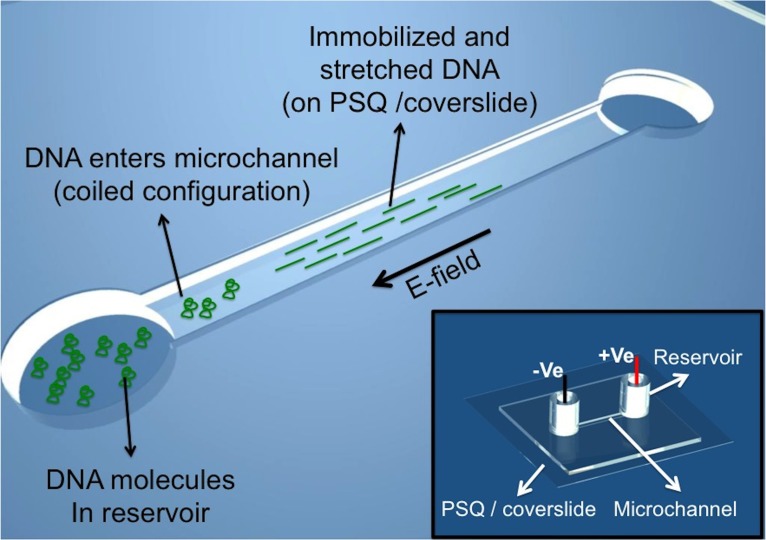
Based on our observations, simple microfluidic channels bonded with LP O2 plasma treated PSQ coverslides may be used for effective combing of DNA molecules. Solution containing DNA molecules was loaded into one reservoir of the microfluidic device and a small electric field (2.5 V/cm) was applied between reservoirs connecting a straight microchannel, driving the DNA molecules from the loading reservoir into the microchannel (Figure 2). Within 5 min, we observed immobilized and stretched DNA molecules along the microchannel, with increased amounts of combed DNA molecules with time (Figures 3(a)–3(c)). The amount of combed DNA molecules along the microchannel depends on DNA concentration in solution, 0.1 μg/ml used in our experiments showed no overcrowding of DNA molecules along the microchannel (Movie M1 in the supplementary material).29 On the contrary, experiments conducted in microchannels bonded with HP O2 plasma treated PSQ did not show any DNA immobilization (Movie M2 in the supplementary material).29
FIG. 2.
Schematics showing DNA combing in microchannels bonded with LP O2 plasma treated PSQ/coverslide. Inset shows a complete device.
FIG. 3.
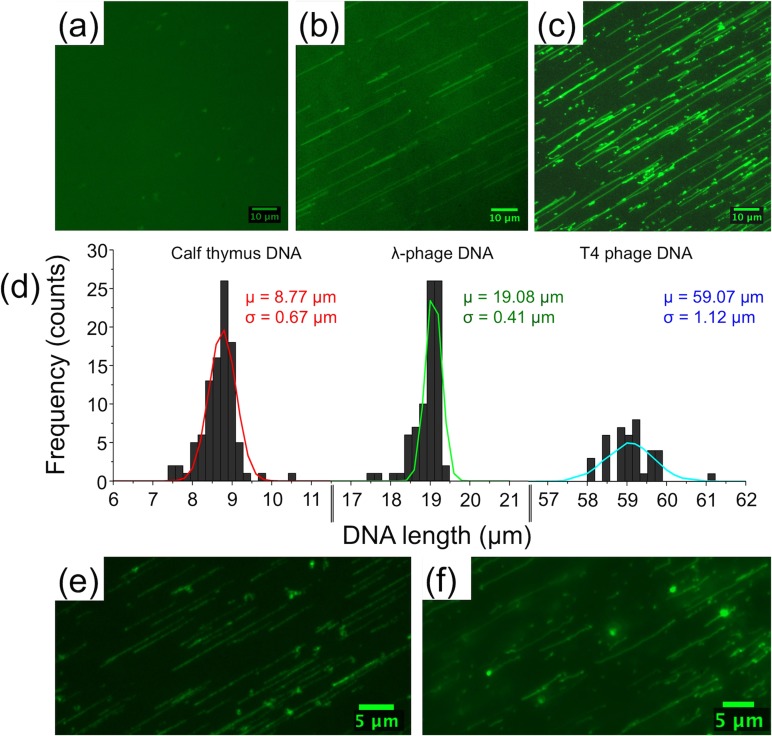
T4 DNA molecules in microchannel bonded with LP O2 plasma treated PSQ (1.5 min): (a) immediately after being driven from the reservoirs, (b) 5–10 min after applying a constant applied electric field (2.5 V/cm), T4 DNA molecules show one end immobilization and stretching, and (c) after 20–30 min, more DNA molecules get combed on the PSQ surface. (d) Gaussian fitted histogram plot showing the length distribution of calf thymus (mean length μ = 8.77 μm, standard deviation σ = 0.67 μm), λ-phage (μ = 19.08 μm and σ = 0.41 μm), and T4 phage (μ = 59.07 μm and σ = 1.12 μm) in microchannel bonded with LP O2 plasma treated PSQ. λ-phage DNA immobilized at (e) both ends in 3-min LP O2 plasma treated PSQ and (f) multiple points in 5-min LP O2 plasma treated PSQ. (Multimedia view) [URL: http://dx.doi.org/10.1063/1.4892515.1]
Combing experiments were conducted with calf thymus, lambda, and T4 DNA, and the corresponding histograms were plotted, showing the length distribution of these DNA molecules (Figure 3(d)). DNA molecules fluorescently labeled with YOYO-1 dye to assist imaging showed contour lengths of 8.77, 19.08, and 59.07 μm, respectively (Figure 3(d)).
O2 plasma exposure for longer durations (3 and 5 min), while keeping other parameters fixed, showed DNA molecules pinning to the surface at multiple points (Figures 3(e) and 3(f), Multimedia view). This makes our device versatile as one could choose different plasma treatment times to alter DNA-surface interactions, depending on experimental needs. DNA pinned at two or more points will be useful for kinetic studies involving DNA-drug or DNA-protein interactions, as no external force is required to keep the molecules stretched. On the other hand, unlike open-top flow stretching or conventional combing devices, microfluidic channels assist buffer exchange and provide better control over experimental conditions.
LP O2 plasma exposure for 1.5 min showed DNA molecules immobilized at only one point (almost always at one of its ends). The small crests observed in the roughened PSQ surface may function as traps to hook and stretch DNA molecules (Movies M1 and M3 in the supplementary material).29 This phenomenon is observed for all three types of DNA molecules (calf thymus, lambda, and T4) irrespective of their ends being sticky (lambda) or blunt (calf thymus and T4), though the origin of this observation is not clear at the moment. Ideally, DNA molecules immobilize at the ends rather than at other regions along the backbone. Moreover, end immobilization is preferred in our experiments for ease and accuracy in length measurements and differentiating false-positives in DNA-protein and DNA-drug interactions (Figures 4–6, Multimedia view). For longer exposure times (3 and 5 min), surface charge density increases showing adsorption at multiple points along the DNA backbone. To be noted, DNA immobilization was also tried on chips prepared one or two days before experiments, confirming that the effect is not a short time-scale one. One advantage of PSQ over PDMS is that, after oxygen plasma treatment, the surface remains hydrophilic for extended period of time, so fluidic channels can be filled easily even after few days of bonding to conduct our experiments.22,23
FIG. 4.
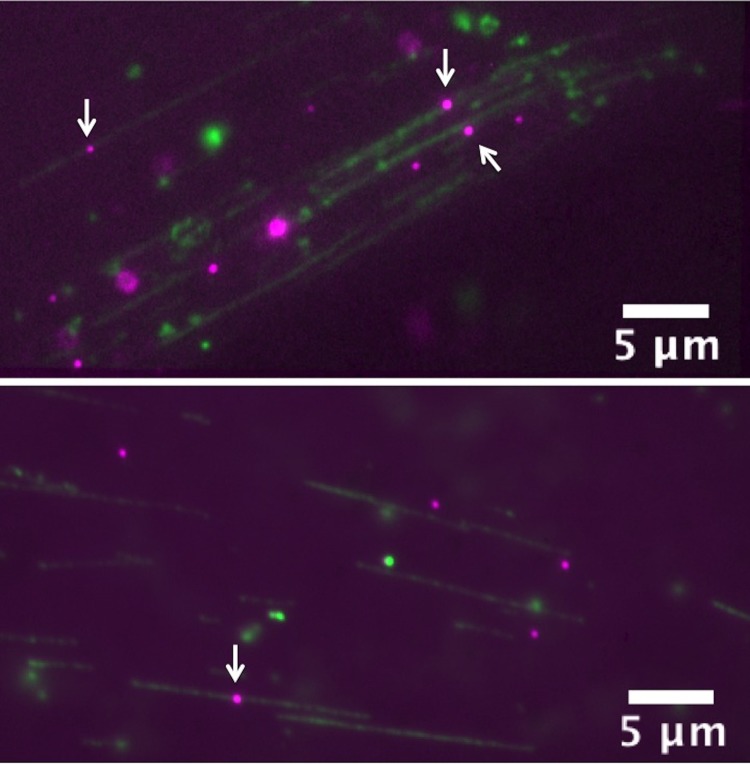
λ-phage DNA (green)—E. coli RNAP complexes (magenta) combed on LP O2 plasma treated PSQ surface. White arrows indicate the candidate DNA-RNAP complexes. DNA molecules were labeled with YOYO-1 nucleic acid stain (1/5 of dye/base-pair ratio) and RNAP with primary-secondary antibody coupling scheme.28 Some QDs also non-specifically immobilized on the surface (bottom), but false positives can be identified using electric-field assisted stretching/recoiling of DNA-protein complexes. (Multimedia view) [URL: http://dx.doi.org/10.1063/1.4892515.5]
FIG. 5.
Cisplatin-induced calf thymus DNA condensation at 0.05 Pt/nucleotide ratios. (a) After 12 h incubation at 37 °C, small kinks appeared along the DNA backbone, (b) after 24 h, more kinks appeared along the DNA backbone, and (c) after 36 h, large condensed globules appeared along the DNA chain. White box represents enlarged images to the right in each case. (d) Intensity-weighted histogram plots comparing contour lengths of untreated and cisplatin treated calf thymus DNA molecules, indicating no considerable change in contour lengths after treating DNA molecules with cisplatin. (Multimedia view) [URL: http://dx.doi.org/10.1063/1.4892515.7]
FIG. 6.
Cisplatin-induced λ-phage DNA condensation at 0.5 Pt/nucleotide ratios. (a) After 4 h incubation at 37 °C, small kinks appeared along the DNA backbone, (b) after 8 h, large condensed globules appeared along the DNA backbone, and (c) after 12 h, complete condensation was observed. White box represents enlarged images to the right in each case. (Multimedia view) [URL: http://dx.doi.org/10.1063/1.4892515.8]
Observation of DNA-RNAP complexes
Mapping transcription factor binding sites (TFBS) along stretched DNA molecules help to understand regulatory circuits that control cellular processes.45 As single molecule methods are developing into a useful platform to map TFBS, a simple DNA immobilization and stretching device might fit very well for such studies.10,12,15,28 Fluorescently labeled DNA-RNAP complexes were allowed to be combed on the LP plasma treated PSQ surface. We observed QDs localized on DNA backbone in many instances, confirming that our device could well be used for such studies (Figure 4). QDs also show adsorption to LP plasma treated PSQ surfaces, but we can differentiate real complexes from false positives using electric-field assisted DNA stretching/recoiling (Figure 4, Multimedia view).
Cisplatin-induced DNA condensation
Although significantly different from live cell studies, single molecule DNA-drug interaction studies are of considerable interest, as they may shed light on possible interaction mechanisms between DNA and the drug involved.46 Methods, such as DNA combing, involve strong DNA-surface interactions might influence the results. Hence, we adopted the one end immobilization scheme here, where contact between DNA and the surface is only at one end of the molecule. Moreover, we carried out DNA-cisplatin reactions in an eppendorf tube, though not an in vivo study, to ensure that DNA molecules are in their physiological random coil configuration during the interactions.
First, cisplatin-induced DNA condensation was studied using calf thymus DNA at 0.05 Pt/nucleotide ratios. Under such conditions, no visible condensation was observed up to 12 h. Samples incubated for 12 h showed visible kinks along the DNA backbone (Figure 5(a), Multimedia view). The number of kinks increased with incubation time for 24 h (Figure 5(b)). At 36 h, we observed large condensed globules along the DNA backbone (Figure 5(c)). Longer incubation times were attempted (up to 96 h), but it did not show complete DNA condensation. This is expected due to the low cisplatin concentration used here. Fluorescence intensity weighted contour length measurements were carried out on cisplatin condensed calf thymus DNA (0.05 Pt/nucleotide) [see supplementary material and Figure S3].29 Briefly, images were subjected to background subtraction and cropped to obtain individual molecules for measurements. Then, fluorescence intensity profile along the DNA backbone was obtained using ImageJ (NIH) and each pixel weighted based on the most probable intensity along the DNA chain.13 DNA contour lengths obtained from ∼50 molecules were plotted into a histogram (Figure 5(d)). Our measurements show no considerable change in contour lengths with and without condensation, which is in accordance with observations by other groups.30 Contour length measurements for 0.05 Pt/nucleotide cisplatin condensed calf thymus DNA showed ∼15%, 35%, and above 50% condensation for 12, 24, and 36 h incubation time, respectively. Next, we increased the cisplatin concentration to 0.2 Pt/nucleotide. Our results showed that enhanced DNA condensation may be observed for shorter reaction times. Visible kinks first appeared around 6 h incubation time and large condensed globules were observed just after 12 h of incubation (data not shown).
Further, we have performed experiments using λ-phage DNA, ∼2.5 times longer than calf thymus DNA. For these experiments, we used 0.5 Pt/nucleotide of cisplatin/DNA ratios. Once again, we observed multiple kinks along the DNA backbone for only 4 h of incubation (Figure 6(a)), which starts to become a large globule around 8 h of incubation (Figure 6(b)), and complete condensation occurred after 12 h of incubation (Figure 6(c)). Enlarged images clearly show small condensed globules that later condensed into a larger globule (Figure 6(c), Multimedia view). Our observations show DNA condensation starts at the central regions of the DNA backbone and the DNA ends are the least affected regions during the process. These results are in accordance with cisplatin-induced condensation mechanism proposed by Hou et al.30 Increase in cisplatin concentration by one order of magnitude (0.5 Pt/nucleotide ratio) showed reduction in condensation times by nearly one third. Moreover, complete condensation was observed only for higher cisplatin concentrations (≥0.5 Pt/nucleotide ratio).
Unlike AFM experiments,30 our platform helps to effectively observe tens of DNA molecules at the same time (higher throughput). Additionally, in AFM experiments, the whole DNA backbone was adsorbed to the substrate surface, which might have adverse influence on the results. In our case, DNA molecules are immobilized at one end for cisplatin-induced condensation study thus eliminating such problems. It is noted that the YOYO-1 staining step was done at the final step, after cisplatin-induced DNA condensation was terminated; hence we expect the staining poses no effect on the condensation process. Further, our platform can easily be extended for DNA condensation studies using other agents such as ethanol, spermine, and spermidine.
Earlier work using differential scanning calorimetry (DSC) helped in understanding temporal behavior of DNA thermal stability during cisplatin mediated DNA condensation.31 While DSC relies on bulk measurements, our microfluidic platform could provide complementary single molecule results through the observation of real time melting profile of condensed single DNA molecules. In addition to the melting profiles, condensation kinetics may well be studied using our device. However, it is cautioned that YOYO-1 dye may have interference against cisplatin binding to DNA in kinetic experiments, as both YOYO-1 and cisplatin being intercalators.47 This issue may be alleviated by using very low amounts of YOYO-1 stain, e.g., 1:100 in dye: base-pair ratio.
CONCLUSIONS
We have developed a simple platform using LP O2 plasma treated PSQ-coated coverslides, compatible to the bonding of microfluidic channels, to effectively immobilize, and stretch DNA molecules (essentially a new molecular combing scenario) without the need of complex surface modification procedures. We demonstrated DNA immobilization at one, two or multiple points with increasing LP O2 plasma exposure time, providing more flexibility in using the PSQ surface for different experimental needs. To interpret our results and explore the working mechanism, we further performed AFM and KPFM measurements indicating increased surface roughness and surface potential of LP O2 plasma treated PSQ/coverslides, compared to that of HP O2 plasma treatment, which are suggested to be responsible for effective DNA combing. The variation of LP O2 plasma treated PSQ reserves the flexibility to immobilize DNA molecules in different schemes. For application purposes, we demonstrated the ease of use for this platform in single molecule studies of DNA-RNAP binding and cisplatin-mediated DNA condensation.
ACKNOWLEDGMENTS
We thank Wei-Chiao Lai for AFM surface roughness measurements, Dr. Chin-Kun Hu for his inputs in cisplatin-DNA condensation experiments, Dr. Yit-Tsong Chen for providing the KPFM tool for surface potential measurements, Dr. Andreas Erbe for helpful discussions, and the reviewers for insightful suggestions. This work was supported by AS Nano Science and Technology Program, AS Integrated Thematic Project (AS-103-TP-A01), Ministry of Science and Technology, Taiwan (102-2112-M-001-005-MY3 and 103-2923-M-001-007-MY3), and Asian Office for Aerospace Research and Development (#FA2386-12-1-4002).
References
- 1.Lam E. T., Hastie A., Lin C., Ehrlich D., Das S. K., Austin M. D., Deshpande P., Cao H., Nagarajan N., Xiao M., and Kwok P. Y., Nat. Biotechnol. 30, 771 (2012). 10.1038/nbt.2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada Y., Funatsu T., Murakami K., Nanoyama Y., Ishihama A., and Yanagida T., Biophys. J. 76, 709 (1999). 10.1016/S0006-3495(99)77237-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J. H. and Larson R. G., Nucleic Acids Res. 35, 3848 (2007). 10.1093/nar/gkm332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guthold M., Zhu X., Rivetti C., Yang G., Thomson N. H., Kasas S., Hansma H. G., Smith B., Hansma P. K., and Bustamante C., Biophys. J. 77, 2284 (1999). 10.1016/S0006-3495(99)77067-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilemann M., Hwang L. C., Lymperopoulos K., and Kapanidis A. N., Methods Mol. Biol. 543, 503 (2009). 10.1007/978-1-60327-015-1_29 [DOI] [PubMed] [Google Scholar]
- 6.Deniz A. A., Mukhopadhyay S., and Lemke E. A., J. R. Soc., Interface 5, 15 (2008). 10.1038/nature04268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efcavitch J. W. and Thompson J. F., Annu. Rev. Anal. Chem. 3, 109 (2010). 10.1146/annurev.anchem.111808.073558 [DOI] [PubMed] [Google Scholar]
- 8.Kim J. H., Dukkipati V. R., Pang S. W., and Larson R. G., Nanoscale Res. Lett. 2, 185 (2007). 10.1007/s11671-007-9057-5 [DOI] [Google Scholar]
- 9.Dukkipati V. R., Kim J. H., Pang S. W., and Larson R. G., Nano Lett. 6, 2499 (2006). 10.1021/nl0617484 [DOI] [PubMed] [Google Scholar]
- 10.Ebenstein Y., Gassman N., Kim S., Antelman J., Kim Y., Ho S., Samuel R., Michalet X., and Weiss S., Nano Lett. 9, 1598 (2009). 10.1021/nl803820b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins T. T., Smith D. E., Larson R. G., and Chu S., Science 268, 83 (1995). 10.1126/science.7701345 [DOI] [PubMed] [Google Scholar]
- 12.Leonardo L. R., Sriram K. K., Liao K. T., Lai S. C., Kuo P. C., Chu M. L., and Chou C. F., Biomicrofluidics 8, 016501 (2014). 10.1063/1.4861435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh J. W., Taloni A., Chen Y. L., and Chou C. F., Nano Lett. 12, 1597 (2012). 10.1021/nl2045292 [DOI] [PubMed] [Google Scholar]
- 14.Bensimon A., Simon A., Chiffaudel A., Croquette V., Heslot F., and Bensimon D., Science 265, 2096 (1994). 10.1126/science.7522347 [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Gottfried A., Lin R. R., Dertinger T., Kim A. S., Chung S., Colyer R. A., Weinhold E., Weiss S., and Ebenstein Y., Angew. Chem., Int. Ed. 51, 3578 (2012). 10.1002/anie.201107714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H. and Schwartz D. C., Anal. Biochem. 380, 111 (2008). 10.1016/j.ab.2008.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T. and Schwartz D. C., Anal. Biochem. 361, 31 (2007). 10.1016/j.ab.2006.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing J., Reed J., Huang J., Hu X., Clarke V., Edington J., Housman D., Anantharaman T. S., Huff E. J., Mishra B., Porter B., Shenker A., Wolfson E., Hiort C., Kantor R., Aston C., and Schwartz D. C., Proc. Natl. Acad. Sci. U.S.A. 95, 8046 (1998). 10.1073/pnas.95.14.8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y. and Jo K., Chem. Commun. 47, 6248 (2011). 10.1039/c0cc05396a [DOI] [PubMed] [Google Scholar]
- 20.Charlot B., Bardin F., Sanchez N., Roux P., Teixeira S., and Schwob E., Biomicrofluidics 8, 014103 (2014). 10.1063/1.4863575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baney R. H., Itoh M., Sakakibara A., and Suzuki T., Chem. Rev. 95, 1409 (1995). 10.1021/cr00037a012 [DOI] [Google Scholar]
- 22.Gu J., Gupta R., Chou C. F., Wei Q., and Zenhausern F., Lab Chip 7, 1198 (2007). 10.1039/b704851c [DOI] [PubMed] [Google Scholar]
- 23.Leichlé T., Lin Y. L., Chiang P. C., Hu S. M., Liao K. T., and Chou C. F., Sens. Actuators, B 161, 805 (2012). 10.1016/j.snb.2011.11.036 [DOI] [Google Scholar]
- 24.Liao K. T. and Chou C. F., J. Am. Chem. Soc. 134, 8742 (2012). 10.1021/ja3016523 [DOI] [PubMed] [Google Scholar]
- 25.Mcdonald J. C. and Whitesides G. M., Acc. Chem. Res. 35, 491 (2002). 10.1021/ar010110q [DOI] [PubMed] [Google Scholar]
- 26.Friend J. and Yeo L., Biomicrofluidics 4, 026502 (2010). 10.1063/1.3259624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinohara H., Mizuno J., and Shoji S., IEEJ Trans. Electr. Electron. Eng. 2, 301 (2007). 10.1002/tee.20157 [DOI] [Google Scholar]
- 28.Sriram K. K., Yeh J. W., Lin Y. L., Chang Y. R., and Chou C. F., Nucleic Acids Res. 42, e85 (2014). 10.1093/nar/gku254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.See supplementary material at http://dx.doi.org/10.1063/1.4892515E-BIOMGB-8-002492 for movies, roughness measurements, and image analysis.
- 30.Hou X. M., Zhang X. H., Wei K. J., Ji C., Dou S. X., Wang W. C., Li M., and Wang P. Y., Nucleic Acids Res. 37, 1400 (2009). 10.1093/nar/gkn933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lando D. Y., Galyuk E. N., Chang C. L., and Hu C. K., J. Inorg. Biochem. 117, 164 (2012). 10.1016/j.jinorgbio.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 32.Florea A. M. and Busselberg D., Cancers 3, 1351 (2011). 10.3390/cancers3011351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strumberg D., Brugge S., Korn M. W., Koeppen S., Ranft J., Scheiber G., Reiners C., Mockel C., Seeber S., and Scheulen M. E., Ann. Oncol. 13, 229 (2002). 10.1093/annonc/mdf058 [DOI] [PubMed] [Google Scholar]
- 34.Cohen S. A., An Introduction to Plasma Physics for Materials Processing ( Academic Press, UK, Amsterdam, 1989). [Google Scholar]
- 35.Watanabe F. and Ohnishi Y., J. Vac. Sci. Technol., B 4, 422 (1986). 10.1116/1.583347 [DOI] [Google Scholar]
- 36.Yousefi H. R., Ghoranneviss M., Tehrani A. R., and Khamseh S., Surf. Interface Anal. 35, 1015 (2003). 10.1002/sia.1633 [DOI] [Google Scholar]
- 37.Shah F. A., Kim K. Nan, Lieberman M., and Bernstein G. H., J. Vac. Sci. Technol., B 30, 011806 (2012). 10.1116/1.3676054 [DOI] [Google Scholar]
- 38.Bhattacharya S., Datta A., Berg J. M., and Gangopadhyay S., J. Microelectromech. Syst. 14, 590 (2005). 10.1109/JMEMS.2005.844746 [DOI] [Google Scholar]
- 39.Almutairi Z., Ren C. L., and Simon L., Colloids Surf., A 415, 406 (2012). 10.1016/j.colsurfa.2012.10.008 [DOI] [Google Scholar]
- 40.Sun P. and Horton J. H., Appl. Surf. Sci. 271, 344 (2013). 10.1016/j.apsusc.2013.01.199 [DOI] [Google Scholar]
- 41.Lyklema J. J., in Fundamentals of Interface and Colloid Science, edited by Lyklema J. J., Keizer A. de, Bijsterbosch B. H.. et al. ( Elsevier Ltd., Amsterdam, 1995), Vol. 2 [Google Scholar]
- 42.Tandon V., Bhagavatula S. K., Nelson W. C., and Kirby B. J., Electrophoresis 29, 1092 (2008). 10.1002/elps.200700734 [DOI] [PubMed] [Google Scholar]
- 43.Yang B., Dukkipati V. R., Li D., Cardozo B. L., and Pang S. W., J. Vac. Sci. Technol., B 25, 2352 (2007). 10.1116/1.2806975 [DOI] [Google Scholar]
- 44.Benke A., Mertig M., and Pompe W., Nanotechnology 22, 035304 (2011). 10.1088/0957-4484/22/3/035304 [DOI] [PubMed] [Google Scholar]
- 45.Latchman D. S., Int. J. Biochem. Cell Biol. 29, 1305 (1997). 10.1016/S1357-2725(97)00085-X [DOI] [PubMed] [Google Scholar]
- 46.Tegenfeldt J. O., Prinz C., Cao H., Huang R. L., Austin R. H., Chou S. Y., Cox E. C., and Sturm J. C., Anal. Bioanal. Chem. 378, 1678 (2004). 10.1007/s00216-004-2526-0 [DOI] [PubMed] [Google Scholar]
- 47.Hurley L. H., Nat. Rev. Cancer 2, 188 (2002). 10.1038/nrc749 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4892515E-BIOMGB-8-002492 for movies, roughness measurements, and image analysis.