Abstract
Airway allergen exposure induces inflammation among individuals with atopy that is characterized by altered airway gene expression, elevated levels of T helper type 2 cytokines, mucus hypersecretion, and airflow obstruction. To identify the genetic determinants of the airway allergen response, we employed a systems genetics approach. We applied a house dust mite mouse model of allergic airway disease to 151 incipient lines of the Collaborative Cross, a new mouse genetic reference population, and measured serum IgE, airway eosinophilia, and gene expression in the lung. Allergen-induced serum IgE and airway eosinophilia were not correlated. We detected quantitative trait loci (QTL) for airway eosinophilia on chromosome (Chr) 11 (71.802–87.098 megabases [Mb]) and allergen-induced IgE on Chr 4 (13.950–31.660 Mb). More than 4,500 genes expressed in the lung had gene expression QTL (eQTL), the majority of which were located near the gene itself. However, we also detected approximately 1,700 trans-eQTL, and many of these trans-eQTL clustered into two regions on Chr 2. We show that one of these loci (at 147.6 Mb) is associated with the expression of more than 100 genes, and, using bioinformatics resources, fine-map this locus to a 53 kb-long interval. We also use the gene expression and eQTL data to identify a candidate gene, Tlcd2, for the eosinophil QTL. Our results demonstrate that hallmark allergic airway disease phenotypes are associated with distinct genetic loci on Chrs 4 and 11, and that gene expression in the allergically inflamed lung is controlled by both cis and trans regulatory factors.
Keywords: allergic airway disease, systems genetics, gene expression quantitative trait loci
Clinical Relevance
Using a systems genetics approach and a new mouse genetics reference population, we examined correlations between key features of allergic inflammation, namely IgE and airway eosinophilia, and identified distinct genetic loci associated with each trait. We show that the expression of many genes (> 4,500) in the lung after allergen challenge is also under genetic control, in part by distal regulatory factors. These gene expression data provide insight about gene regulation in the context of allergen challenge, and also point to a novel candidate gene for allergic inflammation.
The prevalence of allergic airway diseases (AADs), including asthma and allergic rhinitis, has increased dramatically over the last 50 years, especially in more developed countries (1). These diseases affect some 8–16% of the population in the United States, and cause substantial decrements in quality of life and productivity (2–4). Hallmark phenotypes of AAD include elevated serum IgE, airway eosinophilia, and mucus hypersecretion. The etiology of AAD remains enigmatic and complex, and likely involves complex interactions between genes and environment (5–8). Genome-wide association studies in humans have identified several loci associated with asthma in multiple study populations (9–14), and a genome-wide association study of allergic rhinitis also identified multiple loci, some of which overlap with asthma loci (e.g., LRRC32, HLA-G, and TSLP) (15). Several studies have also identified gene expression profiles that differentiate normal versus AAD in affected tissue (16–21).
We set out to expand on existing knowledge of the genetics and transcriptomics of AAD using a systems genetics approach. To accomplish this, we applied a house dust mite (HDM) mouse model of AAD in the lung (22) to incipient lines of the Collaborative Cross (CC). The CC is composed of a panel of recombinant inbred lines derived from eight-way crosses using five classical inbred strains (C57BL/6J, 129S1/SvImJ, A/J, NOD/ShiLtJ, and NZO/HlLtJ) and three wild-derived inbred strains (WSB/EiJ, PWK/PhJ, and CAST/EiJ) (23), the genomes of which have recently been sequenced (24, 25). Quantitative trait loci (QTL) for many traits have been identified using these incipient CC lines (26–29), which we refer to as “preCC” mice.
In the present work, we report on four types of correlations: between AAD phenotypes, between genotype and AAD phenotypes (i.e., QTL), between gene expression and AAD phenotype, and between genotype and gene expression (expression QTL or “eQTL”). We focused much of our work on eQTL discovery in the allergen-challenged mouse lung, because genetically determined variation in gene expression is thought to contribute to asthma (31, 32), and this has not yet been examined in a mouse model of AAD.
Materials and Methods
Additional details about methods are provided in the online supplement.
Mice
We obtained 151 male preCC mice from Oak Ridge National Laboratory (Oak Ridge, TN). Each mouse was from a distinct CC line that had undergone 5–14 generations of inbreeding. All mice were singly housed with α-dri bedding and fed NIH-31 Open Formula mouse sterilized diet in an Association for Assessment and Accreditation of Laboratory Animal Care–approved facility at the National Institutes of Health (Bethesda, MD) under normal 12-hour light/dark cycles.
Phenotyping Protocol
We adapted our model of HDM-induced AAD (22) to a longitudinal study design in which preCC mice were phenotyped at two different time points, baseline and final, as follows. On Day 9 of the study, we took serum samples from 8- to 12-week-old preCC mice for baseline measurements of serum IgE. We then sensitized mice with 10 μg of the immunodominant allergen from the Dermatophagoides pteronyssinus species of HDM, Der p 1, by intraperitoneal injection on Days 22 and 30, followed by challenge with 50 μg of Der p 1, administered by oropharyngeal aspiration, on Day 37. On Day 40, mice were killed, followed by collection of blood and whole-lung lavage fluid. After lavage, lung tissue was snap frozen. Cytokines in lavage fluid were measured using a multiplex bead assay from Millipore (Billerica, MA).
Gene Expression Analysis and eQTL Mapping
Total lung RNA was isolated from lung tissue and then processed and hybridized to Illumina WG-6v2 arrays (San Diego, CA). Normalization was conducted using robust multi-average normalization with quantile normalization and log2 transformation. Array data have been deposited to Gene Expression Omnibus (GSE51768). eQTL mapping was performed in essentially the same fashion as QTL mapping, with some modifications, as described in the online supplement.
Quantitative RT-PCR
We measured gene expression of a long, noncoding RNA (9030622O22Rik) in preCC lung RNA samples using quantitative RT-PCR with locked nucleic acid primers designed by Exiqon (Woburn, MA) that amplify one isoform of 9030622O22Rik, ENSMUST00000136555. For normalization, we measured the expression of Rpl22 and calculated expression using the delta Ct method (33).
Conditional Correlation Analysis
To identify eQTL that may underlie the QTL for eosinophils, we used conditional correlation. Specifically, we ran chromosome (Chr) 11 QTL scans for eosinophils or gene expression, with covariates, either gene expression or eosinophils, as follows:
A large drop in the logarithm of the odds (LOD) score (“delta LOD”) between models 1 and 2 provides evidence to support a causal relationship (QTL→gene expression→eosinophils) (34). Conversely, a large LOD drop between models 3 and 4 is indicative of gene expression that is reactive to the phenotype (i.e., QTL→eosinophils→gene expression) (34). For these analyses, we had complete data from 132 mice, and we permuted the gene expression data (n = 1000 permutations) data to assess the statistical significance of the LOD drops that we observed.
Results
Phenotypes
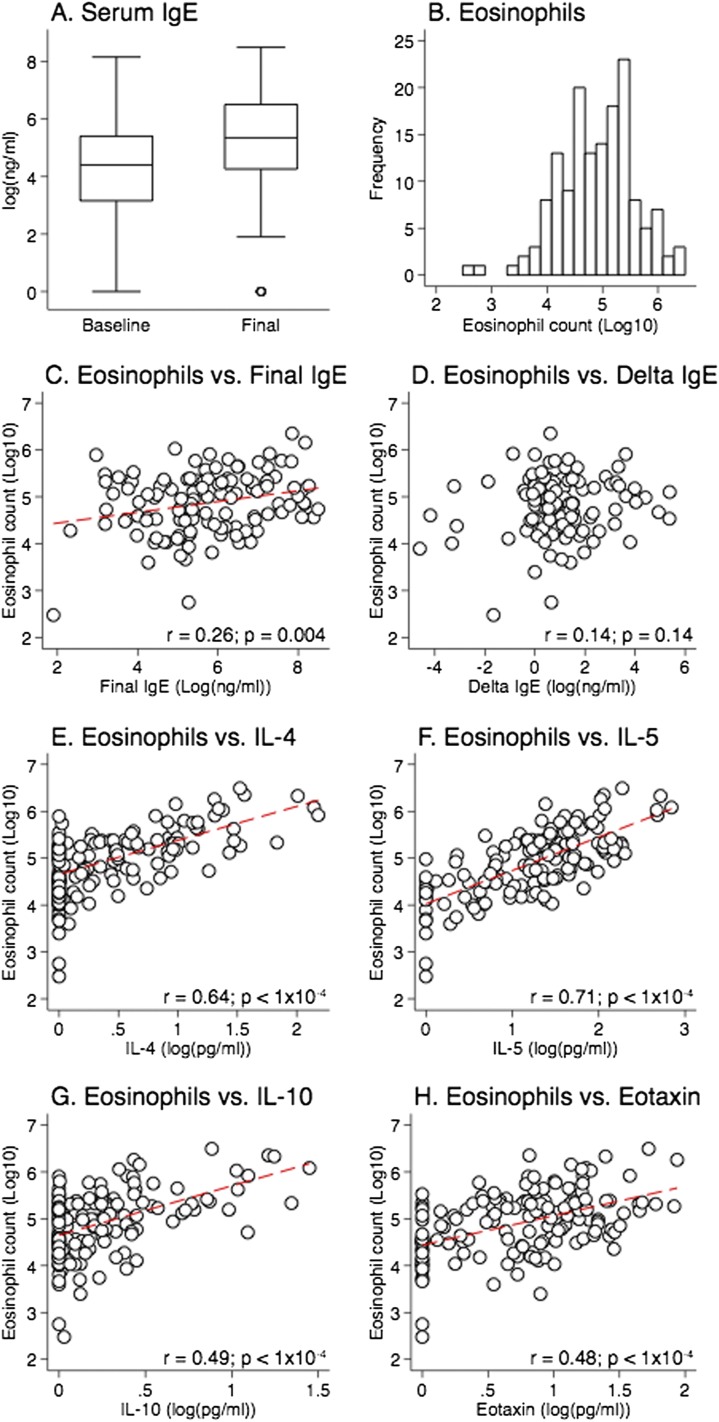
We employed longitudinal study design in which preCC mice were phenotyped for serum IgE before and after sensitization and challenge with the HDM allergen Der p 1 at two different time points (“baseline” and “final”). Total IgE increased over the course of the experiment (Figure 1A), with a mean increase (“delta IgE”) of 347 ng/ml (P = 2.0 × 10−4), demonstrating expected effects of allergen sensitization and challenge across the population. Interestingly, having high baseline IgE generally resulted in little or no increase in antibody concentrations with sensitization and challenge (Figure E1), suggesting that IgE concentration has a plateau value. Eosinophil counts in lung lavage fluid spanned four orders of magnitude (Figure 1B; range, 3.00 × 102 − 3.10 × 106; mean = 8.00 × 104), demonstrating a wide range of responses among the preCC population. Eosinophil counts and final IgE were weakly correlated (Figure 1C; Pearson’s r = 0.26, P = 0.004), but there was no correlation between eosinophil counts and delta IgE (Figure 1D). These results argue against a predominant role for IgE in inflammation in this model of AAD. We did, however, detect very strong correlations between eosinophils and IL-4, -5, and -10 and eotaxin in lavage fluid (Figures 1E and 1F), as expected, based on the known role of these cytokines in T helper type 2 inflammation.
Figure 1.
Allergic airway disease (AAD) phenotypes in pre–Collaborative Cross (preCC) mice. (A) Serum IgE before and after house dust mite (HDM) sensitization and challenge. A total of 12 mice had undetectable IgE levels at the final time point. Each data point represents one mouse. (B) Eosinophil counts in lung lavage fluid. (C–H) Pairwise correlations between traits. Dashed red lines represent linear best fit.
Genetic Analyses
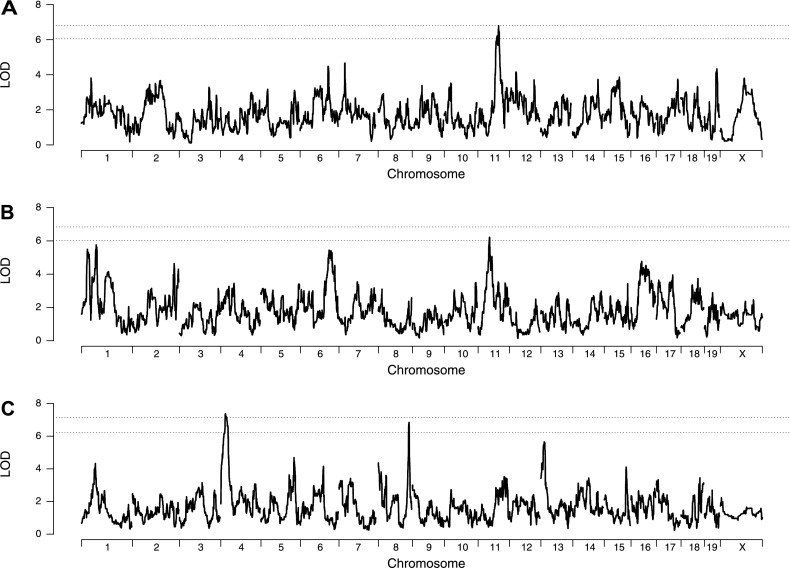
We conducted genome scans using the preCC data and found a single QTL for eosinophil counts on Chr 11 (Dpe1, Der p 1–induced pulmonary eosinophils; 72.904–87.098 megabases [Mb], LOD = 7.0, P < 0.05), as shown in Figure 2A. This QTL accounted for 19% of total variation in phenotype. We found a suggestive peak (P < 0.2) for final IgE on Chr 11 (42.350–51.087 Mb, LOD = 6.2; Figure 2B) that is distinct from the eosinophil QTL. For delta IgE, we found one significant QTL on Chr 4 (DsIgE, 13.950–31.660 Mb, LOD = 7.4; Figure 2C), which explained 26% of variation in delta IgE, and one suggestive QTL (P < 0.1) on Chr 8 (119.086–122.313 Mb, LOD = 6.8). The lack of overlap between QTL for IgE and eosinophils further argues against a major role for IgE in this model of AAD. No QTL were detected for any of the cytokines in lavage fluid.
Figure 2.
Genetic analysis of eosinophils and IgE. Genome scans for eosinophils (A), final IgE (B), and delta IgE (C). The dashed lines denote 95 and 80% significance thresholds determined by permutation. LOD, logarithm of the odds.
We focused our subsequent analysis on the eosinophil QTL with the goal of identifying candidate genes. The eosinophil QTL on Chr 11 spans 14.2 Mb (72.9–87.1 Mb), and contains 359 genes, 252 of which are protein coding and 107 of which are noncoding RNAs (see Table E1 in the online supplement). Among these genes, Ccl11 (eotaxin) is the most obvious candidate gene based on its known role in eosinophil chemotaxis. We noted that there is no coding region variation in Ccl11 in CC founder strains, as determined by sequencing data (24); thus, if variation in Ccl11 underlies the eosinophil QTL, the effect must be mediated through mechanisms other than variation in CCL11 protein sequence variation.
We also asked whether syntenic regions in the human genome contain genes related to relevant phenotypes. The syntenic regions in human genome lie on Chr 17p and 17q, specifically 0–16.5 Mb, 25.5–28.9 Mb, 29.1–36.2 Mb, and 45.6–60.3 Mb. A total of 94 genes in the mouse QTL have human orthologs in these regions (Table E2). Of these, CCL11 is the only gene with prior evidence of association with asthma or other allergic inflammation phenotypes.
Gene Expression
We turned to gene expression as a way to identify candidate genes for the eosinophil QTL in an unbiased manner. We measured gene expression in whole-lung samples from 138 preCC mice and used these data in two ways. First, we identified expressed transcripts that were correlated with eosinophils, so called quantitative trait transcripts (QTTs) (30), by linear regression. We identified 2,852 genes that were correlated with eosinophil counts at a false discovery rate of 5% (Table E3). As expected, well known cytokines related to AAD are in this list (e.g., Il4, Il10, and Il33), as are genes previously associated with asthma (e.g., Chi3l4) and genes associated with mucous cell metaplasia (e.g., Muc5ac, Spdef, and Foxa3). On the whole, this set of QTTs is significantly enriched for a number of immune response pathways (Table E4). We also found that this gene set is enriched for genes associated with alternative activation of macrophages (35) (Alox15, Arg1, Ccl17, Ccl22, Ccl24, Ccr5, Chi3l1, Clec7a, Col6a2, Fcer2a, Il10, Il1rn, Mertk, Mgl2, Mmp12, Mrc1, Retnla, Retnlg, Rgs1, Tgfb1; P = 6.47 × 10−6), a pathway of renewed interest in AAD (36). Hence, it is apparent that this gene set reflects contributions from both resident lung cell types, such as airway epithelia and infiltrating leukocytes (e.g., macrophages).
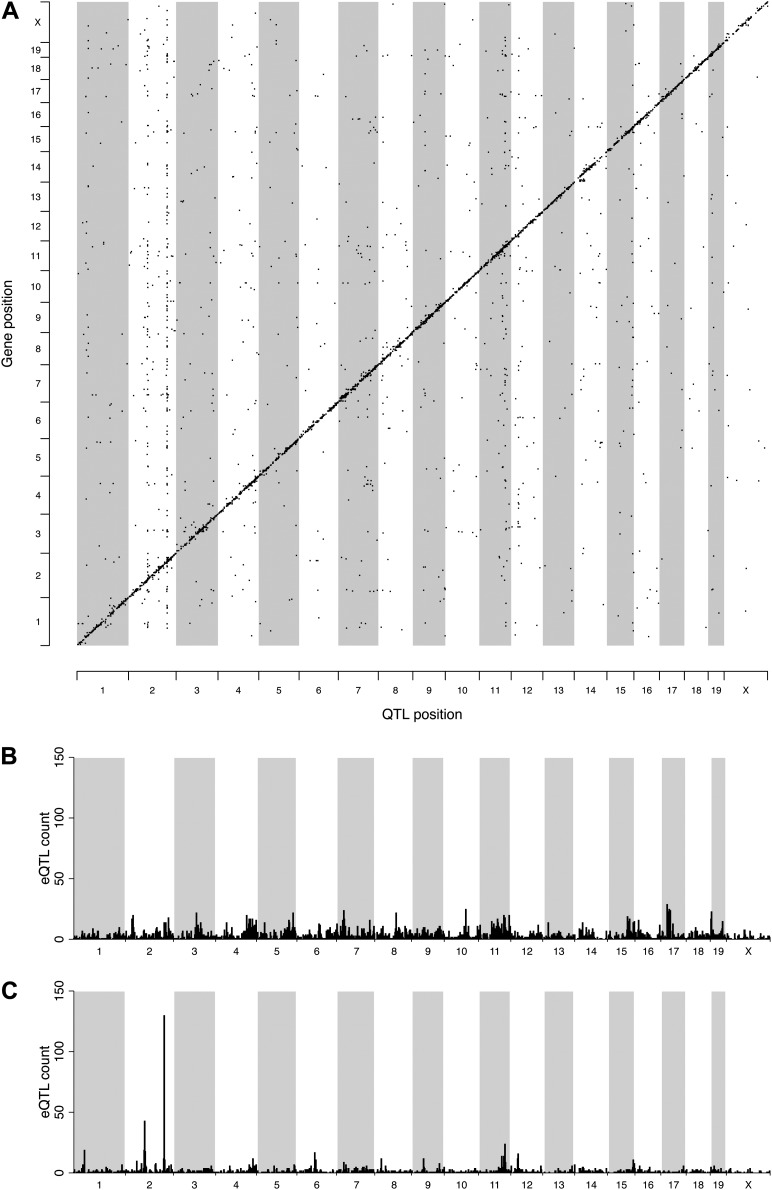
We then used the gene expression data to identify eQTL. As described in the online supplement, we focused our eQTL analysis on genes for which the expression array probe did not contain single nucleotide polymorphisms (SNPs), as these can bias eQTL detection (37, 38). Using these data, we identified 6,457 eQTL associated with 4,556 genes (Figure 3A, Table E5). Of these eQTL, the large majority (n = 4,759, 73.7%) were located on the same chromosome as the cognate gene, whereas 1,698 (26.3%) were located on other chromosomes. The latter are indicative of trans-regulatory variation (e.g., a polymorphic transcription factor located on one chromosome that binds to the promoter of a gene on another chromosome, and are referred to as trans-eQTL).
Figure 3.
Expression quantitative trait locus (eQTL) identification. (A) Genome-wide plot of eQTL (n = 6,457). (B) Distribution of cis-eQTL. (C) Distribution of trans-eQTL. The y axis represents eQTL counts per 2-megabase (Mb) window.
We characterized eQTL located on the same chromosome as the cognate gene in terms of location relative to the corresponding gene and effect size. In most cases, the eQTL confidence intervals contain the cognate gene (n = 4,065 out of 4,759 [85.4%], which equates to 63% of all eQTL); hence, most eQTL act in cis. Among cis-eQTL, there is an intuitive relationship between LOD score and distance to the gene–eQTL with the highest LOD scores tending to be located close to the gene (Figure E2). A small number of eQTL (n = 228 [4.81% of eQTL located on the same chromosome as cognate gene]) were located further than 10 Mb away, and we categorized these eQTL as trans-eQTL (see the online supplement). Overall, our results suggest that regulatory variation located close to genes is a major source of eQTL in the lung.
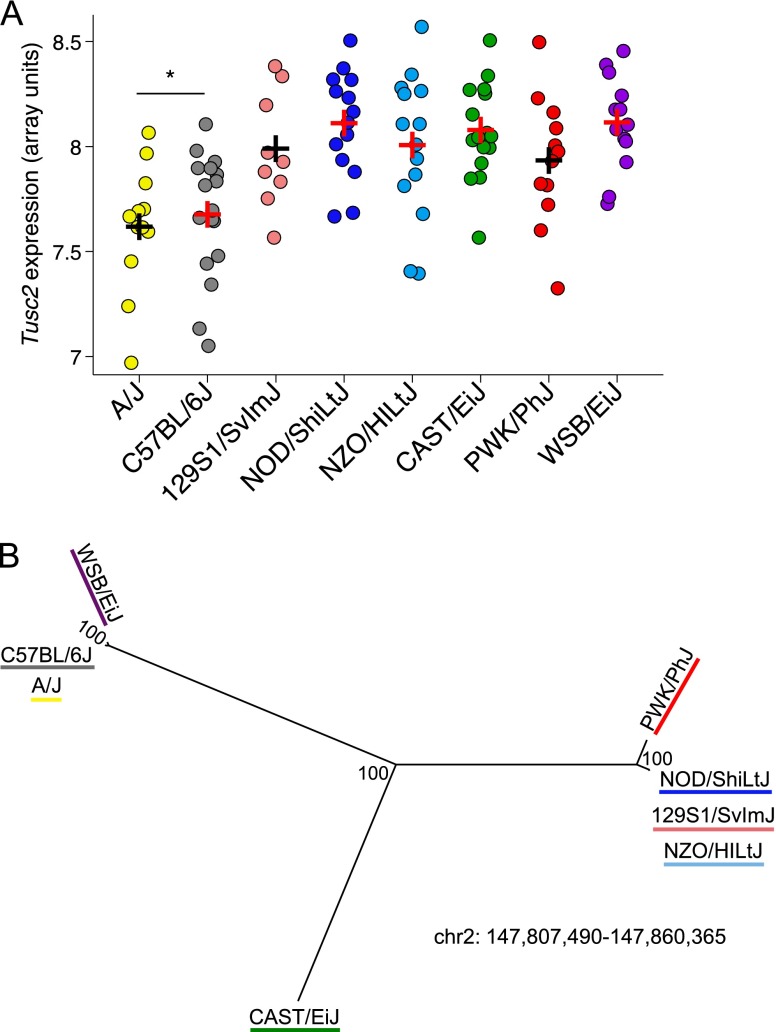
Cis-eQTL were distributed genome wide (Figure 3B), but trans-eQTL clustered primarily into two hotspots (often referred to as “trans-bands”) on Chr 2 (Figure 3C). The existence of trans-bands on Chr 2 suggests that these loci harbor master regulators of gene expression in the allergen-challenged lung. Closer examination of these two regions revealed that each was dominated by a single locus. Specifically, markers at 73.0 Mb and 147.6 Mb were associated with the expression of 22 and 41 genes, respectively. We also found that an additional 12 and 33 genes had trans-eQTL at a lower significance threshold of P less than 0.1 at these two loci, respectively. In addition, when we included probes that contain SNPs in this analysis, these numbers increased to 51 and 102, respectively. We asked whether genes with eQTL at these two loci on Chr 2 were enriched among the set of eosinophil QTTs, and found significant enrichment for genes with trans-eQTL at the distal locus (147.6 Mb, 1.5-fold enriched; P = 0.003), but not for the more proximal locus (73.0 Mb; P > 0.1). This focused our interest on the distal locus as a potential regulator of eosinophilic inflammation. We examined the allele effects for all trans-eQTL at the distal locus and found that, for the majority of genes (95 out of 102 [93%]), gene expression derived from A/J and C57BL/6J alleles was different than expression derived from the other six founder alleles. A representative gene is shown in Figure 4A. These eQTL allele effects suggested that A/J and C57BL/6J share an allele that is distinct from all other CC founder strains, and is associated with altered gene expression. We looked for a shared allele among these two strains in the trans-band confidence interval (Chr2: 146–150 Mb) using sequence data (24) and identified a 53-kb interval spanning 147,807,490–147,860,365 bp in which these two CC founder strains are 99.6% identical. Phylogenetic analysis of this interval shows that A/J and C57BL/6J share a haplotype that is distinct from all other CC founder strains (Figure 4B). A total of 68 SNPs in this region distinguish A/J and C57BL/6J from all other CC founder strains, and thus are potential causal variants for the trans-band (Table E6). All 68 SNPs lie within the putative long noncoding RNA (lncRNA) 9030622O22Rik.
Figure 4.
Fine mapping of a trans-band on Chr 2. (A) Allele effects plot for a representative gene in the trans-band, Tusc2, showing separation of A/J and C57BL/6J allele effects from the other CC founder strain alleles. Array-based measurements of Tusc2 expression from preCC mice partitioned by their founder haplotype at the peak locus for the eQTL. For the sake of clarity, only homozygous mice are shown (n = 99). Each data point represents a preCC mouse and plus symbols (+) represent the means for each genotype. P < 0.05 for contrast between A/J. (B) Maximum likelihood phylogenetic tree for the region of shared ancestry within the trans-band confidence interval. The tree was constructed using single nucleotide polymorphism data from Sanger MGP Wellcome Trust Sanger Mouse Genomes Project (see the online supplement), and the numbers indicate bootstrap support estimates for each branch. Note that C57BL/6J and A/J haplotypes are separated by a short branch (with 100% bootstrap support) from WSB/EiJ. This indicates that the number of genetic variants that causes lower gene expression among CC mice with A/J or C57BL/6J haplotypes at this locus is limited to a small number (n = 68; see Table E6). chr, chromosome.
We then asked whether variation in expression of this putative lncRNA could underlie the trans-band. We measured the expression level of one isoform of the lncRNA (ENSMUST00000136555) using quantitative RT-PCR and found that it had an eQTL that overlapped with the region on Chr 2 where A/J and C57BL/6J share a haplotype (147,417,620–150,318,592 bp, LOD = 11.45). We found that alleles derived from A/J and C57BL/6J were also associated with lower lncRNA eQTL expression (Figure E3), but also that the CAST/EiJ allele was associated with relatively low lncRNA expression. Hence, the allele effects for lncRNA eQTL and genes in the trans-band are not fully congruent. In addition, we examined pairwise correlations between lncRNA expression and the expression of genes in the trans-band, and found no correlations. Collectively, these results indicate that, even though the lncRNA has a colocalized eQTL, it is unlikely to be the source of the trans-band, at least by means of a direct relationship between its expression and the expression of genes in the trans-band.
Identification of Candidate Genes for the Eosinophil QTL (Dpe1) Using Gene Expression Data
Finally, we used our gene expression to identify candidate genes for the eosinophil QTL. Several studies have found that variants associated with disease risk in humans are often eQTL and imply that disease risk is a function of variation in gene expression, and one study used lung eQTL to identify gene networks associated with asthma (32). Therefore, we asked whether lung eQTL could explain eosinophil recruitment responses to allergen. First, we identified genes that met two criteria: (1) their expression was linearly correlated with eosinophils (i.e., the gene is a QTT); and (2) they had an eQTL within the eosinophil QTL confidence interval (Chr 11: 72.904–87.098 Mb). A total of 18 genes met these criteria: 2610507B11Rik, Atad5, Ccl11, Ccl7, Coil, Inpp5k, Lig3, Mmp28, Poldip2, Ppm1d, Ppm1e, Rpa1, Slc6a4, Ssh2, Supt4h1, Tax1bp3, Tlcd1, and Tlcd2.
We then used conditional correlation to evaluate whether the variation in expression in any of these genes could underlie the eosinophil QTL. Accounting for Ccl11 expression did not reduce the LOD score as would be expected if there were a direct relationship between the QTL, Ccl11 expression, and eosinophil recruitment. In fact, the LOD score increased from 5.6 to 8.2 (P < 0.001, Table 1), which is indicative of a more complex (indirect) relationship. We found the greatest drop in the LOD score was associated with the gene Tlcd2 (LOD dropped from 5.6 to 3.1; P < 0.001; Table 1). Because Tlcd2 expression was negatively correlated with eosinophil counts (Table 1), our results suggest that Tlcd2 is a candidate negative regulator of eosinophil recruitment responses to allergen.
Table 1.
Results of Conditional Correlation Analyses to Identify Candidate Genes for the Eosinophil Quantitative Trait Locus (Dpe1)
| Eosinophil Correlation |
eQTL |
Conditional Analysis† |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Entrez Gene ID | Transcript | Probe | Pearson’s r | P Value | Peak (Mb) | LOD | EOS LOD* | EOS|GEX | GEX|EOS |
| Tlcd2 | 380,712 | NM_027249 | ILMN_2814385 | −0.40 | 1.91E−06 | 73.353 | 35.90 | 5.60 | 3.12 | 32.13 |
| Tlcd2 | 380,712 | NM_027249 | ILMN_1249828 | −0.41 | 8.23E−07 | 76.537 | 22.38 | 5.60 | 3.27 | 16.89 |
| Tlcd2 | 380,712 | NM_027249 | ILMN_1213090 | −0.27 | 1.84E−03 | 75.073 | 27.72 | 5.60 | 3.88 | 23.89 |
| Ppm1d | 53,892 | NM_016910 | ILMN_1226504 | 0.33 | 1.18E−04 | 83.453 | 16.53 | 5.60 | 3.99 | 15.65 |
| Mmp28 | 118,453 | NM_172797 | ILMN_1218211 | −0.24 | 5.00E−03 | 83.408 | 16.24 | 5.60 | 4.17 | 14.06 |
| Ccl7 | 20,306 | NM_013654 | ILMN_2835117 | 0.32 | 1.65E−04 | 83.257 | 15.21 | 5.60 | 4.90 | 15.21 |
| Tax1bp3 | 76,281 | NM_029564 | ILMN_2746086 | −0.26 | 2.89E−03 | 73.353 | 10.57 | 5.60 | 4.97 | 10.42 |
| Slc6a4 | 15,567 | NM_010484 | ILMN_2713714 | −0.26 | 2.90E−03 | 75.265 | 17.94 | 5.60 | 4.99 | 17.27 |
| Poldip2 | 67,811 | NM_026389 | ILMN_2543314 | −0.24 | 4.76E−03 | 76.453 | 10.63 | 5.60 | 5.16 | 10.60 |
| Supt4h1 | 20,922 | NM_009296 | ILMN_1247505 | −0.31 | 3.73E−04 | 87.098 | 30.71 | 5.60 | 5.47 | 30.75 |
| Ssh2 | 237,860 | NM_177710 | ILMN_1228331 | −0.24 | 4.78E−03 | 83.444 | 10.37 | 5.60 | 5.49 | 10.24 |
| Lig3 | 16,882 | NM_010716 | ILMN_1256377 | 0.33 | 8.96E−05 | 82.222 | 6.87 | 5.60 | 5.52 | 8.60 |
| Inpp5k | 19,062 | NM_008916 | ILMN_2584558 | −0.33 | 1.41E−04 | 73.872 | 9.76 | 5.60 | 5.53 | 9.98 |
| 2610507B11Rik | 72,503 | NM_001002004 | ILMN_3014965 | 0.30 | 4.74E−04 | 77.338 | 9.15 | 5.60 | 5.65 | 8.21 |
| Coil | 12,812 | NM_016706 | ILMN_2746210 | −0.25 | 3.89E−03 | 84.286 | 11.46 | 5.60 | 5.65 | 11.51 |
| Ccl7 | 20,306 | NM_013654 | ILMN_2771176 | 0.31 | 2.67E−04 | 81.456 | 10.47 | 5.60 | 5.86 | 9.82 |
| Tlcd1 | 68,385 | NM_026708 | ILMN_2781458 | −0.24 | 6.02E−03 | 78.007 | 11.01 | 5.60 | 5.87 | 11.73 |
| Ppm1e | 32,0472 | NM_177167 | ILMN_2702374 | 0.30 | 4.33E−04 | 83.474 | 10.67 | 5.60 | 5.99 | 11.44 |
| Rpa1 | 68,275 | NM_026653 | ILMN_2750801 | 0.32 | 1.71E−04 | 75.265 | 9.63 | 5.60 | 6.51 | 11.68 |
| Atad5 | 237,877 | NM_001029856 | ILMN_2534259 | 0.38 | 5.94E−06 | 81.302 | 18.06 | 5.60 | 7.21 | 19.12 |
| Ccl11 | 20,292 | NM_011330 | ILMN_2647757 | 0.30 | 5.12E−04 | 82.000 | 17.07 | 5.60 | 8.21 | 19.33 |
Definition of abbreviations: EOS, eosinophils; eQTL, expression quantitative trait locus; GEX, gene expression; LOD, logarithm of the odds; Mb, megabases.
LOD score for EOS on chromosome 11. This analysis was based on complete data from 132 mice. In this population, the LOD score was 5.6 (versus 7.0 from the analysis of 151 mice with eosinophil data).
See Materials and Methods for description of conditional models.
Discussion
HDM sensitization and challenge produced a wide range eosinophil recruitment responses in preCC mice, and this diversity facilitated the identification of a QTL for eosinophil counts. It is interesting that despite the fact the distribution of eosinophil counts is suggestive of a polygenic basis of inheritance in which many loci contribute to the phenotype, we only found one QTL associated with eosinophil counts. Additional QTL may be detectable with greater power, and the availability of biological replicates for inbred CC lines will facilitate this. In addition, we detected significant changes in IgE over the course of the experiment. However, these changes were not correlated with eosinophilic inflammation. These results are similar to those of a human genome-wide association study that detected loci associated with asthma and loci associated with IgE, but the two were not the same (14).
We identified a small set of priority candidate genes for the eosinophil QTL by merging the phenotype, gene expression, and eQTL data. We tested whether the expression of any single gene could underlie eosinophil recruitment responses using a simple linear model. Unexpectedly, we did not find evidence for a direct relationship between the QTL, Ccl11 (eotaxin) expression, and eosinophil counts. Rather, our results point to a more complex relationship that opposes the direct effect of Tlcd2, which we suggest is a novel candidate gene. Tlcd2 is an intriguing candidate gene in that TLC domain–containing proteins are thought to act as sphingolipid sensors (39) that are involved in lung inflammation (40, 41), but little else is known about this gene at present. We did not observe changes in Tlcd2 gene expression due to allergen sensitization and challenge in our previous study (22), nor is there evidence of differential expression in other lung disease models for which gene expression data is available in National Center for Biotechnology Information’s Gene Expression Omnibus database. Thus the manner by which variation in Tlcd2 gene expression affects allergic inflammation is an important question to address in future studies, as is the question of which lung cell type(s) express this gene.
We detected a large number of lung eQTL (n = 6,457, corresponding to 4,556 genes). This indicates that the expression of many genes in the lung is controlled in part by genetic variation. Based on our study design, we cannot estimate what fraction of variation in gene expression is due to allergen challenge versus genetic variation or a combination thereof (i.e., gene-by-HDM interactions). In the future, it will be worthwhile to address this, as well as categorizing eQTL by whether they affect gene expression only in the naive/unchallenged lung, only in the allergen-challenged lung, or both (“dynamic” versus “static” eQTL [43]). It will be particularly interesting to investigate which type of eQTL are more important in determining response to allergen. Intuitively, we would expect that the eQTL that regulate the induction/repression of gene expression after allergen challenge are more important, but this hypothesis should be tested directly, as this does not appear to be the case for Tlcd2 and eosinophilia.
Whole lung is, of course, a heterogeneous tissue, and contains numerous cell types, particularly in the context of AAD in which leukocytes are recruited. This has both positive and negative ramifications for eQTL identification and characterization. On the positive side, it is likely that we detected many eQTL that would not have been detected in naive or unchallenged lung, because they are expressed in infiltrating leukocytes. On the other hand, identifying which specific cell type(s) is the source of any particular eQTL remains a significant challenge, but is crucial to gaining further understanding of the biology related to AAD.
One of the most interesting features or our eQTL study was the identification of numerous trans-eQTL. The relative abundance of trans- versus cis-eQTL may be a function of the allergen model, as others have observed that trans-eQTL tend be more common in models where an environmental perturbation is applied (44). We also found that trans-eQTL clustered to specific genomic regions, most notably a locus on Chr 2 that was associated with expression of approximately 100 genes, many of which were correlated with eosinophil counts. We note, however, that the locus on Chr 2 was not itself QTL for eosinophil counts.
Some have expressed concern that trans-bands may be an artifact of hidden confounding in the data (45), but given that we employed randomization procedures to avoid such confounding, we do not think this is the case for this locus. We conducted post hoc analysis to test whether potential confounders or batch effects (RNA extraction date, hybridization date, or enrichment of a particular genotype and batch) could have resulted in spurious associations, and found no evidence of such biases. Based on the consistent allele effects that we observed, we narrowed the confidence interval for the trans-band to a 53-kb region containing a single gene, a putative lncRNA. lncRNAs are known to regulate gene expression, both in cis and in trans (46), and affect lung phenotypes (42); hence, we evaluated whether the lncRNA could underlie the trans-band. Although we found that this lncRNA also has an eQTL at the same locus, its expression did not correlate with genes in the trans-band, and thus it appears unlikely that the lncRNA explains the trans-band exclusively through variation in expression. It remains to be determined, however, whether the 68 SNPs in this putative lncRNA that distinguish A/J and C57BL/6J alleles from the other six CC founder alleles could give rise to structurally different RNAs that differentially control gene expression. Alternatively, this locus may contain other regulatory elements that control gene expression in trans.
It is also interesting to note that the trans-band on Chr 2 lies in a region that appears to be a QTL hotspot for other phenotypes. Previous studies have found QTL in this region for metabolism-related phenotypes, including blood glucose level (47), plasma levels of sterol (48), and growth and body fat parameters (49, 50). In addition, QTL for immune-related phenotypes, including natural killer T cell number in the thymus (51), experimentally induced induced colitis (52), and chemically induced bladder tumorigenesis (53), also map to this locus. It will be interesting to see whether the genes with trans-eQTL at this locus may be related to those phenotypes. A QTL for bronchial hyperresponsiveness also contained this locus (54). However, the bronchial hyperresponsiveness QTL was identified using C57BL/6J and A/J strains, the two strains that share a haplotype at the locus that we identified. Therefore, the trans-band locus cannot explain the bronchial hyperresponsiveness QTL.
In summary, we used a genetically diverse mouse population to quantify correlations between AAD phenotypes and identify genetic loci that control AAD phenotypes, including gene expression in the lung. We identified a candidate gene for eosinophilic inflammation caused by HDM using the gene expression data. Finally, our eQTL data also provide insights about gene regulation in the context of allergen challenge.
Acknowledgments
Acknowledgments
The authors thank Darla R. Miller (University of North Carolina [UNC]) for providing research support that facilitated the mouse experiments, and John P. Didion (UNC) for his assistance with array probe single nucleotide polymorphism mapping. We also thank the reviewers for helpful comments and suggestions.
Footnotes
This work was supported in part by the intramural program of the National Human Genome Research Institute (NIH) ZIA-HG200361 (F.S.C.), the U.S. Department of Energy, Office of Biological and Environmental Research (E.J.C.), and by NIH grants U01CA134240 and U01CA105417 (for production of Collaborative Cross mice), F32GM090667 (D.L.A.), GM070683 (G.A.C.), and a National Institutes of General Medical Sciences Centers of Excellence in Systems Biology program grant GM076468 (G.A.C.).
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2013-0501OC on April 2, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Malone DC, Lawson KA, Smith DH, Arrighi HM, Battista C. A cost of illness study of allergic rhinitis in the united states. J Allergy Clin Immunol. 1997;99:22–27. doi: 10.1016/s0091-6749(97)70296-3. [DOI] [PubMed] [Google Scholar]
- 3.Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma–united states, 1980–1999. MMWR Surveill Summ. 2002;51:1–13. [PubMed] [Google Scholar]
- 4.Mannino DM, Homa DM, Pertowski CA, Ashizawa AA, Nixon LL, Johnson CA, Ball LB, Jack EE, Kang DS. Surveillance for asthma—United States, 1960–1995. MMWR CDC Surveill Summ. 1998;47:1–27. [PubMed] [Google Scholar]
- 5.Cookson W. Genetics and genomics of asthma and allergic diseases. Immunol Rev. 2002;190:195–206. doi: 10.1034/j.1600-065x.2002.19015.x. [DOI] [PubMed] [Google Scholar]
- 6.Ober C, Thompson EE. Rethinking genetic models of asthma: the role of environmental modifiers. Curr Opin Immunol. 2005;17:670–678. doi: 10.1016/j.coi.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Vercelli D. Gene-environment interactions: the road less traveled by in asthma genetics. J Allergy Clin Immunol. 2009;123:26–27. doi: 10.1016/j.jaci.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 8.Tamari M. Genome-wide association studies of allergic diseases. Allergol Int. 2013;62:21–28. doi: 10.2332/allergolint.13-RAI-0539. [DOI] [PubMed] [Google Scholar]
- 9.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, Wilk JB, Willis-Owen SAG, Klanderman B, Lasky-Su J, et al. Genome-wide association analysis identifies pde4d as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers RA, Himes BE, Gignoux CR, Yang JJ, Gauderman WJ, Rebordosa C, Xie J, Torgerson DG, Levin AM, Baurley J, et al. Further replication studies of the eve consortium meta-analysis identifies 2 asthma risk loci in european americans. J Allergy Clin Immunol. 2012;130:1294–1301. doi: 10.1016/j.jaci.2012.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, Tsai YJ, Yang M, Campbell M, Foster C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–346. doi: 10.1016/j.jaci.2009.08.031. e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse north American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sleiman PMA, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SAG, Wang K, Rafaels NM, Michel S, Bonnelykke K, et al. Variants of dennd1b associated with asthma in children N Engl J Med 2010363988–989.; author reply 989 [DOI] [PubMed] [Google Scholar]
- 14.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WOCM. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulay ME, Morin A, Laprise C, Boulet LP. Asthma and rhinitis: what is the relationship? Curr Opin Allergy Clin Immunol. 2012;12:449–454. doi: 10.1097/ACI.0b013e328357cc32. [DOI] [PubMed] [Google Scholar]
- 16.Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA. Effects of allergen challenge on airway epithelial cell gene expression. Am J Respir Crit Care Med. 2005;171:579–586. doi: 10.1164/rccm.200404-532OC. [DOI] [PubMed] [Google Scholar]
- 17.Levine M, Ensom MHH. Post hoc power analysis: an idea whose time has passed? Pharmacotherapy. 2001;21:405–409. doi: 10.1592/phco.21.5.405.34503. [DOI] [PubMed] [Google Scholar]
- 18.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Arron JR, Koth LL, Fahy JV. T-helper type 2–driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattila P, Renkonen J, Toppila-Salmi S, Parviainen V, Joenväärä S, Alff-Tuomala S, Nicorici D, Renkonen R. Time-series nasal epithelial transcriptomics during natural pollen exposure in healthy subjects and allergic patients. Allergy. 2010;65:175–183. doi: 10.1111/j.1398-9995.2009.02181.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang IV, Tomfohr J, Singh J, Foss CM, Marshall HE, Que LG, McElvania-Tekippe E, Florence S, Sundy JS, Schwartz DA. The clinical and environmental determinants of airway transcriptional profiles in allergic asthma. Am J Respir Crit Care Med. 2012;185:620–627. doi: 10.1164/rccm.201108-1503OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelada SNP, Wilson MS, Tavarez U, Kubalanza K, Borate B, Whitehead GS, Maruoka S, Roy MG, Olive M, Carpenter DE, et al. Strain-dependent genomic factors affect allergen-induced airway hyperresponsiveness in mice. Am J Respir Cell Mol Biol. 2011;45:817–824. doi: 10.1165/rcmb.2010-0315OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium CC. The genome architecture of the collaborative cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, et al. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottomly D, Ferris MT, Aicher LD, Rosenzweig E, Whitmore A, Aylor DL, Haagmans BL, Gralinski LE, Bradel-Tretheway BG, Bryan JT, et al. Expression quantitative trait loci for extreme host response to influenza a in pre-collaborative cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelada SNP, Aylor DL, Peck BCE, Ryan JF, Tavarez U, Buus RJ, Miller DR, Chesler EJ, Threadgill DW, Churchill GA, et al. Genetic analysis of hematological parameters in incipient lines of the collaborative cross. G3 (Bethesda) 2012;2:157–165. doi: 10.1534/g3.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passador-Gurgel G, Hsieh W-P, Hunt P, Deighton N, Gibson G. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet. 2007;39:264–268. doi: 10.1038/ng1944. [DOI] [PubMed] [Google Scholar]
- 31.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 32.Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative Ct method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Tesson BM, Churchill GA, Jansen RC. Critical reasoning on causal inference in genome-wide linkage and association studies. Trends Genet. 2010;26:493–498. doi: 10.1016/j.tig.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey B-G, O’Connor TP, Crystal RG. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183:2867–2883. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byers DE, Holtzman MJ. Alternatively activated macrophages and airway disease. Chest. 2011;140:768–774. doi: 10.1378/chest.10-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberts RR, Terpstra PP, Li YY, Breitling RR, Nap JPJ-P, Jansen RCRC. Sequence polymorphisms cause many false cis eQTLs. PLoS One. 2007;2:2. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang G-J, Shifman S, Valdar W, Johannesson M, Yalcin B, Taylor MS, Taylor JM, Mott R, Flint J. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 2009;19:1133–1140. doi: 10.1101/gr.088120.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter E, Ponting CP. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- 40.Nixon GF. Sphingolipids in inflammation: pathological implications and potential therapeutic targets. Br J Pharmacol. 2009;158:982–993. doi: 10.1111/j.1476-5381.2009.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlig S, Gulbins E. Sphingolipids in the lungs. Am J Respir Crit Care Med. 2008;178:1100–1114. doi: 10.1164/rccm.200804-595SO. [DOI] [PubMed] [Google Scholar]
- 42.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincrnas are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ackermann M, Sikora-Wohlfeld W, Beyer A. Impact of natural genetic variation on gene expression dynamics. PLoS Genet. 2013;9:e1003514. doi: 10.1371/journal.pgen.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang HM, Ye C, Eskin E. Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics. 2008;180:1909–1925. doi: 10.1534/genetics.108.094201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muto A, Ochiai K, Kimura Y, Itoh‐Nakadai A, Calame KL, Ikebe D, Tashiro S, Igarashi K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorman J, Rudd R, Johnson C, King M, Minor P, Bailey C, Scalia M, Akinbami L. National surveillance for asthma—United States, 1980–2004. MMWR Surveill Summ. 2007;56:1–54. [PubMed] [Google Scholar]
- 48.Baines KJ, Simpson JL, Bowden NA, Scott RJ, Gibson PG. Differential gene expression and cytokine production from neutrophils in asthma phenotypes. Eur Respir J. 2010;35:522–531. doi: 10.1183/09031936.00027409. [DOI] [PubMed] [Google Scholar]
- 49.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133:16–26. doi: 10.1016/j.jaci.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan BS, Kirby A, Shebzukhov YV, Daly MJ, Kramnik I. Genetic architecture of tuberculosis resistance in a mouse model of infection. Genes Immun. 2006;7:201–210. doi: 10.1038/sj.gene.6364288. [DOI] [PubMed] [Google Scholar]
- 51.Walsh ER, Sahu N, Kearley J, Benjamin E, Kang BH, Humbles A, August A. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med. 2008;205:1285–1292. doi: 10.1084/jem.20071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mähler M, Bristol IJ, Sundberg JP, Churchill GA, Birkenmeier EH, Elson CO, Leiter EH. Genetic analysis of susceptibility to dextran sulfate sodium-induced colitis in mice. Genomics. 1999;55:147–156. doi: 10.1006/geno.1998.5636. [DOI] [PubMed] [Google Scholar]
- 53.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Sanctis GT, Merchant M, Beier DR, Dredge RD, Grobholz JK, Martin TR, Lander ES, Drazen JM. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nat Genet. 1995;11:150–154. doi: 10.1038/ng1095-150. [DOI] [PubMed] [Google Scholar]