Abstract
Background
Trimethylation of histone H3 lysine 4 (H3K4me3) accumulates at promoters in a gene activity-dependent manner. The Set1 complex is responsible for most H3K4me3 in somatic cells and contains the conserved subunit Cfp1, which is implicated in targeting the Set1 complex to CpG islands in mammals. In mouse embryonic stem cells, Cfp1 is necessary for H3K4me3 accumulation at constitutively active gene promoters, but is not required to maintain steady-state transcription of the associated gene.
Results
Here we show that Cfp1 is instrumental for targeting H3K4me3 to promoters upon rapid transcriptional induction in response to external stimuli. Surprisingly, H3K4me3 accumulation is not required to ensure appropriate transcriptional output but rather plays gene-specific roles. We also show that Cfp1-dependent H3K4me3 deposition contributes to H3K9 acetylation genome-wide, suggesting that Cfp1-dependent H3K4me3 regulates overall H3K9 acetylation dynamics and is necessary for histone acetyl transferase recruitment. Finally, we observe increased antisense transcription at the start and end of genes that require Cfp1 for accurate deposition of H3K4me3 and H3K9ac.
Conclusions
Our results assign a key role for Cfp1 in establishing a complex active promoter chromatin state and shed light on how chromatin signaling pathways provide context-dependent transcriptional outcomes.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-014-0451-x) contains supplementary material, which is available to authorized users.
Background
In eukaryotes, specialized chromatin structures contribute to multiple DNA-related processes, including transcription, replication and repair. Combinations of specific histone post-translational modifications correlate well with the functional status of the underlying DNA sequence - for example, at sites of transcriptional initiation, elongation or at distal regulatory elements [1-4]. Transitions between chromatin states accompany differentiation, cellular reprogramming, and disease processes [2,4]. However, it is unclear whether histone modification patterns are set up as a consequence of ongoing dynamic processes such as transcription or if they perform instructive roles. It is therefore crucial to systematically address the role of individual histone modifications in different contexts. As chromatin marks usually arise in reproducible groups comprising the same set of modifications, it is important to decipher their interdependence to help determine the biological significance of complex, potentially redundant, chromatin states.
H3K4me3 is a mark associated with eukaryotic gene promoters. In yeast, it is a feature of actively expressed genes [5,6], suggesting that it positively influences transcription. In mammals, H3K4me3 is found at active and inactive promoters at a level dependent on gene activity [7,8]. Most promoters in mouse and human are associated with CpG islands (CGIs), which are DNA elements showing high G + C and CpG content that are usually free of DNA methylation [9,10]. CGIs possess a characteristic chromatin structure thought to predispose them towards promoter activity [9,11]. For example, CGIs can directly recruit H3K4me3, favoring transcriptional competence [12]. In mammalian stem cells, H3K4me3 is found together with H3K27me3 at bivalently marked CGI promoters [13,14], which are poised for activation by developmental signals upon lineage commitment.
H3K4 methylation is achieved by conserved enzymatic complexes related to the yeast COMPASS (Complex associated with Set1) [15,16]. Mammalian COMPASS complexes vary in their catalytic component (Setd1A and Setd1B, Mll1 to Mll4) as well as in specific subunits that contribute to their functional diversity (reviewed in [17]). Set1-containing COMPASS is the main H3K4 histone methyltransferase in most organisms [18-21]. Mll1/Mll2 COMPASS-like have gene-specific roles in H3K4me3 deposition [22-24], while Mll3/Mll4 COMPASS-like complexes mainly contribute to H3K4me1 at enhancers [25,26].
CxxC finger protein 1 (Cfp1, CXXC1 or CGBP) is a specific component of Set1-containing complexes [17,27]. Cfp1 binds unmethylated CpGs and targets Set1 and H3K4me3 to most CGIs in somatic cells, regardless of their transcriptional activity [12]. In embryonic stem cells, Cfp1 plays a fundamental role in genome-wide H3K4me3 organization [28]. It is required for strong H3K4me3 enrichment at constitutively active gene promoters, but plays little role in depositing this mark at poised genes, including bivalent promoters [28]. Surprisingly, in stem cells, reduced H3K4me3 deposition at active promoters does not drastically affect steady-state transcription [28,29]. On the other hand, loss of the Cfp1 gene in mice results in early embryonic lethality [30] and Cfp1-deficiency in somatic cell lines is toxic [12,31]. Thus, it is possible that Cfp1-deficiency impairs the proper induction of transcription programs in response to differentiation signals or to external stimuli like stress, potentially explaining why Cfp1−/− embryonic stem (ES) cells are unable to differentiate in vitro [32].
In this study, we ask how Cfp1 affects H3K4me3 dynamics in rapid, regulated gene expression, using the transcriptional response to DNA damage as a model. We show that in addition to its role in regulating steady-state H3K4me3 deposition in ES cells, Cfp1 is instrumental in targeting this modification to gene promoters upon rapid transcriptional induction. We also observe that the Cfp1-dependent H3K4me3 accumulation that follows gene induction is not strictly required to ensure appropriate transcriptional output but rather plays gene-specific roles. We also identify a strong co-dependency between H3K4me3 and H3K9ac deposition upon transcriptional induction as well as in normally cycling ES cells. Our results suggest that Cfp1-dependent H3K4me3 regulates overall H3K9 acetylation dynamics and is necessary for histone acetyltransferase (HAT) recruitment. Finally, we describe elevated antisense transcription at the start and end of those genes that require Cfp1 for accurate H3K4me3 and H3K9ac deposition.
Results
Cfp1 is required for H3K4me3 deposition following transcriptional induction
To determine the role of Cfp1 in H3K4me3 deposition upon rapid transcriptional activation, we analyzed the transcriptional response to doxorubicin, a DNA damaging agent that triggers sudden p53-dependent gene expression changes in mouse ES cells [29,33,34]. Doxorubicin treatment (1 μM, 6 hours) in wild-type (WT ) ES cells rapidly caused increased binding of phosphorylated p53 (at serine 18 in mouse) at the Cdkn1a (p21) locus, leading to a robust induction of the Cdkn1a gene (Figure S1A-C in Additional file 1). This was accompanied by increased H3K4me3 levels at the Cdkn1a locus (Figure S1D in Additional file 1) as previously described [29]. To identify genes whose expression is regulated in response to doxorubicin treatment, we performed RNA-Seq experiments using cDNA libraries from ribosomal RNA-depleted total RNA in doxorubicin-treated and untreated WT ES cells. We identified 1,264 genes that significantly changed expression upon doxorubicin treatment (P-value <0.05, fold change ≥2.5) comprising 775 upregulated and 489 downregulated genes (Figure 1A; Additional file 2). The list included many genes previously implicated in the p53-dependent transcriptional response to doxorubicin [33,34] in both the upregulated (Wnt9, Wnt3, Btg2, Cdkn1a, Mdm2, Bbc3) and downregulated (Sox2, Esrrb, Sall4, Zic3, n-Myc, Nanog) gene sets (Figure 1A,B; Figure S2A in Additional file 3). These data confirm the validity of our data analysis pipeline.
Figure 1.
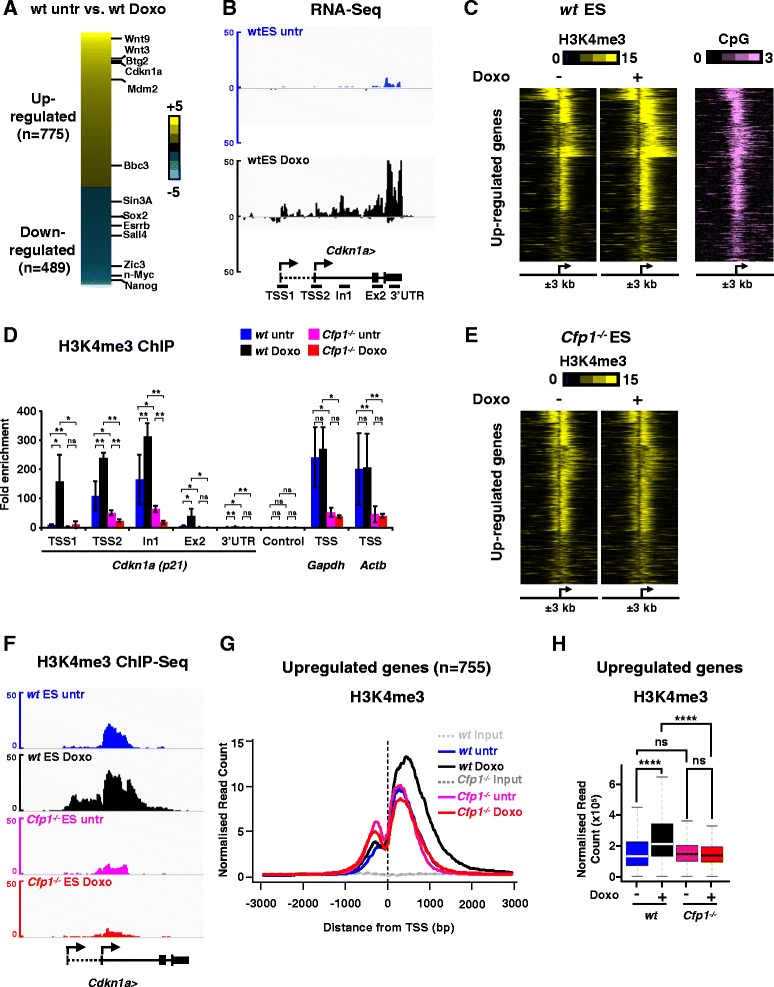
Cfp1 is crucial for H3K4me3 deposition at promoters of genes regulated upon DNA damage. (A) Heatmap representing gene expression changes induced by doxorubicin treatment (P-value <0.05, fold change ≥ 2.5). Genes highlighted were identified in previous studies [33,34]. Untr, untreated. (B) Genome browser screenshots representing RNA-Seq normalized read count in WT ES cells at the Cdkn1a locus. Positions for quantitative PCR (qPCR) primers used during the study are also represented. TSS, transcription start site. (C) Heatmap showing H3K4me3 normalized read count 3 kb upstream and downstream of the TSS of genes induced by doxorubicin in WT ES cells (n = 755). Unmethylated CpGs are represented by CAP-Seq enrichment in WT ES cells [28]. (D) ChIP-qPCR analysis of H3K4me3 enrichment at the Cdkn1a locus for untreated or doxorubicin-treated WT and Cfp1 −/− ES cells. Results are expressed as fold enrichment relative to input DNA and a control intergenic region on chromosome 15. Control corresponds to an intergenic region on chromosome 5. Gapdh and Actb promoters are also shown. Data are represented as mean ± standard deviation, n = 3. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01; P > 0.05 is not significant (ns). (E) Same as (C) for Cfp1 −/− ES cells. (F) Genome browser screenshots representing H3K4me3 ChIP-Seq normalized read count in WT (untreated in blue, treated in black) and Cfp1 −/− (untreated in pink, treated in red) ES cells at the Cdkn1a locus. (G) Average profile showing H3K4me3 normalized read count at TSSs of genes induced by doxorubicin treatment in WT ES cells. Input DNA is shown as a control. Signal is displayed from −3 kb to +3 kb surrounding each annotated TSS. (H) Comparison of H3K4me3 normalized read density at TSSs of upregulated genes, from 3 kb upstream to 3 kb downstream. P-values were calculated using two-tailed unpaired Wilcoxon tests: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
H3K4me3 associates with active or poised promoters at a level dependent on gene activity [7,8]. Furthermore, transcriptional induction can lead to increased H3K4me3 at the activated gene promoter [35]. We asked how expression changes induced by doxorubicin affect H3K4me3 patterns at cognate promoters by performing H3K4me3 chromatin immunoprecipitation (ChIP) coupled to massively parallel DNA sequencing (ChIP-Seq) in WT ES cells that were untreated or treated with doxorubicin. Most promoters of upregulated genes are CpG-rich and displayed H3K4me3 enrichment before induction (Figure 1C), which is compatible with the notion that they are poised for rapid activation in response to external stimuli [36]. As expected, transcriptional induction by doxorubicin led to increased H3K4me3 levels at those promoters (Figure 1C). On the other hand, genes downregulated by doxorubicin treatment were characterized by decreased H3K4me3 (Figure S2A,B in Additional file 3), emphasizing the correlation between H3K4me3 and transcriptional activity.
In ES cells, Cfp1 plays a key role in establishing H3K4me3 at constitutively active loci, while its general contribution to H3K4me3 deposition at poised genes is minimal [28]. To explore how Cfp1 contributes to H3K4me3 dynamics in regulated gene expression, we analyzed H3K4me3 status at the Cdkn1a locus in response to doxorubicin in Cfp1−/− ES cells by ChIP-quantitative PCR (ChIP-qPCR; Figure 1D). While we observed a robust increase in H3K4me3 at transcription start site (TSS)1, TSS2 and intron 1 upon doxorubicin treatment in WT ES cells, it was apparent that Cfp1-deficiency led to a severe defect in H3K4me3 accumulation at the Cdkn1a locus (Figure 1D). ChIP-qPCR using an antibody recognizing total histone H3 and a control IgG confirmed the specificity of this result (Figure S1E,F in Additional file 1). As expected, Cfp1-deficiency also led to decreased H3K4me3 near the TSS of constitutively active Actb and Gapdh gene loci (Figure 1D). We extended the analysis by performing H3K4me3 ChIP-Seq analyses in Cfp1−/− ES cells treated or not with doxorubicin. Indeed, H3K4me3 did not increase at promoters of genes normally upregulated by doxorubicin treatment, but instead remained at basal levels in both untreated or doxorubicin-treated Cfp1−/− ES cells (Figure 1E-H). This establishes that Cfp1 is instrumental in H3K4me3 deposition in response to an external stimulus in ES cells. On the other hand, genes downregulated by doxorubicin showed reduced H3K4me3 in untreated Cfp1−/− compared to untreated WT ES cells and these levels did not drastically change upon doxorubicin treatment (Figure S2B-F in Additional file 3). Down-regulated genes are highly expressed in untreated ES cells (Figure S2G in Additional file 3) and therefore require Cfp1 for accurate H3K4me3 deposition at their promoters in normally proliferating mouse ES cells [28]. We conclude that upon transcriptional activation, H3K4me3 levels are enhanced at induced promoters in a Cfp1-dependent manner. Thus, Cfp1 not only plays a key role in determining proper H3K4me3 patterns in normally proliferating ES cells [28], but is also instrumental in targeting this modification to gene promoters upon transcriptional induction in response to external stimuli. These findings favor a model whereby the Cfp1/Set1-containing complex acts mainly downstream of transcriptional activation.
Low H3K4me3 at promoters only mildly affects transcriptional induction output
Previous work established that lack of Cfp1-dependent H3K4me3 at constitutively active gene promoters does not drastically alter steady-state transcription in ES cells [28]. However, H3K4me3-mediated recruitment of Taf3 is reported to be crucial for transcriptional induction upon doxorubicin treatment in HCT116 cells [37]. We therefore tested whether altered H3K4me3 patterns affect the transcriptional response to doxorubicin. First, we checked that early response to doxorubicin was intact in Cfp1−/− ES cells by confirming that p53 stabilization, phosphorylation and binding to the Cdnkn1a locus was indistinguishable between WT and Cfp1−/− ES cells (Figure S3A,B in Additional file 4). We then performed RNA-Seq experiments in Cfp1−/− ES cells treated or untreated with doxorubicin and looked for differential expression of genes that are inducible in WT ES cells. Many genes, including Mdm2, Btg2 and Wnt9a, were induced appropriately in the mutant cells (Figure S3C in Additional file 4). Others appeared to be affected in their transcriptional response, but this effect was generally mild and gene-dependent, with very few genes showing complete lack of activation or repression (Figure 2A-C). We also observed minor defects in genes repressed by doxorubicin (Figure 2A,B), but confirmed that Nanog responded properly in both treated WT and treated Cfp1−/− ES cells (Figure 2D). To exclude the possibility that Cfp1-deficiency affects global transcription in response to doxorubicin treatment, we performed absolute quantification RT-qPCR experiments. For this, we normalized transcript abundance to a spiked-in standard instead of an endogenous transcript (see Materials and methods for details) as described in [38]. In this setting, we also observe efficient Cdkn1a and Mdm2 induction, as well as Nanog and Sox2 repression in doxorubicin-treated WT and Cfp1−/− ES cells (Figure 2E,F). Our results suggest that Cfp1-dependent H3K4me3 plays a minor role in ensuring appropriate expression of inducible genes. The biological significance of modest deregulation at specific genes is, however, uncertain.
Figure 2.
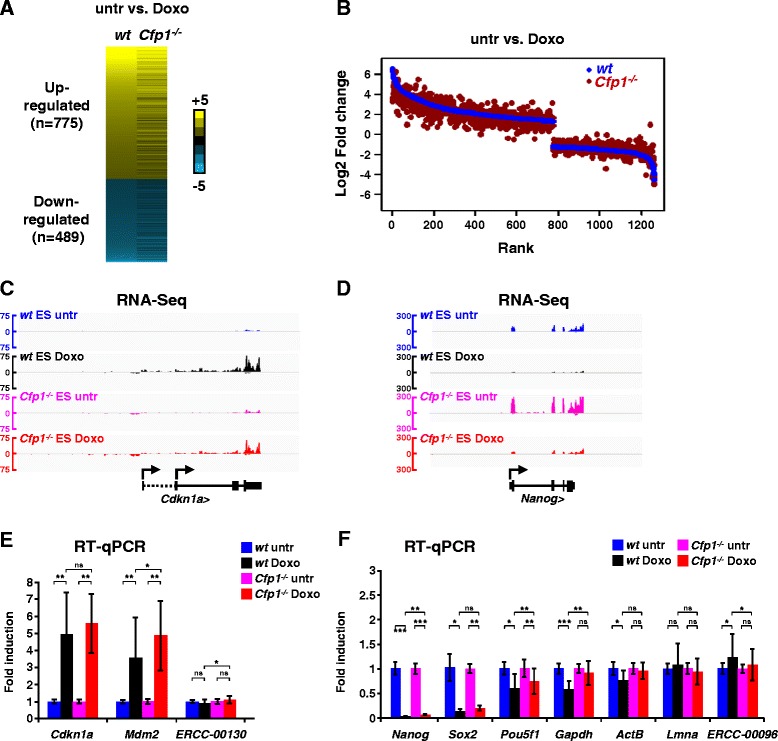
Decreased H3K4me3 at regulated promoters mildly affects transcriptional output. (A) Heatmap comparing gene expression changes (as log2 fold change) induced by doxorubicin treatment in WT and Cfp1 −/− ES cells as determined by RNA-Seq. (B) Dot plot comparing gene expression differences in WT and Cfp1 −/− ES cells. Genes were sorted on the basis of their log2 fold change upon doxorubicin treatment in WT ES cells (most upregulated gene on the far left, most downregulated gene on the far right). The log2 fold changes (untreated (untr) versus doxorubicin treatment (Doxo)) are plotted for WT (blue) and Cfp1 −/− ES cells (red). (C) Genome browser screenshots representing RNA-Seq signal (as normalized read count) in WT ES cells (untreated in blue, treated in black) and Cfp1 −/− (untreated in pink, treated in red) ES cells at the Cdkn1a locus. (D) Same as (C) for the Nanog locus. (E) Absolute quantification of Cdkn1a and Mdm2 mRNA levels in WT (untreated in blue, doxorubicin-treated in black) and Cfp1 −/− ES cells (untreated in pink, doxorubicin-treated in red) by RT-qPCR. Expression levels are normalized using an RNA spike-in standard (ERCC-00074) and then expressed as fold induction compared with the untreated value. The ERCC-00130 standard is shown as control. Data are represented as mean ± standard deviation, n = 4. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01; P > 0.05 is not significant (ns). (F) Same as (E) for Nanog, Sox2, Pou5f1, Gapdh, and Actb mRNA levels. Lmna and ERCC-00096 are shown as controls.
Cfp1 is crucial for H3K9 acetylation at regulated promoters
As lack of Cfp1 led to impaired H3K4me3 accumulation at doxorubicin-regulated promoters, we investigated whether other aspects of the chromatin landscape were also affected. Several reports have linked H3K4me3 levels at TSSs with histone H3 acetylation [29,35,39,40]. Using an antibody directed against H3K9,K14ac, we observed increased histone acetylation at the Cdkn1a locus upon doxorubicin treatment in WT ES cells, but this effect was absent in Cfp1−/− cells (Figure 3A). A possible explanation is that histone H3 acetylation requires Cfp1-dependent H3K4me3 deposition at the induced TSSs. This effect appears specific to H3K9,K14 as H3K27 acetylation levels were unaffected by absence of Cfp1 (Figure 3B). Furthermore, antibodies specifically directed against H3K9 or H3K14 acetylation revealed that Cfp1-deficiency more severely impacts H3K9 rather than H3K14 acetylation (Figure S4A,B in Additional file 5). To test whether Cfp1-dependent H3K4me3 at doxorubicin-regulated promoters is generally associated with H3K9,K14 acetylation, we performed ChIP-Seq experiments. In WT ES cells we observed increased TSS histone acetylation at genes induced by doxorubicin, but acetylation was severely reduced in Cfp1−/− ES cells (Figure 3C,E; Figure S4C in Additional file 5). At down-regulated promoters, H3K9,K14 acetylation was decreased in treated WT ES cells, but its levels appear low in both untreated and doxorubicin-treated Cfp1−/− cells (Figure 3D,F; Figure S4D in Additional file 5). There is therefore a strong similarity between H3K4me3 and H3K9,K14ac patterns at doxorubicin-regulated promoters, as levels of both are directly related to transcriptional activity. Interestingly, lack of Cfp1 impacts both modifications at this subset of genes, strongly suggesting co-dependency.
Figure 3.
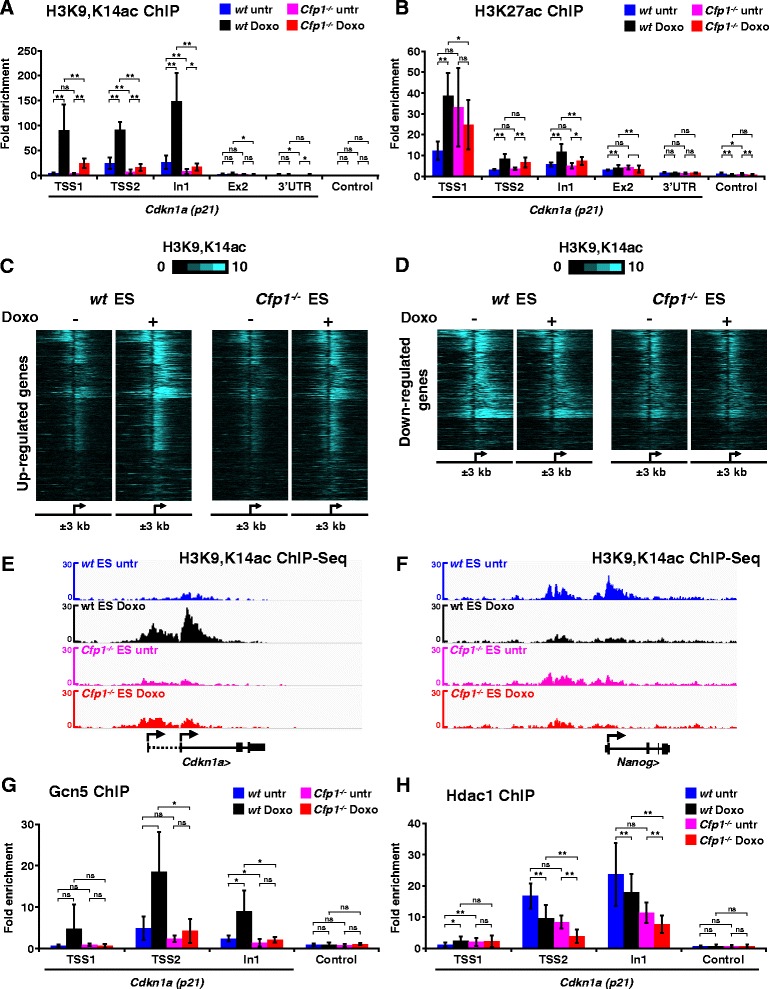
Cfp1 is crucial for H3K9 acetylation at regulated promoters. (A) ChIP-qPCR analysis of H3K9,K14ac enrichment at the Cdkn1a locus for untreated (untr) or doxorubicin-treated (Doxo) WT and Cfp1 −/− ES cells. Results are expressed as fold enrichment relative to input DNA and a control intergenic region on chromosome 15. Control corresponds to an intergenic region on chromosome 5. Data are represented as mean ± standard deviation (SD), n = 3. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01; P > 0.05 is not significant (ns). (B) Same as (A) for H3K27ac enrichment. Data are represented as mean ± SD, untr n = 2, Doxo n = 4. (C) Heatmap representation showing H3K9,K14ac normalized read count 3 kb upstream and downstream of TSSs of genes upregulated by doxorubicin treatment in WT ES cells (n = 755). (D) Same as (C) for downregulated genes (n = 489). (E) Genome browser screenshots representing H3K9,K14ac ChIP-Seq signal (as normalized read count) in WT (untreated in blue, treated in black) and Cfp1 −/− (untreated in pink, treated in red) ES cells at the Cdkn1a locus. (F) Same as (E) for the Nanog locus. (G) Same as (A) for Gcn5 enrichment at the Cdkn1a locus. Data are represented as mean ± SD, n = 3. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01, P > 0.05 (ns). (H) Same as (G) for Hdac1 enrichment at the Cdkn1a locus. Data are represented as mean ± SD, n = 3. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01, P > 0.05 (ns).
H3K4me3 can act as a binding platform that attracts HAT complexes. In particular, the Gcn5-containing SAGA complex can be targeted to H3K4me3 nucleosomes by the adaptor subunit Sgf29 [41]. Consistent with this model, we observed that decreased H3K4me3 at the Cdkn1a promoter is associated with decreased Gcn5 binding at the same genomic location (Figure 3G), which is likely to account for decreased H3K9 acetylation. Furthermore, we observed that reduced histone acetylation at the Cdkn1a locus in Cfp1−/− ES cells was not due to increased binding of histone deacetylase 1 (Hdac1; Figure 3H). We conclude that H3K4me3 defects caused by Cfp1-deficiency at promoters affect overall H3K9 acetylation dynamics, probably by directly impairing H3K4me3-mediated HAT recruitment.
H3K9ac levels follow H3K4me3 at Cfp1-regulated regions independent of gene induction
Cfp1 influences H3K4me3 deposition at many genomic locations spread throughout the mouse ES cell genome and particularly at active gene promoters [28]. We tested whether Cfp1-deficiency affects histone acetylation at non-doxorubicin-inducible promoters by examining H3K4me3 patterns at the most active genes in WT ES cells as identified in our RNA-Seq dataset (n = 2,500; Additional file 6). The results confirm that lack of Cfp1 leads to severely reduced H3K4me3 levels, mainly located downstream of the TSS (Figure 4A,B,E) and that this is accompanied by a reduction in H3K9,K14ac as determined by ChIP-Seq (Figure 4A,B,E). Similar results were obtained for this gene set when comparing doxorubicin-treated WT and Cfp1−/− ES cells (Figure S5A,B,E in Additional file 7). The findings further support a mechanistic link between Cfp1-dependent H3K4me3 and H3K9ac at TSSs in normally cycling mouse ES cells.
Figure 4.
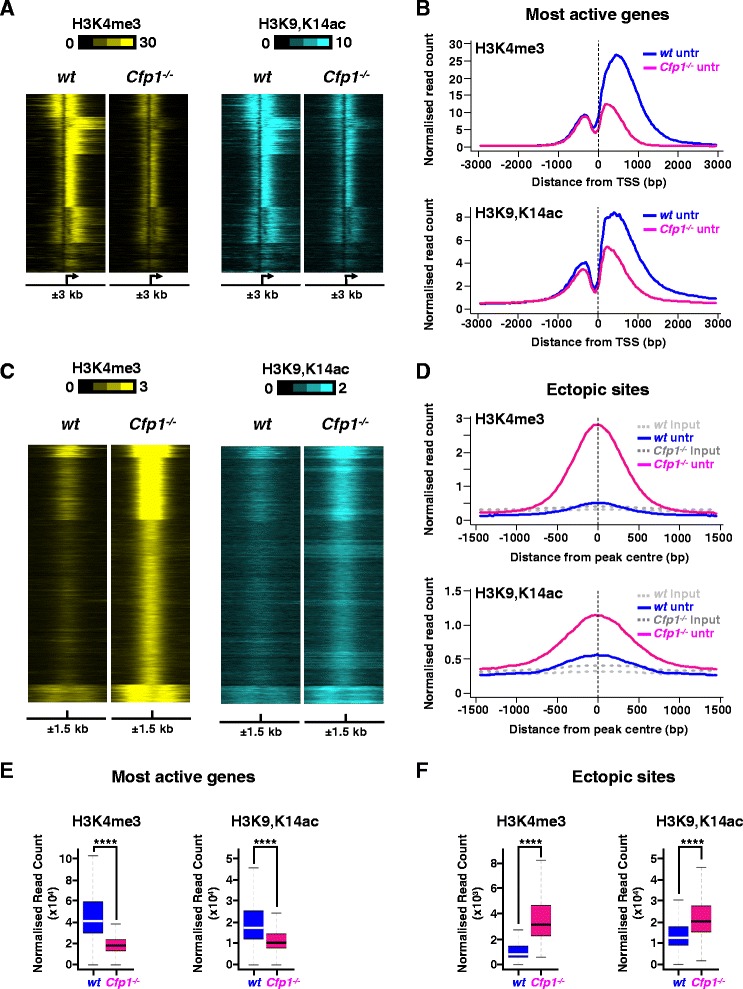
H3K9 acetylation is altered at Cfp1-regulated regions. (A) Heatmap showing H3K4me3 (yellow) or H3K9,K14ac (blue) normalized read count at TSSs for the most active genes in WT ES cells (n = 2,500). Signal is displayed from −3 kb to +3 kb surrounding each annotated TSS for untreated WT or Cfp1 −/− ES cells. (B) Average profile showing H3K4me3 or H3K9,K14ac normalized read count at TSSs for the most active genes in WT ES cells (n = 2,500). Signal is displayed from −3 kb to +3 kb surrounding each annotated TSS for untreated WT or Cfp1 −/− ES cells. (C) Same as (A) for regions aberrantly accumulating H3K4me3 in absence of Cfp1 (n = 14,208). Signal is displayed from −1.5 kb to +1.5 kb surrounding the center of the peak for each region for untreated WT or Cfp1 −/− ES cells. (D) Average profile showing H3K4me3 or H3K9,K14ac normalized read count for regions aberrantly accumulating H3K4me3 in absence of Cfp1 (n = 14,208). Signal is displayed from −1.5 kb to +1.5 kb surrounding the centre of the peak. Input DNA is plotted as a reference. (E) Comparison of H3K4me3 or H3K9,K14ac normalized read density at TSSs of the most active genes (n = 2,500), from 3 kb upstream to 3 kb downstream. Box plots show the central 50% of the data (filled box), the median (central bisecting line) and 1.5× the interquartile range (whiskers). P-values were calculated using two-tailed unpaired Wilcoxon tests: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (F) Same as (E) for H3K4me3 or H3K9,K14ac normalized read density at regions aberrantly accumulating H3K4me3 in absence of Cfp1 (n = 14,208), from 1.5 kb upstream to 1.5 kb downstream.
Absence of Cfp1 in mouse ES cells has been shown to cause aberrant accumulation of H3K4me3 at many non-promoter regions [28]. We tested the possibility that these ectopic sites of H3K4me3 accumulation are also associated with increased H3K9ac. For this, we compared H3K4me3 and H3K9,K14ac density in WT and Cfp1−/− ES cells at those sites where H3K4me3 accumulates in the absence of Cfp1 (Additional file 8). The results confirmed that, on average, increased H3K4me3 at those regions is matched by increased H3K9,K14ac levels (Figure 4C,D,F). Furthermore, it was obvious that regions that accumulated the most H3K4me3 in Cfp1−/− ES cells also showed highest H3K9,K14ac enrichment (Figure 4C,D,F). Again, this was confirmed when comparing doxorubicin-treated WT and Cfp1−/− ES cells (Figure S5C,D,F in Additional file 7). Some of the regions that accumulate H3K4me3 in cells lacking Cfp1 correspond to sites occupied by Ctcf or Cohesin [28]. We therefore analyzed both H3K4me3 and H3K9,K14ac distributions at selected binding sites for those two factors in WT and Cfp1−/− ES cells [42] (Additional files 9 and 10). Lack of Cfp1 leads to increased H3K4me3 and this is associated with an increase in H3K9,K14ac (Additional files 11 and 12). We conclude that the altered H3K4me3 pattern observed in mouse ES cells in the absence of Cfp1 is accompanied by an alteration in H3K9ac deposition. This suggests a mechanistic co-dependency for these two histone marks, both of which are consistently associated with transcriptional activity.
Decreased H3K4me3 and H3K9ac impact transcriptional accuracy upon induction
Given the subtle effect of altered promoter H3K4me3 and H3K9ac on the transcriptional output of inducible genes, we tested whether other aspects of gene expression are affected by the altered chromatin landscape. First we examined RNA processing, as H3K4me3 is proposed to facilitate splicing factor loading [43]. We compared splicing efficiency for the Cdkn1a transcript upon induction in WT and Cfp1−/− ES cells. We detected neither accumulation of the unspliced pre-mRNA nor reduction in a particular splicing product when comparing doxorubicin-treated WT and Cfp1−/− ES cells (Figure 5A,B). This agrees with a previous observation that decreased H3K4me3 at constitutively active genes does not drastically alter splicing in normally cycling ES cells [28].
Figure 5.
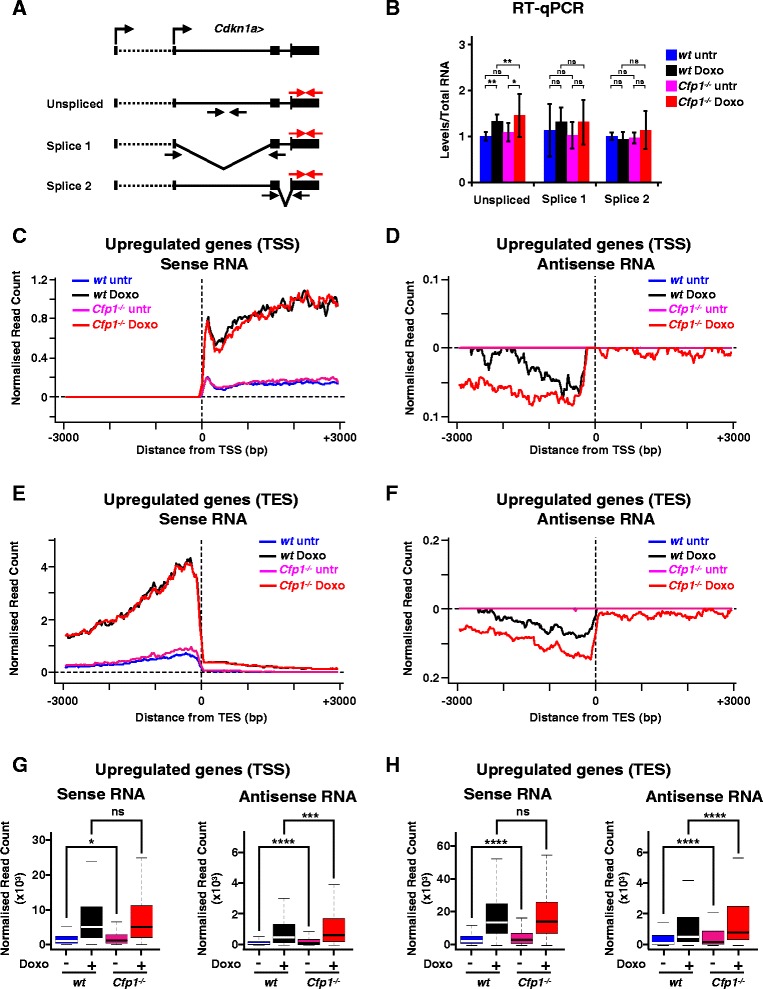
Decreased H3K4me3 and H3K9ac influence transcriptional accuracy upon induction. (A) Schematic representation of the qPCR strategy used to quantify various Cdkn1a mRNA splicing products. Total RNA is detected using primers on 3’ UTRs (shown in red). (B) RT-qPCR analysis of Cdkn1a splicing products. P-values were calculated using two-tailed Student’s t-tests: *P < 0.05, **P < 0.01, P > 0.05 is not significant (ns). (C) Average profile showing sense RNA-Seq normalized read count at TSSs of genes induced by doxorubicin treatment in WT ES cells (n = 755). Signal is displayed from −3 kb to +3 kb surrounding each annotated TSS. (D) Same as (C) for antisense RNA-Seq normalized read count (note that sense and antisense transcripts are plotted on a different scale). (E) Average profile showing sense RNA-Seq normalized read count at transcription end sites (TESs) of genes induced by doxorubicin treatment in WT ES cells (n = 755). Signal is displayed from −3 kb to +3 kb surrounding each annotated TES. (F) Same as (E) for antisense RNA-Seq normalized read count (note that sense and antisense transcripts are plotted on a different scale). (G) Comparison of sense or antisense RNA-Seq normalized read density at TSSs of upregulated genes (n = 755), from 3 kb upstream to 3 kb downstream, in WT or Cfp1 −/− ES cells, treated or not with doxorubicin as indicated. Box plots show the central 50% of the data (filled box), the median (central bisecting line) and 1.5× the interquartile range (whiskers). P-values were calculated using two-tailed unpaired Wilcoxon tests: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (H) Same as (G) for TESs of upregulated genes (n = 755).
Antisense transcription is also a feature of gene expression thought to be affected by H3K4 methylation [44-46]. Active mammalian transcription units produce small sense and antisense RNA molecules at their TSSs and transcription end sites (TESs) [47,48]. We investigated levels of these transcripts at genes regulated by doxorubicin in WT and Cfp1−/− ES cells using strand-specific RNA-Seq datasets. As expected, upon induction sense RNA accumulated both downstream of the TSS and upstream of the TES (Figure 5C,D,G,H) and profiles were strongly similar when comparing doxorubicin-treated WT and Cfp1−/− ES cells. Induced genes also displayed increased divergent antisense transcription at both TSSs and TESs, albeit at much lower abundance, but this was strikingly elevated in Cfp1−/− ES cells (Figure 5C,D,G,H). The results suggest that Cpf1-dependent increases in H3K4me3 and H3K9ac can play a role in regulating TSS- and TES-associated small RNA transcription upon gene induction. The effect is marginal or non-significant at genes downregulated by doxorubicin (Additional file 13). We propose that Cfp1-dependent H3K4me3 and H3K9ac depositions contribute to accurate RNA production at the start and end of genes upon transcriptional induction.
Discussion
Understanding H3K4me3 deposition at gene promoters
Cfp1 is required for accurate H3K4me3 deposition at constitutively active gene promoters in mouse ES cells [28]. Here we show that Cfp1 also plays an important role in targeting this modification upon transcriptional induction. This favors a model in which the Cfp1/Set1 complex acts mainly downstream of the transcriptional apparatus, similar to yeast [5,6,49]. The data suggest a scenario whereby high H3K4me3 levels at promoters of genes regulated by external stimuli result from a multistep process. In the unstimulated state H3K4me3 is low and the promoter is poised for activation. Cfp1 seems to play little role in this 'priming' process. When transcription is induced, however, Cfp1 is instrumental in causing the striking increase in H3K4me3 levels at the TSS. Similarly, Cfp1 is dispensable for H3K4me3 at bivalent genes [28], which are kept silenced in ES cells but are thought to be poised for activation by developmental cues [13,14,50]. The histone methyl transferase Mll2 is largely responsible for H3K4me3 deposition at bivalent promoters [23]. Thus, different H3K4 methyl-transferases play distinct roles in shaping the chromatin landscape, suggesting that histone modification patterns are the results of an intricate network of enzymatic activities. Variation in these histone modifications may create alternative binding surfaces for potential histone tail readers, leading to context-dependent outcomes. According to this scenario, histone modifications do not act as simple on/off switches, but instead create molecular signaling gradients, allowing for subtle regulation of particular genomic loci in various conditions. More quantitative measurements of chromatin features are needed to fully appreciate their complexity and relationship to cell commitment and disease.
H3K4me3 and H3K9ac deposition are linked in mouse embryonic stem cells
A striking finding from our study is that alterations in Cfp1-dependent H3K4me3 deposition are accompanied by parallel defects in H3K9ac deposition. This is true for regions that either lose or gain H3K4me3 in the absence of Cfp1. An attractive possibility is that H3K9 acetylation is downstream of H3K4me3, and, together with previous reports, points to a molecular function for H3K4me3 in targeting H3K9 acetylation. H3K4me3 and H3K9ac coincide at many gene promoters [8,51-53], but we observe that these two marks are mechanistically co-dependent. H3K4me3 was suggested to facilitate H3K9 acetylation [40] and Sgf29, a subunit of the SAGA complex, can bind H3K4me3 and facilitate recruitment of the Gcn5 HAT [41]. Accordingly, we find that decreased H3K4me3 in Cfp1−/− ES cells leads to reduced Gcn5 recruitment at the Cdkn1a promoter. Furthermore, aberrant H3K4me3 accumulation at ectopic sites in the absence of Cfp1 coincides with enhanced H3K9ac deposition at the same loci. These findings do not favor a hypothesis where Cfp1 functions in an as yet uncharacterized HAT complex, but this possibility cannot be formally excluded. We favor a simpler model whereby Cfp1-dependent enhancement of H3K4me3 levels at TSSs contributes to recruitment of HATs, which increase H3K9 acetylation. Interestingly, the inverse relationship, whereby acetylation of histone H3 targets the Set1 complex to chromatin, has been observed biochemically, although the acetylation affects residues other than K9 [54]. Further studies of the functional interplay between histone marks will undoubtedly shed valuable light on the biological significance of histone modifications.
H3K4me3 and H3K9ac function in a context-dependent manner
Reduced H3K4me3 at a TSS does not affect steady-state transcription in ES cells [28,29]. We now extend this observation and show that H3K4me3 is largely dispensable for regulated gene expression in this cell type. In vitro, H3K4me3 stimulates assembly of the pre-initiation complex as well as p53- and p300-dependent transcription [37,54]. Furthermore, Taf3, a TBP-associated protein that can bind H3K4me3, selectively regulates a p53 target gene induced by doxorubicin in colon cancer cells [37]. On the other hand, depletion of Mll2 in ES cells, which is responsible for H3K4me3 deposition at bivalent genes, has little effect on their rapid transcriptional induction [23]. Cfp1/Set1 associates with most CGI promoters in mouse brain and is required for regulated gene expression in colon cancer cells [12,54]. In pluripotent cells, however, Cfp1 is only necessary for H3K4me3 deposition at active genes and does not strongly affect either steady-state or regulated gene expression ([28] and this study). We conclude that the impact of H3K4me3 deposition on nucleosomes is highly context-dependent and that this modification may play gene- and/or cell type-specific roles.
Our data also imply that high level H3K9 acetylation is not required for either steady-state or regulated gene expression in ES cells. The association between histone acetylation and active transcription is well established [55,56], but it has been reported that histone deacetylases, which remove acetylation, are also enriched at active gene promoters [40]. It appears that two of the marks most prominently associated with transcriptional initiation are not necessary for either steady-state or regulated gene expression in mouse ES cells. It is possible that their functional significance is gene-specific and/or context-dependent. Alternatively, there may be general functions for these marks that have so far escaped detection. In yeast, Set1 deletion has effects on regulating antisense transcription [44-46]. We observed aberrant antisense transcription at TSSs and TESs of genes induced by doxorubicin, suggesting that H3K4me3 and/or H3K9ac can regulate complex aspects of mRNA biogenesis. These transcripts are low in abundance, however, and do not seem to critically influence mRNA levels. A second possibility is that H3K4me3 and/or H3K9ac are required to help resolve potentially toxic R-loops that arise at transcriptionally active CpG rich promoters in mammals [57]. Thirdly, the presence of these marks may fine-tune RNA polymerase II dynamics in ways that are not easily caught by genome-wide studies, which provide only an averaged snapshot of the cell population.
Our data could also fit with the idea that histone modifications mainly modulate nucleosome dynamics [58,59]. ES cells reportedly have an intrinsically accessible, dynamic chromatin structure, which is believed to contribute to their pluripotency [4,60]. Such an atypical structure could be robust enough to sustain key processes such as transcription, for which histone modification only acts redundantly. Upon lineage commitment, however, a more restricted chromatin state may be acquired [4] for which histone modification-mediated processes become necessary. One also needs to stay open to the idea that histone marks strongly associated with promoters or coding regions can play roles outside transcription. For example, H3K4me3, together with insulator proteins, demarcates the border of physical domains and might thus be important for shaping the three-dimensional organization of the nucleus [61] and H3K36me3 has recently been implicated in regulation of DNA repair [62]. There is still much to understand regarding the role of chromatin in complex genome organization and function.
Conclusions
We show that Cfp1 is required for H3K4me3 accumulation at promoters upon rapid transcriptional induction, linking chromatin structure and transcriptional activity. We also uncover a strong relationship between Cfp1-dependant H3K4me3 accumulation and accurate H3K9ac deposition. As defects in H3K4me3 and H3K9ac accumulation at promoters do not drastically alter steady-state or regulated expression of the associated gene, our results suggest that those two histone modifications function downstream of the transcriptional apparatus and could potentially regulate small RNA levels around transcription unit boundaries.
Materials and methods
Embryonic stem cell culture
ES cells were grown in gelatinized dishes in Glasgow MEM (Gibco Life Technologies Ltd, Paisley, UK) supplemented with 10% fetal bovine serum (Hyclone GE Healthcare Bio-Sciences AB, Uppsala, Sweden), 1× MEM nonessential amino acids, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol (Gibco) and LIF (Leukemia Inhibitory Factor). E14TG2a ES cells were used as WT ES and Cfp1−/− ES cells have been described [32]. Doxorubicin was purchased from Sigma (Gillingham, UK) and used at a final concentration of 1 μM for 6 hours.
Chromatin immunoprecipitation
ES cells were crosslinked with 1% formaldehyde at room temperature for 10 minutes, followed by 5-minute incubation with 125 mM glycine at room temperature. Cells were washed in phosphate-buffered saline, flash frozen in liquid nitrogen and stored at −80°C prior to use. Cell pellets were thawed on ice, resuspended in lysis buffer 1 (50 mM Hepes KOH, pH 7.9, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and incubated for 10 minutes on a rotator at 4°C. Cells were washed in TE (10 mM Tris–HCl, pH 8, 1 mM EDTA).
For histone modifications and RNA polymerase II ChIP, cells were resuspended in TE containing 0.5% SDS, incubated on ice for 10 minutes and sonicated using a Bioruptor (Diagenode, Seraing, Belgium) on high setting for 18 cycles of 30 s (30 s pause between pulses). The sonicated extract was centrifuged at 20,000 g for 10 minutes at 4°C and the supernatant diluted 5-fold with ChIP dilution buffer (20 mM Tris–HCl, pH 8, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA).
For Gcn5, CBP, Hdac1 and p53S15P ChIP, cells were sonicated in 20 mM Tris–HCl, pH 8, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS for 60 cycles of 30 s (30 s pause between pulses).
Sonicated chromatin was incubated overnight with the appropriate antibody and immune complexes were precipitated using protein A-Sepharose (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) pre-blocked with bovine serum albumin and yeast tRNA, for 30 minutes at 4°C. Complexes were washed three times with 20 mM Tris–HCl, pH 8, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS, once with 20 mM Tris–HCl, pH 8, 500 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS, and once with TE 1X. Bound complexes were eluted from the beads with TE containing 1% SDS by heating at 65°C for 1 h. Samples were diluted to 0.5% SDS with TE, then RNase A and NaCl (final concentration 0.3 M) were added and samples incubated for 4 h to overnight at 65°C to reverse crosslinking. Samples were treated with proteinase K (100 μg/ml) for 1 to 2 h at 55°C, phenol extracted and ethanol precipitated.
Strand-specific RNA-Seq
Cells were lysed in TRI reagent (Sigma). Total RNA was then extracted with chloroform, precipitated with isopropanol and DNAse treated (DNA-free, Ambion Life Technologies Ltd, Paisley, UK). Ribosomal RNA from 5 μg of total RNA was depleted using Ribo-Zero kit (Epicentre, Madison, WI, USA). Strand specific RNA-Seq libraries were prepared using Script-Seq V2 RNA-Seq library preparation kit (Epicentre) according to the manufacturer’s instructions.
Preparation of ChIP DNA for Illumina sequencing
ChIP DNA was end repaired by incubation at 20°C for 30 minutes with 3 U of T4 DNA Polymerase (NEB, Hitchin, UK), 10 U of Polynucleotide Kinase (NEB), 2 U DNA Polymerase I Large (Klenow) fragment (NEB), 1× T4 DNA ligase reaction buffer (NEB) and 400 nM dNTPs. The enzymes were then heat inactivated at 75°C for 20 minutes, after which the DNA was ethanol precipitated. A tail of 'A' bases was added to the 3’ ends of the DNA by incubation with 5 U Klenow Fragment (3’-5’ exo-; NEB), 200 nM dATP and 1X buffer 2 (NEB) at 37°C for 30 minutes. The enzymes were heat inactivated and cleaned up as before. Barcoded Ilumina paired end adaptors were then ligated to the processed ChIP DNA by incubation with 300 U of T4 DNA ligase (NEB), 1× T4 DNA ligase buffer (NEB), 7.5% PEG-6000 and 2 pmol of annealed Illumina adaptors for 3 h at room temperature. Ligated DNA was purified using MinElute PCR columns (Qiagen, Hilden, Germany) and eluted in 10 μl water.
Library preparation and Illumina sequencing
Ligated DNA was amplified by 16 to 18 cycles of PCR with primers complementary to the adaptor sequences and Phusion 2x premix (Finnzymes, Waltham, MA, USA). The DNA was purified using QIAquick PCR Purification columns (Qiagen). The purified DNA was captured on an Illumina flow cell for cluster generation. Libraries were sequenced using an Illumina HiSeq to generate paired-end 75 bp reads. For ChIP-Seq, six samples were multiplexed. Paired-end sequence reads were demultiplexed and mapped to the mouse genome (NCBIm37) using BWA version 0.5.9-r16 [63]. Uniquely aligned reads were selected for further analysis and duplicate reads were removed for H3K9,K14ac ChIP-Seq samples (duplication was very low in H3K4me3 samples). For H3K4me3, sequencing from two biological replicates was combined in a single file. For RNA-Seq, eight samples were multiplexed. Paired-end sequence reads were mapped to the mouse genome (NCBIm37) using STAR version 2.20c_r200 [64] to allow spliced alignments. Biological replicates (two for each condition) were kept separate for differential expression analysis but were merged to generate average profiles.
RNA expression analysis
A GTF file containing ensEMBL mouse transcript information (release 67) was used to generate a count table with HTSeq-count [65] with the following parameters: −-stranded yes -mode union -type exon. These options summarize unambiguous read counts, on the sense strand, over entire gene units using all exons. Genes differentially expressed upon doxorubicin treatment in WT ES cells were identified using the Bioconductor package DESeq [66]. Genes with adjusted P-value <0.05 and fold change ≥2.5 were identified as differentially expressed. Genes with a base mean count <200 (empirically determined) were removed from the list as they correspond to genes weakly expressed in both conditions. The corresponding differential expression heatmap (Figure 2A) was generated using Java Treeview [67]. A normalized count table was used to determine expression values (Figure S2G in Additional file 3) and identify the 2,500 most active genes in mouse ES cells (Figure 4) by correcting WT raw read counts for gene length. Average profile plots of sense and antisense transcription (Figure 5C-F) were generated in an identical manner to ChIP-Seq occupancy plots (see below) using normalized bigwig files for each strand (based on direction of paired end alignment).
Average profile and heatmap analysis of ChIP-seq occupancy
All read depths were normalized to the total number of uniquely mapped reads in each experiment. ChIP-seq density was determined by calculating the average number of hits per base in 100 bp windows with a 20 bp slide using normalized bigwig files over a 6 kb interval centered on the TSS (for promoters) or a 3 kb interval over the center of each peak (for non-promoter regions) using custom Perl and R scripts interfaced with the Galaxy server [68,69], which are available upon request. Average profiles were generated by plotting the median value in each window for each sample. For heat maps, the sliding window file obtained from WT ES cells was subjected to k-means clustering using Cluster 3.0 [70]. Results from clustering were used to rank sliding windows files for all other samples accordingly. Clustered heatmaps were generated using Java Treeview.
RNA extraction and RT-qPCR
Cells were lysed in TRI reagent (Sigma). RNA was then extracted with chloroform, precipitated with isopropanol and DNAse treated (DNA-free, Ambion). Total RNA (2 μg) was used per reverse transcription reaction. RNA was denatured in the presence of 2 μg of random hexamers (Invitrogen Life Technologies Ltd, Paisley, UK) for 5 minutes at 75°C, and reverse transcribed in a final volume of 40 μl with 200 U of SuperScript II (Invitrogen) at 42°C overnight followed by heat inactivation at 70°C for 15 minutes. Synthesized complementary DNAs were diluted in 300 μl of water and stored at −20°C until used.
Absolute quantification experiments were performed essentially as described in [39]. Briefly, 5.105 cells were lysed in 1 ml TRI reagent (Sigma) and 1 μl of a 1:20 dilution of ERCC Spike-in Mix#1 (Life Technologies Ltd, Paisley, UK) was added to each sample. After RNA extraction and reverse transcription, cDNAs were analysed by RT-qPCR. Transcript abundance were normalized to ERCC-00074 spike-in standard. ERCC-00096 and ERCC-00130 were also used as secondary controls.
Protein extraction and western blotting
Cells were lysed in RIPA buffer (25 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) plus protease inhibitors and sonicated with a Bioruptor (Diagenode) for 10 cycles (30 s each cycle). Whole cell lysate were subjected to SDS-PAGE and transferred on nitrocellulose.
Antibodies
All antibodies are listed in Additional file 14.
Oligonucleotides
Oligonucleotides used for ChIP-qPCR, RT-qPCR and splicing analysis are listed in Additional file 15.
Data access
High throughput sequencing data (ChIP-Seq and RNA-Seq) have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE53492.
Acknowledgements
We thank Sophie Messager and Nathalie Smerdon at the Wellcome Trust Sanger Institute for managing and performing the DNA sequencing. We also thank Gaëlle Legube and Luisa Di Stefano for a critical reading of the manuscript. This work was funded by the Wellcome Trust, The Medical Research Council UK and Cancer Research UK.
Abbreviations
- bp
base pair
- CGI
CpG island
- ChIP
chromatin immunoprecipitation
- ES
embryonic stem
- HAT
histone acetyltransferase
- qPCR
quantitative polymerase chain reaction
- TES
transcription end site
- TSS
transcription start site
- UTR
untranslated region
- WT
wild type
Additional files
Embryonic stem cell response to doxorubicin.
Differentially expressed genes comparing WT versus WT doxorubicin-treated cells (P > 0.05, fold change ≥2.5).
Cfp1 regulates H3K4me3 at promoters of genes responding to doxorubicin in ES cells.
Impact of decreased H3K4me3 at regulated promoters on transcriptional output.
Cfp1-deficiency associates with decreased H3 acetylation at regulated promoters.
The 2,500 most expressed gene in WT ES cells.
H3K9 acetylation is altered at Cfp1-regulated H3K4me3 binding sites in doxorubicin-treated cells.
Coordinates for ectopic H3K4me3 peaks.
Subset of Ctcf binding sites (from [42]).
Subset of Smc1a binding sites (from [42]).
H3K9 acetylation is altered at Ctcf binding sites in the absence of Cfp1.
H3K9 acetylation is altered at cohesin binding sites in the absence of Cfp1.
Transcription at TSSs and TESs of genes downregulated by doxorubicin.
List of antibodies used in the study.
List of oligonucleotides used in the study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TC conceived of the study, designed and carried out the experiments, analyzed sequencing data, and drafted the manuscript. SW analyzed the sequencing data and drafted the manuscript. AB participated in the design of the study, supervised and coordinated the study and drafted the manuscript. All authors have read and approved the final version of this manuscript.
Contributor Information
Thomas Clouaire, Email: thomas.clouaire@univ-tlse3.fr.
Shaun Webb, Email: swebb1@staffmail.ed.ac.uk.
Adrian Bird, Email: a.bird@ed.ac.uk.
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 6.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2010;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illingworth RS, Bird AP. CpG islands–'a rough guide'. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Blackledge NP, Klose R. CpG island chromatin: a platform for gene regulation. Epigenetics. 2011;6:147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 23.Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol. 2013;20:1093–1097. doi: 10.1038/nsmb.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branch of the COMPASS family is a major H3K4 monomethylase at enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 28.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, Lee JH, Skalnik D, Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlone DL, Skalnik DG. CpG binding protein is crucial for early embryonic development. Mol Cell Biol. 2001;21:7601–7606. doi: 10.1128/MCB.21.22.7601-7606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young SR, Skalnik DG. CXXC finger protein 1 is required for normal proliferation and differentiation of the PLB-985 myeloid cell line. DNA Cell Biol. 2007;26:80–90. doi: 10.1089/dna.2006.0535. [DOI] [PubMed] [Google Scholar]
- 32.Carlone DL, Lee JH, Young SR, Dobrota E, Butler JS, Ruiz J, Skalnik DG. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol Cell Biol. 2005;25:4881–4891. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, Hockemeyer D, Mitalipova M, Lewis CA, Vander Heiden MG, Sur M, Young RA, Jaenisch R. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Geli V, Holstege FC. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3'-end antisense transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dijk EL, Chen CL, D'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 47.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW, Roeder RG. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 56.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 60.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burrows-Wheeler Aligner. [http://bio-bwa.sourceforge.net/]
- 64.Rna-star [http://code.google.com/p/rna-star/]
- 65.HTSeq-count [http://www-huber.embl.de/users/anders/HTSeq/doc/count.html]
- 66.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Java TreeView [http://www.jtreeview.sourceforge.net]
- 68.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10:Unit 10 15. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galaxy [http://main.g2.bx.psu.edu/]
- 70.Cluster 3.0 [http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm]


