Abstract
The Pax6 transcription factor is expressed in cerebellar granule cells and when mutated, as in the Sey/Sey mouse, produces granule cells with disturbed survival, migration, and defects in neurite extension. The impact of Pax6 on other genes in the context of cerebellar development has not been identified. In this study, we performed transcriptome comparisons between wildtype and Pax6-null whole cerebellar tissue at embryonic day (E) 13.5, 15.5 and 18.5 using Affymetrix arrays (U74Av2). Statistical analyses identified 136 differentially regulated transcripts (FDR 0.05, 1.2 fold change cut-off) over time in Pax6-null cerebellar tissue. In parallel we examined the Math1-null granuloprival cerebellum and identified 228 differentially regulated transcripts (FDR 0.05, 1.2 fold change cut-off). The intersection of these two microarray datasets produced a total of 21 differentially regulated transcripts. For a subset of the identified transcripts, we used qRT-PCR to validate the microarray data and demonstrated the expression in the rhombic lip lineage and differential expression in Pax6-null cerebellum with in situ hybridization analysis. The candidate genes identified in this way represent direct or indirect Pax6-downstream genes involved in cerebellar development.
Keywords: Cerebellum, Development, Granule cell, Transcription factor, Sey mutant, Transcriptome profiling
Introduction
The rhombic lip (RL) gives rise to the excitatory neurons of the cerebellum: first glutamatergic cerebellar nuclear neurons and then granule cell precursors and unipolar brush cells (Wang et al., 2005; Englund et al., 2006; Fink et al., 2006). The key transcription factors Math1 and Pax6 are expressed in this region as well as the external germinal layer (EGL)(Ben-Arie et al., 1997; Engelkamp et al., 1999). Cerebellar granule cells go through several epochs of development from their origins in the rhombic lip around E12.5 to the trans-migratory cells that establish the EGL, to the highly proliferative and then migratory population that produces the largest cohort of neurons in the brain (Hatten & Heintz, 1995; Goldowitz & Hamre, 1998; Machold & Fishell, 2005). In spite of numerous studies on granule cell development, the understanding of the genetic underpinnings of the establishment of the EGL is limited. Previous studies of Pax6 mutant mice (Pax6Sey/Sey) had indicated a defect in the colonization of the EGL which could be observed as early as embryonic day (E) 15 (Engelkamp et al., 1999; Swanson et al., 2005; Swanson & Goldowitz, 2010). We used the timing of the appearance of the Sey mutant trait to hone-in on genes important for granule cell development.
Pax6 belongs to the Paired box gene (PAX) family of transcription factors (TFs) and has been the focus of intensive study for its role in eye development (Hill et al., 1991; Halder et al., 1995), pancreatic islet cell development (St-Onge et al., 1997), brain patterning (Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001), neurogenesis (Estivill-Torrus et al., 2002; Heins et al., 2002), as well as other key events in neurodevelopment (Quinn et al., 2007). In neural stem cells, it has recently been shown that the level of Pax6 is essential for the balance between stem cell self-renewal and neurogenesis (Sansom et al., 2009). In the cerebellum, Pax6 is highly expressed in the RL region, which gives rise to the granule cell precursors in the EGL. Pax6 plays an important role in the organization of EGL as loss of Pax6 function results in aberrant organization of this structure in terms of foliation, disorganization of post-mitotic granule cells and neuronal migration (Engelkamp et al., 1999; Swanson et al., 2005).
In this study, we combined the mutant mouse strategy with genome-wide gene expression profiling technology to identify genes that could be part of Pax6 regulatory gene networks involved in the development of cerebellar granule cells. As part of this strategy we compared transcriptome profiles of whole cerebellar tissue at E13.5, E15.5 and E18.5 from littermates of wildtype and Pax6-null mice. We also applied a biological filter by comparing the changes in gene expression in the Pax6-null cerebellum with that of a the Math1 (Atoh1, atonal homolog 1)-null cerebellum that lacks glutamatergic rhombic lip lineage neuronal precursors such as granule cells, unipolar brush cells and glutamatergic cerebellar nuclear neurons (Ben-Arie et al., 1997; Englund et al., 2006). Through these approaches, we identified discrete cohorts of genes that are related to developmental processes perturbed in the mutant cerebellum. We find that the intersection of genes that are significantly down-regulated in the Math1-null and significantly altered in the Pax6-null cerebellum are excellent candidates for a role in Pax6-dependent gene networks that underlie granule cell development.
Materials and methods
Mice
Three strains of mice were used in this study: two strains of Pax6Sey mice, the original Pax6Sey strain (obtained from Drs. Robert Grainger and Marilyn Fisher, University of Virginia, maintained on a mixed genetic background) and the Pax6Sey-Neu strain (obtained from Brigid Hogan, Duke University, maintained on an ICR background for more efficient animal production (Grindley et al., 1997), and Math1β-Gal (Ben-Arie et al., 1997; Wang et al., 2005) on a mixed C57-129SvEv background. Mice were housed in a room with 12/12 hr light/dark controlled environment. Male and female heterozygous mice from the Pax6Sey and Pax6Sey-Neu strains were mated to generate double heterozygous (i.e., non-functional homozygous knock-out) Pax6-null embryos for microarray, RNA collection, and in situ work. Heterozygous Math1β-Gal male and female mice were mated to generate homozygous Math1-null and littermate control embryos. Embryos were obtained from timed pregnant females at midnight of the day when a vaginal plug was detected; this was considered embryonic day 0 (E0). Pregnant females were cervically dislocated and embryos were harvested from the uterus and identified as Pax6Sey/Sey, Pax6Sey/+, or Pax6+/+ by eye phenotype. For in situ hybridization (ISH), E15.5 embryos were decapitated and whole heads were immersion fixed for 1–2 hours in 4% paraformaldehyde made with DEPC treated water. Tissues were cryopreserved in 20% sucrose, and then quick-frozen in OCT (Fisher) in preparation for cryosectioning. The use of mice was in strict accordance with SFN guidelines and was based on IACUC and CCAC approved protocols at the University of Tennessee HSC and the University of British Columbia.
Gene expression microarray
The cerebellum was isolated from each embryo, pooled with littermates of like genotype, and snap-frozen in liquid nitrogen. 3–4 replicate pools of 6–10 whole cerebella samples were collected from E13.5, E15.5 and E18.5 embryos of Pax6+/+ and Pax6Sey/Sey genotypes and processed for total RNA extraction. Care was taken to avoid thawing prior to homogenization of the tissue in RNASTAT60 (Tel-Test Inc., Friendswood, TX). The tissue was disrupted by passage through a 23G syringe needle and then passed over a Qiashredder column to ensure complete disruption. The homogenate was cleared by centrifugation and the RNA precipitated from the supernatant with isopropanol. RNA pellets were sent to Genome Explorations (Memphis, TN; URL: http//www.genome-explorations.com) for quality control (QC) and preparation for expression analysis. The RNA was resuspended, quantified, and the RNA integrity was verified using an Agilent Bioanalyzer 2100. We estimate that each E15.5 cerebellum has a wet weight of approximately 0.50–0.75mg and produced on average 3.5–6.0μg of total RNA. Biotinylated cRNA probes were generated from total RNA, tested for quality and hybridized using an Affymetrix platform. For the Pax6 data, the available format at the time of experiment was the Affymetrix U74Av2 GeneChip, which contains 12,488 probe sets (mgu74av2.cdf). During the progress of these studies, when we were ready to examine Math1 gene expression, Affymetrix upgraded the platform to the GeneChip Mouse Expression Array 430AB, which contained 45,037 probe sets (MOE430A.cdf and MOE430B.cdf). The cross-platform comparison of differentially regulated genes was performed at the gene level. The details of common probe sets and details are provided by the manufacturer (Affymetrix) and available for download on the web (www.affymetrix.com/support/technical/comparison_spreadsheets.affx). Other than using different microarray chips from Affymetrix, all experimental procedures for the E15.5 Math1 sample were done in an identical manner as the Pax6 samples described above.
Microarray data preprocessing
Each chip served as an independent hybridization experiment and four pooled samples were analyzed for each genotype. CEL files were read in the R environment using the package ‘affy’(Gautier et al., 2004; Team, 2011). Background adjustment and quantile normalization was done using RMA function justRMA and fold changes were calculated on normalized raw intensity values. In subsequent principal component analysis, the first principal component which represents the major variances across all samples was correlated to each sample and plotted to check for outliers. One sample, a wildtype E15.5 from the Pax6 analysis, was excluded as its correlation to first principle component (0.95) was beyond 3 standard deviations (0.0086) from the median (0.994). The remaining set was renormalized and further analyzed in R environment. For single time point, mutant vs wildtype comparisons, the Significance Analysis of Microarrays (SAM) program was used (Tusher et al., 2001).
General linear modeling and statistical methods
For expression analysis we applied general linear modeling followed by planned linear contrasts suitable for low-density time series analyses. The application of this technique to microarray data was described by others previously (Li et al., 2006). For each transcript we fitted a two-way ANOVA model with an interaction for embryonic age and genotype (i.e. Pax6 mutation) using ‘anova’ and ‘lm’ functions in R. The Age factor had 3 levels: E13.5, E15.5 E18.5 and the Genotype factor had 2 levels: mutant and wildtype. Based on the results of the fits we classified the genes into three separate groups as genes with an interaction effect (Set 1, Genotype*Age False Discovery Rate (FDR) <=0.05 and fold change > 1.2), genes with genotype effects (Set 2, Genotype*Age FDR >0.05, Genotype FDR <=0.05, and fold change > 1.2) and genes with age effects only (Set 3, Genotype*Age FDR >0.05, Genotype FDR >0.05, and Age FDR <=0.05). FDR is defined as the expected proportion of false positives among the rejected hypotheses and we used the FDR method of Benjamini and Hochberg to control for FDR (Benjamini & Hochberg, 1995).
Post-hoc contrast analysis
Once the data had been classified in one of the 3 models, post-hoc contrast analysis was performed. The contrasts were constructed based only on the Age effect for genes for which Age was significant (Set 3). A set of orthogonal contrast vectors was constructed using the R function “contr.poly” to test specific hypotheses regarding the temporal pattern of the age effect. For example, for each probe set the best fit for linear temporal pattern (constant increase (↗↗) or decrease over time (↘↘)) or polynomial temporal pattern (inflection points at E15.5 (↗↘ or ↘↗)) was determined. The coefficients for each polynomial contrast are listed here.
| E13.5 | E15.5 | E18.5 | |
|---|---|---|---|
| Linear | −0.707 | 0 | 0.707 |
| Quadratic | 0.408 | −0.817 | 0.408 |
For each gene in Set 3, we chose the contrast (linear or polynomial) with the smallest p-value and based on the sign of the estimates we classified the contrasts as linear increasing, linear decreasing, upward parabola and downward parabola. In addition, we computed the power to detect age and type effects using R function power.anova.test.
Real-time PCR
Real-time PCR was performed as a validation tool for the microarray data. An independent set of RNA samples from corresponding littermates of wildtype and mutant mice were used for the cDNA synthesis. As an additional control, heterozygote cDNA samples were prepared for the Pax6 sample set. cDNA samples were produced with random hexamers using the High Capacity cDNA Archive kit (Applied Biosystems). cDNA products were diluted and aliquoted to 100 ng total RNA input. Sequences of the transcript of interest were loaded into Primer Express® software (Applied Biosystems) and primers designed (see Supplemental Table 4 for sequences). Amplicon lengths were between 75–125 bp. The qPCR was performed with the FAST SYBR Green PCR Master Mix (Applied Biosystems) on an ABI StepOne Plus Sequence Detection System (Applied Biosystems). The linear dynamic range of the input RNA from 1 ng to 10 pg was determined. All primer sets were tested for optimal dissociation curves with amplification efficiencies between 85–100%. Any primer sets not meeting those standards were redesigned. All runs were normalized to 18s RNA, our endogenous control. The relative delta-delta-CT method (Gibson et al., 1996) was employed for analysis and an expression ratio calculated for each sample pair. All runs were performed in triplicate and replicated at least three times per transcript.
In situ hybridization (ISH) for analysis of candidate gene expression
The cRNA probes were generated from a cDNA library generated from the E15.5 and P0 mouse brain using Invitrogen cDNA synthesis kit (Invitrogen). We used M13F and M13R primers to PCR amplify templates directly from plasmids (pGemT, Promega) that contained the target gene. The obtained PCR yield was directly used as a template to generate the Dig-labelled cRNA probe. A total of 16 differentially expressed genes selected from DNA micro-arrays were used for the ISH study.
Embryonic tissues were sagittally cryosectioned, mounted on Superfrost™ slides (Fisher), air-dried at room temperature and stored at −80 °C until use. Control wildtype or Pax6Sey/+ and experimental Pax6Sey/Sey mutant tissues were mounted on a single slide. The ISH was performed at 55°C overnight in a hybridization buffer containing about 300ng/ml of Dig-labelled cRNA probe. After hybridization, the slides were washed and gradually desalted in 4X SSC, 2X SSC, 1X SSC and 0.5 X SSC at 55°C, then incubated with an anti-Dig antibody (Roche) for 2 hrs at room temperature at a concentration of 1:300. After washing, slides were colorized with NTP/BCIP (Roche) and mounted with 1.5% gelatin containing 15% glycerol.
Bioinformatic tools for gene ontology categorization and transcription factor binding site analysis
For Gene Ontology category analysis of identified differentially regulated genes from microarray data analysis, DAVID bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) was used (Dennis et al., 2003). The filtered gene list was entered via a web interface and the results were downloaded directly from the website. For Pax6 binding site analysis, we used oPOSSUM (http://burgundy.cmmt.ubc.ca/oPOSSUM/), a bioinformatics tool that was designed to detect over-represented, conserved TF binding sites in human and mouse genes (Ho Sui et al., 2005). We used the recommended settings for the analysis.
Results
Time series microarray comparison of Pax6 mutant vs wildtype cerebellum and post-hoc contrast analysis
In order to identify the genes directly and indirectly regulated by Pax6 in cerebellar development, we employed genome-wide gene expression analysis on the Pax6 and Math1 mutant mouse cerebella (Fig. 1). We compared transcriptome profiles of Pax6-null mutant and wildtype littermate cerebellum at E13.5, E15.5 and E18.5 using the Affymetrix U74Av2 GeneChip platform. We first performed wildtype vs Pax6-null comparisons at each timepoint with Significance Analysis of Microarrays (SAM). With FDR<0.05 and 1.2 raw intensity value fold change cut-offs, the number of differentially regulated genes was 9 at E13.5 (9 down and 0 up), 23 at E15.5 (17 down and 6 up) and 344 at E18.5 (148 down and 196 up) (Fig 2A). The 2 genes that were common for all 3 time points were Neurod1 (neurogenic differentiation 1) and Calb2 (calbindin 2). This trend of increasing transcriptome changes over embryonic development closely followed the reported histological findings of Pax6-null mutant cerebellar phenotypes as the mutant phenotype becomes observable at E15.5 and more pronounced at E18.5 (Engelkamp et al., 1999; Swanson et al., 2005). In light of this finding, we selected E15.5 as the key timepoint of enquiry based on the number of differentially expressed genes and the high overlap of the differentially expressed genes at E15.5 (87%) with the other time points (Fig 2A).
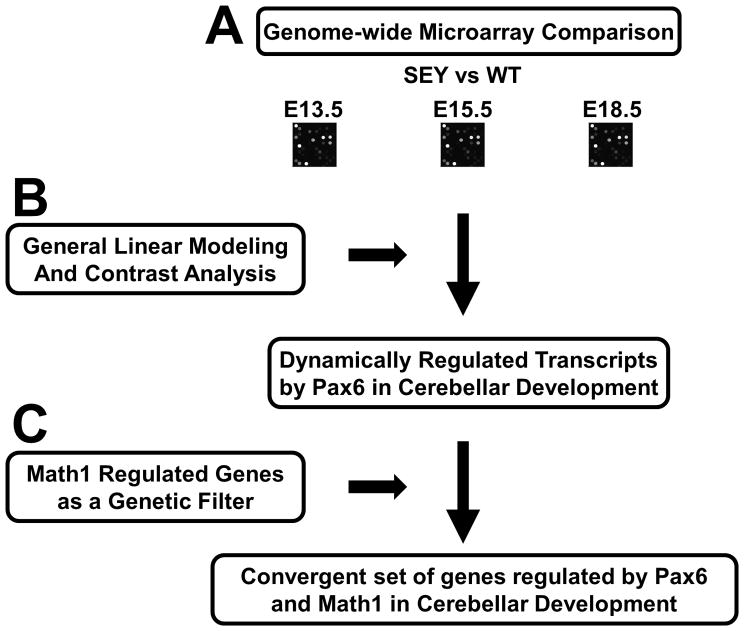
Fig 1.
Strategy for identification of Pax6 differentially regulated genes in cerebellar development. (A) An embryonic mini-time series transcriptome analysis was performed on RNAs of whole cerebellar tissues from Pax6-null and littermate wildtype mice. (B) Dynamically regulated transcripts based upon expression in the Pax6-null cerebellum in the time series expression data were identified by general linear modeling. (C) We then used Math1-downregulated transcripts as a filter for Pax6 differentially regulated genes to get a convergent set of genes important for cerebellar development.
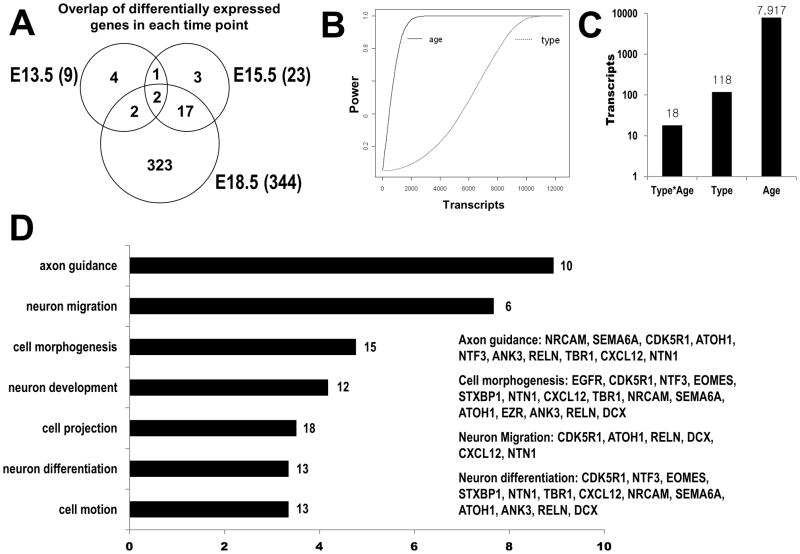
Fig 2.
Bioinformatic analysis of Pax6-null mini-time series microarray data. (A) Overlap of differentially expressed genes in each time point of Pax6-null mini-time series microarray data. Wildtype vs Pax6-null comparison was performed at each time point. The number of differentially regulated genes was 9 at E13.5, 23 at E15.5 and 344 at E18.5 at q<0.05 and 1.2 fold change cut-offs. The E15.5 showed the highest overlap with the other two time points (87%). (B) Power analysis of one-way ANOVA. For each transcript, one way ANOVA model (either age or type) was fitted. Between and within variance components were estimated and then used to calculate the power. The x-axis is the transcripts and the y-axis is the sorted power for age (left side) and sorted power for genotype (right side). The analysis demonstrates that there is more power to detect age than type. (C) The Pax6-null mini-time series microarray data were subjected to ANOVA with a two-way model using two factors, embryonic age and genotype. Based on the best fits to the model, we classified genes into three major groups (FDR<0.05): the interaction effect (Genotype*Age, 18 transcripts), genotype effect (118 transcripts) and age only effect (7,917 transcripts). The age only group significantly outnumbered other groups. These results suggest Pax6 regulated genes are a small subset of the whole cerebellar developmental program. (D) The set of Pax6-regulated genes was compared to the reference Affymetrix U74Av2 set with DAVID tools to identify any functionally or biologically over-represented GO categories in the set. The enriched GO categories (p<0.001) are listed with labeling of number of genes in the category. x-axis: enrichment fold.
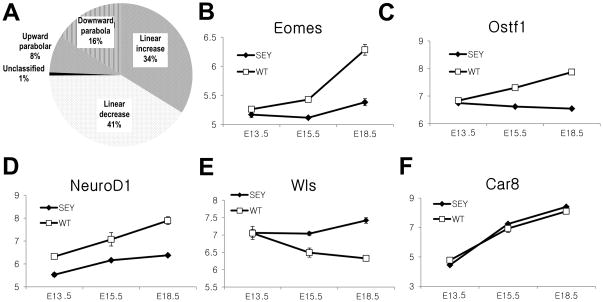
The microarray data were further subjected to general linear modeling using ANOVA to fit a two-way (3 × 2) model using 2 factors: embryonic age (E13.5, E15.5, and E18.5), genotype (Pax6-null vs wildtype) and their interaction. Based on the results of the fits, we classified the genes into three major groups (Fig 2C): Set 1 --18 transcripts with a genotype-specific temporal pattern (Genotype*Age FDR<0.05 and fold change > 1.2, Fig 3B and 3C), Set 2 --118 transcripts with genotype effects (Genotype*Age FDR>0.05, Genotype FDR<0.05, and fold change > 1.2, Fig 3D and 3E), and Set 3 -- 7,917 transcripts with temporal variation only (Genotype*Age FDR>0.05, Genotype FDR>0.05, and Age FDR<0.05) (Fig 3F). The power analysis of one way ANOVA revealed that the developmental effects on gene expression were of much greater magnitude and abundance than the effect of the mutation (Fig 2B, 136 transcripts in Set 1 and 2 vs. 7,917 transcripts in Set 3). Based on the post-hoc contrast analysis we classified each transcript by fitting linear contrast patterns (linear increase (↗↗) or decrease (↘↘)) or polynomial (upward (↗↘) or downward parabola (↘↗)) and selected the pattern with the smallest p-value (Fig 3A). We found that linear patterns significantly outnumber parabolic patterns suggesting the majority of transcripts have a linear mode of gene regulation.
Fig 3.
Dynamic expression of Pax6-regulated transcripts in the developing cerebellum. (A) Polynomial contrast analysis. The transcripts were classified to either fitting linear (linear increase (↗↗) or decrease(↘↘)) or polynomial (upward (↗↘) or downward parabola (↘↗)) contrast patterns based on the polynomial contrast analysis. Linear patterns significantly outnumber parabola patterns indicating that the majority of transcripts have a linear mode of gene regulation. (B and C) Eomes and Ostf1 exemplifies Set 1 transcripts with a mutation-specific temporal pattern (Age*Genotype q<0.05). (D and E) Neurod1 and Wls are examples of Set 2 transcripts with mutation effects only (Age*Genotype q>0.05 and Genotype q<0.05). (F) Car8 (carbonic anhydrase 8) is an example of Set 3 transcripts with temporal variation only (Age q<0.05, Age*Genotype q>0.05 and Genotype q>0.05).
Functional annotation of Pax6 differentially expressed genes (136 transcripts in Set 1 and 2) with Gene Ontology revealed a significant enrichment for biological processes related to nervous system development (Fig 2D). Notable examples for enriched GO biological process terms with p-value < 0.001 include axon guidance (fold enrichment- 8.93, p = 1.31E-06), neuron migration (7.67, p = 9.90E-04), cell morphogenesis (4.77, p = 2.18E-06), neuron development (4.18, p = 1.14E-04) and neuron differentiation (3.35, p = 4.17E-04). GO enrichment analysis affirms the important role of Pax6 as a regulator of neuronal development in the cerebellum and the identified genes in Sets 1 and 2 (ie, those affected by mutant genotype) are candidates for Pax6 regulation. The data and statistical analyses can be viewed in detail online (http://www.cbgrits.org/ and ftp://cbgrits.org/pub/supplemental/HA2012EJN/). The CbGRiTS site is a publicly accessible relational database that provides for the mining of these data and a growing number of expression profiles from wildtype and mutant cerebella at various developmental times. Lists of differentially expressed transcripts by pattern are also stored in Gene Weaver (Baker et al., 2009; Baker et al., 2012) where they may be compared directly to user submitted and other published gene sets.
Math1 microarray data as a means to filter the Pax6 gene list to identify genes involved in cerebellar glutamatergic neuron development
Because we used the whole cerebellum for the microarray experiments, the differentially regulated genes included all the genes that are influenced by Pax6 in all cell types of the cerebellum. Therefore, we made use of the differentially regulated genes from Math1 mutant mice as a filter to enrich the Pax6 data for the genes relevant for the development of rhombic lip lineage neurons such as cerebellar nuclear neurons and granule cells (Fig 1).
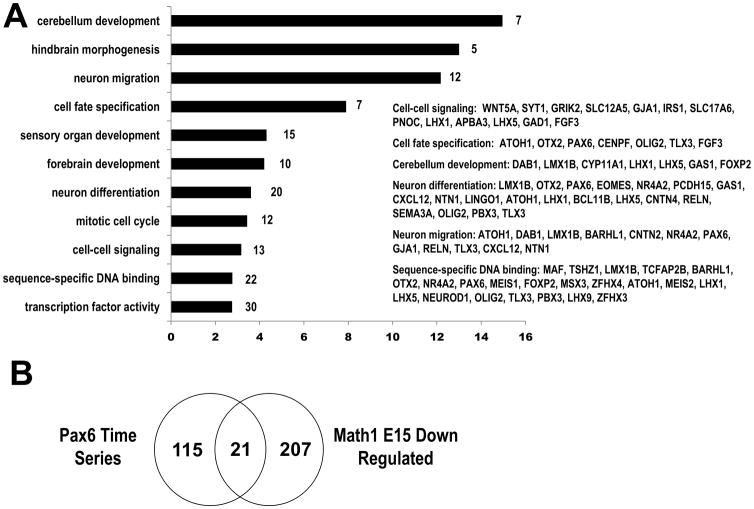
We generated gene expression profiles for wildtype and Math1 mutant cerebellum at E15.5 using the Affymetrix GeneChip Mouse Expression Array 430AB platform for four biological replicates for each genotype. The Significance Analysis of Microarrays identified 307 transcripts (430A- 148 down and 76 up, 430B- 80 down and 3 up) differentially regulated at the q<0.05 level of FDR and 1.2 fold change cut-off in the Math1 mutant cerebellar tissue. Functional annotation of 307 Math1 differentially expressed genes with Gene Ontology revealed a significant enrichment for biological processes related to nervous system development and transcriptional regulation (Fig 4A). Notable enriched GO terms include cerebellum development (fold enrichment- 14.96, p = 5.30E-06), neuron migration (12.17, p =2.62E-09), cell fate specification (7.90, p = 2.29E-04), cell-cell signaling (3.17, p = 7.67E-04) and sequence-specific DNA binding (2.78, p = 3.47E-05). GO enrichment analysis shows the important roles of Math1 in transcriptional control and regulation of cerebellar development. The down-regulated genes from Math1-null expression profiling were compared to the Pax6-null mini-time series set and the intersection resulted in a discrete set of 21 genes, which, theoretically, would reflect genes more specifically involved in the development of rhombic lip derivatives (Table 1, Fig 4B).
Fig 4.
GO enrichment analysis of Math1 differentially expressed genes. (A) The differentially expressed genes of Math1-null cerebellum were compared to the reference Affymetrix 430AB set with DAVID tools to identify any functionally or biologically over-represented GO categories in the set. The enriched GO categories (p<0.001) are listed with labeling of number of genes in the category. x-axis: enrichment fold. (B) The intersection between the Pax6-null time series and Math1-null down-regulated microarray dataset produced 21 genes common between the two dataset. The 21 genes represent converging set of genes that are under the downstream of the transcription factor Pax6 and Math1 and their effect on cerebellar development.
Table 1.
Genes differentially expressed in the Pax6-null and down-regulated in the Math1-null cerebellum.
| Probe Set ID | Gene Symbol | Set | Pax6 ANOVA FDR | E13 FC | E15 FC | E18 FC | Math1 E15.5 FC | Math1 FDR |
|---|---|---|---|---|---|---|---|---|
| 161028_at | Bmp6 | 2 | 0.00 | −1.02 | −1.17 | −1.39 | −1.30 | 0.02 |
| 160375_at | Car3 | 1 | 0.04 | 1.00 | 1.09 | 1.36 | −1.41 | 0.00 |
| 92932_at | Cbln1 | 1 | 0.00 | −1.08 | −1.33 | −1.91 | −1.77 | 0.00 |
| 98782_at | Cplx2 | 2 | 0.00 | −1.19 | −1.27 | −1.70 | −2.33 | 0.00 |
| 160511_at | Cxcl12 | 2 | 0.00 | −1.03 | −1.10 | −1.39 | −1.37 | 0.04 |
| 103532_at | Eomes | 1 | 0.02 | −1.11 | −1.35 | −1.75 | −1.93 | 0.00 |
| 98863_at | Grik2 | 2 | 0.03 | −1.03 | −1.15 | −1.40 | −1.21 | 0.05 |
| 95671_at | Hey1 | 2 | 0.00 | −1.05 | −1.19 | −1.33 | −1.23 | 0.00 |
| 103654_at | Hmgn5 | 2 | 0.03 | 1.22 | 1.16 | 1.30 | −1.22 | 0.05 |
| 100493_at | Hsd11b2 | 2 | 0.00 | 1.08 | 1.47 | 1.94 | −2.10 | 0.00 |
| 99010_at | Islr | 2 | 0.00 | −1.06 | 1.52 | 1.87 | −1.23 | 0.02 |
| 102569_at | Math1 | 2 | 0.02 | 1.04 | 1.28 | 1.39 | −2.54 | 0.05 |
| 161962_f_at | Mfap4 | 2 | 0.03 | −1.01 | −1.07 | −1.32 | −1.70 | 0.00 |
| 161050_at | Nav2 | 2 | 0.02 | −1.16 | −1.16 | −1.21 | −1.89 | 0.00 |
| 92717_at | Neurod1 | 2 | 0.00 | −1.77 | −2.28 | −2.87 | −2.33 | 0.00 |
| 97977_at | Ntn1 | 2 | 0.00 | 1.04 | 1.11 | 1.30 | −1.36 | 0.00 |
| 99158_at | Ostf1 | 1 | 0.00 | −1.07 | −1.59 | −2.55 | −1.68 | 0.00 |
| 97967_at | Plxnd1 | 1 | 0.00 | 1.03 | 1.15 | 1.60 | −1.27 | 0.00 |
| 96719_i_at | Pvalb | 1 | 0.01 | 1.03 | 1.32 | 1.53 | −1.56 | 0.00 |
| 96591_at | Reln | 2 | 0.00 | −1.08 | −1.94 | −1.65 | −2.08 | 0.00 |
| 93411_at | Sema7a | 2 | 0.04 | 1.00 | 1.13 | 1.54 | −1.34 | 0.01 |
“Probe ID” is the probe set ID number from the Affymetrix U74Av2 array platform. The “Set” refers to the groupings from general linear modeling analysis; 1 – Genotype*Age, 2- Genotype. The fold changes (FC) are represented in normalized raw intensity values of mutant expression compared to wildtype; negative values mean downregulation in the mutant while positive values indicate upregulation in the mutant cerebellum. FDR: False Discovery Rate
Validation of the microarray data with qRT-PCR
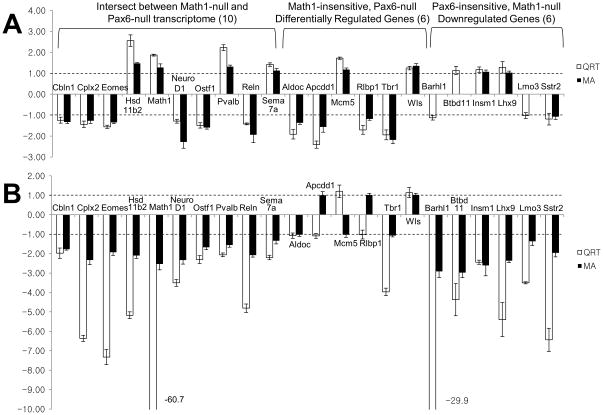
In order to validate the differential expression effects from microarray analyses, we performed quantitative real-time PCR (qRT-PCR) (Fig 5). First, we selected 10 genes from the intersect between Pax6-null mini-time series microarray data and Math1-null down-regulated gene lists (Fig 4B) and examined their relative expression level in cDNA samples from wildtype and Pax6-null and Math1-null cerebellum at E15.5 time point. The qRT-PCR analysis revealed that all genes tested were consistent with the array data (Fig 5). For qRT-PCR analysis of E15.5 wildtype vs Pax6-null cerebellum samples, the expression levels of 4 genes were increased 1.42 – 2.56 fold (Hsd11b2 (hydroxysteroid 11-beta dehydrogenase 2), Math1, Sema7a (semaphorin 7A), and Pvalb (parvalbumin)) and 6 decreased 1.24 – 1.56 fold (Cbln1 (cerebellin 1), Cplx2 (complexin 2), Eomes (eomesodermin homolog), Neurod1, Ostf1 (osteoclast stimulating factor 1), and Reln (reelin)), respectively (Fig5A). In the qRT-PCR analysis of E15.5 wildtype vs Math1-null cerebellum, all 10 genes showed a decrease, within the range of 1.98 – 60.71 folds in expression levels (note: these were selected from Math1 down-regulated gene set). Notably, qRT-PCR analysis on the expression level of Math1 transcript in Math1-null tissue revealed a decrease of 60.7 fold compared to the wildtype (Fig 5B).
Fig 5.
Validation of microarray data by qRT-PCR. Twenty-two genes from a total of 343 were selected for qRT-PCR analysis from the 3 groups in Fig 4B; 10 genes from the intersect between Math1-null and Pax6-null transcriptome, 6 genes that were differentially expressed in the Pax6-null mini-time series data, but not in Math1-null data, and 6 genes that were down-regulated in Math1-null data, but not in Pax6-null data. qRT-PCR analysis was performed on cDNAs derived from wildtype and Pax6-null cerebella at E15.5 (A) and wildtype and Math1-null at E15.5 (B). Values on the Y-axis represent the ratios of expression levels between the mutant and the wildtype with positive values corresponding to an up-regulation in the mutant and negative values to a down-regulation in the mutant. The white bars are the ratio from qRT-PCR and black bars are from the microarray data. The dotted line indicates no change in expression (±1 fold change). The Pax6-null microarray data for three genes, Barhl1, Btbd11 and Lmo3, are missing due to absence of the probes in the U74Av2 chip. While the ratio differences were more pronounced in qRT-PCR than in microarray in certain cases, all differentially expressed transcripts tested showed the same trend (up- or down-regulation) between microarray results and qRT-PCR except one discrepancy for Tbr1 between qRT-PCR and microarray in Math1-null expression data.
Then we selected 12 additional genes, 6 that were differentially expressed in the Pax6-null mini-time series data, but not in Math-null1 data (115 from Fig 4B) and 6 that were down-regulated in Math1-null, but not in Pax6-null data (207 from Fig 4B) (Fig 5). We examined the expression level of the 6 genes from the Pax6-null data in cDNA samples from E15.5 wildtype and Pax6-null cerebellum and found that qRT-PCR results were consistent with the array data (1.70 – 2.41 decrease and 1.25 – 1.72 fold increase, Fig 5A). However, when the qRT-PCR analysis was performed on E15.5 wildtype and Math1-null cerebellum for these 6 genes, there was no difference in expression levels between wildtype and Math1-null cerebellum except for Tbr1. The discrepancy between qRT-PCR and microarray data for Tbr1 could be due to very low signal intensity for Tbr1 probe in the Math1 microarray data. In contrast, the expression level of the 6 down-regulated genes from the Math1-null data determined by qRT-PCR showed down-regulation in cDNA samples from E15.5 wildtype and Math1-null cerebellum (2.44 – 29.9 fold decrease, Fig 5B), however, there was no difference in cDNA samples from E15.5 wildtype and Pax6-null cerebellum. Therefore, there was high concordance between the microarray and qRT-PCR data.
Expression analysis of identified genes in cerebellar tissue
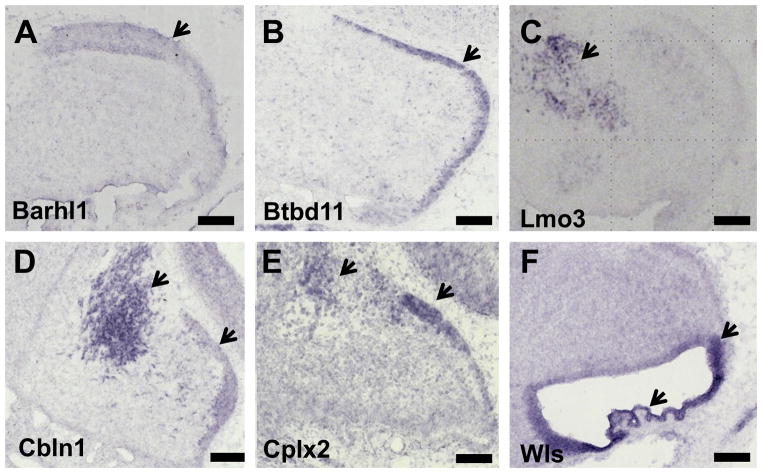
In order to examine our assumption that the down-regulated genes from Math1-null transcriptome will be preferentially expressed in the rhombic lip lineages, we performed ISH on 12 genes selected from the down-regulated genes from Math1-null transcriptome and examined their expression on cerebellar tissue at E15.5 (Fig 6). All 12 genes examined had specific expression either in the EGL (N=8, Fig 6A and B) or cerebellar nuclei (N=2, Fig 6C) or both (N=2, Fig 6D and E). The in situ expression analysis of these 12 selected genes from the Math1 down-regulated gene set supported the use of Math1-null expression data as a filter to obtain genes that were expressed in the rhombic lip lineage. Additional in situ analysis was performed for Wls, a mammalian homologue of dedicated Wnt secretion factor Evi/Wntless (Fig 6E). Wls was specifically expressed in the rhombic lip and choroid plexus at E15.5 in wildtype cerebellum, which is important for sustained activity of Wnt signaling in rhombic lip.
Fig 6.
In situ hybridization analysis of Math1 down-regulated genes and Wls in the E15.5 cerebellum. To test the hypothesis that Math1 down-regulated genes are expressed in rhombic lip derivatives such as EGL and the cerebellar nucleus, ISH was performed on E15.5 wildtype cerebellum for 12 selected genes from the Math1 down-regulated set. All 12 genes examined had specific expression either in the EGL [N=8: (A) Barhl1 (BarH-like 1) and (B) Btbd11(BTB domain containing 11)] or cerebellar nuclei [N=2 (C) Lmo3 (LIM domain only 3)] or both [N=2: (D) Cbln1and (E) Cplx2]. The in situ expression analysis of these 12 selected genes from Math1 down-regulated genes supports the use of the Math1-null expression data as a filter to obtain genes that are expressed in the rhombic lip lineage. (F) Additional in situ analysis was performed for Wls, which is specifically expressed in the rhombic lip and choroid plexus at E15.5 (arrow heads). Arrowheads point to the EGL and/or the cerebellar nuclei. Scale bar: 100 um.
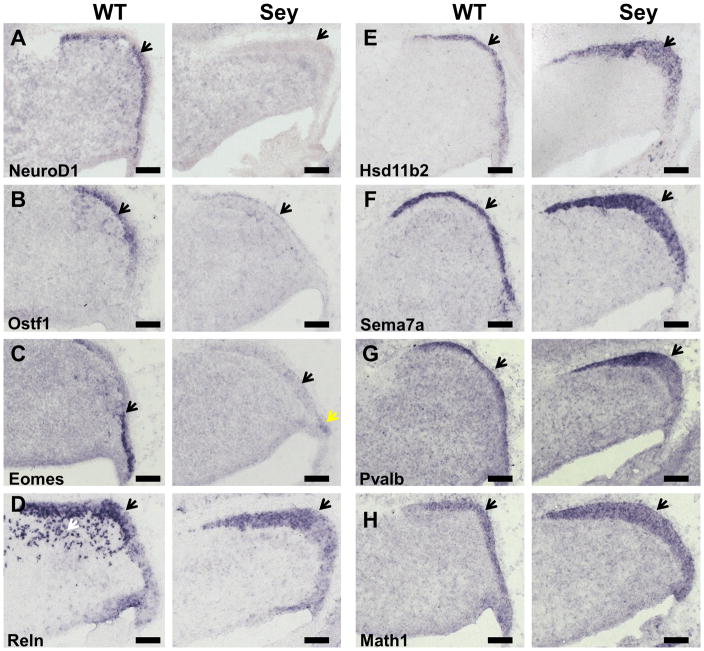
Our Pax6-null/Math1-null transcriptome analysis identified a set of 21 genes whose expression was disrupted upon Pax6 and Math1 mutation. These genes include several genes known to be involved in cerebellar development (Cbln1, Eomes, Neurod1, Reln) as well as novel genes (Cplx2, Ostf1 and Sema7a) that have not previously been implicated in cerebellar development. Interestingly, transcription factor binding site analysis showed that 9 out of the 21 genes including Cplx2, Eomes, Hsd11b2, Neurod1, Ostf1, Reln and Pvalb had one or more Pax6 binding sites. We selected 8 genes and studied the expression of these genes by ISH in wildtype and Pax6-null cerebellar tissue (Fig 7). Neurod1, Eomes, Ostf1 and Reln showed a markedly weaker signal in the EGL of Pax6-null cerebellar tissue, whereas Hsd11b2, Sema7a, Math1 and Pvalb showed a stronger signal in the mutant cerebellum. In case of Reln, the signals present in cerebellar anlage in wild type disappeared in Sey cerebellum and the EGL of Sey cerebellum was wider due to the loss of division and disorganization of EGL cells (Engelkamp 1999). These results are consistent with the microarray and qRT-PCR data and lend support for their involvement in the Pax6-null granule cellular phenotypes. The identified genes would also be candidates as direct targets of Pax6 transcription factor regulation and part of a gene regulatory network involved in the cerebellar development.
Fig 7.
In situ hybridization analysis of Pax6 differentially regulated genes in the wildtype (WT) and Pax6-null (Sey) E15 cerebellum. We performed in situ hybridization analysis on selected transcripts for cellular localization and the effect of Pax6 mutation on their expression. (A–D) Neurod1, Ostf1, Eomes, and Reln are down-regulated in the Pax6-null cerebellum and their expression in the Pax6-null EGL is significantly reduced. (A) Neurod1in situ signal is absent from EGL of Pax6-null cerebellum. (B) Ostf1 in situ signal is absent from the EGL of the Sey cerebellum. (C) A slight signal for Eomes remains in the rhombic lip of Sey cerebellum (yellow arrow). (D) The Reln signals present in cerebellar anlage in wild type (white arrows) are absent in Sey cerebellum and also the Reln signals in the EGL are less intense compared to the wild type cerebellum. (E–H) Hsd11b2 (E), Sema7a (F), Pvalb (G), Math1 (H) are up-regulated in the Pax6-null cerebellum and their in situ signals in the Pax6-null EGL is stronger compared to wildtype. Also note that the EGL of Sey cerebellum looks wider due to the loss of division and disorganization of cells in the EGL in the mutant cerebellum (Engelkamp 1999). All images are from E15.5 cerebellum and black arrow heads indicate EGL expression. Left panel – wildtype (WT). Right panel – Pax6Sey/Sey(Sey). Scale bar: 100 um.
Discussion
Identification of Pax6 regulated genes involved in cerebellar development
We utilized Affymetrix array platform to identify differentially regulated genes in the Pax6-null mutant cerebella at 3 different time points. Previous reports characterizing the mutant phenotype of Pax6-null cerebellum have identified E15.5 as the earliest time point when the SEY phenotype is apparent and presage the foliation defects and lack of fissurization seen at E18.5 (Engelkamp et al., 1999; Swanson et al., 2005). Therefore, we bracketed that time (examining E13.5, E15.5, and E18.5 cerebella) to inquire into the early molecular changes that would precede, coincide or proximally follow phenotypic changes due to loss of Pax6 function in the cerebellar granule cells. Indeed, when we made individual comparisons between wild-type vs Pax6 mutant at E13.5, E15.5 or E18.5 timepoints, the number of differentially regulated genes was the smallest at E13.5 (9), modest at E15.5 (23) and largest at E18.5 (344). These data corroborate the phenotypic data and indicate E15.5 as the time point when the molecular sequelae of the Pax6 mutation may first become most obvious in the mutant cerebellum; and are candidate genes most proximal to the cellular phenotypes that have been described (Engelkamp et al., 1999; Swanson et al., 2005). The use of Math1 dataset to identify genes involved in the development of rhombic lip derivatives was rewarded by finding a discrete set of 21 genes (Table 1). These 21 genes represent convergences between the transcription factor Pax6 and Math1 and their effect on cerebellar development. Further studies with a closer look at these genes are warranted from our findings.
While we employed the filtering approach by using the Math1-null transcriptome as a way of acquiring genes with rhombic lip derivative expression, alternative methods for obtaining cell-type specific transcriptome are available such as fluorescence-activated cell sorting (FACS) or laser capture microdissection (Cantor et al., 1975; Kamme et al., 2003). For example, by applying FACS, Glassman et al. (2009) obtained the transcriptome profile of inhibitory interneurons in cerebellum and Arlotta et al. (2005) acquired pure populations of corticospinal motor neurons for developmental stage specific transcriptome analysis. Kamme et al. (2003) and Tietjen et al., (2003) used laser capture microdissection methods for transcriptomic analyses of individual hippocampal neurons and neuronal progenitors. We are undertaking such approaches for the cerebellar granule cell progenitor population.
Discovery of novel signaling molecules in cerebellar development
Bioinformatic analysis using Gene Ontology annotation has identified several significantly over-represented GO categories related to neuronal development that reaffirm the crucial roles of Pax6 and Math1 in cerebellar development (Fig 2D and 4A). The 136 differentially regulated genes in the Pax6-null transcriptome would be the primary candidates for association with observed Pax6 mutant cerebellar phenotypes. In Math1-null transcriptome data, considering that the Math1-null cerebellum lacks glutamatergic rhombic lip lineage neuronal precursors such as granule cells and glutamatergic cerebellar nuclear neurons (Ben-Arie et al., 1997), the 228 down-regulated genes are likely to be important for the development of the rhombic lip neuronal lineage (Fig 6), whereas the 79 up-regulated genes are likely to be involved for the development of GABAergic neuroepithelium neuronal lineage (Hoshino et al., 2005). Recently, microarray analysis on Math1-lineage cells at E14.5 using a transgenic labeling strategy has been reported (Machold et al., 2011). Machold et al. labelled E10.5 and E13.5 Math1-lineage cells with tamoxifen, isolated these cells at E14.5 through FACS and identified genes enriched for early labeled extra-cerebellar and cerebellar nuclear cells and late born granule cell precursors. Using SAM, we identified 3,072 transcripts enriched in granule cell precursors compared to early born cells. We find that 64% (145 out of 228) of our Math1-null downregulated genes are common with the granule cell precursor-enriched transcripts. While these are two different approaches to transcriptome profiling, the large overlap between these studies provides co-validation and interesting comparative datasets.
The 21 genes that are common between Pax6- and Math1-null transcriptome include such well-characterized cerebellar development genes as Reln (D’Arcangelo et al., 1995), Neurod1 (Miyata et al., 1999), Eomes (Fink et al., 2006) and Cbln1(Hirai et al., 2005), and also novel genes that have not been implicated previously in cerebellar development or linked to Pax6, such as Ostf1 (Szymkiewicz et al., 2004) and Cplx2 (Glynn et al., 2003). Analysis of these genes could yield clues on the identity of molecules responsible for Pax6 mutant phenotypes. For example, one of the major phenotypes of the Pax6 mutation is the disorganisation and broadening of the EGL (Engelkamp 1999). This could be caused by the dysregulation of genes affecting neuronal development and differentiation in the EGL such as the downregulation of NeuroD1 and Eomes and/or upregulation of Math1 in Pax6-null cerebellum. The functional studies of these genes will advance our knowledge of the role of Pax6 and its transcriptional network in cerebellar granule cell development.
One of the most interesting genes on the Pax6-null transcriptome list is Wls (Banziger et al., 2006; Bartscherer et al., 2006). The expression of Wls, a mammalian homologue of the recently characterized dedicated Wnt secretion factor Evi/Wntless (Banziger et al., 2006; Bartscherer et al., 2006) does not decline over time in the Pax6-null cerebellum as it does in wildtype (Fig 3E). We find the expression of Wls in the developing cerebellum is localized to the RL (Fig 6F) where Wnt1 (wingless-type MMTV integration site family member 1) and Wnt3a are expressed (GenePaint). Considering the role of Wls as a dedicated Wnt secretion factor and the potency of Wnt-family members as morphogens (Logan & Nusse, 2004), the prolonged and upregulated expression of Wls could have a significant effect on the development of cerebellar granule cells. For example, the sustained presence of Wnt signals in the RL may prolong progenitor characteristics and inhibit and/or delay differentiation. Together with downregulation of NeuroD1 and Eomes and upregulation of Math1, these molecular changes are likely to set up the Pax6-null granule cell progenitors at E15.5 for the manifestation of SEY mutant phenotype in later embryonic and postnatal development. Thus, the identification of Pax6 downstream genes and affected signalling molecules could be the first step toward the identification of molecular accomplices for the observed mutant phenotypes.
Interestingly, Unc5c, the netrin receptor, which was shown to mediate the migration disruption of Pax6-null granule cells (Engelkamp et al., 1999) is absent from 136 genes from Pax6-null mini-time series ANOVA analysis. However, Unc5c is down-regulated 1.3 fold in wildtype vs Pax6-null comparison at E18.5 and there was no significant difference in expression of Unc5c between wildtype and Pax6-null cerebellum at E15.5 or E13.5. This kind of time series analysis may be able to provide information regarding the time line and/or sequences of molecular events stemming from the mutation of Pax6. Another notable observation was that there were some cases of different homologs belonging to the same protein family showing differential regulation either by Pax6 or Math1. For example, in wildtype vs Pax6-null comparison at E18.5, Sema6a was down-regulated, whereas Sema7a was up-regulated. In the Math1-null transcriptome analysis, Lhx9 (LIM homeobox protein 9) was down-regulated, whereas Lhx1 and Lhx5 were up-regulated. Other examples include homologs of Zic (zinc finger protein of the cerebellum), Plxn (plexin) and Grik (ionotropic kainate glutamate receptor). In these cases, the different homologs are likely to be either expressed in different cell-types or may have opposing roles in the same cell.
The Pax6 downstream genes in brain development
Several transcriptome profiling studies have been carried out to identify downstream genes of Pax6 on the eyes and cortex from Pax6 mutant and overexpressing mice (Chauhan et al., 2002a; Chauhan et al., 2002b; Holm et al., 2007; Sansom et al., 2009). The present study is the first transcriptome analysis on the effect of loss of Pax6 function in cerebellar development and provides a comparative dataset to examine the downstream effecter genes of Pax6 in the eye and brain. Holm et al. identified a set of Pax6-downstream genes in developing cortex at E12 and E15 and compared it to the genes regulated by Pax6 in the development of the heterozygous mutant eye and found little to no overlap between the two datasets (Holm et al., 2007). We also find that Pax6-dependent genes in cerebellar development have no overlap with the Pax6-dependent genes reported in the development of the heterozygous mutant eye, but a notable overlap with the Pax6 cortex gene set such as Neurod1 and Eomes that were represented in the cortex microarray data from Pax6sey/sey and D6-Pax6 transgenic mice (Sansom et al., 2009). One possible explanation for the low overlap is that Pax6 may interact with different activator/repressor cofactors in different tissues and the transcriptional network involving Pax6 may be tissue- or cell-type specific. Thus, our list of Pax6 differentially regulated genes provides a valuable reference point to gauge the tissue and cell-type specific transcriptional network involving Pax6 in other regions of the developing brain.
CONCLUDING REMARKS
As more genomic information regarding transcriptional networks and their regulation accumulates, we will have a better understanding of gene regulatory networks responsible for development and how they are regulated as development progresses. The present study has identified novel genes with functional implications for cerebellar development and present comparative transcriptome data for tissue and cell-type specific regulation of transcriptional networks involving Pax6.
Supplementary Material
Acknowledgments
Supported by NIH grant R01 HD 52472. Authors thank Dr. Ramin Homayouni and Dr. Matt Larouche for their insightful discussions. We also thank Randy Glenn for his informatics support.
Footnotes
The authors have no conflict of interest to declare.
References
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ. GeneWeaver: a web-based system for integrative functional genomics. Nucleic acids research. 2012;40:D1067–1076. doi: 10.1093/nar/gkr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Philip VM, Zhang Y, Li Z, Kirova R, Langston MA, Chesler EJ. Ontological discovery environment: a system for integrating gene-phenotype associations. Genomics. 2009;94:377–387. doi: 10.1016/j.ygeno.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Cantor H, Simpson E, Sato VL, Fathman CG, Herzenberg LA. Characterization of subpopulations of T lymphocytes. I. Separation and functional studies of peripheral T-cells binding different amounts of fluorescent anti-Thy 1.2 (theta) antibody using a fluorescence-activated cell sorter (FACS) Cell Immunol. 1975;15:180–196. doi: 10.1016/0008-8749(75)90174-4. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Yang Y, Cermak L, Reneker L, Duncan MK, Cvekl A. A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens. Genes Cells. 2002a;7:1267–1283. doi: 10.1046/j.1365-2443.2002.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW, Cvekl A. Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. The Journal of biological chemistry. 2002b;277:11539–11548. doi: 10.1074/jbc.M110531200. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development (Cambridge, England) 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development (Cambridge, England) 2002;129:455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics (Oxford, England) 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Glynn D, Bortnick RA, Morton AJ. Complexin II is essential for normal neurological function in mice. Human molecular genetics. 2003;12:2431–2448. doi: 10.1093/hmg/ddg249. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends in neurosciences. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Hargett LK, Hill RE, Ross A, Hogan BL. Disruption of PAX6 function in mice homozygous for the Pax6Sey-1Neu mutation produces abnormalities in the early development and regionalization of the diencephalon. Mechanisms of development. 1997;64:111–126. doi: 10.1016/s0925-4773(97)00055-5. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science (New York, N Y. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annual review of neuroscience. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nature neuroscience. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nature neuroscience. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic acids research. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Molecular and cellular neurosciences. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wood CL, Liu Y, Getchell TV, Getchell ML, Stromberg AJ. Identification of gene expression patterns using planned linear contrasts. BMC bioinformatics. 2006;7:245. doi: 10.1186/1471-2105-7-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Machold R, Klein C, Fishell G. Genes expressed in Atoh1 neuronal lineages arising from the r1/isthmus rhombic lip. Gene Expr Patterns. 2011;11:349–359. doi: 10.1016/j.gep.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes & development. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Developmental biology. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS genetics. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DJ, Goldowitz D. Experimental Sey mouse chimeras reveal the developmental deficiencies of Pax6-null granule cells in the postnatal cerebellum. Developmental biology. 2010;351:1–12. doi: 10.1016/j.ydbio.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Swanson DJ, Tong Y, Goldowitz D. Disruption of cerebellar granule cell development in the Pax6 mutant, Sey mouse. Brain research. 2005;160:176–193. doi: 10.1016/j.devbrainres.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Szymkiewicz I, Destaing O, Jurdic P, Dikic I. SH3P2 in complex with Cbl and Src. FEBS letters. 2004;565:33–38. doi: 10.1016/j.febslet.2004.03.100. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development (Cambridge, England) 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development (Cambridge, England) 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.