Abstract
Effector-triggered immunity (ETI) was originally termed gene-for-gene resistance and dates back to fundamental observations of flax resistance to rust fungi by Harold Henry Flor in the 1940s. Since then, genetic and biochemical approaches have defined our current understanding of how plant “resistance” proteins recognize microbial effectors. More recently, proteomic approaches have expanded our view of the protein landscape during ETI and contributed significant advances to our mechanistic understanding of ETI signaling. Here we provide an overview of proteomic techniques that have been used to study plant ETI including both global and targeted approaches. We discuss the challenges associated with ETI proteomics and highlight specific examples from the literature, which demonstrate how proteomics is advancing the ETI research field.
Keywords: Effector-Triggered Immunity, Proteomics, Type III effector, NB-LRR proteins, Pseudomonas syringae
Introduction
The plant immune system can be triggered by the recognition of two broad classes of microbial molecules. PRR-triggered immunity (PTI) is activated following the recognition of conserved microbe-associated molecular patterns (MAMPs) by cell surface plant pattern recognition receptors (PRRs), while effector-triggered immunity (ETI) is activated by recognition of pathogen effector proteins by intracellular nucleotide binding, leucine-rich repeat (NB-LRR) proteins. Both PTI and ETI result in similar immune responses, although the amplitude of the ETI-induced response is often substantially higher and frequently associated with a localized programmed cell death (PCD) response around the site of infection called the hypersensitive response (HR). The recognition of effectors by NB-LRR proteins can be direct where the NB-LRR protein directly binds the effector to trigger ETI, or indirect where the NB-LRRs interact with the host target of pathogen effectors and monitor them for perturbations.1,2 An exception to the effector/NB-LRR paradigm for ETI activation is the recognition of transcription activator-like (TAL) effectors which can transcriptionally activate expression of non NB-LRR genes to activate ETI, but proteomic analyses have yet to be conducted on these TAL ETI “executors.”3
Genetic approaches have been successful at identifying numerous genes required for ETI and have largely built our current understanding of the ETI network. More recently, proteomic analyses of ETI have emerged as a powerful complement to genetic methods by expanding the repertoire of proteins and PTMs responsible for ETI signaling, identifying protein complexes that contain novel ETI signaling components, and providing an overview of the molecular events responsible for manifesting the cellular responses associated with ETI (Table 1 and 2). Additionally, proteomic approaches have provided important insights into the physiology of the ETI response, such as alterations in photosynthesis, lipid metabolism and redox potential. The co-regulation of antagonistic aspects of these physiological responses reveals that they are tightly regulated during ETI.
Table 1. Summary of Global Proteomic Analyses of ETI-Regulated Proteins.
| T3SE | NB-LRR-protein | Cellular Fraction/PTM enrichment |
Proteomics Approach | Protein IDsa | Reference | |
|---|---|---|---|---|---|---|
| AvrRpm1 | RPM1 | Chloroplast,Mitochondria,Cytosol | 2D-PAGE | GSTF8 (At2g47730) GST79 (At2g30860) OEC33 (At5g66570) OEC23 (At1g06680) |
17 , 18 | |
| AvrRpm1 | RPM1 | Secreted Proteins | SDS-PAGE iTRAQ LC-MS/MS |
Ubiquitin GAPDH(At1g13440/At3g04120) Phosphoglucomutase (At1g23190) Enolase (At2g36530) |
19 | |
| AvrPto/HopAB2 | Prf | All soluble proteins | iTRAQ 2D-LC-MS/MS |
14–3-3 protein (TC217464_3, TC217870_1, TC233482_2) Acyl-binding carrier protein (TC238115_2) Acyl-carrier protein transacylase (TC219533_3) |
20 | |
| GVG:AvrRpm1 | RPM1 | Microsome, Cytosol | 2D-PAGE MALDI-TOF |
AtRem1.2 (At3g61260) PP2C PIA1 (At2g20630) C2-domain containing protein (At4g34150) |
44 , 45 | |
| GVG:AvrRpt2 | RPS2 | Plasma Membrane | SDS-PAGE LC-MS/MS Label free quantitation |
SOBIR1 (AT2G31880) RIPK (AT2G05940) PIA1 (At2g20630) SYP122 (AT3G52400) SOBER1(At4g22300) PLDγ1 (AT4G11850) DGK5 (AT2G20900) |
PEPR1 (AT1G73080) WAK1 (AT1G21250) BIK1 (AT2G39660) SNAP33 (AT5G61210) PEN1/SYP121 (AT3G11820) PLDα1 (AT3G15730) |
49 |
| AvrRpm1 | RPM1 | Phosphorylated Proteins | iTRAQ LC-MS/MS |
Rubsico large subunit (AtCg00490) | 60 | |
| AvrB | RPM1 | S-nitrosylated cysteine | Biotin-Switch SDS-PAGE MALDI-TOF/TOF | Rubsico large subunit (AtCg00490) GAPDH (At3g26650) Triosephosphate isomerase (At3g55440) PsbQ (At4g21280) |
65 | |
| AvrB | RPM1 | Tyrosine nitration | Immunoaffinity enrichment 2D-PAGE LC-MS/MS |
Rubsico large subunit (AtCg00490) PsbO2 (At3g50820) PsbO1 (At5g66570) Fructose-bisphosphate aldolase 1 (At2g39730) Fructose-bisphosphate aldolase 2 (At4g38970) |
66 | |
Note: aTomato gene indices as annotated by DFCI – LeGI
Table 2. Summary of Targeted Proteomic Analyses of ETI Protein Complexes.
| Target | Tag | System | Purification Strategya | Proteins Identified | Reference | |
|---|---|---|---|---|---|---|
| HopN1 (T3SE) | 6xHIS | N. benthamiana | In vitro pulldown, Ni2+-IMAC | PsbQ | 72 | |
| RPS2 (NB-LRR-protein) | HA-PreScission-Biotin (HBP) | A. thaliana | Biotin/Streptavidin AP | RIN4 (At3g25070) AtHIR1 (At1g69840) AtHIR2 (At3g01290) |
BSK1 (At4g35230) BSK8 (At5g41260) |
73 |
| Prf (NB-LRR-protein) | SBP-FLAG | Tomato | TAP, Streptavidin AP/FLAG IAP | Pto Fen |
Pth2 Pth3 |
80 |
| Pto (Monitored by Prf) | FLAG | Tomato | FLAG IAP | Prf Fen |
Pth3 Pth 5 |
|
| RIN4 (Monitored by RPM1/RPS2) | A. thaliana | Native IAP | RIPK (AT2G05940) | 50 | ||
| MOS4 (mediates NB-LRR protein signaling) | HA | A. thaliana | HA IAP | AtCDC5 (At1g09770) PRL1 (At4g15900) |
MAC3A (At1g04510) MAC3B (At2G33340) |
86 |
Note: aAP: affinity purification, IAP: immunoaffinity purification
The technical progress made in analyzing plant proteomes,4 general proteomic strategies, theory and instrumentation have been reviewed elsewhere and we will focus on proteomic approaches used to study ETI induced by type III secreted effectors (T3SEs) of Pseudomonas syringae.5,6 We use the term “global” to refer to studies aiming to identify and quantify proteins from a total cellular extract, whereas “targeted” approaches identify the components of specific protein complexes. This review highlights the contributions that both approaches have made to our understanding of the plant ETI response (Table 1 and Table 2).
Proteomics of ETI
Despite significant technical advances, plant proteomics remains challenging. Plant cells generally have low cytoplasmic volume relative to cell wall mass, with high protease and phosphatase content.7 As a result, careful consideration must be paid to the isolation of proteins from plant tissues to preserve both their integrity and PTMs. Further, the predominance of ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco), which catalyzes the first major step of carbon fixation and is generally considered the most abundant protein on earth, makes it difficult to concentrate plant protein extracts, and necessitates a large dynamic range for protein separation, detection and identification.4
An additional challenge in the proteomic study of ETI simply involves the induction of the ETI response. Many signaling events in plants occur downstream of perception of small molecules or peptides (i.e., elicitors) by their cognate receptors. These systems can generally be readily induced under experimental conditions by the application of elicitors such as the bacterial flagellin peptide flg22, which is a potent activator of PTI,8-11 or the elicitation of phytohormone signaling by the application of exogenous hormones.12,13 In contrast, receptors mediating ETI are intracellular and generally detect the activity of proteinaceous effectors translocated from pathogens into the host cytoplasm.14 Therefore, ETI is difficult to induce experimentally, and requires either the delivery of effectors by pathogens or their transgenic expression.
While pathogen delivery of effectors represents the most biologically and physiologically relevant stimulus, these interactions involve a broad range of host response, not all of which are directly associated with ETI. For example, P. syringae strains can inject dozens of T3SE,15,16 which can alter host physiology and inhibit immune responses, including ETI. Consequently, it can be difficult to deconvolute ETI signaling from other T3SE functions. Consequently, three experimental treatments are typically compared with identify proteins regulated by ETI signaling,17-19 including plants inoculated with: (1) P. syringae (with its endogenous complement of T3SEs); (2) P. syringae + an exogenous T3SE that triggers ETI; and (3) P. syringae lacking a functional type III secretion system that is unable to secrete any T3SEs. Further, since ETI often culminates in PCD, its proteomic study requires careful coordination of inoculation and tissue collection to ensure a consistent stage of ETI in all tissues and limited protein loss from PCD. Despite these complications, bacterial delivery of T3SE is usually easily implemented in crop species20 and cell culture,19 and therefore represents a viable strategy for translational work in non-model plant systems.
ETI can also be induced by transgenic expression of T3SE in planta. Since ETI culminates in PCD, T3SE expression is typically controlled by an inducible promoter. A popular system for transgenic delivery of effectors is the dexamethasone (Dex) inducible GAL4 / VP16 / glucocorticoid receptor domain (GVG) system,21 which provides a transcription factor that drives expression of the transgene in a corticosteroid sensitive manner. While the GVG system produces substantially higher levels of the transgenic T3SE than would be expected under natural conditions, ETI induced by these constructs generally recapitulates phenotypes seen with bacterial T3SE delivery22,23 and has been successfully used to identify genes required for ETI.24 The GVG system allows simple and synchronized ETI induction by small molecule (Dex) application. Furthermore, because only one T3SE is induced, as opposed to the entire complement carried by a P. syringae strain, it simplifies experimental design and downstream analysis.
Global Analysis of Protein Abundance and PTMs during ETI
Pathogen Delivery of Effectors
In Arabidopsis thaliana (hereafter Arabidopsis), the membrane associated NB-LRR protein RPM1 mediates ETI triggered by the P. syringae T3SE AvrRpm1 (Fig. 1a).25,26 Jones et al.17,18 provided the first insight into proteins differentially regulated during RPM1-mediated ETI. Ultimately, 19 proteins were identified as upregulated in response to P. syringae pv tomato DC3000 (PtoDC3000) expressing AvrRpm1 (PtoDC3000(avrRpm1)), but not PtoDC3000 or PtoDC3000 lacking a functional TTSS (PtoDC3000(hrpA)), and therefore uniquely regulated by RPM1-mediated ETI signaling. In particular, proteins involved in redox regulation, lipid metabolism and photosynthesis were identified.
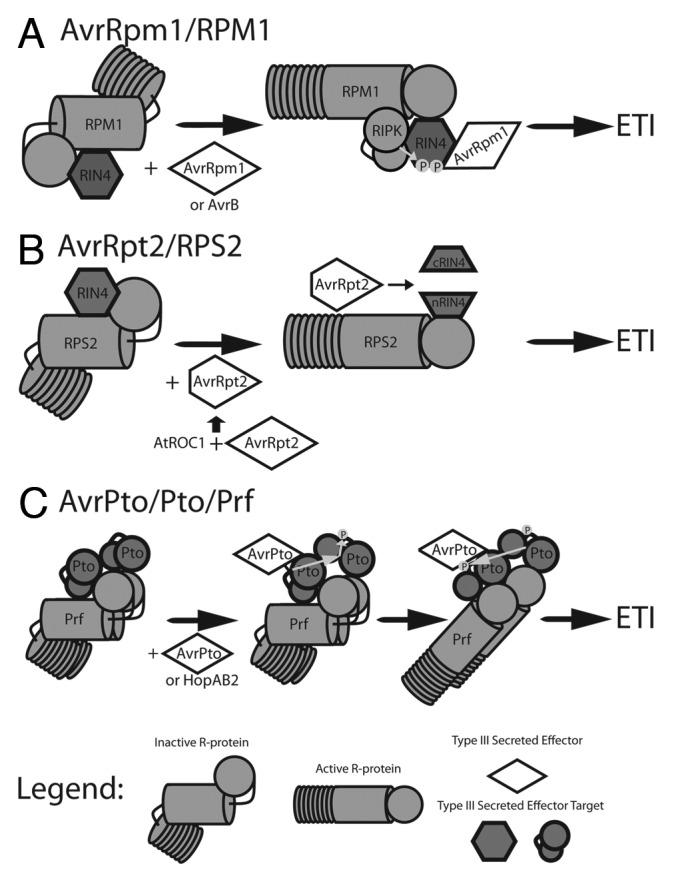
Figure 1. Models of T3SE recognition by NB-LRR proteins to trigger ETI. NB-LRR proteins monitor host proteins targeted by T3SE for modification. Detection of modified host proteins causes NB-LRR conformational change, initiating ETI signaling.14 (A) The NB-LRR RPM1 monitors the host protein RIN4.25,26 Interaction of RIN4 with the T3SE AvrRpm1 or AvrB triggers RIN4 phosphorylation by the host kinase RIPK, activating RPM1.50,83 (B) RIN4 is also monitored by the NB-LRR RPS2. The T3SE AvrRpt2 is activated by the host cyclophillin AtROC1, and causes proteolytic cleavage of RIN4 resulting in activation of RPS2.48 The RIN4 complexes in A and B are membrane associated. (C) Oligomeric complexes of Pto family host kinases80 and the NB-LRR Prf recognize the T3SEs AvrPto35,36 and HopAB2.37 Interaction of AvrPto or HopAB2 with a Pto monomer activates Pto kinase activity, causing transphosphorylation and activation of the second Pto monomer. This causes a second transphosrphorylation event resulting in phosphorylation of both Pto kinases and activation of Prf.81
The glutathione-s-transferases (GST) GSTF8 and GSTF9 were AvrRpm1-upregulated while the NADPH quinone reductase (NQR) and 2 cysteine peroxiredoxin PRxB were downregulated. The concomitant up and downregulation of radical detoxifying enzymes suggests a tight regulation of redox status during ETI. The downregulation of NO turnover proteins such as peroxiredoxin is likely associated with the rapid build-up of NO preceding HR.27 The accumulation of ROS during ETI can directly oxidize fatty acids leading to lipid peroxidation, which may be detoxified by NQRs.25,28 The downregulation of NQRs suggests that the accumulation of peroxidated lipids may play a role in ETI signaling.
Proteins involved in photosynthesis also respond to PtoDC3000(avrRpm1) providing a link between ETI and altered photosynthetic function. Two members of the oxygen evolving complex (OEC) of Photosystem II (PSII), OEC33 K protein and OEC23 K protein, accumulated in response to AvrRpm1 triggered ETI. Additionally, AvrRpm1 also induces the cytosolic accumulation of ferredoxin reductase which normally catalyzes the terminal photosynthetic electron transfer to NADP(+).18 Together, these suggest ETI might alter in photosynthetic electron transport around PSII. Consistent with altered regulation of photosynthesis during ETI, PtoDC3000(avrRpm1) causes a reduction in carbon fixation,29 altered photosynthetic electron transport,30,31 and over-activation of chlorophyll associated with ROS production and lipid peroxidation.31
In another study, Kaffarnik et al.19 characterized the secretome of Arabidopsis suspension cells in response to incubation with PtoDC3000, PtoDC3000(AvrRpm1), PtoDC3000(hrpA), and identifying 13 proteins specifically upregulated by RPM1 mediated ETI. Surprisingly, only three ETI induced secreted proteins have predicted signal peptides. Non-canonical secretion during ETI could result from loss of cell integrity, autophagy and/or membrane trafficking processes that contribute to ETI.32-34
In tomato, the P. syringae T3SEs AvrPto and HopAB2 (formerly AvrPtoB) carried by PtoDC3000 induce ETI mediated by the NB-LRR protein Prf and the host kinase Pto (Fig. 1C).35-37 The tomato cultivar Rio Grande (RG) consists of two homogenic genotypes: RG-PtoR, which expresses Pto and recognizes PtoDC3000, and RG-prf3, which lacks Prf and is susceptible to PtoDC3000. Parker et al.20 performed a proteomic analysis of Prf mediated ETI by inoculating the tomato cultivars RG-PtoR and RG-prf3 with PtoDC3000. Ultimately, 550 proteins were identified as uniquely regulated by Prf mediated ETI signaling. Many proteins regulated by Prf were consistent with similar experiments in Arabidopsis challenged with PtoDC3000(AvrRpm1), including proteins involved in photosynthesis, redox regulation and lipid metabolism.18,20 Similarities in the proteomes induced by AvrRpm1/RPM1 in Arabidopsis and AvrPto/HopAB/Prf in tomato strongly imply conserved early signaling events and proteomic changes during the ETI response.
This study also identified three 14–3-3 proteins that were upregulated by Prf mediated ETI.20 14–3-3 proteins mediate protein-protein interactions in a phosphorylation dependent manner, strongly implying that early ETI signaling events are regulated by phosphorylation and protein-protein interactions mediated by 14–3-3 proteins. Further, 14–3-3 proteins are known to mediate Prf signaling.38 Interestingly, 14–3-3 proteins are emerging as targets of T3SE that inhibit both PTI39,40 and ETI suggesting a central role for 14–3-3 proteins in mediating plant immune signaling.41
Both studies in Arabidopsis18 and tomato20 were designed to complement previous transcriptome studies.42,43 Interestingly, both studies revealed that there was little correlation between protein and transcript abundance in early ETI signaling (4 h post inoculation). However, correlations were observed at later time points (24 h post inoculation) with increases in transcript and protein abundance for Pathogenesis-Related protein 1 (PR1), peroxidases and lipoxygenases.20 It is perhaps not surprising that early ETI signaling events are likely mediated by proteome changes including PTMs such as phosphorylation or S-nitrosylation, protein-protein interactions or by signaling events such as lipid peroxidation, redox, and ROS/reactive nitrogen species generation that precede transcriptional re-programming.
Transgenic Expression of Effectors
Proteins regulated by RPM1-mediated ETI were also investigated using the GVG system to express AvrRpm1.44,45 Total protein and proteins from microsomal preparations were used for proteomic analysis. Despite limited protein identification, the accumulation of OEC complex members, GSTs and PRXs, were observed, demonstrating that proteomic changes resulting from transgenic T3SE expression resemble those produced by bacterially delivered T3SE.17,18 Similar to previous studies, protein discovery was confounded by high concentrations of rubisco.18,44 In order to improve protein discovery, varying concentrations of polyethylene glycol (PEG) were used to selectively precipitate Rubisco from total protein and microsomal preparations. This approach yielded a greater proportion of putative signaling proteins including the remorin AtREM1.2, a C2 domain containing protein and the protein phosphatase 2C (PP2C) induced by AvrRpm1 (PIA1).
Follow-up studies of PIA1 have demonstrated that pia1 plants show increased resistance to PtoDC3000(avrRpm1).44 The RPM1 interacting protein 4 (RIN4) is phosphorylated in response to AvrRpm1 and is required for AvrRpm1 recognition by RPM1 (Fig. 1A).46 However, pia1 plants do not display altered RIN4 phosphorylation during RPM1 mediated ETI implying that the phosphorylation status of an unknown protein participates in regulating RPM1 mediated ETI. The TTSE AvrB also triggers ETI requiring RPM1 and RIN4.24 While PtoDC3000(avrRpm1) induces the accumulation of PIA1, PtoDC3000 expressing AvrB does not.45 Disrupting PIA1 also has no effect on PtoDC3000(avrB) growth in planta, indicating that multiple signaling pathways exist for RPM1-mediated ETI.
The NB-LRR protein RPS2 also interacts with RIN4 and monitors RIN4 for cleavage by the T3SE AvrRpt2 (Fig. 1B).47 Both RPS2 and RIN4 are peripheral membrane proteins (Fig. 1).46,48 Recently, Elmore et al. characterized the proteomic responses of the plasma membrane during ETI mediated by RPS2 using the GVG system to express AvrRpt2 in Arabidopsis.49 Ultimately, 235 proteins increasing in abundance and 188 decreasing in abundance were identified during RPS2 mediated ETI. This was the first proteomic study of sufficient depth in Arabidopsis to identify the finer details of ETI signaling. For example, protein kinases and phosphatases were overrepresented among proteins upregulated by RPS2 ETI. Kinases promoting ETI (e.g., RPM-1 induced protein kinase,50 and suppressor of BIR1) were upregulated alongside PP2Cs, including PIA1, hinting at shared regulation of ETI signaling by RPM1 and RPS2. Interestingly, the pattern recognition receptors PEPR151 and WAK152 were upregulated during ETI, as was the PTI co-receptor BIK1,53 highlighting the potential crosstalk between ETI and PTI.
Proteins upregulated by RPS2 during ETI strongly support the role of membrane dynamics during ETI. Proteins involved in membrane trafficking were upregulated in response to RPS2 signaling, including multiple SNARE complex members involved in vesicle fusion.32,54 The increased association of normally cytosolic glycolytic enzymes (pyruvate kinase, PEP carboxylase,55 GAPDH56) with purified plasma membranes would be consistent with autophagy and membrane trafficking turning over cytosolic enzymes during ETI. Phospholipases (PL) upregulated by RPS2 mediated ETI are likely involved in membrane turnover and signaling. Both PLCs and PLDs associated with the release of phosphatidic acid (PA) during ETI were upregulated,57 while a PLA2 known to decrease PA levels and inhibit ETI triggered by the Xanthomonas campestris T3SE AvrBsT58 was also upregulated. Therefore, antagonistic branches of phospholipase signaling are concomitantly upregulated by ETI. This is similar to upregulation of antagonistic enzymes in protein phosphorylation and ROS signaling pathways and highlights the tight regulation of molecular events during ETI.59
ETI Induced Post-Translation Modifications
Given the evidence of altered phosphorylation in response to ETI mediated by RPM1, Jones et al.60 examined the quantitative changes in the phosphoproteome of Arabidopsis infiltrated with PtoDC3000, PtoDC3000(avrRpm1), or PtoDC3000(hrpA) by phosphopeptide enrichment. Five proteins were identified as differentially regulated by PTI and one protein by ETI. The large subunit of rubisco showed increased phosphorylation in response to RPM1 mediated ETI, which may contribute to altered photosynthetic activity during ETI.29
A rapid buildup of NO, such as that observed during ETI, can modify cysteine and tyrosine residues by S-nitrosylation or nitration, respectively. Enzyme classes known to mediate ETI signaling can be regulated by these redox sensitive PTMs, including peroxidoxins61 and metacaspases.62 Further, turnover of S-nitrosylated glutathione compromises ETI63 and is essential for modulation of plant immune responses by salicylic acid.64 Given the potential role of differential S-nitrosylation or tyr-nitration during ETI signaling, Romera-Puertas et al.65 and Cecconi et al.66 investigated the NO redox modified subproteome during AvrB triggered ETI. Most proteins differentially modified by NO redox reactions belong to primary metabolism including components of the PSII OEC, the large subunit of rubisco and three glycolytic enzymes: triose-phosphate isomerase, phosphoglycerate kinase and GAPDH. The carboxylase activity of rubisco and turnover are modulated by S-nitrosylation67,68 and S-nitrosylation of GAPDH’s catalytic cysteine is known to inhibit enzyme activity, reducing glycolytic flux.69 Therefore, accumulation of NO and other ROS might act as a regulator of plant metabolism during ETI.
Targeted Proteomics of ETI Complexes
Proteomics of ETI protein complexes has also been an invaluable tool for the in planta study of ETI signaling (Table 2). One common strategy has been the fusion of affinity tags to T3SEs or genetically characterized components of ETI. Affinity chromatography is then used for purification of protein complexes associated with the tagged “bait,” followed by protein identification by mass spectrometry.70 This approach can identify proteins that participate in the same biological process as the “bait” and identify immune components that would be recalcitrant to genetic analyses due to redundancy or essentiality.
Proteomics of Effector complexes
The P. syringae T3SE HopN1 is a cysteine protease that inhibits cell death in tomato and Nicotiana tabacum71 and ROS production in Arabidopsis.72 Proteomic investigation of its host targets was performed by in vitro pulldown of HopN1. Recombinant HopN1-His6 was immobilized and incubated with tomato extract, ultimately identifying photosystem b Q, the water oxidizing complex of PSII, as an interacting protein.72 Recombinant HopN1 demonstrated proteolytic activity against PsbQ in thylakoid membranes, while chloroplasts isolated from leaves inoculated with bacteria expressing HopN1 had reduced PSII activity. Silencing of PsbQ in N. benthamiana inhibited ROS production and HR during ETI, suggesting that HopN1 inhibits PCD by decreasing PSII activity required for ETI signaling.72
Proteomics of NB-LRR protein complexes
RPS2
In order to identify RPS2 associated proteins, microsomal preparations of transgenic Arabidopsis expressing affinity tagged RPS2 were treated with a protein chemical crosslinker prior to affinity purification of RPS2 complexes and protein identification by mass spectrometry.73 Chemical cross-linking of protein complexes allowed for use of harsh detergent conditions to solubilize microsomal pellets, and has been beneficial in purification of transient protein complexes in yeast74 and Arabidopsis.75 While RIN4 was identified as a member of RPS2 complexes, RPM1 was absent. Interestingly, immunoaffinity purification of RIN4 in an independent study identified RPS2 but not RPM1, suggesting that either different pools of RIN4 interact with each NB-LRR-protein76 or that RPM1/RIN4 interactions are transitory based on RIN4 phosphorylation.50
Two members of the hypersensitive induced reaction gene family, AtHIR1 and AtHIR2, were also identified as RPS2 complex members. AtHIR proteins are oligomeric plasma membrane proteins that are associated with detergent resistant plasma membrane microdomains.77 These proteins are known to increase in abundance during RPS2 mediated ETI.50 Disruption of AtHIR2 or AtHIR3 compromises AvrRpt2 induced ETI, while overexpression of AtHIR1, AtHIR2 or AtHIR3 enhances ETI and reduces PtoDC3000(AvrRpt2) growth. This supports the role of AtHIR proteins in RPS2 signaling and suggests that RPS2 complexes may be organized in plasma membrane microdomains.
The RPS2 complex also includes kinases associated with brassinosteroid (BR) signaling, namely the BR-signaling kinases (BSK) 1 and 8.77 These kinases also interact with the PRR FLS277,78 supporting a functional link between ETI and PTI. Further confirmation of ETI/PTI receptor association was also provided by the co-immunoprecipitation of RPM1, RPS2 and RPS5 with FLS2.79 Altogether, this demonstrates that receptors from both branches of plant immunity can interact and are likely co-localized in large protein complexes organized into plasma membrane subdomains.
Prf /Pto
A targeted proteomic approach was also used to identify Prf-associated proteins using transgenic tomato expressing affinity tagged Prf.80 Multiple Pto kinase family members including Pto, Fen, Pth2, and Pth3 were identified as Prf complex members. Proteomic analysis of affinity tagged Pto also identified Prf, Fen, Pth5 and either Pth2 or Pth3 as members of Pto/Prf complexes.80 This implies that Prf forms heterocomplexes containing both Pto and Pth family members, that may allow Prf to recognize T3SEs that potentially target Pto-related kinases.
Characterization of Pto phosphorylation sites during Prf mediated ETI was accomplished through immunoaffinity purification of Pto transiently expressed with AvrPto and Prf in N. benthemiana.81 Double phosphorylation events on Ser198 and Thr199 within the kinase activation domain of Pto were identified as ETI specific. Mutation of either site to residues that do not accept phosphor-transfer had no effect on Prf signaling while a double mutant PtoS198A/T199A inhibits Prf signaling. Nevertheless, expression of a phosphomimetic PtoS198D/T199D does not trigger T3SE independent ETI, indicating that Pto phosphorylation alone is not sufficient for ETI signaling, rather, Prf mediated ETI requires a combination of Pto phosphorylation and T3SE interaction. Consistent with this, the kinase inactive, double phosphomimetic mutant PtoD164N/S198D/T199D can still recognize AvrPto, whereas the kinase inactive PtoD164N does not. Therefore, both Pto phosphorylation and T3SE interaction are required for Prf mediated ETI. Currently, T3SEs are thought to trigger Prf signaling by interacting and perturbing one Pto kinase, resulting in transphosphorylation of the perturbed kinase by the reciprocal Pto kinase of the Prf/Pto oligomeric complex. Critically, such a model provides a mechanism for signaling through Prf heterocomplexes containing Pto kinase family members lacking kinase activity.82 Theoretically, inactive family members Pth2–5 may act as a molecular “trap” or decoy for T3SEs in the Prf heterocomplex. Interaction of T3SEs and the inactive Pto family member may induce transphosphorylation by the active family members Pto or Fen to trigger Prf signaling.
Proteomics of NB-LRR associated proteins
RIN4
Targeted proteomics of RIN4 complexes have provided insight into the mechanism underlying ETI signaling. Complexes containing RIN4 were isolated from transgenic Arabidopsis expressing DEX inducible AvrRpm1, identifying the RPM1-induced protein kinase (RIPK) as a RIN4 interacting protein.50 Disruption of RIPK compromises ETI triggered by AvrB, and to a lesser extent AvrRpm1.50,83 RIPK phosphorylation sites on RIN4 were mapped by mass spectrometry to Thr21, Ser160 and T166. Transient co-expression of RPM1 and phosphomimetic RIN4T21D/S160D/T166D or RIN4T166D induces HR in N. benthamiana, while disruption of RIPK in Arabidopsis reduces RIN4 phosphorylation in response to AvrRpm1 and AvrB.50 These studies show that phosphorylation of RIN4 by RIPK at T166 positively regulates RPM1 mediated ETI.
MOS4
Arabidopsis plants with constitutively active forms of the NB-LRR protein SNC1 display enhanced disease resistance.84 In addition, constitutive SNC1 activity also results in smaller stature of Arabidopsis plants and constitutive expression of defense-related genes. Signaling components downstream of SNC1 have been identified by forward genetic screens for suppression of small stature and constitutive defense gene expression, termed Modifier of snc1 (MOS) genes. One SNC1 suppressor, MOS4, interacts with a three proteins by yeast-two-hybrid that form a MOS4-associated complex (MAC).85 The MAC protein AtCDC5 is a transcription factor homologous to yeast and human proteins involved in RNA processing and splicing. In order to determine if the MAC complex contains more than the three core members, proteomic analysis of transgenic Arabidopsis expressing affinity tagged MOS4 was used to identify MAC interacting proteins86 In addition to the three core proteins, another 22 proteins were identified as MAC members including 19 proteins that showed homology to members of the Nineteen Complex (NTC) in yeast and humans. This complex plays roles in spliceosome assembly, DNA repair and cell-cycle progression. Two MAC complex members, MAC3A and MAC3B, are homologous to the founding member of the NTC in yeast. Arabidopsismac3a/mac3b plants have compromised immunity to PtoDC3000 and PtoDC3000 expressing the ETI inducing T3SEs AvrRps4 or AvrPphB, implying a role for the MAC in both PTI and ETI. Further, mac3a/mac3b mutants suppress constitutive SNC1 activation, confirming its role downstream of SNC1 in mediating ETI.86 Currently, it is unclear if MAC mediates ETI through altered transcription or splicing. However, the role of MAC in ETI signaling suggests that RNA processing and/or splicing play crucial roles in ETI.
Conclusions
Our current molecular understanding of plant ETI has been shaped by genetic studies. More recently, proteomic approaches have expanded our knowledge of the players and mechanisms involved in ETI using both global and targeted proteomic approaches (Table 1 and Table 2). Despite these advances, the proteomics of ETI still faces important challenges. ETI complexes are often membrane associated and their study incurs the challenges faced by researchers studying membrane proteins, including extraction and maintenance of complex integrity during purification. It is unclear if complexes isolated from plasma membrane microdomains represent protein complexes maintained by protein-protein interactions or bridged together by common lipid properties maintained during extraction procedures. Although significant advances have been made in global analyses, abundant proteins still confound deep protein discovery in many studies. Consequently, global proteomic analyses have failed to identify components of ETI that have been uncovered using genetic approaches. These limitations are likely due to their low abundance, subcellular localization and/or their lack of changes in abundance during ETI signaling. Rather, global analyses to date have provided insight into the output of ETI including changes in photosynthesis, redox regulation and lipid metabolism rather than signaling mechanisms. On the other hand targeted proteomics approaches, including affinity purification of known immune components, have been more successful at advancing our knowledge of ETI signaling, but lack the breadth of responses revealed by global approaches.
Future advances in plant proteomics will require continued application of the most advanced mass spectrometry technologies and sample preparation techniques. This may involve the development of a repository of contaminant plant proteins identified using affinity purification-mass spectrometry analogous to the contaminant repository for affinity purification (the CRAPome) developed for humans and yeast.87 Despite current limitations, there is no doubt that proteomic approaches form a crucial component for future advances in the study of ETI signaling. These will include the characterization of more NB-LRR complexes, comparative proteomic analyses of ETI induced by different T3SE as well as the temporal and spatial dynamics of the ETI response. The proteomic age of ETI research should continue to increase in momentum as access to proteomic facilities, latest technologies and expertise continues to increase. Currently, the field is perfectly positioned to build on the foundation provided by classical genetics, and to complement advanced genomics approaches provided by next-generation sequencing technologies.
Acknowledgments
Proteomics and ETI work in the Desveaux and Guttman labs is supported by Natural Sciences and Engineering Research Council of Canada awards; a Canada Research Chair in Plant-Microbe Systems Biology (Desveaux D) or Comparative Genomics (Guttman DS); the Centre for the Analysis of Genome Evolution and Function.
Glossary
Abbreviations:
- ETI
Effector-triggered immunity, T3SE: Type III secreted effector, NB-LRR: nucleotide-binding and leucine-rich repeat
References
- 1.van der Biezen EA, Jones JDG. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:R226–7. doi: 10.1016/S0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 2.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–33. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal GK, Sarkar A, Righetti PG, Pedreschi R, Carpentier S, Wang T, Barkla BJ, Kohli A, Ndimba BK, Bykova NV, et al. A decade of plant proteomics and mass spectrometry: translation of technical advancements to food security and safety issues. Mass Spectrom Rev. 2013;32:335–65. doi: 10.1002/mas.21365. [DOI] [PubMed] [Google Scholar]
- 5.Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 6.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 7.Quirino BF, Candido ES, Campos PF, Franco OL, Krüger RH. Proteomic approaches to study plant-pathogen interactions. Phytochemistry. 2010;71:351–62. doi: 10.1016/j.phytochem.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Nühse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2:1234–43. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Nühse TS, Bottrill AR, Jones AME, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–40. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem. 2010;285:39140–9. doi: 10.1074/jbc.M110.160531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Hoehenwarter W, Weckwerth W. Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 2010;63:1–17. doi: 10.1111/j.1365-313X.2010.04218.x. [DOI] [PubMed] [Google Scholar]
- 13.Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci U S A. 2010;107:15986–91. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takken FLW, Tameling WIL. To nibble at plant resistance proteins. Science. 2009;324:744–6. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 15.Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–6. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 16.Chang JH, Urbach JM, Law TF, Arnold LW, Hu A, Gombar S, Grant SR, Ausubel FM, Dangl JL. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci U S A. 2005;102:2549–54. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones AME, Thomas V, Truman B, Lilley K, Mansfield J, Grant M. Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochemistry. 2004;65:1805–16. doi: 10.1016/j.phytochem.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Jones AME, Thomas V, Bennett MH, Mansfield J, Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006;142:1603–20. doi: 10.1104/pp.106.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaffarnik FA, Jones AM, Rathjen JP, Peck SC. Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics. 2009;8:145–56. doi: 10.1074/mcp.M800043-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Parker J, Koh J, Yoo M-J, Zhu N, Feole M, Yi S, Chen S. Quantitative proteomics of tomato defense against Pseudomonas syringae infection. Proteomics. 2013;13:1934–46. doi: 10.1002/pmic.201200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–12. doi: 10.1046/j.1365-313X.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 22.McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, Staskawicz BJ. Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 1998;14:247–57. doi: 10.1046/j.1365-313X.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 23.Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6:e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eitas TK, Nimchuk ZL, Dangl JL. Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci U S A. 2008;105:6475–80. doi: 10.1073/pnas.0802157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci U S A. 1998;95:15849–54. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–6. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 27.Groß F, Durner J, Gaupels F. Nitric oxide, antioxidants and prooxidants in plant defence responses. Front Plant Sci. 2013;4:419. doi: 10.3389/fpls.2013.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mano J, Torii Y, Hayashi S, Takimoto K, Matsui K, Nakamura K, Inzé D, Babiychuk E, Kushnir S, Asada K. The NADPH:quinone oxidoreductase P1-zeta-crystallin in Arabidopsis catalyzes the alpha,beta-hydrogenation of 2-alkenals: detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 2002;43:1445–55. doi: 10.1093/pcp/pcf187. [DOI] [PubMed] [Google Scholar]
- 29.Freeman BC, Beattie GA. Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol Plant Microbe Interact. 2009;22:857–67. doi: 10.1094/MPMI-22-7-0857. [DOI] [PubMed] [Google Scholar]
- 30.Alméras E, Stolz S, Vollenweider S, Reymond P, Mène-Saffrané L, Farmer EE. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003;34:205–16. doi: 10.1046/j.1365-313X.2003.01718.x. [DOI] [PubMed] [Google Scholar]
- 31.Mur LA, Aubry S, Mondhe M, Kingston-Smith A, Gallagher J, Timms-Taravella E, James C, Papp I, Hörtensteiner S, Thomas H, et al. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 2010;188:161–74. doi: 10.1111/j.1469-8137.2010.03377.x. [DOI] [PubMed] [Google Scholar]
- 32.Teh O-K, Hofius D. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J Exp Bot. 2014;65:1297–312. doi: 10.1093/jxb/ert441. [DOI] [PubMed] [Google Scholar]
- 33.Hackenberg T, Juul T, Auzina A, Gwizdz S, Malolepszy A, Van Der Kelen K, Dam S, Bressendorff S, Lorentzen A, Roepstorff P, et al. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell. 2013;25:4616–26. doi: 10.1105/tpc.113.117192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J, Chen W. The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. PLoS One. 2013;8:e73091. doi: 10.1371/journal.pone.0073091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol. 1992;174:1604–11. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell. 1994;6:511–20. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–98. doi: 10.1016/S0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 38.Oh C-S, Martin GB. Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J Biol Chem. 2011;286:14129–36. doi: 10.1074/jbc.M111.225086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozano-Durán R, Bourdais G, He SY, Robatzek S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014;202:259–69. doi: 10.1111/nph.12651. [DOI] [PubMed] [Google Scholar]
- 40.Taylor KW, Kim J-G, Su XB, Aakre CD, Roden JA, Adams CM, Mudgett MB. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 2012;8:e1002768. doi: 10.1371/journal.ppat.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teper D, Salomon D, Sunitha S, Kim J-G, Mudgett MB, Sessa G. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 2014;77:297–309. doi: 10.1111/tpj.12391. [DOI] [PubMed] [Google Scholar]
- 42.Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB. Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32:299–315. doi: 10.1046/j.1365-313X.2002.01424.x. [DOI] [PubMed] [Google Scholar]
- 43.Truman W, de Zabala MT, Grant M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 2006;46:14–33. doi: 10.1111/j.1365-313X.2006.02672.x. [DOI] [PubMed] [Google Scholar]
- 44.Widjaja I, Naumann K, Roth U, Wolf N, Mackey D, Dangl JL, Scheel D, Lee J. Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics. 2009;9:138–47. doi: 10.1002/pmic.200800293. [DOI] [PubMed] [Google Scholar]
- 45.Widjaja I, Lassowskat I, Bethke G, Eschen-Lippold L, Long H-H, Naumann K, Dangl JL, Scheel D, Lee J. A protein phosphatase 2C, responsive to the bacterial effector AvrRpm1 but not to the AvrB effector, regulates defense responses in Arabidopsis. Plant J. 2010;61:249–58. doi: 10.1111/j.1365-313X.2009.04047.x. [DOI] [PubMed] [Google Scholar]
- 46.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–54. doi: 10.1016/S0092-8674(02)00661-X. [DOI] [PubMed] [Google Scholar]
- 47.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–89. doi: 10.1016/S0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 48.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–77. doi: 10.1016/S0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 49.Elmore JM, Liu J, Smith B, Phinney B, Coaker G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics. 2012;11:014555. doi: 10.1074/mcp.M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Elmore JM, Lin Z-JD, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–46. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci U S A. 2006;103:10104–9. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci U S A. 2010;107:9452–7. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18:257–73. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Leary B, Park J, Plaxton WC. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J. 2011;436:15–34. doi: 10.1042/BJ20110078. [DOI] [PubMed] [Google Scholar]
- 56.Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell. 2012;24:2200–12. doi: 10.1105/tpc.111.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M. Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J. 2006;47:947–59. doi: 10.1111/j.1365-313X.2006.02844.x. [DOI] [PubMed] [Google Scholar]
- 58.Kirik A, Mudgett MB. SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci U S A. 2009;106:20532–7. doi: 10.1073/pnas.0903859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vellosillo T, Vicente J, Kulasekaran S, Hamberg M, Castresana C. Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol. 2010;154:444–8. doi: 10.1104/pp.110.161273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones AME, Bennett MH, Mansfield JW, Grant M. Analysis of the defence phosphoproteome of Arabidopsis thaliana using differential mass tagging. Proteomics. 2006;6:4155–65. doi: 10.1002/pmic.200500172. [DOI] [PubMed] [Google Scholar]
- 61.Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AME, Perazzolli M, Vandelle E, Dietz K-J, Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell. 2007;19:4120–30. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inzé D, Delledonne M, Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem. 2007;282:1352–8. doi: 10.1074/jbc.M608931200. [DOI] [PubMed] [Google Scholar]
- 63.Feechan A, Kwon E, Yun B-W, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci U S A. 2005;102:8054–9. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–6. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romero-Puertas MC, Campostrini N, Mattè A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–69. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- 66.Cecconi D, Orzetti S, Vandelle E, Rinalducci S, Zolla L, Delledonne M. Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis. 2009;30:2460–8. doi: 10.1002/elps.200800826. [DOI] [PubMed] [Google Scholar]
- 67.Abat JK, Mattoo AK, Deswal R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008;275:2862–72. doi: 10.1111/j.1742-4658.2008.06425.x. [DOI] [PubMed] [Google Scholar]
- 68.Abat JK, Deswal R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics. 2009;9:4368–80. doi: 10.1002/pmic.200800985. [DOI] [PubMed] [Google Scholar]
- 69.Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013;4:450. doi: 10.3389/fpls.2013.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunham WH, Mullin M, Gingras A-C. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–90. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 71.López-Solanilla E, Bronstein PA, Schneider AR, Collmer A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol Microbiol. 2004;54:353–65. doi: 10.1111/j.1365-2958.2004.04285.x. [DOI] [PubMed] [Google Scholar]
- 72.Rodríguez-Herva JJ, González-Melendi P, Cuartas-Lanza R, Antúnez-Lamas M, Río-Alvarez I, Li Z, López-Torrejón G, Díaz I, Del Pozo JC, Chakravarthy S, et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 2012;14:669–81. doi: 10.1111/j.1462-5822.2012.01749.x. [DOI] [PubMed] [Google Scholar]
- 73.Qi Y, Katagiri F. Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 2009;57:932–44. doi: 10.1111/j.1365-313X.2008.03736.x. [DOI] [PubMed] [Google Scholar]
- 74.Kaake RM, Wang X, Huang L. Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol Cell Proteomics. 2010;9:1650–65. doi: 10.1074/mcp.R110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–84. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qi Y, Tsuda K, Nguyen V, Wang X, Lin J, Murphy AS, Glazebrook J, Thordal-Christensen H, Katagiri F. Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J Biol Chem. 2011;286:31297–307. doi: 10.1074/jbc.M110.211615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25:1143–57. doi: 10.1105/tpc.112.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi Y, Tsuda K, Glazebrook J, Katagiri F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol. 2011;12:702–8. doi: 10.1111/j.1364-3703.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AME, Rathjen JP. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–18. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 81.Ntoukakis V, Balmuth AL, Mucyn TS, Gutierrez JR, Jones AME, Rathjen JP. The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog. 2013;9:e1003123. doi: 10.1371/journal.ppat.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang JH, Tai Y-S, Bernal AJ, Lavelle DT, Staskawicz BJ, Michelmore RW. Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol Plant Microbe Interact. 2002;15:281–91. doi: 10.1094/MPMI.2002.15.3.281. [DOI] [PubMed] [Google Scholar]
- 83.Chung E-H, da Cunha L, Wu A-J, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–36. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–46. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, Zhang Y, Li X. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21:1484–93. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Monaghan J, Xu F, Gao M, Zhao Q, Palma K, Long C, Chen S, Zhang Y, Li X. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 2009;5:e1000526. doi: 10.1371/journal.ppat.1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–6. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]


