Summary
Acid sensing ion channels (ASICs) detect extracellular protons produced during inflammation or ischemic injury and belong to the super family of degenerin/epithelial sodium channels. Here, we determine the cocrystal structure of chicken ASIC1a with MitTx, a pain-inducing toxin from the Texas coral snake, to define the structure of the open state of ASIC1a. In the MitTx-bound open state and in the previously determined low pH desensitized state, TM2 is a discontinuous α-helix in which the Gly-Ala-Ser selectivity filter adopts an extended, belt-like conformation, swapping the cytoplasmic one-third of TM2 with an adjacent subunit. Gly 443 residues of the selectivity filter provide a ring of 3 carbonyl oxygen atoms with a radius of ~3.6 Å, presenting an energetic barrier for hydrated ions. The ASIC1a-MitTx complex illuminates the mechanism of MitTx action, defines the structure of the selectivity filter of voltage-independent, sodium-selective ion channels and captures the open state of an ASIC.
Introduction
Six distinct ASIC subtypes, (1a, 1b, 2a, 2b, 3 and 4) are expressed throughout vertebrate central and peripheral nervous systems, where they assemble and function as homo- or heteromeric complexes exhibiting a broad range of kinetic, steady-state, pharmacological and ion selectivity properties (Deval et al., 2010; Gründer and Chen, 2010; Hesselager et al., 2004; Sherwood et al., 2012). ASICs detect extracellular protons (Krishtal and Pidoplichko, 1980) and are activated under injury conditions that are accompanied by local tissue acidosis, such as inflammation, muscle ischemia, and stroke (Wemmie et al., 2006; Xiong et al., 2004). ASIC1 and 3 are expressed by primary afferent sensory neurons (Alvarez de la Rosa et al., 2002; Molliver et al., 2005; Waldmann et al., 1997a), where they initiate acute or persistent pain signals upon transient or sustained drops in pH, such as in cardiac ischemia or chronic inflammatory syndromes (Deval et al., 2010; Krishtal and Pidoplichko, 1981; Lingueglia, 2007).
ASICs are sodium-selective, voltage-independent and amiloride-blockable ion channels that activate and desensitize on a millisecond to second time-scale (Gründer and Chen, 2010). Early studies on epithelial sodium channels (ENaCs), the namesake founder of the ASIC/ENaC/degenerin superfamily, defined their ionic selectivity (Palmer, 1982) and subsequent experiments on ENaCs and ASICs mapped a ‘(Gly/Ser)-X-Ser’ tripeptide in the second transmembrane segment (TM2) – deemed the ‘GAS’ motif – as central to ion selectivity (Carattino and Vecchia, 2012; Kellenberger et al., 2001; Kellenberger et al., 1999; Li et al., 2011a; Sheng et al., 2000; Sheng et al., 2001; Sheng et al., 2005; Snyder et al., 1999). Amino acids implicated in amiloride block are also located on TM2, several residues on the extracellular side of the ‘GAS’ motif (Kellenberger et al., 2003; Schild et al., 1997). Despite crystal structures of the chicken ASIC1a in a low pH desensitized state (Gonzales et al., 2009), psalmotoxin (PcTx1) - bound ion selective and non selective (Baconguis and Gouaux, 2012), or inactive states (Dawson et al., 2012), together with the initial high resolution structure of an inactive form of the ion channel (Jasti et al., 2007), elucidation of the mechanism of sodium selectivity, the role of the ‘GAS’ motif in ion selectivity, and binding sites for amiloride have proven elusive, as has been stabilization of an ASIC in a physiologically relevant open channel conformation.
Recently, snake (Bohlen et al., 2011; Diochot et al., 2012), spider (Escoubas et al., 2000) and sea anemone toxins (Diochot et al., 2004; Karczewski et al., 2010) have been identified that elicit or suppress pain by selectively activating or blocking ASICs (Bohlen and Julius, 2012; Chen et al., 2005; Karczewski et al., 2010). In addition to validating a role for ASICs in nociception and pain sensation, peptide toxins provide powerful tools to arrest ASICs in specific conformational states for pharmacological, biophysical, and structural studies (Baconguis and Gouaux, 2012; Baconguis et al., 2013; Bohlen et al., 2011; Chen et al., 2006). The Texas coral snake toxin, MitTx, is a hetero-dimeric polypeptide toxin that activates ASIC1a channels at nanomolar concentrations in a pH independent manner (Bohlen et al., 2011). MitTx consists of two, non-covalently associated α and β subunits that resemble Kunitz and phospholipase-A2 proteins (Huber et al., 1970; Scott et al., 1990), respectively, and which together function as a potent and selective ASIC1a agonist.
Here we determine the crystal structure and probe the function of the chicken ASIC1a – MitTx complex, illuminating how the toxin binds to and stabilizes ASIC1a in a physiologically relevant, open channel, sodium-selective state. We elucidate the structure of the ‘GAS’ motif, showing that it adopts an extended conformation that not only defines the selectivity filter, but that also enables the cytoplasmic portion of TM2 to undergo a swap between subunits. Furthermore, we map ion sites in the selectivity filter and in the ion channel pore and define binding sites for amiloride in the acidic pocket of the extracellular domain and within the fenestrations of the extracellular vestibule.
RESULTS AND DISCUSSION
Architecture of channel-toxin complex
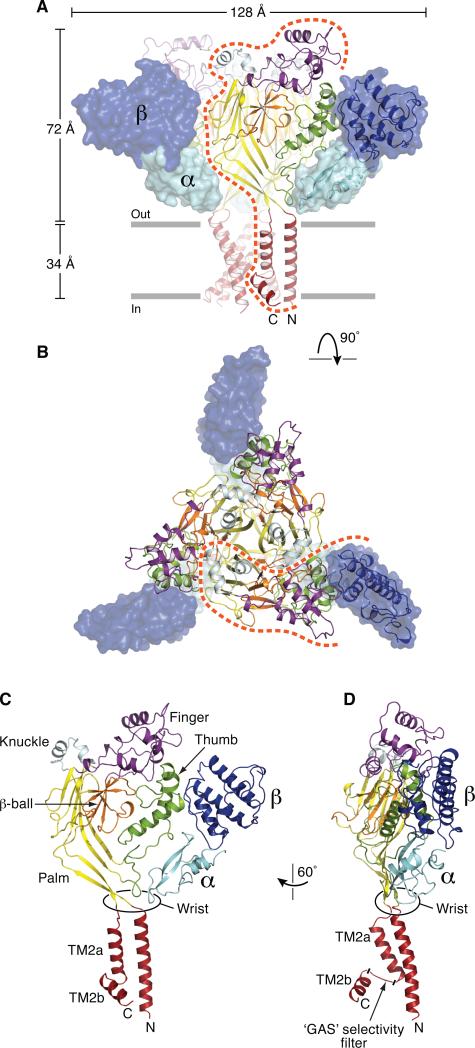
The heterodimeric MitTx activates the wild type-like chicken Δ13 ASIC1a construct (Baconguis and Gouaux, 2012) employed in this study. At pH 7.4, application of MitTx to cells expressing the Δ13 construct induces a slow inward current that reaches a steady-state amplitude that is ~15% of the peak amplitude resulting from a step to low pH (Figure S1A). In addition, MitTx slows the rate of channel closure and increases the magnitude of steady state current upon step to pH 5.5 (Figure S1B). We crystallized the Δ13-MitTx complex at pH 5.5 and determined crystal structures of the complex in the presence of Na+, amiloride or cesium at resolutions of 2.1, 2.3 and 2.6 Å, respectively (Figures 1 and S2 and Table S1) by molecular replacement and iterative rounds of manual rebuilding and crystallographic refinement. In the Δ13-MitTx crystals there is one Δ13 subunit and a single MitTx heterodimer in the asymmetric unit of the R3 unit cell and thus the physiological trimer hews to ideal crystallographic 3-fold symmetry.
Figure 1.
Architecture of the Δ13-MitTx complex. (A) Subunits of Δ13 and MitTx are color-coded by domain. The Δ13 ion channel is in cartoon and MitTx subunits are in surface representation with one heterodimer also shown in cartoon representation. The orange dashed line defines the region of one subunit. (B) View along the 3-fold axis of symmetry, from the extracellular side of the membrane. (C, D) Cartoon representation of a single subunit derived from the Δ13-MitTx complex. See also Figure S1 and Table S1.
The toxin-channel complex has a triskelion-like shape when viewed down the 3-fold axis of symmetry with one toxin heterodimer radiating from each Δ13 subunit (Figures 1A, 1B and Figure 2A). Toxin subunits protrude from the ‘edges’ of the channel trimer, with each heterodimer interacting almost exclusively with a single subunit, thus explaining how the toxin can activate ASIC1a/2 heteromeric channels (Bohlen et al., 2011). The toxin forms extensive contacts from the ion channel ‘wrist’, proximal to the extracellular boundary of the membrane bilayer and to the knuckle and thumb domains, as far as 60 Å from the membrane surface. Consistent with the high affinity of the toxin for the channel (Bohlen et al., 2011), one toxin heterodimer buries 1350 Å2 of solvent accessible surface area in the channel-toxin complex, of which 800 Å2 and 550 Å2 are derived from the α and β subunits respectively (Figure 2B).
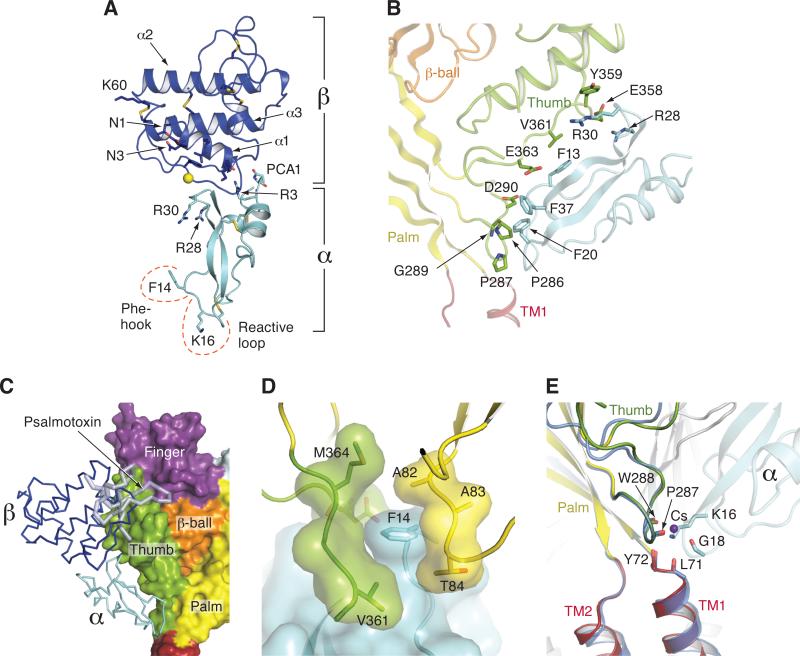
Figure 2.
Structures of MitTx and illustration of key residues and interactions. (A) Structure of MitTx derived from the complex with Δ13. In the heterodimeric α/β toxin complex, each subunit buries ~500 Å of solvent accessible surface area at the subunit interface. Residues near the N and C- termini of the α subunit play particularly important roles in the heterodimeric complex, forming helix-capping contacts to the C-terminus of the α1 helix of the β subunit, as well as mediating interactions with the β-wing domain. The α subunit, depicting the key Phe 14 and Lys 16 residues, along with the 3 disulfide bonds. (B) Illustration of residues that participate in extensive interactions between the α subunit of MitTx (cyan) and residues on the thumb domain (green) with portions of the Δ13 palm domain (light yellow) and β-ball (light orange) also shown. Phe 13, 20 and 37 supplement the interactions between α and the thumb of Δ13, and together with Arg 28 and 30, make interactions with residues on the α4 and α5 helices. (C) The binding site of MitTx (blue and cyan) overlaps with the psalmotoxin (light blue) binding site. Δ13 is shown in surface representation and toxins are in ribbon representation. (D) View of the subunit interface separated by Phe 14 of the α subunit, which serves as a ‘flange’ of the MitTx ‘churchkey.’ (E) View of the ‘wrist’ region following superposition of Cα positions of residues 285-290 and 70-74 of Δ13-MitTx and low pH Δ13-PcTx1 soaked in Cs+. Coordination of the ammonium group of Lys 16 is similar to the carbonyl oxygen coordination with Cs+ in the Δ13-PcTx1 structure. See also Figure S2.
The high resolution diffraction data derived from the Δ13-MitTx crystals allowed us to reinterpret the electron density in the transmembrane domain and now we find that TM2 is not a continuous α-helix but rather has a break in helical structure approximately 2/3 of the distance across the membrane, thus dividing the second transmembrane domain into segments TM2a and TM2b (Figure 1C). Remarkably, the ‘GAS’ motif bridges TM2a and TM2b by adopting an extended polypeptide conformation aligned approximately parallel to the membrane plane, allowing the TM2b element of one subunit to interact with TM1a of an adjacent subunit. The swapping of TM2b elements between subunits effectively generates a continuous TM2 helical segment with the ‘GAS belt’ reinforcing the structure of the ion channel pore.
Mechanism of MitTx binding
MitTx, held together by extensive interactions between the α and β subunits (Figure 2A, S2A and S2B), acts like a ‘churchkey’ bottle opener on Δ13, forming extensive interactions with the wrist, palm and thumb domains, supporting the notion that these domains play major roles in gating the ion channel (Figures 1A,B and 2). The overlap of the binding sites between PcTx1 and the β subunit of MitTx also explains why the binding and biological activity of these toxins are mutually exclusive (Figure 2C) (Bohlen et al., 2011). The α subunit is the ‘flange’ of the toxin ‘churchkey’ positioned on the underside of the thumb domain and insinuating a Phe 14 ‘hook’ into a Δ13 subunit interface and a Lys 16 ‘barb’ into the wrist, at the juncture of the extracellular ends of the transmembrane domains and at the base of the palm and thumb domains (Figure 2D and 2E). The aromatic ring of Phe 14 splays subunits apart and forms extensive interactions with residues Ala 82 through Thr 84 of the β1-β2 linker, a region of ASICs central to channel gating (Li et al., 2010b; Roy et al., 2013; Springauf et al., 2011) and, on the adjacent subunit, with residues Val 361 to Met 364. Lys 16 of α, situated on the ‘reactive loop’ of protease inhibitor homologs, juts into the wrist-TM1 interface of Δ13 with the ammonium group coordinated by 4 main chain carbonyl oxygens from Δ13 and one from the α subunit, thereby coupling the base of the wrist domain to the extracellular end of TM1. Remarkably, the ammonium group occupies the same position as Cs+ ions occupy in the open-state of the Δ13-PcTx1 complexes (Baconguis and Gouaux, 2012) (Figure 2E).
Repurposing of MitTx interaction surfaces
Despite relatively low sequence conservation between MitTx-α and other Kunitz-type proteins, or between MitTx-β and other PLA2 homologues, these toxin subunits retain canonical folds and 3D structures characteristic of their respective families, offering insights into evolutionary repurposing of conserved structural elements (Figure S2C and S2D). In the case of MitTx-α, this can be appreciated by comparing its structure with that of bovine trypsin inhibitor (BPTI) (Rühlmann et al., 1973), a classic Kunitz domain protein (Kunitz and Northrop, 1936). Moreover, just as Lys 16 in MitTx-α forms critical interactions with carbonyl oxygens in ASIC1a, the equivalent residue (Lys 15) in BPTI protrudes deep into the catalytic pocket of trypsin to mediate inhibition (Rühlmann et al., 1973). Thus, the ‘business end’ of these proteins has been structurally conserved, despite their meager sequence similarity of 37% and dissimilar physiological targets (Figure S2C).
Functional specification of Kunitz family members is also mediated by residues on the opposite end of the protein, at the N-terminus. In the case of δ-dendrotoxin and other Kunitz domain potassium channel antagonists, residues within the N-terminus are primary determinants of channel inhibition, suggesting that this region specifies interaction with the physiologic target (Imredy and MacKinnon, 2000) (Figure S2C). For MitTx-α, the N-terminus fulfills a different, but nonetheless critical structural role by serving as the primary surface for inter-subunit interactions. Indeed, a similar arrangement is seen with β-bungarotoxin, the other known example of a Kunitz-PLA2 heterodimeric snake toxin, wherein subunit interactions involve contacts between the N-terminus of the Kunitz partner and residues within and around the β-wing region of the PLA2 component (Kwong et al., 1995). Again, structural conservation is seen between MitTx and β-bungarotoxin subunits, despite limited sequence similarity of 34 and 42% for Kunitz and PLA2 subunits, respectively, and minimal sequence conservation within binding interfaces (Figure S2A, S2B and S2D).
Functional re-purposing of structural regions is also apparent in interactions between MitTx-β and ASIC1a. Here, the main interface corresponds to the region that in functional PLA2 enzymes contains the catalytic site and a surrounding hydrophobic patch that facilitates interaction with membrane lipids (Figure S2E-G). Thus, equivalent zones are devoted to functional activities of these proteins, irrespective of their divergent physiologic targets (Figure S2D).
Duplication and hypermutability of toxin genes accelerates the neo-functionalization of protein scaffolds commonly found in animal venoms, such as those belonging to the prolific Kunitz, PLA2, and inhibitor cysteine knot families (Bohlen and Julius, 2012). The comparison of MitTx with other known Kunitz and PLA2 structures illustrates the repeated use of regions peripheral to their compact and rigid cores in specifying interactions with subunits or physiological targets. The malleability of two established protein scaffolds, such as Kunitz and PLA2 proteins, provides a mechanism for enhanced functional diversification in venomous organisms, whose chemical armamentarium must evolve to keep pace with adaptive changes in predator-prey relationships.
‘GAS’ motif allows transmembrane helix swap
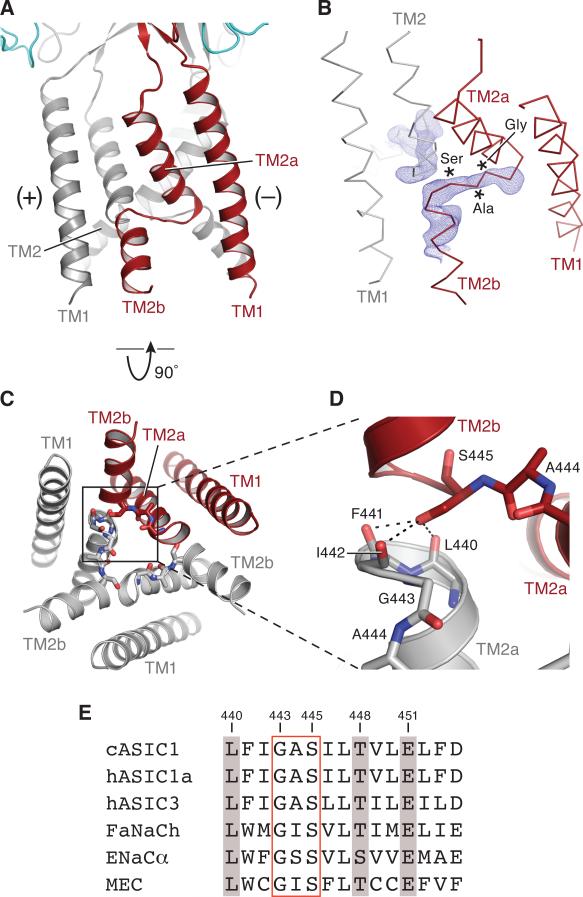
To solve the Δ13-MitTx crystal structures we employed the extracellular and transmembrane domains derived from the desensitized-state cASIC1 crystal structure (Gonzales et al., 2009) (PDB code 3HGC). In the latter structure, TM1 and TM2 are continuous α-helices. Upon inspection of 2Fo-Fc and Fo-Fc electron density maps of the Δ13-MitTx complexes, however, we noticed residual positive electron density located between TM1 and TM2, near Leu 50 (TM1) and Ile 442 (TM2) and weak density at Gly 443. These features, also seen upon reanalysis of the desensitized state structure (Gonzales et al., 2009), led us to alter the trace of TM2 (Figure 3A). Analysis of ‘omit’ electron density maps using diffraction data to ~2.1 and 2.3 Å resolution from both the Δ13-MitTx and the Δ13-MitTx-amiloride complexes (Figure 3B) provided convincing evidence for ending the helical structure of TM2 at Gly 443, for building residues Gly 443, Ala 444 and Ser 445 of the ‘GAS’ motif in an extended conformation, and for re-initiating an α-helix at Ile 446 to the last ordered residue, Ala 456 (Figure 3C). Reanalysis of the cASIC1 desensitized state structure, determined at 3 Å resolution, indicates that a similar break in TM2 and a swapping of TM2b between subunits is present. We therefore rebuilt and refined the desensitized state structure and now use that structure in our comparisons to the Δ13-MitTx structures. A careful review of electron density maps of the initial cASIC1a structure (Jasti et al. 2007) and the PcTx1 complexes (Baconguis and Gouaux, 2012) provides no evidence for swapping of the TM2 helices as we observe in the low pH desensitized state and in the MitTx complex. Furthermore, for the TM2 segments in the 3-fold symmetric high pH PcTx1 complex, and for TM2 segments of chains A-C and A-B in the asymmetric, low pH PcTx1 complex, the distance that the ‘GAS’ motif would be required to span is greater than 22 Å, which is too large for the tripeptide unit. In the TM2-swapped structures, the comparable distance is ~9 Å between the α-carbons of I442-S445 residues on adjacent TM2 helices. Thus we conclude that the swap in TM2 does not occur in the low pH and high pH PcTx1 structures or in the initial cASIC1a structure.
Figure 3.
Transmembrane domain swap mediated by the ‘GAS’ belt. (A) Cartoon representation of Δ13 transmembrane domains. One subunit is red and the region above and below the ‘GAS’ belt is labeled as ‘TM2a’ and ‘TM2b,’ respectively. (B) Fo-Fc ‘omit’ map generated with a model lacking residues Ile 442 to Leu 450 contoured at 3.0 σ. The Δ13 is shown as an α-carbon trace with one subunit omitted for clarity. (C) View of the transmembrane domain from the intracellular side and shown in cartoon representation. Residues near or in the ‘GAS’ belt are shown as sticks. (D) Close-up view of the boxed region in (C) illustrating how the hydroxyl group of Ser 445 caps the TM2a helix of an adjacent subunit.
The ‘GAS’ motif acts like a peptide ‘belt’ around the waist of the channel, breaking TM2 into TM2a and TM2b and allowing for the TM2b helical element to interact with the cytoplasmic portion of TM1 from the (−) subunit (Figure 3A). The ‘GAS’ motif is flanked by Ile 442 and Ile 446 which brace the ‘GAS’ motif by intrasubunit and intersubunit interactions with TM1 and TM2, respectively. Within the ‘GAS’ motif, Gly 443 exploits its conformational flexibility to end TM2a and initiate the extended structure of the motif while Ser 445 utilizes the hydrogen bonding capacity of its hydroxyl group to ‘cap’ the carbonyl oxygens atoms exposed at the beginning of TM2B of the (−) subunit (Figure 3D). Thus, the ‘GAS’ motif positions the carbonyl oxygen of Gly 443 toward the ion channel pore, in an ideal position to interact with permeant ions, and it also crosslinks subunits, thereby reinforcing the architecture of an ion channel pore composed of only 6 transmembrane segments. Inspection of amino acid sequences underscores the extent to which the (Gly/Ser)-X-Ser selectivity filter has been conserved across evolution and suggests that a discontinuous TM2 together with an extended belt is present in ASIC/ENaC/DEG family members (Fig. 3E).
MitTx expands the extracellular vestibule
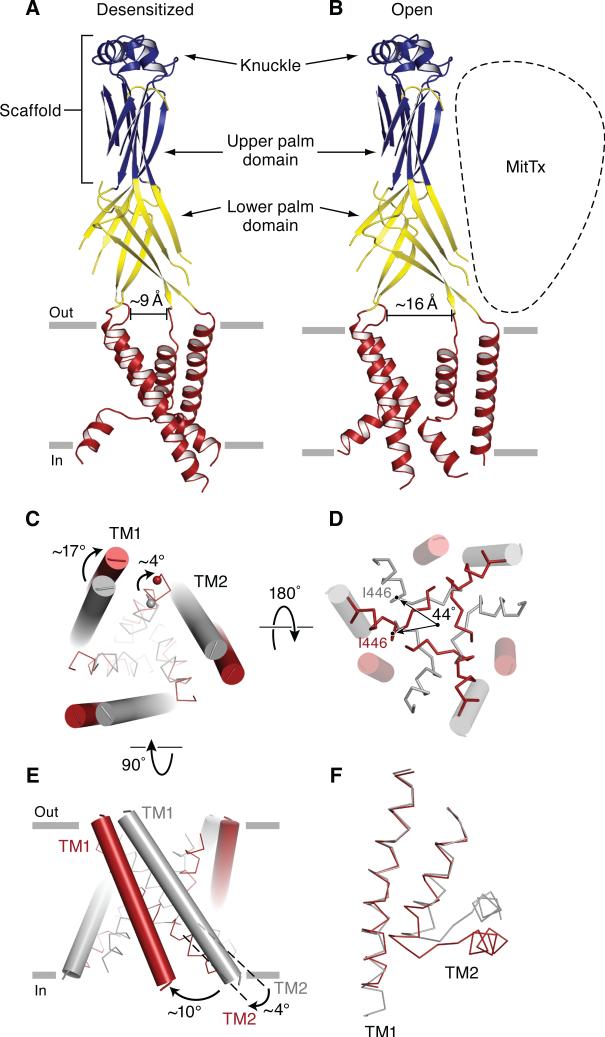
Superposition of the desensitized state and Δ13-MitTx structures demonstrates a conserved structural scaffold defined by the upper palm and knuckle domains (Baconguis and Gouaux, 2012). Whereas the knuckle and upper palm domains adopt the same conformation between the desensitized and MitTx-bound states, the lower palm domain flexes at the juncture of the β-strands spanning the extracellular and central vestibules, leading to an expansion of the extracellular vestibule (Figure 4A,B and Figure S3A,B). Using Ala 424 as a reference, the intersubunit separation increases from 9 Å in the desensitized state to 16 Å in the Δ13-MitTx complex. The flexing of the palm β-strands also decreases the length of the constriction, along the 3-fold axis, that occludes a direct, solvent accessible pathway between the central and extracellular vestibules (Figure S3A,B). In the desensitized state, the occlusion is flanked by Val 75 and Leu 78 and is a ~10 Å barrier between the central vestibule and the extracellular vestibule. By contrast, in the MitTx-bound state, Val 75 has shifted away from the 3-fold axis, giving rise to a constriction primarily mediated by Leu 78. These conformational changes are in accord with studies that have previously implicated the central vestibule in the modulation of gating in ASICs (Cushman et al., 2007; Li et al., 2011b; Roy et al., 2013; Yu et al., 2010).
Figure 4.
Gating movements in the extracellular and transmembrane domains deduced from comparison of the desensitized state and MitTx-bound structures. Cartoon representation of the desensitized state (A) and the Δ13-MitTx state (B) where the structurally conserved scaffold is blue, the malleable lower palm domain is yellow, and the transmembrane domains are red. Measurements use the Cα atoms Ala 424 on adjacent subunits as landmarks. (C) View of the transmembrane domains from the extracellular side illustrates the iris-like rotation of TM1 and TM2a. TM1 and TM2a are in cylinders and ribbon representation, respectively. In panels C-F the desensitized and Δ13-MitTx structures are gray and red, respectively. (D) View of the transmembrane domains from the intracellular side shows that TM2a rotates by ~44°, in the plane of the membrane. (E) View of the transmembrane domains perpendicular to the bilayer illustrating tilts of ~10° and ~4° by TM1 and TM2, respectively. (F) Close-up view of the transmembrane helices of one subunit. The TM1 domain of the Δ13-MitTx structure is superimposed onto the TM1 domain of the desensitized state structures using Cα positions of residues Cys 50 to Phe 70. Both TM1 and TM2 are shown in ribbon representations. See also Figure S3.
By contrast with the graded flexing of the lower palm domain β-strands, the binding of MitTx to the Δ13 ion channel promotes discrete ‘switching’ in the conformation of the β1-β2 and β11-β12 linkers, short regions of polypeptide implicated in ion channel gating (Li et al., 2010a; Li et al., 2010b; Springauf et al., 2011) and residing near the ‘pivot points’ of the lower palm β-strands. Relative to the desensitized state, residues 414 and 415 of the β11-β12 linker in the MitTx complex undergo a dramatic conformational change where hydrophobic Leu and polar Asn swap positions (Figure S3A,B). In the desensitized state, the Leu and Asn side chains are oriented toward the central vestibule and the β1-β2 linker, respectively, yet in the MitTx complex their orientations switch, with the Leu side chain packing against the β1-β2 loop and the Asn side chain facing the vestibule and making hydrogen bonds with the main chain amide of Ala 82 in the β1-β2 linker and a water molecule (Figure S3C).
MitTx stabilizes a symmetric open pore
The expansion of the extracellular vestibule opens the ion channel by way of an iris-like motion of the transmembrane α-helices. Comparison of the transmembrane domains between the desensitized and the MitTx-bound conformations demonstrates differential rotational movements of TM1, TM2a and TM2b by ~17°,~4°, and ~44°, respectively, around the 3-fold axis (Figure 4C,D). The trapping of Δ13 in a symmetric, open channel conformation by MitTx provides several insights into the molecular principles underpinning gating in ASICs. First, the extensive TM2-mediated intersubunit contacts that define the ~10 Å occlusion of the desensitized, shut ion channel are ruptured. Second, within each subunit TM1 and TM2a largely move together as a consequence of the iris-like expansion of the lower palm domain, although there is a scissor-like intrasubunit movement of TM1 relative to TM2 that alters interhelical contacts on both the extracellular and intracellular portions of the TM domains and that is manifested in the tilting of TM1 and TM2 by ~10° and ~4°, respectively (Figure 4E and S3D-G). Third, the role of TM1 in opening the pore and stabilizing a 3-fold symmetric conformation of the ion channel is underscored by the fact that either an alkali cation (Baconguis and Gouaux, 2012) or the ammonium group of a lysine residue provided by Lys 16 of the MitTx α subunit reinforces the contacts between the extracellular end of TM1 and the base of the thumb domain. Fourth, superposition of TM1 of one subunit of the desensitized state with a subunit of the Δ13-MitTx complex reveals a rearrangement of TM2b (Figure 4E,F), showing how the cytoplasmic regions of the TM segments could modulate ion channel gating.
In comparing the desensitized state and Δ13-MitTx structures, there is a structurally immobile pivot point at the ‘GAS’ selectivity filter. On the extracellular side of the ‘GAS’ belt, Phe 441 is clasped by Leu 57 and Leu 58 in both the desensitized and Δ13-MitTx structures and forms one set of interactions at the TM1-TM2 pivot. Similarly, Leu 447, a key residue that forms a leucine zipper-like interaction in the low pH PcTx1-bound structure (Baconguis and Gouaux, 2012), forges interactions with Cys 50 and Phe 51 of TM1 in both the open-state MitTx complex and in the desensitized state structure, thus forming another locus of interactions at the pivot, on the cytoplasmic side of the ‘GAS’ belt (Figure S3D,E).
By contrast, interactions ‘above’ and ‘below’ the ‘GAS’ belt exhibit large rearrangements disrupting multiple interactions observed in the desensitized state. Buried by van der Waals interactions with Val 61 and Cys 62 in the desensitized state, Gln 437 breaks away from Cys 62 and only maintains contacts with Val 61 in the Δ13-MitTx complex. Similarly, the contacts between Trp 47 and Asp 454 in the desensitized state are ruptured, allowing Trp 47 to make a hydrogen bond to Glu 451 in the MitTx state (Figure S3F, G). The role of the multiple interactions formed by Gln 437 in stabilizing the desensitized state is best shown in the Q437A mutant, which ablates the interactions of the amide side chain and results in an increase in the ratio of MitTx to pH induced current (Figure S3H, I). Eliminating interactions that favor the desensitized state do not appear to modify the pore conformation, based on measurements of amiloride block, but more likely alter the probability of the channel to transition between the open and desensitized states.
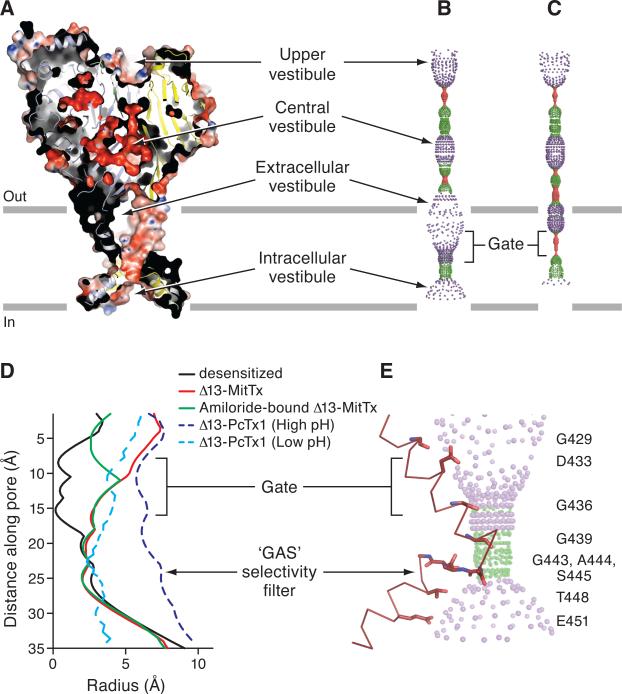
Gate and selectivity filter
The MitTx-bound state of the Δ13 construct demonstrates an open ion channel pore and access to the ion channel by way of the fenestrations of the extracellular vestibule (Figure 5A-C). Electrostatic mapping of the solvent accessible surface illustrates that the ion channel pore harbors a modest negative potential conferred by the presence of Asp 433, Gln 437, carbonyl oxygen atoms of Gly 436, 439 and 443, as well as by Thr 448 and Glu 451. The central vestibule has a strongly negative potential, due to the presence of a number of acidic residues, thus providing an explanation for the binding of Cs+ ions within this chamber (Figures 5A, S4A and S4B).
Figure 5.
The Δ13-MitTx complex harbors an open pore with a constriction located below the gate near the ‘GAS’ belt. (A) A section of an electrostatic potential of Δ13. Pore-lining surface down the threefold axis of the Δ13-MitTx (B) and the desensitized state structures (C). The plots in (B) and (C) were generated using the HOLE software (pore radius: red < 1.15 Å < green < 2.3 Å < purple). (D) Plot of radius as a function of longitudinal distance along the pore for Δ13-MitTx (red), Δ13-MitTx (amiloride, green), desensitized state (black), Δ13-PcTx1 (high pH, dark blue), and Δ13-PcTx1 (low pH, light blue). (E) Close-up view of the pore domain. Only one TM2 domain is shown for clarity and is in ribbon representation. Residues lining the pore are shown as sticks.
Comparison of the ion channel pore between the MitTx complex and the desensitized state shows that the extracellular vestibule undergoes a large expansion upon channel opening, rupturing the shut gate of the desensitized state and revealing an hourglass-shaped pore with upright and inverted ‘funnel shaped’ extracellular and intracellular vestibules in the open state (Figure 5B, C). The selectivity filter regions of the desensitized and MitTx states do not undergo large conformational changes, demonstrating that the ion channel gate is spatially distinct from the selectivity filter (Figure 5B-D). Further comparisons of the ion channel pores between the desensitized state, the PcTx1-bound selective (low pH) and non selective (high pH) states, and the MitTx-bound open state (Baconguis and Gouaux, 2012), show how the non-selective PcTx1 conformation has a large pore while that of the low pH PcTx1 bound conformation has a pore with dimensions that are similar to that of the MitTx-state and with a constriction that is also near the ‘GAS’ selectivity filter (Figure 5D). Finally, the pore of the Δ13-MitTx complex is primarily lined by residues contributed by TM2a and TM2b, in general agreement with accessibility studies (Carattino and Vecchia, 2012; Li et al., 2011a) but in contrast with earlier studies on FaNaCh (Pöet et al., 2001), and with structures of Δ13 in complex with psalmotoxin (Baconguis and Gouaux, 2012).
Mechanism of ion permeation and selectivity
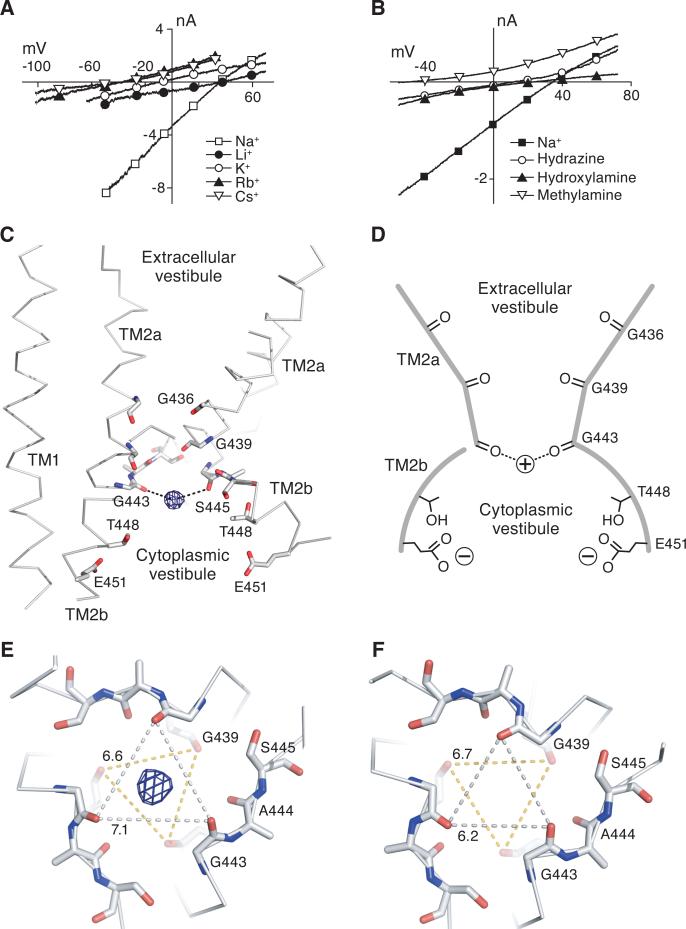
To probe the ion selectivity properties of the Δ13-MitTx complex, we performed biionic experiments using alkali metal and organic cations. The Δ13-MitTx complex is selective for Li+ and Na+ over K+ (~4:1) and nearly impermeable to the larger cations Rb+ and Cs+, similar to the wild-type ASIC1a upon low pH application (Baconguis and Gouaux, 2012) (Figure 6A). These observations reinforce the conclusion that the transmembrane pore of the Δ13-MitTx complex is a faithful representation of the proton-activated, wild-type ASIC1a pore. Because organic cations have proven to be valuable probes for voltage-gated Na+ channels, we measured the reversal potentials of the Δ13-MitTx ion channel using external solutions composed of hydrazine, hydroxylamine or methylamine. Like voltage-gated Na+ channels (Hille, 1971), hydroxylamine passes through the pore more easily than hydrazine while methylamine is impermeable (Figure 6B and Table S2), consistent with the conclusion that ions are hydrated as they permeate through the pore.
Figure 6.
Selectivity and structure of the ‘GAS’ selectivity filter. Whole cell, patch-clamp current/voltage analysis of the Δ13-MitTx complex in the presence of alkali metal cations (A) or organic cations (B). (C) Close-up view of the ‘GAS’ belt. Glycine residues 436 and 439, and residues in the ‘GAS’ belt are shown as sticks. One subunit has been omitted for clarity. A 2Fo-Fc map contoured at 1.5 σ illustrates the monovalent Cs+ site in the ‘GAS’ belt. This peak overlaps with an electron density peak calculated using anomalous difference amplitudes as coefficients. (D) Schematic illustration showing how the conductive pore is lined with carbonyl oxygens. (E) View of the ‘GAS’ belt from the intracellular side, perpendicular to the membrane plane, from the Cs+ soaked crystals, showing the 2 Fo-Fc peak for Cs+ and distances between the Gly 443 (7.1 Å) and Gly 439 (6.6 Å) carbonyl oxygen atoms. (F) Same view as in (E) of the ‘GAS’ belt derived from the Δ13-MitTx complex in the presence of Na+, showing how the size of the selectivity contracts in the presence of Na+, as measured by the distance between Gly 443 carbonyl oxygen atoms (6.2 Å). See also Table S2.
Using the diffraction data of Cs+ - soaked crystals, we observed a strong electron density peak in both Fo-Fc and anomalous difference maps located on the 3-fold axis and situated on the same plane as residues within the ‘GAS’ selectivity filter sequence (Figure 6C). The carbonyl oxygens of Gly 443 residues form a 3-fold symmetric triangle, 7.1 Å on a side, with each of the three carbonyl oxygens 4.1 Å from the Cs+ peak. We suggest that a hydrated Cs+ ion, with a radius of ~4.5 Å (Mähler and Persson, 2011), interacts with the carbonyl oxygens of Gly 443 by way of water-mediated contacts. The coordination of the Cs+ ion by the Gly 443 carbonyl oxygen atoms, together with the striking ‘belt-like’ conformation of the ‘GAS’ motif, support the long standing proposal that the ‘GAS’ motif is the selectivity filter of ASICs and ENaCs. Directly ‘above’ the plane of the GAS selectivity motif reside the 3-fold symmetric carbonyl oxygen atoms of Gly 439, which together with the carbonyl oxygen atoms of Gly 443, form a triangular antiprism. The carbonyl groups of Gly 439 are tipped off the helix axis by ~20°, protruding into the ion channel pore and in a position to interact with hydrated ions. We do not find evidence in electron density maps for a Cs+ ion in the plane defined by Gly 439 carbonyl oxygen atoms, perhaps because the radial distance from the 3-fold axis to the carbonyl oxygen atoms is 3.8 Å and thus too small to accommodate a hydrated Cs+ ion. Even though Cs+ is a weakly permeant ion, occupancy of the site in the selectivity filter occurs in crystals because the ion has access to both sides of the ion channel pore. In a membrane environment, Cs+ is a poorly permeant ion because the constrictions at residues Gly 439 and Gly 443 present high energy barriers to ion permeation.
Inspection of the electron density maps using diffraction data measured from either the Δ13-MitTx or the Δ13-MitTx-amiloride crystals, both of which were grown in the presence of Na+, did not reveal electron density peaks in the selectivity filter, suggesting that either the affinity for Na+ is weak or that bound ion(s) are disordered, despite the presence of 100 – 150 mM Na+ in the crystallization solution. Nevertheless, there is a change in the local structure of the selectivity filter between crystals grown in Na+ and those soaked in Cs+. For the Na+ containing crystals, the distance between Gly 443 carbonyl oxygen atoms is 6.2 Å, compared to 7.1 Å for the Cs+ soaked crystals. This contraction of the selectivity filter in the presence of Na+ reduces the filter's radius, as defined by the distance from the 3-fold axis to the carbonyl oxygen atoms of Gly 443, to 3.6 Å. The ring of carbonyl oxygens at Gly 439, immediately above the selectivity filter and with a radius of ~3.8 Å, matches the size of a hydrated Na+ (~3.8 Å) (Mähler and Persson, 2011). Moreover, the radius of a hydrated potassium ion is ~4.2 Å and thus too large to be accommodated within the selectivity filter (Mähler and Persson, 2011). Thus the mechanism of ion selectivity in ASICs is best described by a barrier mechanism (Hille, 2001), where selectivity is achieved by discrimination between cations on the basis of the size of the hydrated ion. Consistent with this mechanism, we note that the strongly hydrated lithium ion, with a radius of 3.5 Å (Mähler and Persson, 2011), is accommodated within the selectivity filter and is a highly permeable ion in ASICs and ENaCs (Palmer, 1982; Waldmann et al., 1997b).
Blocker and ion entry to ion channel pore
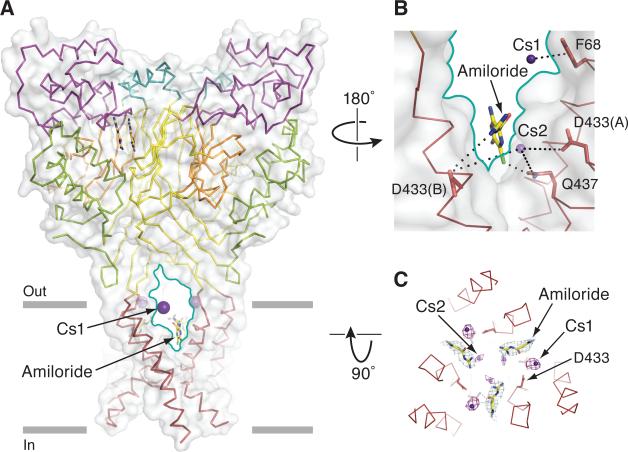
To understand how small molecule blockers and ions enter the ASIC ion channel pore, we soaked crystals in amiloride, the classical blocker of ENaCs and ASICs, or in Cs+-containing solutions, respectively. Upon analysis of electron density maps derived from crystals soaked in amiloride, we found density that could be well fit by amiloride in the acidic pocket of the extracellular domain (Figure S4C). While the presence of the two, head-to-tail amiloride molecules in the acidic pocket is striking and might define how amiloride stimulates ASICs (Adams et al., 1999), thorough testing of this conjecture requires additional experiments. Further inspection of ‘omit’ and 2Fo-Fc maps revealed prominent electron density features nestled between TM2 helices at the base of the ‘V’-shaped fenestrations of the extracellular vestibule, within the membrane-spanning region of the ion channel (Figure 7A-C). The three symmetry-related amiloride molecules partially occlude the pore in the extracellular vestibule with the guanidino groups positioned toward the three-fold axis (Figure 5B, 7A-C) at a site similar to that found in a recent computational study (Qadri et al., 2010). Forming hydrogen bonds and van der Waals interactions with amiloride are Asp 433 of one subunit and Gln 437 of the adjacent subunit, respectively (Figure 7B). Because the amiloride molecules are not occluding the ion channel pore, and mutagenesis studies of ENaC suggest that residues deeper in the pore, at positions equivalent to Gly 439, are implicated in amiloride binding (Kellenberger et al., 2003), we suggest that this site represents a binding site for amiloride as it enters the pore. We speculate that a single amiloride molecule blocks ion conduction by dipping its amidino group into the pore, binding at or near the carbonyl oxygens of Gly 436, occluding the ion conduction pathway. Perhaps the voltage-dependence of amiloride block (Adams et al., 1999) makes it energetically unfavorable for amiloride to occupy its blocking site in crystals.
Figure 7.
Fenestrations allow cations and amiloride access to the pore. (A) View of Δ13-MitTx complex bound to amiloride or Cs+. The structures of the amiloride-soaked and the Cs+ soaked Δ13 were superimposed. One fenestration is highlighted by a solid teal line, Cs+ and amiloride are in sphere and sticks representation, respectively. Δ13 is shown in both surface (gray) and ribbon representation and colored as in Figure 1A. (B) Close-up view of the fenestration. Residues near the Cs+ sites and amiloride are in sticks representation. Dashed lines indicate that interactions are mediated by water. (C) View of Cs+ and amiloride sites from the extracellular side showing how the two types of sites are near one another. The anomalous difference map showing Cs+ sites is contoured at 3.0 σ and shows one strong Cs+ site (5.0 σ) above amiloride and a weaker site (3.7 σ) near the guanidine group of amiloride. See also Figure S4.
To define how hydrated cations access the pore, we measured anomalous diffraction data of Δ13-MitTx crystals soaked in Cs+-containing solutions (Figure S7A). Prominent Cs+ sites (5 σ) were observed at the extracellular/transmembrane interface and below the pore constriction (Figure S7A). There is a strong Cs+ site within the triangle-shaped fenestrations ~5 Å from the aromatic ring of Tyr 68. Importantly, both 2Fo-Fc maps of Cs+-soaked and native Na+ crystals show density at this site, demonstrating that cations exploit a cation-π interaction to enter the extracellular vestibule (Figure 7A-C and S4E-G) (Dougherty, 1996). This cation binding site may also play a role in channel gating, because in the desensitized state the guanidinium group of Arg 65 forms a cation-π interaction with Tyr 68 (Figure S4H) (Gonzales et al., 2009).
Mechanism of gating, ion-selectivity, and block in ASICs
The ASIC1a-toxin structures demonstrate that the elements of gating involve a structural scaffold composed of the upper palm and knuckle domains, positioned at the core of the channel, in combination with a cluster of surrounding flexible domains that include the lower palm, finger, thumb, and wrist. As shown by the structures of ASIC1a in the desensitized state and in complexes with PcTx1 and MitTx, flexible domains undergo varying extents of conformational change which, in turn, are transduced into distinct conformational and functional properties of the ion channel. The MitTx complex shows that ions exploit cation-π interactions within the lateral fenestrations to enter and exit the extracellular vestibule. The selectivity filter of ASICs is defined by the lateral GAS belt which forms a triangular ring, allows for swapping of TM2b between subunits, and positions the carbonyl oxygen atom of Gly 443 directly into the pore. The size of the filter, with a radius of ~3.6 Å, is precisely matched to the radius of a hydrated Na+, and thus selectivity in ASICs is achieved by a barrier mechanism in which ions are discriminated on the basis of the size of the hydrated species. Lastly, the MitTx complex defines the mechanism by which a complex toxin allosterically modulates the activity of an ion channel. Together, the high-resolution structures of the Δ13-MitTx complexes provide the structural bases for pain initiation, ion conduction and selectivity in trimeric Na+-selective ion channels.
Experimental Procedures
Protein expression and purification
The Δ13 cASIC1a protein and native MitTx were expressed and purified as previously described (Baconguis and Gouaux, 2012; Bohlen et al., 2011). Prior to crystallization, the complex was formed by mixing Δ13 with MitTx at an approximate molar ratio of 1.0 Δ13 subunit to 1.5 MitTx, respectively.
Expression, refolding and purification of recombinant MitTxα and MitTxβ
Genes coding for the mature MitTxα and MitTxβ proteins (Genebank accession numbers JN613325 and JN613326, respectively) (Bohlen et al., 2011) preceded by N-terminal enterokinase cleavage sites were synthesized and subcloned into the pET32b vector. The fusion proteins were expressed in E. coli RosettagammiB (DE3) pLysS cells following induction with 0.1 mM isopropylthiogalactopyranoside. Cells were collected by centrifugation and disrupted by sonication in buffer containing 20 mM Tris pH 8, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 5 mM MgCl2, 20 μg/ml DNase-I and 0.4 mg lysozyme. The cell extracts were centrifuged at 8000 g for 15 min, inclusion bodies were collected, washed and solubilized as previously described (Chen and Gouaux, 1997) except that inclusion body solubilization buffer was supplemented with 300 mM β-mercaptoethanol. The inclusion bodies were subsequently solubilized in 6 M GuCl, refolded and the tags removed by enterokinase digestion at RT, followed by purification by gel filtration (α-subunit) or ion exchange chromatography (β-subunit). Analysis by mass spectrometry showed that the α and β subunits have masses of 7104 and 13736.6 Da, in close agreement to the predicted masses of 7104 and 13738 Da, respectively.
Crystallization and cryoprotection
Crystals of the Δ13-MitTx complex were grown by vapor diffusion using reservoir conditions containing either 50 mM sodium acetate (pH 5.5), 22-25% PEG 400, and 10 mM magnesium acetate, or 100 mM sodium acetate (pH 5.5) and 6-9% PEG 4000 from drops composed of a ratio of 1:1 and 2:1, protein to reservoir respectively, at 4 °C and an initial protein concentration of ~2.5 mg/ml. The complexes with amiloride or Cs+ were produced by soaking crystals in reservoir solution supplemented with 17.5 mM amiloride and 5% DMSO overnight or with cesium acetate (pH 5.5) and 500 mM CsCl, respectively.
Structure determination
Diffraction data sets collected from crystals of Δ13-MitTx and Δ13-MitTx (amiloride) were indexed, integrated, and scaled using the HKL2000 software and XDS (Kabsch, 2010; Otwinowski and Minor, 1997) or by using XDS, XSCALE and the ‘microdiffraction assembly’ method (Hanson et al., 2012), the latter of which yielded data sets that gave electron density maps with the most detail and thus were used for all analyses. The Δ13-MitTx structure was solved by molecular replacement using the computer program PHASER (McCoy, 2007), with the extracellular domain coordinates of the ΔASIC1 structure (PDB code: 2QTS) (Jasti et al., 2007) as a search probe.
Homology models of the MitTx α and β subunits were built using the ‘Automated Mode’ of the Swiss-Model homology modeling server (http://swissmodel.expasy.org/). Using the molecular replacement solution obtained with the extracellular domain of the ΔASIC1 structure, truncated models of the α subunit (residues 13-38) and the β subunit (helices α1, α2, and α3) were employed in molecular replacement. Coordinates were then subjected to iterative rounds of manual model building and crystallographic refinement using the computer programs COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2002). “Omit” electron density maps were calculated to validate residue registration in the transmembrane region and presence of the swap of TM2 between adjacent subunit. The final structure includes residues Val 45 to Gly 296 and Glu 299 to Ala 456 of the Δ13 construct and residues PCA 1 to Gly 60 and Asn 1 to Cys 118 of the α and β subunits of MitTx, respectively.
The Δ13-MiTx-amiloride structure was solved by molecular replacement. 2Fo-Fc and Fo-Fc electron density maps demonstrated distinct new features best accommodated by amiloride molecules, two in the acidic pocket of the extracellular domain and one in the fenestration of the extracellular vestibule. Iterative rounds of refinement and manual model adjustment were performed until satisfactory model statistics were achieved. The final model of the amiloride-bound Δ13-MitTx structure includes the same residues as the Δ13-MitTx structure.
Diffraction data from the Cs+-soaked crystals and the Δ13-MitTx structure described above were used in molecular replacement, followed by rounds of manual model building and crystallographic refinement. Inspection of the anomalous difference electron density maps was employed to identify Cs+ sites and a Cs+ ion was placed in all peaks that were greater than ~4 σ, resulting in a total of 15 Cs+ ions. Analysis of electron density maps, together with the refined structure, indicated that the most significant differences between the ‘native’ and the Cs+ soaked structures involved an expansion of the TM helices near Gly 443, at a Cs+ ion binding site. The final model consists of residues Val 46 to Gly 296 and Glu 299 to Phe 453.
We revisited the previously determined structure of the desensitized state (PDB code: 3HGC) and examined electron density maps. Indeed, TM2 also demonstrates a swapped conformation. Thus, we rebuilt and refined the structure until satisfactory model statistics were achieved with final Rwork and Rfree values of 20.4% and 24.6%, respectively. RMSD values of bonds and angles from ideal values were 0.007 Å and 1.006°, respectively (PDB code 4NYK).
Electrophysiology
Whole-cell recordings were carried out with CHO-K1 cells 24 hours after transfection by plasmid DNA encoding the Δ13 construct and GFP expressed from an internal ribosome entry site. Pipettes were pulled and polished to 2-3 MΩ resistance and filled with internal solution containing (in mM): 150 KCl, 2 MgCl2, 5 EGTA and 10 HEPES (pH 7.35). External solution contained (in mM): 150 NaCl, 2 MgCl2, 2 CaCl2, 8 Tris and 4 MES. For ion selectivity experiments, NaCl was substituted with equimolar concentrations of LiCl, KCl, RbCl, CsCl, hydrazine, hydroxylamine, or methylamine in the external solution. Recombinant α and β MitTx were applied at 600 nM and 300 nM concentrations, respectively. Reversal potentials were measured for 100 ms using voltage ramps from −100 to 40 mV for experiments using RbCl and CsCl, and −60 mV to 70 mV for the remaining cations.
Supplementary Material
Highlights.
ASIC1 structure trapped in open state by MitTx, a heterodimeric snake toxin
MitTx subunits harbor Kunitz and phospholipase A2 folds
Discontinuous transmembrane helix 2 creates novel selectivity filter
Selectivity achieved by barrier mechanism discriminating on basis of hydrated ion size
Acknowledgements
We thank members of the Gouaux lab and C. Jahr for helpful discussions, W. Lu for data processing, L. Vaskalis for figures and H. Owen and J. Dieter for manuscript preparation. This work was supported by National Research Service Awards from the National Institute of Neurological Disorders and Stroke (I.B. and C.B.) and grants from the NIH (E.G. and D.J.). E.G. is an investigator with the Howard Hughes Medical Institute. The authors declare no conflict of interest. The x-ray coordinates and structure factors are available in the Protein Data Bank, entries 4NTW, 4NTX and 4NTY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
I.B. purified the Δ13 ion channel and carried out the crystallographic and electrophysiological studies of the Δ13-MitTx complex. C.B. purified native MitTx from Texas coral snake and A.G. and E.G. prepared recombinant MitTx. I.B. wrote the manuscript and all authors edited the manuscript.
References
- Adams CM, Snyder PM, Welsh MJ. Paradoxical stimulation of a DEG/ENaC channel by amiloride. J Biol Chem. 1999;274:15500–15504. doi: 10.1074/jbc.274.22.15500. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alvarez de la Rosa D, Zhang P, Shao D, White F, Canessa CM. Functional implications of the localization and activity of acid-sensing channels in rat peripheral nervous system. Proc Natl Acad Sci USA. 2002;99:2326–2331. doi: 10.1073/pnas.042688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid- sensing ion channel-spider toxin complexes. Nature. 2012;489:400–405. doi: 10.1038/nature11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I, Hattori M, Gouaux E. Unanticipated parallels in architecture and mechanism between ATP-gated P2X receptors and acid sensing ion channels. Curr Opin Struct Biol. 2013 doi: 10.1016/j.sbi.2013.04.005. doi: 10.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Chesler AT, Sharif-Naeini R, Medzihradszky KF, Zhou S, King D, Sanchez EE, Burlingame AL, Basbaum AI, Julius D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Julius D. Receptor-targeting mechanisms of pain-causing toxins: How ow? Toxicon. 2012;60:254–264. doi: 10.1016/j.toxicon.2012.04.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino MD, Vecchia MCD. Contribution of residues in second transmembrane domain of ASIC1a protein to ion selectivity. J Biol Chem. 2012;287:12927–12934. doi: 10.1074/jbc.M111.329284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Gouaux E. Overexpression of a glutamate receptor (GluR2) ligand binding domain in Escherichia coli: Application of a novel protein folding screen. Proc Natl Acad Sci USA. 1997;94:13431–13436. doi: 10.1073/pnas.94.25.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kalbacher H, Gründer S. The tarantula toxin psalmotoxin 1 inhibits acid-sensing ion channel (ASIC) 1a by increasing its apparent H+ affinity. J Gen Physiol. 2005;126:71–79. doi: 10.1085/jgp.200509303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kalbacher H, Gründer S. Interaction of acid-sensing ion channel (ASIC) 1with the tarantula toxin psalmotoxin 1 is state dependent. J Gen Physiol. 2006;127:267–276. doi: 10.1085/jgp.200509409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman KA, Marsh-Haffner J, Adelman J, McCleskey EW. A conformational change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J Gen Physiol. 2007;129:345–350. doi: 10.1085/jgp.200709757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RJP, Benz J, Stohler P, Tetaz T, Joseph C, Huber S, Schmid G, Hügin D, Pflimlin P, Trube G, et al. Structure of the acid-sensin ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nature Commun. 2012;3:936. doi: 10.1038/ncomms1917. [DOI] [PubMed] [Google Scholar]
- Deval E, Gasull X, Noel J, Salinas M, Baron A, Diochot S, Lingueglia E. Acid-sensing ion channels (ASICs): pharmacology and implication in pain. Pharmacol Ther. 2010;128:549–558. doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Diochot S, Baron A, Rash LD, Deval E, Escoubas P, Scarzello S, Salinas M, Lazdunzki M. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 2004;23:1516–1525. doi: 10.1038/sj.emboj.7600177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diochot S, Baron A, Salinas M, Douguet D, Scarzello S, Dabert-Gay A-S, Debayle D, Friend V, Alloui A, Lazdunzki M, Lingueglia E. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature. 2012;490:552–555. doi: 10.1038/nature11494. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Escoubas P, De Weille JR, A., L., Diochot S, Waldmann R, Champigny G, Moinier D, Menez A, Lazdunski M. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J Biol Chem. 2000;275:25116–25121. doi: 10.1074/jbc.M003643200. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer S, Chen X. Structure, function, and pharmacology of acid-sensing ion channels (ASICs): focus on ASIC1a. Int J Physiol Pathophysiol Pharmacol. 2010;2:73–94. [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, et al. Crystal structure of a lipid-G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselager M, Timmermann DB, Ahring PK. pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- Hille B. The permeability of the sodium channel to organic cations in myelinated nerve. J Gen Physiol. 1971;58:599–619. doi: 10.1085/jgp.58.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion channels of excitable membranes. 3rd ed. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- Huber R, Kukla D, Ruehlmann A, Epp O, Formanek H. The basic trypsin inhibitor of bovine pancreas. 1. Structure-analysis and conformation of polypeptide-chain. Naturwissenschaften. 1970;57:389–392. doi: 10.1007/BF00599976. [DOI] [PubMed] [Google Scholar]
- Imredy JP, MacKinnon R. Energetic and structural interactions between delta-dendrotoxin and a voltage-gated potassium channel. J Mol Biol. 2000;296:1283–1294. doi: 10.1006/jmbi.2000.3522. [DOI] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales E, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski J, Spencer RH, Garsky VM, Liang A, Leitl MD, Cato MJ, Cook SP, Kane S, Urban MO. Reversal of acid-induced and inflammatory pain by the selective ASIC3 inhibitor, APETx2. Br J Pharmacol. 2010;161:950–960. doi: 10.1111/j.1476-5381.2010.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Auberson M, Gautschi I, Schneeberger E, Schild L. Permeability properties of ENaC selectivity filter mutants. J Gen Physiol. 2001;118:679–692. doi: 10.1085/jgp.118.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Gautschi I, Schild L. Mutations in the epithelial Na+ channel ENaC outer pore disrupt amiloride block by increasing its dissociation rate. Mol Pharmacol. 2003;64:848–856. doi: 10.1124/mol.64.4.848. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Gautschi K, Schild L. A single point mutation in the pore region of the epithelial Na+ channel changes ion selectivity by modifying molecular sieving. Proc Natl Acad Sci USA. 1999;96:4170–4175. doi: 10.1073/pnas.96.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishtal OA, Pidoplichko VI. A receptor for protons in the nerve cell membrane. Neuroscience. 1980;5:2325–2327. doi: 10.1016/0306-4522(80)90149-9. [DOI] [PubMed] [Google Scholar]
- Krishtal OA, Pidoplichko VI. A receptor for protons in the membrane of sensory neurons may participate in nociception. Neuroscience. 1981;6:2599–2601. doi: 10.1016/0306-4522(81)90105-6. [DOI] [PubMed] [Google Scholar]
- Kunitz M, Northrop JH. Isolation from beef pancreas of crystalline trypsinogen, trypsin, a trypsin inhibitor, and an inhibitor-trypsin compound. J Gen Physiol. 1936;19:991–1007. doi: 10.1085/jgp.19.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, McDonald NQ, Sigler PB, Hendrickson WA. Structure of b2- bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action. Structure. 1995;3:1109–1119. doi: 10.1016/s0969-2126(01)00246-5. [DOI] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. Asn415 in the beta11-beta12 linker decreases proton-dependent desensitization of ASIC1. J Biol Chem. 2010a;285:31285–31291. doi: 10.1074/jbc.M110.160382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. Leu85 in the beta1-beta2 linker of ASIC1 slows activation and decreases the apparent proton affinity by stabilizing a closed conformation. J Biol Chem. 2010b;285:22706–22712. doi: 10.1074/jbc.M110.134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat Commun. 2011a;2:399. doi: 10.1038/ncomms1409. [DOI] [PubMed] [Google Scholar]
- Li WG, Yu Y, Huang C, Cao H, Xu TL. Nonproton ligand sensing domain is required for paradoxical stimulation of acid-sensing ion channel 3 (ASIC3) channels by amiloride. J Biol Chem. 2011b;286:42635–42646. doi: 10.1074/jbc.M111.289058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E. Acid sensing ion channels in sensory perception. J Biol Chem. 2007;282:17325–17329. doi: 10.1074/jbc.R700011200. [DOI] [PubMed] [Google Scholar]
- Mähler J, Persson I. A study of the hydration of the alkali metal ions in aqueous solution. Inorg Chem. 2011;51:425–438. doi: 10.1021/ic2018693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliver DC, Immke DC, Fierro L, Paré M, Rice FL, McCleskey EW. ASIC3, an acid sensing ion channel, is expressed in metaboreceptive sensory neurons. Mol Pain. 2005;1:35–47. doi: 10.1186/1744-8069-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palmer LG. Ion selectivity of the apical membrane Na channel in the toad urinary bladder. J Membr Biol. 1982;67:91–98. doi: 10.1007/BF01868651. [DOI] [PubMed] [Google Scholar]
- Pöet M, Tauc M, Lingueglia E, Cance P, Poujeol P, Lazdunzki M, Counillon L. Exploration of the pore structure of a peptide-gated Na+ channel. EMBO J. 2001;20:5595–5602. doi: 10.1093/emboj/20.20.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri YJ, Song Y, Fuller CM, Benos DJ. Amiloride docking to acid-sensing ion channel-1. J Biol Chem. 2010;285:9627–9635. doi: 10.1074/jbc.M109.082735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Boiteux C, Alijevic O, Liang C, Berneche S, Kellenberger S. Molecular determinants of desensitization in an ENaC/degenerin channel. FASEB J fj.13. 2013;230680:1–12. doi: 10.1096/fj.13-230680. [DOI] [PubMed] [Google Scholar]
- Rühlmann A, Kukla D, Schwager P, Bartels K, Huber R. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. Crystal structure determination and stereochemistry of the contact region. J Mol Biol. 1973;77:417–436. doi: 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- Schild L, Schneeberger E, Gautschi I, Firsov D. Identification of amino acid residues in the alpha, beta, and gamma subunits of the epithelial sodium channel (ENaC) involved in amiloride block and ion permeation. J Gen Physiol. 1997;109:15–26. doi: 10.1085/jgp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DL, White SP, Otwinowski Z, Yuan W, Gelb MH, Sigler PB. Interfacial catalysis: the mechanism of phospholipase A2 action. Science. 1990;250:1541–1546. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S, Li J, McNulty KA, Avery D, Kleyman TR. Characterization of the selectivity filter of the epithelial sodium channel. J Biol Chem. 2000;275:8572–8581. doi: 10.1074/jbc.275.12.8572. [DOI] [PubMed] [Google Scholar]
- Sheng S, Li J, McNulty KA, Kieber-Emmons T, Kleyman TR. Epithelial sodium channel pore region. Structure and role in gating. J Biol Chem. 2001;276:1326–1334. doi: 10.1074/jbc.M008117200. [DOI] [PubMed] [Google Scholar]
- Sheng S, Perry CJ, Kashlan OB, Kleyman TR. Side chain orientation of residues lining the selectivity filter of epithelial Na+ channels. J Biol Chem. 2005;280:8513–8522. doi: 10.1074/jbc.M413880200. [DOI] [PubMed] [Google Scholar]
- Sherwood TW, Frey EN, Askwith CC. Structure and activity of the acid sensing ion channels. Am J Physiol Cell Physiol. 2012;303:C699–710. doi: 10.1152/ajpcell.00188.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PM, Olson DR, Bucher DB. A pore segment in DEG/ENaC Na+ channels. J Biol Chem. 1999;274:28484–28490. doi: 10.1074/jbc.274.40.28484. [DOI] [PubMed] [Google Scholar]
- Springauf A, Bresenitz P, Gründer S. The interaction between two extracellular linker regions controls sustained opening of acid-sensing ion channel 1. J Biol Chem. 2011;286:24374–24384. doi: 10.1074/jbc.M111.230797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann R, Bassilana F, de Weille JR, Champigny G, Heurteaux C, Lazdunski M. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997a;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997b;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Price MP, Welsh MJ. Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 2006;29:578–586. doi: 10.1016/j.tins.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, MacDonald JF, Wemmie JA, Price MP, Welsh MJ, Simon RP. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Z, Li WG, Cao H, Feng EG, Yu F, Liu H, Jiang H, Xu TL. A nonproton ligand sensor in the acid-sensing ion channel. Neuron. 2010;68:61–72. doi: 10.1016/j.neuron.2010.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.