Abstract
DNA CpG methylation has been associated with chromatin compaction and gene silencing. Whether DNA methylation directly contributes to chromatin compaction remains an open question. In this study, we used fluorescence fluctuation spectroscopy (FFS) to evaluate the compaction and aggregation of tetra-nucleosomes containing specific CpG patterns and methylation levels. The compactness of both unmethylated and methylated tetra-nucleosomes is dependent on DNA sequences. Specifically, methylation of the CpG sites located in the central dyad and the major grooves of DNA seem to have opposite effects on modulating the compactness of tetra-nucleosomes. The interactions among tetra-nucleosomes, however, seem to be enhanced because of DNA methylation independent of sequence contexts. Our finding can shed light on understanding the role of DNA methylation in determining nucleosome positioning pattern and chromatin compactness.
Introduction
DNA methylation of a CpG dinucleotide is the most frequent epigenetic modification that occurs on the genome. DNA methylation has been associated with chromatin compaction and gene silencing via different pathways (1,2). For example, DNA methylation can recruit methyl binding domain proteins (MBDP) and protein complexes such as SIN3A (3–5). These proteins act together to facilitate chromatin compaction and subsequently transcriptional suppression. The direct effect of DNA CpG methylation on chromatin structure, however, remains to be understood. Whether DNA methylation can directly modulate chromatin compactness is still an open question.
Recent genome-wide-association-studies (GWAS) have revealed interesting correlations among DNA sequences, DNA methylation, and nucleosome positioning patterns. Specifically, unmethylated CpG sites are found to preferentially locate in the major grooves of nucleosomal DNA, facing away from histone octamers (6,7). These preferences have been consistently observed in GWAS conducted in yeast, plants, invertebrates, and mammals (6–9). Methylated CpG sites, on the other hand, have been found to exist predominantly at 10 bp periodicity in the minor grooves of nucleosomal DNA facing histone octamers. These observations seem to suggest that methylated DNA fragments preferentially occur at certain nucleosomal locations because of either the timing of DNA methylation reaction or the sequence-encoded nucleosome positioning pattern. Additionally, in vitro studies have shown that besides the 10 bp periodicity of CpG sites, other CpG pattern, such as a stretch of CpG dinucleotides, can also have a significant effect on nucleosome positioning and chromatin conformation depending on their methylation state (10–13). Understanding how DNA methylation at those different nucleosomal locations contributes to the formation and stability of chromatin fibers holds the key to deciphering the direct impact of DNA methylation on chromatin.
The effect of DNA methylation on chromatin structure has been challenging to determine given the hierarchical structure of the complex. At DNA level, DNA methylation has been found to decrease the binding affinity of DNA to histone octamers (7,13–15) and contribute to determining the nucleosome positioning pattern of in vitro reconstituted chromatins (10,16,17). Recently, the effects of DNA methylation on nucleosome structure have been well studied (11,18–20). DNA methylation was found to affect the compactness of mono-nucleosomes based on its sequence contexts, and more importantly the relative location of the methylated CpG sites (11). Although there is extensive evidence suggesting that hyper-methylated chromatin tend to adopt a more compact folding in vivo (21), fewer studies exist as to the detailed role of DNA methylation in altering the inter- and intranucleosomal array interactions.
In this paper, we evaluated the effect of different CpG methylation patterns, selected based on GWAS and previous studies (6,7,10,11), on the compaction and the oligomerization of tetra-nucleosomes. Tetra-nucleosomes, consisting of 147 bp DNA repeats connected by linker DNA of 30 bp, were used as a model nucleosome array in our study. Tetra-nucleosomes have been broadly used to study the structural and functional properties of chromatin (23–29). They are preferred over native chromatin because of their well-defined features (linker DNA length, and types of histone octamers) and homogenous sample quality (30). Although tetra-nucleosomes may not capture all essential interactions required for forming a chromatin fiber (31–33), they provide a minimum system to elucidate the methylation effects on inter- and intranucleosomal array interactions.
In this study, we used fluorescence fluctuation spectroscopy (FFS) to quantitatively assess the compactness and oligomerization states of tetra-nucleosomes containing different CpG patterns and methylation levels. Our results showed that the compactness of tetra-nucleosomes is dependent on both CpG patterns and DNA methylation states. The sequence-dependence of tetra-nucleosome compactness is inversely related to what we have previously observed for mono-nucleosomes (11). DNA methylation can either facilitate or suppress the compaction of tetra-nucleosomes depending on the location of CpG sites. The oligomerization of tetra-nucleosomes, measured as the particle size increases with increasing Mg2+ concentrations, is positively affected by increase in DNA methylation levels, independent of DNA sequence content. Our results suggest that DNA methylation contributes to short-range intranucleosomal interaction in a sequence dependent manner. Meanwhile, long-range internucleosomal interactions that lead to the formation of tetra-nucleosome oligomers are enhanced by DNA methylation. Our results provide insights as to how DNA methylation modulates chromatin interactions and regulates gene expression.
Materials and Methods
Preparation of DNA repeats containing Widom-601 sequence
Tetra-nucleosomes have been used as model systems to study multiple chromatin features, including chromatin structure (23), inter- and intranucleosome interactions (24), and the effects of histone tails and posttranslational modifications on chromatin structure (24,25). In this study, we prepared DNA fragments containing four repeats of Widom-601 sequence (34) separated by linker DNA of 30 bp in length. The tetra-nucleosomes were reconstituted via a stepwise dialysis procedure (35). Additionally, we embedded three different CpG dinucleotide patterns, namely, 1), five consecutive CpG dinucleotides located in the central dyad ((CG)5), 2), five CpG dinucleotides at 10 bp intervals located in the major grooves ((CGX8)5,major groove), and 3), five CpG dinucleotides at 10 bp intervals located in the minor grooves ((CGX8)5,minor groove), to the original Widom-601 sequences as shown in Fig. 1 A . These three types of DNA methylation patterns were selected because of their distinctive nucleosome locations, and consequently different contacting modes with histone octamers as we detailed in our previous publication (11). For example, based on the crystal structure of nucleosomes, the CpG dinucleotides located in the major grooves do not form direct contacts with histone octamers, whereas CpG dinucleotides located in the minor grooves directly face histone octamers (36).
Figure 1.
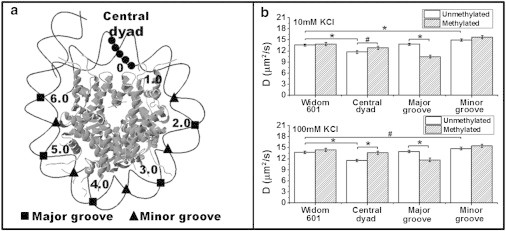
(A) Schematic drawing of the CpG patterns within a nucleosome (adapted from Jimenez-Useche et al. (11)). (B) Translational diffusion coefficient (D) of tetra-nucleosome at 10 mM KCl and 100 mM KCl. Data: mean ± standard error. ∗ p < 0.005; # p < 0.05.
DNA repeats containing Widom-601 sequence were cloned into pUC57 plasmid (Genscript, Piscataway, NJ) and the sequence was verified using DNA sequencing. The detailed DNA sequences of each construct are summarized in Table S1 in the Supporting Material. DNA fragments, free of CpG methylation, were produced in E. coli TOP10 cells (Invitrogen, Grand Island, NY). To produce DNA fragments with methylated CpG dinucleotides, plasmids containing the DNA of interest were co-transformed with pAIT2 plasmids into E. coli ER1821 cells (NEB, Ipswich, MA) (37). Plasmid pAIT2 codes for M.SssI methyltransferase, which catalyzes the addition of methyl groups to cytosines in the context of CpG dinucleotides. DNA produced using E. coli ER1821 strains is heavily methylated at CpG sites as reported in literature (37). Plasmid DNA was purified following a standard protocol (35) and digested to release the DNA repeats. The digested DNA samples were purified using a gel permeation column (Sephacryl S-500 HR, GE Healthcare Life Sciences, Pittsburgh, PA). The quality of the DNA fragments was examined using a polyacrylamide gel as shown in Fig. S1 A.
The methylation level of the DNA samples was assessed using the BstUI digestion pattern. The cleavage ability of BstUI is blocked by CpG methylation. Typically, DNA fragments produced in ER1821 are 85% to 100% methylated based on BstUI digestion patterns (Fig. S1 B). These DNA fragments were incubated with M.SssI (NEB, Ipswich, MA) and SAM (16 μM) at 37°C overnight to increase the methylation level to ∼ 100% (Fig. S1 B). M.SssI was then removed by phenol-chloroform extraction.
Refolding of histone octamers
Recombinant core histone proteins H2A, H2B, H3, and H4 with sequence derived from Xenopus laevis were individually expressed in bacteria as described by Luger et al. (35). Plasmids encoding for mutated histone proteins H2BT112C, H4S47C, and H3C110A were prepared by modifying the expression vector via a site-directed mutagenesis approach (38). The added cysteines provide labeling sites for fluorescent dyes, whereas H3C110A mutant prevents nonspecific fluorescence labeling by removing the only cysteine present in wild-type core histone proteins. The mutations we chose have been shown to minimally disrupt nucleosome conformation in literature (39,40). Two types of fluorescent dyes were used for labeling the proteins, i.e., Alexa Fluor 488 C5 maleimide (Life Technologies, Grand Island, NY) and Fluorescein-5-maleimide (AnaSpec, San Jose, CA). Fluorescent labeling reactions were carried out overnight at 4°C. Reaction mixtures were then dialyzed to water to remove unreacted fluorescent molecules. The quality of the samples was verified using SDS-PAGE. (Fig. S2 A). Histone octamers were refolded and purified by a gel permeation column (Superdex 200, GE Healthcare Life Sciences, Pittsburgh, PA) as described before (35). A typical gel of purified histone octamers was shown in Fig. S2 B.
Reconstitution of tetra-nucleosomes
DNA fragments were mixed with histone octamers at an optimal ratio to reconstitute tetra-nucleosomes (23). The ratio of DNA to histone octamers was optimized to minimize the presence of unbound DNA. The quality of reconstituted tetra-nucleosomes was assessed using agarose gels (Fig. S3 A) and restriction enzyme digestion patterns (Fig. S3 B). EcoRV sites or ScaI sites were embedded in the linker region by design. The saturation level of the tetra-nucleosomes was assessed by the digestion pattern following an established protocol (41). Specifically, undersaturated tetra-nucleosomes with less than four histone octamers per DNA molecule will yield free DNA and mono-nucleosomes after enzyme digestion. Meanwhile, saturated tetra-nucleosomes with all four sites occupied will only exhibit mono-nucleosome bands after digestion (Fig. S3 B). All prepared tetra-nucleosomes were found to have a saturation level > 95% estimated as (INCP/(INCP + IDNA)), where IDNA and INCP are the intensities of free DNA and mononucleosome band, respectively. The recovered reconstitution products were further purified using a Mg2+ (4.4 mM) precipitation step to remove any unbound histone octamers (23).
Fluorescence fluctuation spectroscopy
FFS was used to assess the compaction and oligomerization of tetra-nucleosomes. All measurements were carried out using fluorescently labeled tetra-nucleosomes in TEK buffer (10 mM Tris pH 7.9, 0.1 mM EDTA, and 10 mM or 100 mM KCl) with varying Mg2+ concentrations (0 to 1.5 mM). The nucleosome concentration was kept at ∼ 0.50 μM (corresponding to a tetra-nucleosome concentration of 0.125 μM with 3% labeling efficiency and a labeled tetra-nucleosome concentration of ∼ 4 nM) to obtain a good signal-to-noise ratio and prevent array dissociation. Fluctuations in fluorescence intensity data were collected using a dual-channel confocal spectrometer (ALBA FCS system, ISS, Champaign, IL) similar as described previously (14).
The FFS data were correlated to obtain correlation curves (G(τ)). The fluorescence correlation spectroscopy (FCS) curves were analyzed using the VistaVision Software (ISS, Champaign, IL) to reveal the translational diffusion coefficient (D) of the tetra-nucleosomes in the following equation:
| (1) |
where G(0) is the inverse of the number of molecules in the observation volume, and wo and zo are characteristic dimensions of the observation volume. wo and zo were calculated using a standard solution of Rhodamine 110 with known concentration and translational diffusion coefficient (430 μm2/s) (42). A typical correlation curve of tetra-nucleosomes and its fitting are shown in Fig. S4. We also followed the changes in G(0) as a function of Mg2+ concentrations. Increases in G(0) corresponds to decreases in the number of molecules in the observation volume. Significant changes in G(0) thus suggest the formation of oligomers or aggregates in the sample volume.
To eliminate the possibility that the dissociation of H2A/H2B dimers contribute to the observed translational diffusion coefficient change, we compared the diffusion coefficient of tetra-nucleosomes with fluorescent dyes attached to H4S47C and H2BT112C, respectively. As shown in Fig. S5, the measured diffusivities of two arrays are comparable with each other. To minimize the system noise of FFS experiments, each data point as shown in Figs. 2 and 3 was collected using tetra-nucleosomes from more than two batches of reconstitutions and more than three independent measurements of each reconstitution batch.
Figure 2.
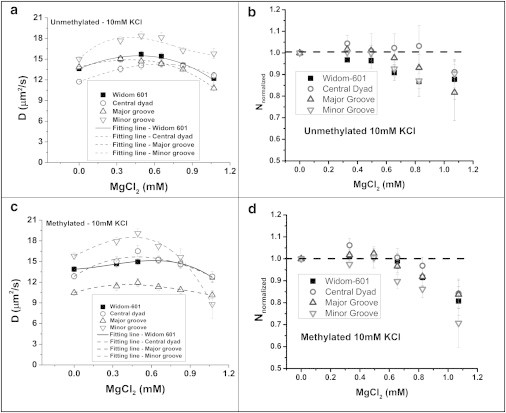
Effect of increasing concentration of Mg2+ cations on (A,B) translational diffusion coefficient (D) and (C,D) normalized particle numbers of unmethylated and methylated tetra-nucleosome arrays within the detection volume, respectively. The tetra-nucleosome concentration was kept at ∼ 0.125 μM. Data: mean ± standard error.
Figure 3.
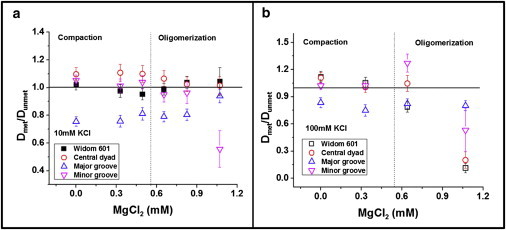
Effect of CpG methylation on the translational diffusion coefficients of tetra-nucleosomes with varying Mg2+ concentrations at (A) KCl = 10 mM and (B) KCl = 100 mM. The dotted line indicates the transition between tetra-nucleosome compaction and oligomerization. To see this figure in color, go online.
Results
DNA sequence modulates compaction of tetra-nucleosomes
DNA sequence has been shown to affect the conformation of nucleosomes (11,43). Here we evaluated the effect of three different CpG patterns on the compaction of tetra-nucleosomes. The translational diffusion coefficient (D) of tetra-nucleosomes was measured by FCS. Fig. 1 B shows the translational diffusion coefficients of unmethylated tetra-nucleosomes at 10 and 100 mM KCl. At 10 mM of KCl, the addition of CpG sites in the minor grooves results in a 10% increase (p < 0.005) in the translational diffusion coefficients, equivalent to a 9% reduction in the calculated hydrodynamic size based on Stoke-Einstein equation. In contrast, CpG sites in the central dyad lead to a decrease of ∼ 14% in the translational diffusion coefficient (p < 0.005). No significant difference is observed between tetra-nucleosomes with CpG sites in major groove and original Widom-601 repeats. Addition of KCl to 100 mM slightly increases the compaction of all tetra-nucleosomes, but the relative compactness of each type of tetra-nucleosomes remains unchanged (Fig. 1 B).
Divalent cations (Mg2+) mediate the compaction and oligomerization of tetra-nucleosomes
Nucleosome arrays assume dynamic structures and their compactness can be modulated by the presence of cations (44). In particular, Mg2+ cations are known to be effective in facilitating the compaction of nucleosome arrays by contributing to both short-range and long-range interaction forces (24,44). In this study, we varied the Mg2+ concentration from 0 to 1.1 mM to study the compaction and oligomerization of tetra-nucleosomes.
The translational diffusion coefficient of all tetra-nucleosomes under different MgCl2 concentrations are summarized in Fig. 2 A and C. For unmethylated tetra-nucleosomes, we observed an increase in translational diffusivities at low Mg2+ concentrations (∼ 0 to 0.40 mM) (Fig. 2 A). The maximum tetra-nucleosome compaction, measured as the highest translational diffusivity, was achieved at Mg2+ concentrations ranging from 0.40 to 0.65 mM as summarized in Table 1. The translational diffusivity increases by ∼ 8% to 22% for all unmethylated tetra-nucleosomes, corresponding to a hydrodynamic size reduction of 7% to 18%. The tetra-nucleosome with the CpG pattern embedded in the minor grooves exhibits the largest compaction induced by Mg2+ cations. Because G(0) is inversely related to the average number of fluorescent particles within the focal volume, we calculated the relative number of particles (Nnormalized) under different Mg2+ concentrations as G(0)control/G(0)sample where G(0)sample and G(0)control are the G(0) value of tetra-nucleosomes in TEK buffers with and without Mg2+, respectively. The values of Nnormalized remain almost constant around 1, at Mg2+ < 0.6 mM, as shown in Fig. 2 B. This result suggests that tetra-nucleosomes do not form multimers under this Mg2+ concentration range. Interestingly, although the presence of divalent cations compacts each type of tetra-nucleosomes, the relative compactness of each construct remains unaffected by the additional Mg2+ cations ([Mg2+] < 0.6mM).
Table 1.
The Mg2+ concentration (mM) at which tetra-nucleosomes assume the maximum compaction.
| Unmethylated (mM) | Methylated (mM) | |
|---|---|---|
| Widom 601 | 0.48 ± 0.01 | 0.62 ± 0.01 |
| Central dyad | 0.65 ± 0.01 | 0.49 ± 0.01 |
| Major groove | 0.41 ± 0.01 | 0.42 ± 0.01 |
| Minor groove | 0.40 ± 0.01 | 0.46 ± 0.01 |
This concentration is estimated by fitting the data with cubic polynomial function and estimating the Mg2+ concentration at the maximum of the function. All data = mean ± standard error.
Further increases in Mg2+ concentration are accompanied by decreases in the diffusion coefficients and significant changes in the normalized number of particles (Nnormalized). Both decompaction of tetra-nucleosomes and oligomerization of tetra-nucleosomes can contribute to the decreased diffusivity. Combining both pieces of evidence, it suggests the formation of tetra-nucleosome oligomers is the more likely cause. The oligomerization trends as demonstrated by different unmethylated tetra-nucleosomes are quite comparable with each other (Fig. 2 A and B). Our results suggest that additional CpG dinucleotides do not significantly contribute to the long-range interactions that lead to the formation of tetra-nucleosome oligomers.
It should be noted that although the translational diffusion coefficients of tetranucleosomes in the absence of Mg2+ may be similar to those obtained at a certain Mg2+ concentration (for example, wild-type tetranucleosomes at 0 and ∼ 0.7 mM of MgCl2), we expect that the conformations of tetranucleosomes at these two concentrations are quite different from each other. Specifically, tetra-nucleosomes exist as individual molecules with relatively open conformations without Mg2+. The measured diffusivities are thus fairly low. After adding Mg2+, tetra-nucleosomes increase their compactness and exhibit higher diffusion coefficients. Further increases in Mg2+ concentrations can lead to the onset of the oligomerization process. The sample mixture contains individual compact tetranucleosomes and tetranucleosome oligomers. As a result, the diffusion coefficient of the sample decreases and may result in diffusivities with similar values of tetranucleosomes without Mg2+.
DNA CpG methylation affects tetra-nucleosome compactness dependent on DNA sequences
We examined the effects of methylation on the compaction of tetra-nucleosomes. In the absence of Mg2+ ions, DNA methylation differentially affects the compactness of tetra-nucleosomes dependent on sequence contexts, as shown in Fig. 1 B top and bottom panels. The most significant changes in tetra-nucleosome compactness were observed for modified Widom-601 sequences with an additional (CG)5 stretch in the central dyad and with additional CpG dinucleotides in the major grooves. Specifically, the DNA construct with (CG)5 stretch exhibits a translational diffusivity 10% higher than the unmethylated counterpart (p < 0.005), suggesting a 9% reduction in the hydrodynamic radius of the tetra-nucleosome. On the other hand, tetra-nucleosomes, with additional (CGX8)5, major groove, shows a 24% decrease (p < 0.005) in diffusivities upon DNA methylation. This change corresponds to 32% increase in the hydrodynamic radii of tetra-nucleosomes. The compactness of tetra-nucleosome with the original Widom-601 sequence and embedded (CGX8)5,minor groove seem to be unaffected by the presence of DNA methylation. A similar trend was observed at 100 mM KCl as shown in Fig. 1 B bottom panel.
Similar to unmethylated samples, methylated tetra-nucleosomes increase their compactness with increasing Mg2+ concentration (∼ 0 to 0.45 mM) as shown in Fig. 2 C. The relative number of particles remains close to 1 within these MgCl2 concentrations as shown in Fig. 2 D. Maximum tetra-nucleosome compaction is observed at 0.42 to 0.62 mM Mg2+ as summarized in Table 1. Similar to unmethylated tetra-nucleosomes, methylated tetra-nucleosomes show maximum compaction with 9% to 22% increase in diffusivity, corresponding to 8% to 18% reduction in hydrodynamic size.
To quantitatively account for the effect of DNA methylation on tetra-nucleosome conformation at various Mg2+ concentrations, we calculated the ratio of the translational diffusivities of methylated and unmethylated tetra-nucleosomes, as Dmet/Dunmet. Our results are summarized in Fig. 3. At low Mg2+ concentrations (0 to 0.55 mM), the values of Dmet/Dunmet remain almost constant. Tetra-nucleosomes with (CG)5 in the central dyad consistently exhibit more compact folding upon DNA methylation, whereas tetra-nucleosomes with (CGX8)5, major groove become less compact with the introduction of DNA methylation. The folding of the other two types of tetra-nucleosomes, i.e., original Widom-601 and (CGX8)5,minor groove, seems to be unaffected by DNA methylation.
DNA CpG methylation facilitates tetra-nucleosome oligomerization
At high Mg2+ concentrations (> 0.42 to 0.62 mM), methylated tetra-nucleosomes show decreased diffusivity with increasing divalent cationic concentrations (Fig. 2 C). Decreases in the values of Nnormalized were also observed in this concentration range similar to unmethylated tetra-nucleosomes (Fig. 2 D). We calculated Dmet/Dunmet using the same approach as described in the previous section. Interestingly, all tetra-nucleosomes have Dmet/Dunmet less than or equal to one, as shown in the right-halves of Fig. 3 A and B. This trend is more significant when KCl = 100 mM. The reduction in diffusivities can be primarily attributed to the oligomers formed in the solution phase in this concentration range. Dmet/Dunmet less than or equal to one can thus be interpreted as formation of larger oligomers and/or more oligomers. This trend is also consistent with the sedimentation experiments as we carried out using both unmethylated and methylated Widom-601 tetra-nucleosomes (Fig. S6).
Discussion
Effects of DNA sequences on the compaction of tetra-nucleosomes
To quantify the effect of embedded CpG patterns on tetra-nucleosome conformation, we calculated the relative compactness of each tetra-nucleosomes as Di/DWidom 601, where Di and DWidom 601 correspond to the translational diffusion coefficients of tetra-nucleosome (i) and Widom 601, respectively. We compared our results with mono-nucleosome compactness as we observed in our previous work (11). Specifically, we calculated the relative compactness of mono-nucleosomes as Ei/EWidom 601, where E is the energy transfer efficiency measured from a FRET dye pair attached to the ends of nucleosomal DNA (Fig. 4 A insert) based on our previous findings (11). In mono-nucleosomes, the FRET results are dominated by the motion of the DNA ends because of our labeling strategy. We observed an increase in nucleosome compaction when a (CG)5 stretch is introduced to the central dyad and a decrease in nucleosome compaction when CpG sites are introduced in the minor grooves of the nucleosomal DNA, as shown in Fig. 4 A. An opposite trend was observed in tetra-nucleosomes with the same CpG patterns. Our results seem to suggest the existence of a correlation between the DNA-end breathing motion of nucleosomes and the compaction of tetra-nucleosome. Surprisingly, the revealed correlation is negative. One possible explanation for this is that linker DNA as observed in the tetra-nucleosome crystal structure adopts a quite open configuration (23). Enhanced DNA-end breathing (manifested as decreased compactness in mono-nucleosomes) permits the existence of different linker DNA trajectories in an array structure and may consequently result in a more compact tetra-nucleosome conformation.
Figure 4.
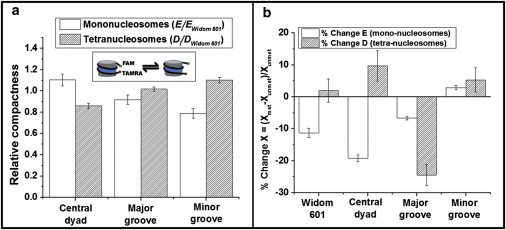
Comparison of the compaction level of (A) unmethylated or (B) methylated nucleosomes and tetra-nucleosomes at 10 mM KCl. Inset: schematic drawing of DNA end breathing motion in nucleosomes. Data: mean ± standard error. ∗ p < 0.005; # p < 0.05. To see this figure in color, go online.
The preferential compaction of tetra-nucleosomes with additional CpG dinucleotides embedded in the minor grooves is consistent with literature report showing that G+C rich DNA strands condense more easily at low Mg2+ concentrations than A+T rich DNA strands (45). Consequently, Mg2+ cations are more likely to reduce the short-range repulsion forces between neighboring nucleosomes and facilitate the compaction process.
Effect of DNA methylation on compaction and oligomerization of tetra-nucleosomes
The effects of DNA methylation on tetra-nucleosome conformation was compared with our previous observations for mono-nucleosomes as shown in Fig. 4 B. Our mono-nucleosome study suggests an overall decrease in mono-nucleosome compaction as demonstrated by a reduction in measured energy transfer efficiency (E) between the ends of nucleosomal DNA, with the exception of DNA fragments containing additional CpG dinucleotides in the minor grooves. The trend observed in tetra-nucleosome diffusivities, however, does not demonstrate any correlation with what was observed for mono-nucleosomes. The discrepancy between these two observed trends (DNA methylation effects on mono- and tetra-nucleosomes) seems to suggest that DNA methylation contributes to the tetra-nucleosome structure by more than affecting the compactness of mono-nucleosomes. DNA methylation may also directly contribute to the intranucleosome interactions that determine the distance between neighboring nucleosomes within the same array.
It is widely accepted that methyl groups of methylated cytosines face into the major grooves (46,47). In the case of tetra-nucleosomes containing methylated CpG sites in the major grooves, the methyl groups will be accommodated at the interior of the nucleosomal structure, facing the histone octamer surface. In this position, the methyl groups pose a hindrance for the nucleosomal DNA to bend and comply with the histone octamer curvature and can also alter the interactions between DNA and histone octamers (48). As a result, a less compact structure of tetra-nucleosomes with (CGX8)5,major groove is to an extent expected. On the other hand, in the case of tetra-nucleosomes with the CpG sites in the minor grooves, the methyl groups will be accommodated on the outside of the nucleosomal structure, facing away from the histone octamer. Because these grooves are already widened because of the bending of nucleosomal DNA, the addition of extra methyl-groups are not expected to pose any hindrance or interfere with DNA-histone octamer contacts. As a result, DNA methylation does not significantly alter the compactness of tetra-nucleosomes. Methylation of the additional (CG)5 located in the central dyad facilitates the tetra-nucleosome compactness by potentially reducing the repulsion forces between DNA fragments located in the linker and the central dyad regions. The presence of methyl-groups in cytosines can potentially reduce the repulsive interactions between negatively charged DNA backbone via the formation of a hydration layer around methyl-groups (49). As a result, the linker DNA can exist in close proximity of neighboring nucleosomes and lead to a more compact array structure.
When DNA methylation was positioned in the major grove of tetranucleosomes, a 32% increase in the hydrodynamic radius was observed. The magnitude of this increase is on scale comparable with what has previously been observed for histone modifications. Analytical centrifugation experiments on nucleosome arrays containing twelve nucleosomes saw increase in hydrodynamic radius of 15%, 19%, and 19% for H2B ubiquitylation (25), H4-K16 acetylation (50), and H4 tail truncation (51), respectively. Additionally, the 15% increase in hydrodynamic radius because of H2B ubiquitylation corresponded to increases in nucleosome accessibility as measured by an in vitro methyltransferase accessibility assay and H2B ubiquitylation was found to be associated with active chromatin in vivo (25). Although these studies were performed using nucleosome arrays containing 12-repeats rather than tetranucleosomes, it has been predicted that tetranucleosome arrays exhibit a lesser extent of compaction as seen 12mer repeat nucleosome arrays (24). This suggests that the increase in hydrodynamic radius is even more significant when seen in tetranucleosomes as compared with 12mer repeat nucleosome arrays. The decrease in array compaction caused by CpG methylation in the major groove of the nucleosome is large enough to play a significant role in the nucleosome accessibility of DNA binding proteins.
Based on our results, DNA methylation facilitates the oligomerization of tetra-nucleosomes independent of DNA sequence context. Significantly, tetra-nucleosomes with additional methyl groups in minor grooves (the preferred location of methyl groups in chromatin, as found in GWAS studies) demonstrate the strongest oligomerization trend as shown in Fig. 2 D. The exact oligomerization/aggregation mechanism of tetra-nucleosomes, however, remains to be clarified. This observation suggests that DNA methylation can contribute to the long-range interactions between tetra-nucleosomes by increasing the effective attraction forces. These forces could be the result of interactions between the attached methyl groups in tetra-nucleosomes. There are literature reports suggesting that the presence of methyl groups in DNA facilitate the interactions between DNA strands (52,53). The nature of this attraction force, however, is controversial, with some literature claiming that these interactions are because of hydrophobicity (52) and other suggesting that they are due to the increased polarizability of the methyl groups (49,54).
Conclusions
In conclusion, our results suggest that the conformation of tetra-nucleosomes is dependent on DNA sequence context just as we observed before for mono-nucleosomes (11). As expected, tetra-nucleosomes exhibit compaction followed by oligomerization with increasing Mg2+ concentrations. Tetra-nucleosomes with additional CpG in the minor grooves exhibit the largest compaction among all four constructs studied in this work, suggesting increased intranucleosome interactions because of the preferential binding of Mg2+ cations. Two of the CpG patterns studied here were able to distinctively modulate the compaction of tetra-nucleosomes upon methylation. Specifically, a stretch of (meCpG)5 dinucleotides favors the compaction of tetra-nucleosomes, whereas meCpG sites in the major grooves induced a less compact structure. Significantly, DNA methylation level seems to promote the oligomerization of tetra-nucleosomes, suggesting increased internucleosome interactions. The results from this study contribute to understanding the detailed mechanistic role of DNA methylation in determining chromatin compaction and regulating gene expression.
Acknowledgments
The authors thank Agnes Mendonca, Nathaphon Yu King Hing, and Robert Khakimov for the helping performing part of the experiments in this study. We also thank New England BioLabs (NEB) for generously sharing with us plasmid pAIT2 and E. coli cells ER1821. We gratefully acknowledge Prof. Tim Richmond in ETH, Zürich, for generously sharing with us the expression plasmids of histone proteins.
This work was supported by the Purdue Engineering School faculty start-up package and the American Chemical Society (Grant No.: PRF# 50918-DNI7).
Supporting Material
References
- 1.Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl.):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 2.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 4.Kass S.U., Pruss D., Wolffe A.P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 5.Nan X., Ng H.-H., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 6.Chodavarapu R.K., Feng S., Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collings C.K., Waddell P.J., Anderson J.N. Effects of DNA methylation on nucleosome stability. Nucleic Acids Res. 2013;41:2918–2931. doi: 10.1093/nar/gks893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan N., Moore I.K., Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valouev A., Ichikawa J., Johnson S.M. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey C.S., Pennings S., Allan J. A determining influence for CpG dinucleotides on nucleosome positioning in vitro. Nucleic Acids Res. 2004;32:4322–4331. doi: 10.1093/nar/gkh749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez-Useche I., Ke J., Yuan C. DNA methylation regulated nucleosome dynamics. Sci. Rep. 2013;3:2121. doi: 10.1038/srep02121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey C., Pennings S., Allan J. CpG methylation remodels chromatin structure in vitro. J. Mol. Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 13.Pérez A., Castellazzi C.L., Orozco M. Impact of methylation on the physical properties of DNA. Biophys. J. 2012;102:2140–2148. doi: 10.1016/j.bpj.2012.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Useche I., Shim D., Yuan C. Unmethylated and methylated CpG dinucleotides distinctively regulate the physical properties of DNA. Biopolymers. 2014;101:517–524. doi: 10.1002/bip.22411. [DOI] [PubMed] [Google Scholar]
- 15.Godde J.S., Kass S.U., Wolffe A.P. Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J. Biol. Chem. 1996;271:24325–24328. doi: 10.1074/jbc.271.40.24325. [DOI] [PubMed] [Google Scholar]
- 16.Drew H.R., McCall M.J. Structural analysis of a reconstituted DNA containing three histone octamers and histone H5. J. Mol. Biol. 1987;197:485–511. doi: 10.1016/0022-2836(87)90560-2. [DOI] [PubMed] [Google Scholar]
- 17.Karymov M.A., Tomschik M., Zlatanova J. DNA methylation-dependent chromatin fiber compaction in vivo and in vitro: requirement for linker histone. FASEB J. 2001;15:2631–2641. doi: 10.1096/fj.01-0345com. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez-Useche I., Yuan C. The effect of DNA CpG methylation on the dynamic conformation of a nucleosome. Biophys. J. 2012;103:2502–2512. doi: 10.1016/j.bpj.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choy J.S., Wei S., Lee T.H. DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J.Y., Lee T.-H. Effects of DNA methylation on the structure of nucleosomes. J. Am. Chem. Soc. 2012;134:173–175. doi: 10.1021/ja210273w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trojer P., Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted in proof.
- 23.Schalch T., Duda S., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 24.Blacketer M.J., Feely S.J., Shogren-Knaak M.A. Nucleosome interactions and stability in an ordered nucleosome array model system. J. Biol. Chem. 2010;285:34597–34607. doi: 10.1074/jbc.M110.140061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierz B., Chatterjee C., Muir T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh R.P., Horowitz-Scherer R.A., Woodcock C.L. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittig B., Wittig S. Nucleosome mono, di, tri-, and tetramers from chicken embryo chromatin. Nucleic Acids Res. 1977;4:3901–3917. doi: 10.1093/nar/4.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson E.C., Butler P.J.G., Thomas J. Higher-order structure of nucleosome oligomers from short-repeat chromatin. EMBO J. 1983;2:1367–1372. doi: 10.1002/j.1460-2075.1983.tb01593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finch J.T., Noll M., Kornberg R.D. Electron microscopy of defined lengths of chromatin. Proc. Natl. Acad. Sci. USA. 1975;72:3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huynh V.A., Robinson P.J., Rhodes D. A method for the in vitro reconstitution of a defined ‘30 nm’ chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 31.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassett A., Cooper S., Travers A. The folding and unfolding of eukaryotic chromatin. Curr. Opin. Genet. Dev. 2009;19:159–165. doi: 10.1016/j.gde.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 35.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 36.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Sandoval A., Ertl H.C. CpG methylation of a plasmid vector results in extended transgene product expression by circumventing induction of immune responses. Mol. Ther. 2004;9:249–261. doi: 10.1016/j.ymthe.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Nurse N.P., Jimenez-Useche I., Yuan C. Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys. J. 2013;104:1081–1088. doi: 10.1016/j.bpj.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park Y.J., Dyer P.N., Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 40.Balasubramanian S., Xu F., Olson W.K. DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences. Biophys. J. 2009;96:2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carruthers L.M., Tse C., Hansen J.C. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- 42.Gendron P.-O., Avaltroni F., Wilkinson K.J. Diffusion coefficients of several rhodamine derivatives as determined by pulsed field gradient-nuclear magnetic resonance and fluorescence correlation spectroscopy. J. Fluoresc. 2008;18:1093–1101. doi: 10.1007/s10895-008-0357-7. [DOI] [PubMed] [Google Scholar]
- 43.Gansen A., Tóth K., Langowski J. Structural variability of nucleosomes detected by single-pair Förster resonance energy transfer: histone acetylation, sequence variation, and salt effects. J. Phys. Chem. B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 44.Poirier M.G., Oh E., Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sitko J.C., Mateescu E.M., Hansma H.G. Sequence-dependent DNA condensation and the electrostatic zipper. Biophys. J. 2003;84:419–431. doi: 10.1016/S0006-3495(03)74862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pennings S., Allan J., Davey C.S. DNA methylation, nucleosome formation and positioning. Brief. Funct. Genomics Proteomics. 2005;3:351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- 47.Heinemann U., Hahn M. C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J. Biol. Chem. 1992;267:7332–7341. [PubMed] [Google Scholar]
- 48.Buttinelli M., Minnock A., Travers A. The exocyclic groups of DNA modulate the affinity and positioning of the histone octamer. Proc. Natl. Acad. Sci. USA. 1998;95:8544–8549. doi: 10.1073/pnas.95.15.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer-Jung C., Moras D., Timsit Y. Hydration and recognition of methylated CpG steps in DNA. EMBO J. 1998;17:2709–2718. doi: 10.1093/emboj/17.9.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shogren-Knaak M., Ishii H., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 51.Dorigo B., Schalch T., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 52.Kaur P., Plochberger B., Lindsay S. Hydrophobicity of methylated DNA as a possible mechanism for gene silencing. Phys. Biol. 2012;9:065001. doi: 10.1088/1478-3975/9/6/065001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer-Jung C., Moras D., Timsit Y. Effect of cytosine methylation on DNA-DNA recognition at CpG steps. J. Mol. Biol. 1997;270:328–335. doi: 10.1006/jmbi.1997.1137. [DOI] [PubMed] [Google Scholar]
- 54.Sowers L.C., Shaw B.R., Sedwick W.D. Base stacking and molecular polarizability: effect of a methyl group in the 5-position of pyrimidines. Biochem. Biophys. Res. Commun. 1987;148:790–794. doi: 10.1016/0006-291x(87)90945-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


