Abstract
Inbreeding increases homozygosity, exposes genome-wide recessive deleterious alleles and often reduces fitness. The physiological and reproductive consequences of inbreeding may be manifested already during gene regulation, but the degree to which inbreeding influences gene expression is unknown in most organisms, including in birds. To evaluate the pattern of inbreeding-affected gene expression over the genome and in relation to sex, we performed a transcriptome-wide gene expression (10 695 genes) study of brain tissue of 10-day-old inbred and outbred, male and female zebra finches. We found significantly lower gene expression in females compared with males at Z-linked genes, confirming that dosage compensation is incomplete in female birds. However, inbreeding did not affect gene expression at autosomal or sex-linked genes, neither in males nor in females. Analyses of single genes again found a clear sex-biased expression at Z-linked genes, whereas only a single gene was significantly affected by inbreeding. The weak effect of inbreeding on gene expression in zebra finches contrasts to the situation, for example, in Drosophila where inbreeding has been found to influence gene expression more generally and at stress-related genes in particular.
Keywords: inbreeding, gene expression, sex chromosome, zebra finch
1. Introduction
It is well established that inbreeding deteriorates the physiological and reproductive performance of most organisms, a phenomenon referred to as inbreeding depression. Because of these negative effects, inbreeding plays a significant role in important evolutionary processes, including the evolution of mating systems and sexual reproduction [1,2]. Despite a long-standing research tradition, many crucial aspects of inbreeding are poorly evaluated. One of these poorly evaluated areas regards the effect of inbreeding on the initial stages of phenotypic development, the expression of genes. Studying the links between inbreeding and gene expression could help in understanding the importance of specific genes, genetic networks and physiological pathways for inbreeding depression [2].
In Drosophila melanogaster, studies of the expression of genes genome-wide have shown that inbreeding affects fundamental molecular processes and in particular stress responses, such as expression of genes related to protein folding and heat shock [3,4]. Similar research in the house mouse (Mus domesticus) and Pacific oyster (Crassostrea gigas) shows different expression patterns as a result of fixation of line-specific alleles [5,6]. However, many of these studies use inbred lines, which have been purebred over several generations in laboratory conditions, and are therefore displaying the long-term consequences of inbreeding. Little is known about short-term, genome-wide expression effects of inbreeding, although such studies could provide important insights into how mating between close relatives, for example, during founder events and rapid population contractions, may affect individuals and populations [2].
Another aspect that has been poorly evaluated is the sex-specific consequence of inbreeding. In species with heteromorphic sex chromosomes (e.g. most mammals and birds), recessive mutations at the sex chromosomes are continuously exposed to selection in the heterogametic sex (XY males, e.g. in mammals; ZW females, e.g. in birds) and can therefore be selected against, or sex-specific counter adaptations may evolve. Therefore, the homogametic sex (XX females; ZZ males) may be more severely affected by inbreeding than the heterogametic sex. Indeed, in several bird species males and females are differently affected by inbreeding at the phenotypic level [7–9] and in D. melanogaster a recent study shows that inbreeding results in stronger net selection on males, with implications for mutation load and female fitness [10]. Still, the sex chromosomes constitute only a small fraction of the total genome, and it remains largely unknown to what extent sex-linked genes contribute to inbreeding depression relative to autosomal genes. The large number of autosomal genes could potentially harbour a vast number of recessive deleterious mutations causing inbreeding effects in both sexes.
In this study, we perform a transcriptome-wide microarray gene expression study in inbred and outbred, 10-day-old, male and female zebra finches (Taeniopygia guttata) to evaluate the overall consequences of inbreeding on gene expression and potential sex-specific effects. Our study was conducted on birds from a captive population that has been kept at a high effective population size and holds substantial genetic variation with a heterozygosity level (0.88) comparable to that of wild zebra finches (0.93–0.94; [11]). Thus, we expect a substantial inbreeding load of harmful recessives segregating in the population. Previous work in zebra finches and other birds [12–14] has established that females have lower gene expression than males at Z-linked genes—a phenomenon referred to as incomplete dosage compensation—and we account for this effect in our analyses of inbreeding and gene expression. We hypothesized that if the sexes were differently affected by inbreeding, the homogametic males would be more sensitive than females due to exposure of recessive deleterious Z-linked alleles during inbreeding. We tested this by analysing how inbreeding influences gene expression at autosomal and Z-linked genes both at the chromosome-wide level and at single genes.
2. Material and methods
We studied captive zebra finches in a population at Lund University (Sweden), which was founded in 2001/2002 by 160 birds from breeders in Denmark [11]. In 2007, we crossed full-sibs and unrelated individuals to produce inbred (f = 0.25) and outbred (control) (f = 0) offspring. Breeding pairs were housed in separate cages (rearing conditions are described elsewhere [14]). Eggs were hatched in an incubator and hatchlings were randomly assigned to foster parents. We selected one offspring of each sex (if available) from each pair, in total 10 inbred and 10 outbred offspring of each sex (i.e. 40 offspring). The offspring were sacrificed by decapitation when 10 days old. Total RNA was extracted from the entire telencephalon of all offspring. Details regarding RNA extraction and quality control are given elsewhere [14,15]. All samples had good-quality RNA with high and comparable RNA integrity numbers.
We used the Lund-zfa array, which is an Affymetrix array based on 22 630 zebra finch ESTs [15]. A detailed description of the array, the hybridization procedure, and normalization and filtering of signal intensities is given in the electronic supplementary material (see also [14,15]). All samples were run on separate arrays (i.e. 40 arrays were used). Redundant ESTs representing the same gene were removed based on their chromosome position in the 3.2.4 zebra finch genome assembly (see the electronic supplementary material for details). Our analyses are based on 10 695 ESTs (10 131 autosomal; 564 Z-linked; Dryad data deposition).
Linear mixed models (R package nlme; http://cran.r-project.org/web/packages/nlme/index.html) were used to analyse gene expression (log2 signal intensity) in relation to three fixed factors (‘sex’: male or female; ‘chromosomal location’: autosomal or Z-linked; ‘inbreeding treatment’: inbred or outbred) and their interactions. To account for potential array effects, we included ‘individual identity’ as a random factor. Significances were assessed with F-tests (see the electronic supplementary material). Differential expression between inbred and outbred individuals at the gene level was analysed with significance analysis of microarray (SAM) [16] using the SAM v. 3.02 Excel plugin. Males and females were analysed separately (because of the pronounced sex difference at Z-linked genes). Two class (unpaired) tests with false discovery rates (FDR) of less than 1% were used (500 permutations). In the same way, we tested for differentially expressed genes between males and females in separate models for inbred and outbred individuals, respectively.
3. Results
The main result from the linear mixed models (table 1) was a significant interaction between sex and chromosomal location (F1,427756 = 224, p < 0.001) caused by females having lower expression at Z-linked genes compared with males (figure 1a). There was also a significant difference between autosomal genes and Z-linked genes, independent of sex (F1,427 756 = 356, p < 0.001). By contrast, there was no significant difference in gene expression between inbred and outbred individuals (p = 0.76; figure 1b), and the inbreeding treatment did not interact significantly with either sex or chromosomal location (p = 0.44 and 0.91; table 1).
Table 1.
Results from a linear mixed model of the association between gene expression (log2 signal) and sex (male versus female), inbreeding treatment (inbred versus outbred) and chromosomal location (autosomal versus Z-linked). Expression was measured at 10 695 genes in 40 young zebra finches. The model also included two random factors: individual identity (to account for array effects) and gene identity (to account for expression differences between genes). Shown are nominator and denominator degrees of freedom (nDf, dDf), F-value (F) and p-value (p).
| variable | nDf, dDf | F | p |
|---|---|---|---|
| intercept | 1, 427 756 | 7 075 060 | <0.0001 |
| sex | 1, 36 | 3 | 0.0739 |
| inbreeding treatment | 1, 36 | ≈0 | 0.7637 |
| chromosomal location | 1, 427 756 | 356 | <0.0001 |
| sex × inbreeding treatment | 1, 36 | 1 | 0.4363 |
| sex × chromosomal location | 1, 427 756 | 224 | <0.0001 |
| inbreeding treatment × chromosomal location | 1, 427 756 | ≈0 | 0.9067 |
| sex × inbreeding treatment × chromosomal location | 1, 427 756 | ≈0 | 0.8849 |
Figure 1.
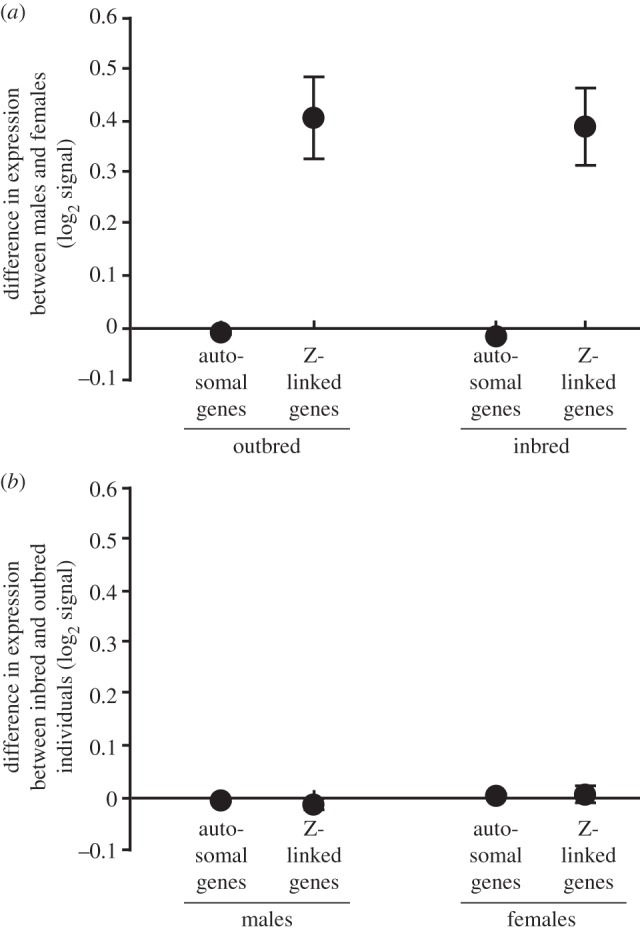
Difference in gene expression (mean ± 99.9% CI log2 signal intensity) (a) between males and females (in outbred and inbred individuals, and at autosomal and Z-linked genes) and (b) between inbred and outbred individuals (in males and females, and at autosomal and Z-linked genes).
The SAM analyses detected a single significantly differentially expressed gene between inbred and outbred individuals: the gene with NCBI Gene ID 100230905 was more highly expressed in inbred than in outbred females (fold change = 1.34). By contrast, a large number of Z-linked genes (330 of 564 in the outbred group; 316 of 564 in the inbred group) and a few autosomal genes (three of 10 131 in the outbred group; three of 10 131 in the inbred group) were significantly differentially expressed between males and females, and the vast majority of those (≈98%) were more strongly expressed in males than in females.
4. Discussion
We found no difference in overall gene expression between inbred and outbred zebra finches, neither in males nor in females, and single-gene analyses detected only a single gene that was differentially expressed between inbred and outbred individuals and then only in females. This gene, a protein-coding gene described as ‘family with sequence similarity 13, member C’ (NCBI Gene ID: 100230905), is located on chromosome 6 and its function is presently unknown. These weak inbreeding effects were surprising given the study population has been kept at a high effective population size, is known to maintain substantial genetic variation [11] and therefore is expected to hold a significant inbreeding load. Moreover, we analysed transcriptome-wide expression on a comparatively large dataset, 10 individuals of each sex in each of the inbreeding and outbreeding groups. Within both the outbred and inbred groups, males and females showed substantial expression differences at Z-linked genes. This sexually differentiated gene expression is caused by incomplete dosage compensation in female birds as previously shown in zebra finch and other species [12–14] and suggests for this study that the lack of inbreeding effects was not caused by low statistical power.
That inbreeding does not influence gene expression in zebra finches contrasts with studies, for example, of D. melanogaster, where inbreeding affected gene expression more widely, and in particular at stress-related genes [3,4]. Moreover, a study of inbred lines and crosses between inbred lines in Pacific oyster has shown genome-wide gene expression responses to inbreeding [6]. There are several potential explanations for why the gene expression response to inbreeding may differ between studies and organisms. First, the inbreeding levels do differ between studies and perhaps the detectable effects would have occurred had the inbreeding been stronger. However, full-sib mating as used in this study is still a quite high level of inbreeding (f = 0.25). Second, there may be intrinsic differences in the genomic distribution of recessive alleles between systems. This could be related to differences in long-term effective population sizes, with more recessive mutations being maintained in large populations, such as in Drosophila. Third, we have studied gene expression in the brain of chicks at a certain time point in early life, and it is possible that other developmental stages are more sensitive to inbreeding [2]. Fourth, inbreeding has been shown to strike more severely under stressful conditions [17,18], and it is therefore possible that stronger inbreeding effects would have occurred had we studied the birds in less benign environments.
To conclude, the surprisingly weak effects of inbreeding on gene expression in zebra finches may suggest that the phenotypic and reproductive consequences of inbreeding that are frequently observed in birds [7,8], including in zebra finches [19], might not be driven primarily by gene regulatory mechanisms. However, the question of whether inbreeding affects gene expression in birds should remain open until other tissues, life-stages and environmental conditions have been investigated.
Ethics statement
The correct ethical approvals for conducting the experiments were obtained from Malmö/Lund's ethical committee.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.ps515.
Supplementary Material
Acknowledgement
We thank SCIBLU genomics and Marcus Ljungqvist for assistance.
Funding statement
This study was financially supported by the Swedish Research Council (B.H. and D.H.) and the Crafoord Foundation (B.H.).
References
- 1.Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. ( 10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 2.Kristensen TN, Pedersen KS, Vermeulen CJ, Loeschcke V. 2010. Research on inbreeding in the ‘omic’ era. Trends Ecol. Evol. 25, 44–52. ( 10.1016/j.tree.2009.06.014) [DOI] [PubMed] [Google Scholar]
- 3.Kristensen TN, Sorensen P, Kruhoffer M, Pedersen KS, Loeschcke V. 2005. Genome-wide analysis on inbreeding effects on gene expression in Drosophila melanogaster. Genetics 171, 157–167. ( 10.1534/genetics.104.039610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen KS, Kristensen TN, Loeschcke V. 2005. Effects of inbreeding and rate of inbreeding in Drosophila melanogaster—Hsp70 expression and fitness. J. Evol. Biol. 18, 756–762. ( 10.1111/j.1420-9101.2005.00884.x) [DOI] [PubMed] [Google Scholar]
- 5.Turk R, Hoen PA, Sterrenburg E, de Menezes RX, de Meijer EJ, Boer JM, van Ommen GJ, den Dunnen JT. 2004. Gene expression variation between mouse inbred strains. BMC Genomics 5, 57 ( 10.1186/1471-2164-5-57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedgecock D, Lin JZ, DeCola S, Haudenschild CD, Meyer E, Manahan DT, Bowen B. 2007. Transcriptomic analysis of growth heterosis in larval Pacific oysters (Crassostrea gigas). Proc. Natl Acad. Sci. USA 104, 2313–2318. ( 10.1073/pnas.0610880104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller LF. 1998. Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia). Evolution 52, 240–250. ( 10.2307/2410939) [DOI] [PubMed] [Google Scholar]
- 8.Jamieson IG, Roy MS, Lettink M. 2003. Sex-specific consequences of recent inbreeding in an ancestrally inbred population of New Zealand takahe. Conserv. Biol. 17, 708–716. ( 10.1046/j.1523-1739.2003.01400.x) [DOI] [Google Scholar]
- 9.Brekke P, Bennett PM, Wang J, Pettorelli N, Ewen JG. 2010. Sensitive males: inbreeding depression in an endangered bird. Proc. R. Soc. B 277, 3677–3684. ( 10.1098/rspb.2010.1144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallet MA, Chippindale AK. 2011. Inbreeding reveals stronger net selection on Drosophila melanogaster males: implications for mutation load and the fitness of sexual females. Heredity 106, 994–1002. ( 10.1038/hdy.2010.148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forstmeier W, Segelbacher G, Mueller JC, Kempenaers B. 2007. Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 16, 4039–4050. ( 10.1111/j.1365-294X.2007.03444.x) [DOI] [PubMed] [Google Scholar]
- 12.Mank JE, Ellegren H. 2009. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102, 312–320. ( 10.1038/hdy.2008.116) [DOI] [PubMed] [Google Scholar]
- 13.Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. 2010. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 20, 512–518. ( 10.1101/gr.102343.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naurin S, Hansson B, Hasselquist D, Kim YH, Bensch S. 2011. The sex-biased brain: sexual dimorphism in gene-expression in two species of songbirds. BMC Genomics 12, 37 ( 10.1186/1471-2164-12-37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naurin S, Bensch S, Hansson B, Johansson T, Clayton DF, Albrekt AS, von Schantz T, Hasselquist D. 2008. A microarray for large-scale genomic and transcriptional analyses of the zebra finch (Taeniopygia guttata) and other passerines. Mol. Ecol. Res. 8, 275–281. ( 10.1111/j.1471-8286.2007.01979.x) [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121. ( 10.1073/pnas.091062498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox CW, Reed DH. 2011. Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution 65, 246–258. ( 10.1111/j.1558-5646.2010.01108.x) [DOI] [PubMed] [Google Scholar]
- 18.Reed DH, Fox CW, Enders LS, Kristensen TN. 2012. Inbreeding–stress interactions: evolutionary and conservation consequences. Ann. NY Acad. Sci. 1256, 33–48. ( 10.1111/j.1749-6632.2012.06548.x) [DOI] [PubMed] [Google Scholar]
- 19.Bolund E, Martin K, Kempenaers B, Forstmeier W. 2010. Inbreeding depression of sexually selected traits and attractiveness in the zebra finch. Anim. Behav. 79, 947–955. ( 10.1016/j.anbehav.2010.01.014) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.ps515.


