Abstract
Learning and memory deficits are early clinical manifestations of Huntington’s disease (HD). These cognitive impairments have been mainly associated with frontostriatal HD pathology; however, compelling evidence provided by several HD murine models suggests that the hippocampus may contribute to synaptic deficits and memory dysfunction in HD. The neurotrophin receptor p75NTR negatively regulates spine density, which is associated with learning and memory; therefore, we explored whether disturbed p75NTR function in the hippocampus could contribute to synaptic dysfunction and memory deficits in HD. Here, we determined that levels of p75NTR are markedly increased in the hippocampus of 2 distinct mouse models of HD and in HD patients. Normalization of p75NTR levels in HD mutant mice heterozygous for p75NTR prevented memory and synaptic plasticity deficits and ameliorated dendritic spine abnormalities, likely through normalization of the activity of the GTPase RhoA. Moreover, viral-mediated overexpression of p75NTR in the hippocampus of WT mice reproduced HD learning and memory deficits, while knockdown of p75NTR in the hippocampus of HD mice prevented cognitive decline. Together, these findings provide evidence of hippocampus-associated memory deficits in HD and demonstrate that p75NTR mediates synaptic, learning, and memory dysfunction in HD.
Introduction
Evidence of cognitive deficits including altered acquisition of new motor skills, paired attention, planning, and memory has been demonstrated in Huntington’s disease (HD) patients before the onset of motor symptoms (1–3). These clinical signs have been mainly attributed to corticostriatal dysfunction (4, 5). However, in recent years the idea has emerged that memory decline in HD is likely a reflection of a widespread brain circuitry defect and not exclusively a dysfunction of the basal ganglia (6–8). Indeed, besides the caudate and putamen, the volume of the hippocampus is reduced in premanifest HD individuals, while no changes in the amygdala, thalamus, or pallidum are observed (9). Interestingly, spatial and recognition memories have been reported to be altered in HD patients. Thus, in moderately advanced HD patients there is a simultaneous impairment of allocentric (hippocampal-dependent) and egocentric (striatal-dependent) spatial navigation, supporting the idea that beyond the atrophy of the striatum and cortex, a more general neurodegenerative process that involves the hippocampus could contribute to HD memory impairment (2, 10, 11). Actually, cytoplasmic and nuclear huntingtin aggregates within the hippocampus have been described in HD individuals (12). Importantly, such behavioral deficits, together with hippocampal long-term potentiation (LTP) disturbances, have been replicated in different HD mouse models (2, 13–15). Although altered synaptic plasticity and aberrant dendritic spine density and morphology have been proposed as underlying mechanisms (14, 16, 17), little is known about the precise molecular pathways involved in HD synaptic and memory disturbances.
In the adult brain, neurotrophins play a critical role in synaptic plasticity regulation. Among the different neurotrophins, brain-derived neurotrophic factor (BDNF) is the best characterized for its role in regulating LTP and long-term depression (LTD) through binding to TrkB and p75NTR receptors (18). It is generally accepted that BDNF via interaction with TrkB receptors modulates synaptic transmission and plasticity in adult synapses by regulating transcription, translation, and trafficking of distinct synaptic proteins (19, 20). Less is known about the role of p75NTR in synaptic plasticity. Null p75NTR mice show improved spatial learning and enhanced LTP (21, 22) while impaired NMDA-dependent LTD (23), which points to an antagonistic role of p75NTR in synaptic plasticity. Moreover, it has been demonstrated that p75NTR is a negative modulator of spine-dendrite morphology and complexity (24), likely by regulation of RhoA activity (25). Consistently, while p75NTR–/– mice exhibit increased hippocampal dendritic spine density, overexpression of p75NTR in hippocampal neurons decreases spine number and branching (24). Interestingly, upregulation of p75NTR levels has been reported in the cortex and hippocampus of Alzheimer’s disease (AD) patients (26), while small-molecule p75NTR ligands prevent both cognitive decline and neuritic dystrophy in AD mouse models (27–29). In this scenario, we were interested to explore whether p75NTR plays a role in different memory deficits in HD. To this aim we have examined the levels of p75NTR in the hippocampus of 2 distinct HD mouse models, exon 1 and full-length, as well as in the hippocampus of HD patients. p75NTR levels were increased at early HD disease stages in the hippocampus of HD mice and also in postmortem samples from HD patients. Increased p75NTR levels were accompanied by long-term memory deficits, altered LTP, reduced levels of synaptic proteins, decreased dendritic spine density, and hyperactivation of the small GTPase RhoA. Such alterations were mimicked by overexpression of p75NTR in WT mice and reversed by genetic normalization of p75NTR levels in HD mutant mice. Finally, specific reduction of p75NTR in the hippocampus of HD mice by intracranial injections of AAV-shp75NTR prevented HD memory impairments. In summary, our findings identify p75NTR as a crucial mediator of synapse loss and cognitive decline in HD and provide mechanistic rationale for p75NTR as a novel target to treat memory deficits in HD.
Results
Increased p75NTR expression in the hippocampus of HD mouse models and HD patients.
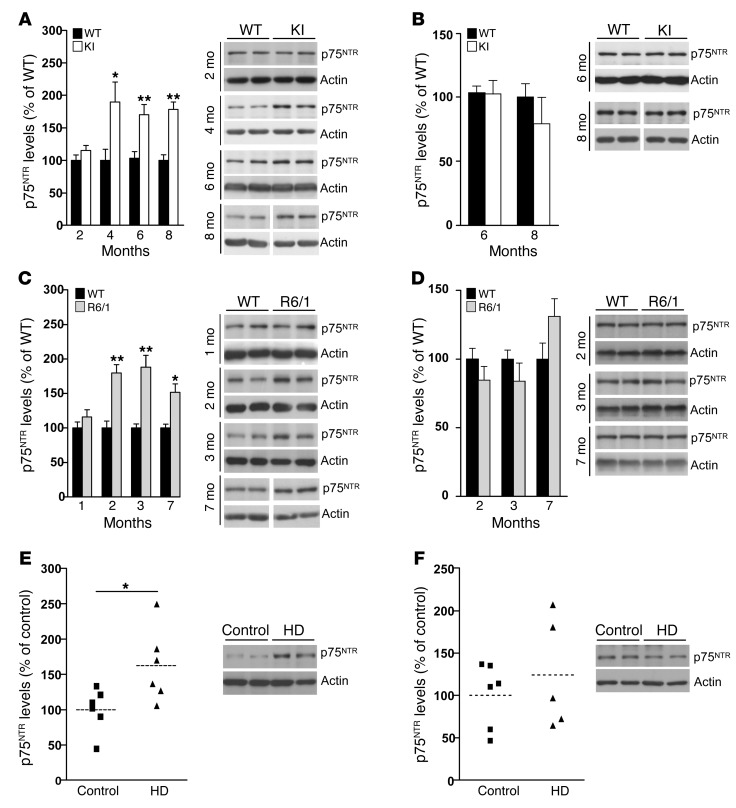
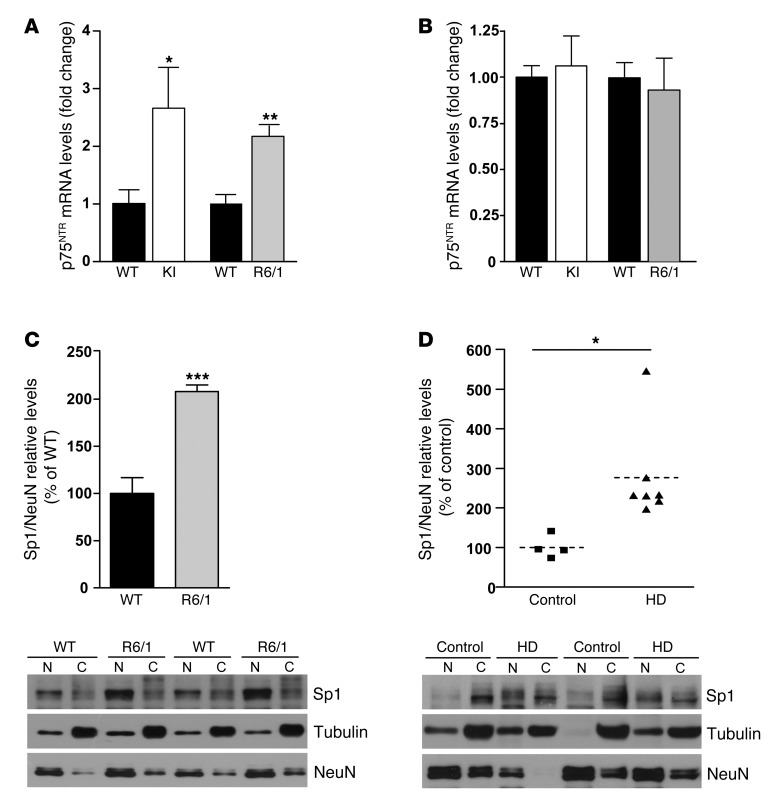
Previous data from our group demonstrated increased levels of p75NTR in the striatum of HD mice as well as in the putamen of HD patients (30). To analyze the role of p75NTR in HD memory dysfunction, we extended these data by evaluating hippocampal and cortical p75NTR levels in 2 HD mouse models at different stages and in postmortem human HD samples. The 2 different HD mouse models, R6/1 and HdhQ7/Q111 knock-in mutant mice (mutant KI mice), differ in the onset and progression of HD phenotypes; R6/1 mice show earlier onset and faster disease progression than mutant KI mice (31). Quantitative Western blot analysis revealed a significant increase in hippocampal p75NTR levels in HD mutant mice at early disease stages (2 months and 4 months in R6/1 and KI mice, respectively), levels that remained elevated at late stages (Figure 1, A and C). Interestingly, cortical p75NTR levels were similar between genotypes (Figure 1, B and D). To analyze whether deregulation of p75NTR levels also occurs in humans, we examined hippocampal and cortical tissue from controls and individuals affected by HD. Consistent with our data in HD mice, levels of p75NTR were found increased in the hippocampus but not in the cortex of HD patients compared with controls (hippocampus: control, 100.1 ± 12.68; HD, 162.4 ± 21.69; cortex: control, 100.02 ± 17.14; HD, 124.3 ± 22.7; Figure 1, E and F, respectively), which supports a pathological role for p75NTR in HD. The increase in hippocampal p75NTR protein levels was likely due to increased p75NTR transcription, since higher mRNA levels measured by RT-PCR were found in the hippocampus but not in the cortex of HD versus WT mice (Figure 2, A and B). We next aimed to determine the molecular mechanism by which mutant huntingtin induces aberrant hippocampal p75NTR expression. The transcription factor Sp1 has been described to drive expression of p75NTR under cellular stress conditions (32, 33), while upregulation of Sp1 has been described in cellular and mouse models of HD (34). These data prompted us to investigate whether increased p75NTR expression was related to higher hippocampal Sp1 levels. Thus, Sp1 levels were determined by Western blot analysis in nuclear and cytosolic enriched fractions obtained from R6/1 and HD human hippocampus (Figure 2, C and D). Interestingly, we found a significant increase in Sp1 levels in the nuclear fraction in R6/1 and HD human hippocampus compared with WT mice and control human samples, suggesting that deregulated Sp1 expression could contribute to aberrant expression of p75NTR in the HD hippocampus.
Figure 1. p75NTR expression is increased in the hippocampus but not in the cortex of 2 different HD mouse models and in HD human brain.
(A–D) Western blot for p75NTR and actin as loading control in total hippocampus and cortex extracts from WT and KI mice (A and B) or WT and R6/1 mice (C and D) at different ages (n = 5–7 per genotype). Right: Representative immunoblots. At 8 and 7 months, lanes were run on the same gel but were noncontiguous (white line). (E and F) Western blot for p75NTR and actin as a loading control in total hippocampus and cortex extracts from control and HD brain samples (n = 6–7). Right: Representative immunoblots. All plots represent mean ± SEM. Student’s 2-tailed t test was performed. *P < 0.05, **P < 0.01 compared with WT mice or control human samples.
Figure 2. Increased hippocampal p75NTR mRNA expression associates with higher levels of Sp1 in the mouse and human HD hippocampus.
(A and B) Histograms showing p75NTR mRNA expression analyzed by RT-PCR in the hippocampus (A) and cortex (B) of 8-month-old WT and KI mice and 12-week-old WT and R6/1 mice. Results were normalized to 18S gene expression. Data represent mean ± SEM and are expressed as fold change. (n = 5–8 for each genotype.) (C and D) Western blot for Sp1 in nuclear and cytosolic enriched fractions from the hippocampus of WT and R6/1 mice at 14 weeks of age and from control and HD patients (n = 4–6). Representative immunoblots showing Sp1 protein levels in cytosolic (loading control α-tubulin) and nuclear (loading control NeuN) fractions. Data were analyzed by Student’s 2-tailed t test. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with WT mice or control human samples.
Altered p75NTR synaptic location in HdhQ7/Q111 knock-in mutant mice.
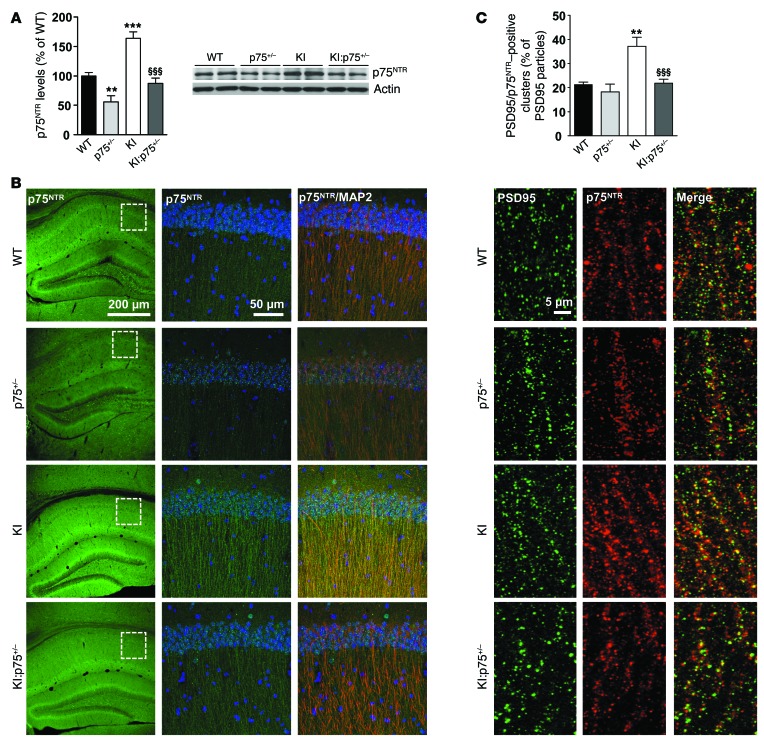
Mutant KI and R6/1 mice develop age-dependent memory deficits (13, 15). Given the role of p75NTR in synaptic plasticity (19, 24, 35), we next analyzed whether the increase in p75NTR levels in the hippocampus of mutant KI mice could be involved in HD memory deficits. To address this question, levels of p75NTR were normalized in KI mice by cross-mating of HdhQ7/Q111 mice with p75NTR/ExonIII+/– mice (p75+/– mice) to obtain double-mutant mice (KI:p75+/– mice). Hippocampal extracts from WT, mutant KI, heterozygous p75+/–, and double-mutant KI:p75+/– mice were analyzed by Western blot. As expected, quantification of band intensities revealed a significant increase of p75NTR in mutant KI mice, an increase that was reversed in double-mutant KI:p75+/– mice (Figure 3A). Confocal analysis of brain sections showed p75NTR immunoreactivity as a punctate staining in pyramidal cell bodies as well as in fibers concentrated in the stratum radiatum and more dispersed in the stratum oriens within the CA1 region of the hippocampus. Similar staining was observed in CA3 and the dentate gyrus (data not shown). According with our biochemical data, p75NTR immunoreactivity in mutant KI mice was higher than in WT animals, whereas no significant differences were detected between double KI:p75+/– and WT mice (Figure 3B). When subcellular localization was analyzed in the CA1 region, we found that in all analyzed genotypes p75NTR immunoreactivity colocalized with MAP2, a dendritic marker, suggesting that p75NTR is expressed by hippocampal neurons. To further analyze the distribution of p75NTR, double immunostaining using p75NTR and PSD95 as a glutamatergic postsynaptic marker was conducted. We found that in apical dendrites of the CA1 region p75NTR staining colocalized with PSD95 immunoreactivity, revealing a postsynaptic distribution of p75NTR (Figure 3C). Interestingly, whereas in WT mice about 20% of PSD95-positive clusters were positive for p75NTR, in mutant KI mice this percentage rose to 40%, suggesting that the increase in p75NTR in mutant KI mice is localized in spines. Importantly, this increase was completely reversed in double-mutant mice. Since p75NTR is also expressed by astrocytes, especially after neuronal damage (36, 37), coimmunostaining with the astrocytic marker GFAP was performed in hippocampal slices from WT and mutant KI mice (see Supplemental Figure 6; supplemental material available online with this article; doi:10.1172/JCI74809DS1). Lack of colocalization between p75NTR and GFAP was found, indicating that astrocytes in WT and KI mutant mice do not overexpress p75NTR. These findings together with our data showing increased p75NTR staining in KI hippocampus colocalizing with MAP2 suggest that neuronal p75NTR upregulation underlies hippocampal HD dysfunction.
Figure 3. Increased postsynaptic localization of p75NTR in mutant KI mice.
(A) Western blot for p75NTR and actin as loading control in total hippocampus extracts from WT, p75+/–, mutant KI, and double-mutant KI:p75+/– mice at 6 months of age (n = 6–7 per genotype) showing normalization of p75NTR levels in double-mutant mice. Right: Representative immunoblots. (B) Representative confocal microscopy images (low magnification) showing the expression pattern of p75NTR in 8-month-old mouse hippocampus. Magnified images (right) show colocalization between p75NTR and MAP2 in the CA1 hippocampal region. (C) Representative confocal microscopy images (high magnification) showing p75NTR and PSD95 colocalization in the CA1 hippocampal region. Quantitative analysis reveals a significant increase in the percentage of PSD95-positive clusters also positive for p75NTR in KI mice. Data are presented as mean ± SEM. One-way ANOVA with Tukey post hoc comparisons was performed; **P < 0.01 and ***P < 0.001 compared with WT mice, §§§P < 0.001 compared with KI mice.
Genetic normalization of p75NTR levels rescues spatial, recognition, and associative memory deficits in HdhQ7/Q111 knock-in mutant mice.
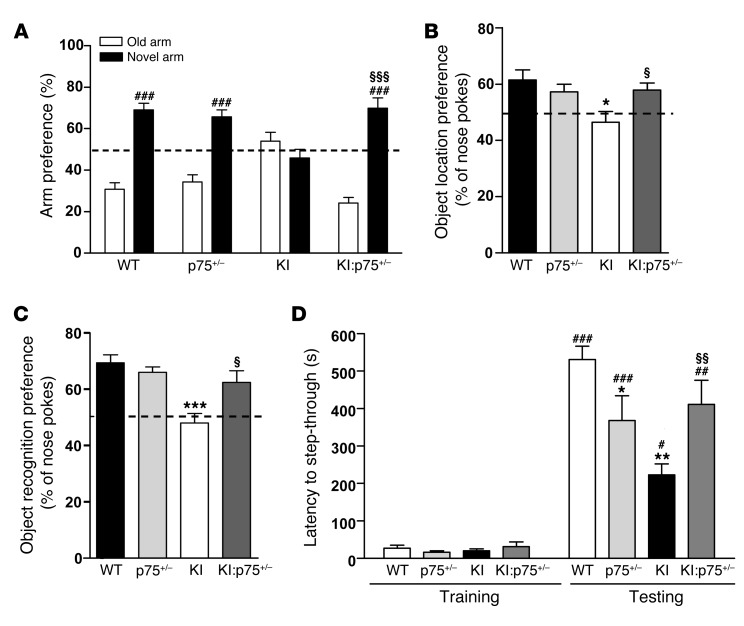
We next assessed whether aberrant hippocampal p75NTR expression was associated with HD memory impairments. Spatial, recognition, and associative memories were evaluated in double-mutant KI:p75+/– mice at 6 and 8 months of age, since previous results from our lab demonstrated lack of recognition memory deficits in mutant KI mice at 4 months of age (15). First, animals were tested in the elevated plus maze and the light-dark exploration paradigms to assess anxiety-related behaviors. These tests are based on the natural aversion of rodents to elevated open spaces and brightly illuminated spaces, respectively (38, 39). In the elevated plus maze paradigm (Supplemental Figure 1A), all genotypes displayed similar exploratory activity measured as the time in the open arms and the distance traveled, suggesting that neither KI, p75+/–, nor KI:p75+/– mice exhibit higher anxiety levels compared with WT mice. As a second test for anxiety-like behaviors we performed the light-dark box test (Supplemental Figure 1B). Consistent with our previous data, we found that all mice, independently of their genotype, made similar transitions to the light compartment and spent similar time in the light. We conclude from these data that nonprominent anxiety-like behaviors are manifested by any of the genotypes studied. Then, spatial, recognition, and associative memories were analyzed (Figure 4). Spatial memory was evaluated by the T-maze spontaneous alternation task (T-SAT) and the object location task (OLT) (40, 41), both behavioral tasks sensitive to hippocampal dysfunction albeit not completely dependent (42–44). In the T-SAT test, mice have the natural tendency to alternate and enter the previously unvisited arm (novel arm) in a T-maze apparatus. At 6 months of age, mutant KI mice showed no preference for the novel versus the old arm, while a significant preference was observed in WT, p75+/–, and double-mutant KI:p75+/– mice (Figure 4A). In the OLT, all mice were first habituated to the open field arena and ambient conditions and then subjected to a training session in the presence of 2 similar objects (A1 and A2). All mice similarly explored both objects, indicating no object or place preferences between genotypes (Supplemental Figure 1C). This test is based on the ability of rodents to recognize when a familiar object has been relocated. When spatial memory was assessed 24 hours after training, KI mice exhibited a significantly lower preference for the object displaced to the new location compared with WT, p75+/–, or double-mutant KI:p75+/– mice (Figure 4B). Overall, these results indicate that normalization of p75NTR levels in KI mice rescue spatial memory deficits. Similar results were obtained at 8 months of age in the T-SAT test, suggesting that reduced p75NTR expression in HD mice prevented, rather than merely delayed, spatial memory deficits (Supplemental Figure 2A). We next evaluated recognition memory by using the novel object recognition test (NORT), based on the natural tendency of mice to spend more time exploring a novel object than a familiar one and known to be dependent on hippocampal and cortical circuits (45–47). No significant differences between genotypes were found during the training period, indicating no object preference (Supplemental Figure 1D). Twenty-four hours after training, mutant KI mice at either 6 months (Figure 4C) or 8 months of age (Supplemental Figure 2B) exhibited a significantly lower preference for the novel object compared with WT, p75+/–, and double-mutant KI:p75+/– mice, indicating preserved long-term recognition memory in double but not in mutant KI mice. Finally, we examined associative memory in the passive avoidance task, based on the association formed between an aversive stimulus (electrical foot shock) and a specific environmental context (light-dark) (Figure 4D), which relies on cortical and hippocampal circuits (48–50). Latency to step-through during the training session was found to be similar between genotypes. However, in the testing session, although all genotypes showed a significant increase in the latencies to enter the dark compartment 24 hours after receiving an electrical shock, the time latency in mutant KI and p75+/– mice was lower than in WT or double-mutant KI:p75+/– mice, indicating that altered levels of p75NTR contribute to associative memory impairments. Altogether, these results demonstrate memory decline in mutant KI mice that was prevented by normalization of p75NTR levels.
Figure 4. Normalization of p75NTR levels in mutant KI mice rescues spatial and nonspatial memory deficits.
(A) Percentage of time spent in arms (old versus novel) from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age (n = 8–12 per genotype). Mutant KI mice exhibit no preference for a previously unexposed (novel) arm of a T-maze. (B) Percentage of nose pokes to the displaced object from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age. Mutant KI mice showed significantly less preference for the novel-object location (n = 7–8 per genotype). (C) Percentage of nose pokes to the new object from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age (n = 8–12 per genotype) in the novel object recognition task. Mutant KI mice display no preference for the novel object. (D) Latency to step-through from light to dark compartment from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age (n = 8–12 per genotype). Mutant KI mice present worse (shorter latency to crossover) retention performance than the other genotypes. One-way ANOVA with Bonferroni post hoc comparisons; *P < 0.05, **P < 0.01, and ***P < 0.001 compared with WT mice; §P < 0.05, §§P < 0.01, and §§§P < 0.001 compared with KI mice. #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with old arm or latency to step-through in the training. Data are presented as mean ± SEM.
Genetic normalization of p75NTR levels rescues hippocampal synaptic dysfunction in HdhQ7/Q111 knock-in mutant mice.
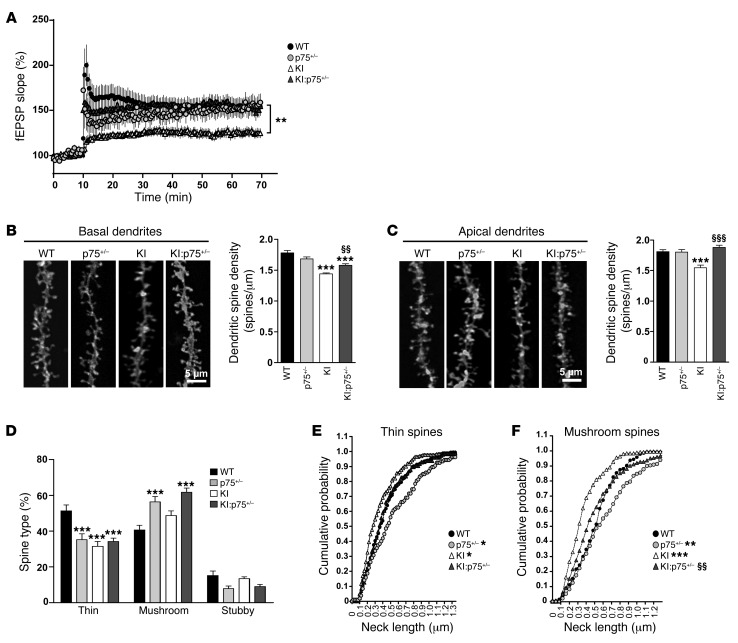
Defective LTP and altered dendritic spine dynamics and morphology are involved in early cognitive and memory deficits in HD (14, 16, 51). Because normalization of p75NTR levels prevented memory impairments in mutant KI mice, we next examined whether this improvement was accompanied by LTP recovery and/or amelioration of dendritic spine loss. High-frequency conditioning tetanus (HFS) to induce LTP was used to investigate synaptic plasticity in CA1 hippocampal slices at 6 months of age. Baseline responses were monitored for 10–30 minutes before conditioning and were found to be stable. Mutant KI mice showed diminished HFS-induced LTP compared with WT animals (Figure 5A). Thus, at 60 minutes after tetanus, potentiation (as mean percentage of baseline) in WT mice was 154.3% ± 9% (n = 8 slices, 3 mice) versus 124.9% ± 5.2% in mutant KI mice (P < 0.01, n = 20 slices, 9 mice). By contrast, tetanic stimulation induced in either double-mutant KI:p75+/– mice or p75+/– mice led to sustained LTP (KI:p75+/–: 154.9% ± 8%, P < 0.01, n = 14 slices, 8 mice; p75+/–: 159.02% ± 10%, P < 0.01, n = 9 slices, 4 mice), which indicates that normalization of p75NTR levels in mutant KI mice restored the LTP impairment.
Figure 5. Normalization of p75NTR levels in mutant KI mice prevents functional and structural synaptic plasticity deficits.
(A) Time course of fEPSP potentiation during HSF-induced LTP in WT (n = 3), p75+/– (n = 4), KI (n = 9), and KI:p75+/– (n = 8) mice at 6 months of age. **P < 0.01 compared with WT mice. (B and C) Representative basal (B) and apical dendrites (C) of CA1 pyramidal neurons from WT, p75+/–, KI, and KI:p75+/– mice at 8 months of age. Right: Quantitative analysis showing dendritic spine density per micrometer of dendritic length. Mutant KI mice exhibit a significant reduction in dendritic spines that was significantly ameliorated in double-mutant mice. One-way ANOVA with Tukey post hoc comparisons was performed (63–83 dendrites; n = 4–5 animals per genotype); ***P < 0.001 compared with WT mice; §§P < 0.01, §§§P < 0.001 compared with KI mice. (D) Percentage of each morphological type of dendritic spine (see Methods and Supplemental Figure 4 for classification criteria) from WT, p75+/–, KI, and KI:p75+/– basal dendrites at 8 months of age. One-way ANOVA with Tukey post hoc comparisons was performed (480 spines from 100 dendrites from 4 animals per genotype were analyzed); ***P < 0.001 compared with WT mice. (E and F) Spine neck length distribution was examined by plotting of the cumulative frequency of neck length of all examined spines and comparison of distributions using the Kolmogorov-Smirnov test. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with WT mice. §§P < 0.01 compared with KI mice.
Next, hippocampal histology and spine density were examined. Stereological estimation of hippocampal volume and CA1 pyramidal cell density revealed no significant differences between genotypes at 8 months of age (Supplemental Figure 3), suggesting no gross anatomical deficiencies in KI or KI:p75+/– mice. To compare spine density and morphology, DiOlistic labeling in fixed brain slices was used and apical and basal dendritic spines of CA1 pyramidal neurons were counted. Mutant KI mice displayed a significant decrease (~20%) in basal and apical dendritic spine density compared with WT mice (Figure 5, B and C). Normalization of p75NTR levels completely (apical dendrites) or partially (basal dendrites) prevented the decay in spine density in mutant KI mice, suggesting that aberrant p75NTR expression in HD could contribute to loss of dendritic spines. To elucidate whether increased p75NTR levels also affect spine morphology, dendritic spine type was assessed on basal dendrites of CA1 pyramidal neurons (Figure 5D). Spines were defined as thin, stubby, or mushroom on the basis of morphology described by Harris and Stevens (ref. 52 and Supplemental Figure 4). Mutant KI mice exhibited altered spine distribution with a significant decrease (~20%) in the proportion of thin spines. Surprisingly, both p75+/– and double-mutant KI:p75+/– mice also presented less thin spines compared with WT mice (~20%). However, in these 2 genotypes the proportion of mushroom spines was significantly higher (~15%–20%), which may indicate a compensatory mechanism. Finally, we assessed the spine neck length, which is known to influence Ca2+-dependent signaling and synaptic plasticity (53). These data were plotted as cumulative distribution for each animal to examine in detail the potential differences (Figure 5, E and F). Cumulative probability indicates that either increased or decreased levels of p75NTR alter spine neck morphology. Thus, while in mutant KI mice both thin and mushroom spines exhibited shorter necks, in p75+/– mice the neck lengths were longer. By contrast, equal graph distributions were found between WT and double-mutant KI:p75+/– mice, indicating that both genotypes display spines with similar neck lengths (Figure 5, E and F). Altogether, these data imply that HD mutant mice exhibited not only reduced dendritic spine density but also a shift in the spine distribution and morphology. Remarkably, these dendritic changes were significantly improved by reduction of aberrant p75NTR levels.
Deregulation of synaptic-related proteins in KI mutant mice is also reversed by genetic normalization of p75NTR levels.
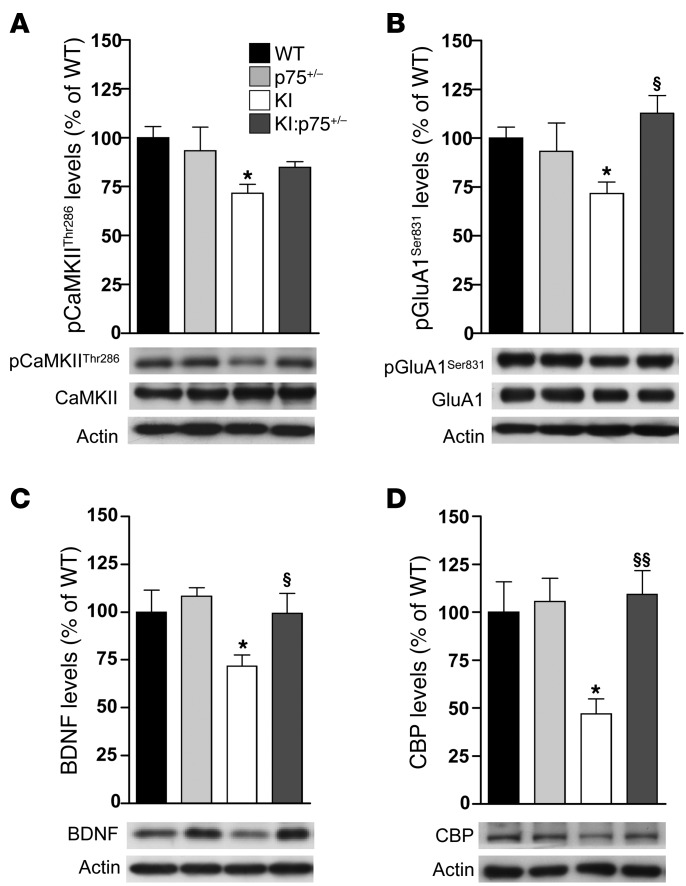
Decreased levels of synaptic-related proteins have been associated with memory impairments and aberrant synaptic plasticity in HD (15, 16, 54, 55). Thus, we next analyzed several synaptic markers and proteins involved in hippocampal synaptic plasticity in the hippocampus of mutant KI mice. Similar levels of the presynaptic marker synaptophysin and the postsynaptic markers GluN1 and GluN2B (NMDA receptor subunits) and GluA1 and GluA2/3 (AMPA receptor subunits) were found between genotypes (Supplemental Table 1). However, levels of phospho-CaMKIIThr286, a key modulator of hippocampal activity through phosphorylation of NMDA and AMPA receptors (56), were significantly reduced in KI mice, a reduction that was prevented by normalization of p75NTR levels in double KI:p75+/– mice (Figure 6A). Consistent with these data, the levels of the CaMKII-mediated GluA1Ser831 phosphorylation were decreased in KI mice, while no significant differences were found in double-mutant mice (Figure 6B). Interestingly, the levels of 2 glutamatergic-scaffolding proteins such as PSD95 and SAP102 were found significantly increased in double KI:p75+/– mice compared with WT mice (Supplemental Table 1), which could be related to the increase in mushroom spines previously shown in these mice (Figure 4D). Finally, levels of BDNF and CREB-binding protein (CBP), previously found to be downregulated in the HD hippocampus (15, 54), were tested. As expected, levels of BDNF and CBP were decreased in the hippocampus of mutant KI mice, whereas no significant differences were found when double KI:p75+/– mice were analyzed (Figure 6, C and D). Thus, altogether these findings suggest that disruption of LTP and memory deficits in KI mutant mice involve deregulation of synaptic-related proteins and that normalization of p75NTR levels prevents such alterations.
Figure 6. Normalization of p75NTR levels in mutant KI mice reverses the altered expression of synaptic-related proteins.
Representative Western blots showing (A) total and phospho-CaMKIIThr286, (B) total and phospho-GluA1Ser831, (C) BDNF, and (D) CBP and actin as loading control in total hippocampus extracts from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age (n = 6–7 per genotype). All histograms represent mean ± SEM. One-way ANOVA with Tukey post hoc comparisons was performed. *P < 0.05 compared with WT mice; §P < 0.05 and §§P < 0.01 compared with KI mice. pCaMKIIThr286, phospho–Ca2+/calmodulin-dependent protein kinase II at threonine 286; pGluA1Ser831, phospho–α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor subunit 1 at serine 831; BDNF, brain-derived neurotrophic factor; CBP, CREB-binding protein.
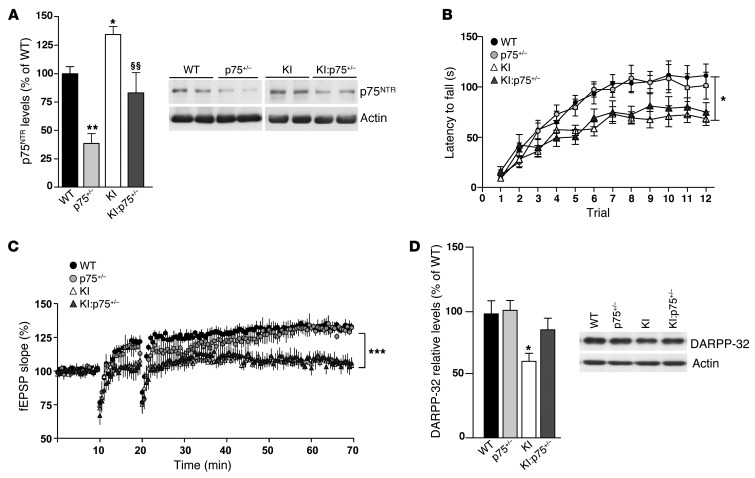
Genetic normalization of p75NTR levels partially restores striatal pathology in HdhQ7/Q111 knock-in mutant mice.
Our data demonstrate that downregulation of p75NTR levels in KI mutant mice fully prevented synaptic and cognitive deficits exhibited by these mice. However, these improvements could be related to the normalization of p75NTR levels in other brain regions known to be affected in HD, such as the striatum, in which p75NTR levels have been demonstrated to be increased (30, 57). Therefore, we next analyzed whether striatal normalization of p75NTR levels in KI mutant mice (Figure 7A) could also ameliorate striatal-dependent behavior, synaptic deficits, and striatal neuropathology. To this aim, motor learning, corticostriatal synaptic transmission, and DARPP-32 expression were analyzed (Figure 7). Compared with WT mice, KI mutant mice at 6 months of age displayed motor learning deficits as evidenced by decreased latency to fall during the accelerating rotarod test (Figure 7B). Importantly, impaired performance in the accelerating rotarod was not prevented or ameliorated by normalization of p75NTR levels in KI:p75+/– mice. Consistent with these findings, when corticostriatal synaptic plasticity was analyzed by induction of LTP through HFS, both KI and KI:p75+/– mice displayed impaired LTP induction and maintenance (Figure 7C). Finally, the levels of the adult striatal neuronal marker DARPP-32, known to be reduced in HD mice from early stages (58), were determined by Western blot analysis in total striatal extracts obtained from WT, p75+/–, KI, and KI:p75+/– mice (Figure 7D). Interestingly, normalization of p75NTR levels rescued reduction of striatal DARPP-32 expression in KI mice, suggesting that striatal neuronal dysfunction could be slightly ameliorated but not prevented by downregulation of aberrant p75NTR expression in KI mutant mice.
Figure 7. Normalization of p75NTR levels in mutant KI mice slightly restores striatal pathology.
(A) Western blot for p75NTR and actin as loading control in total striatal extracts from WT, p75+/–, mutant KI, and double mutant KI:p75+/–. (B) Latency to fall in the accelerating rotarod task procedure in WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age. Data represent the mean ± SEM (n = 9–14 per genotype). Statistical analysis was performed using 2-way ANOVA with repeated measures. *P < 0.05 and **P < 0.01 compared with WT; §§P < 0.05 compared with KI. (C) Time course of fEPSP potentiation during HSF-induced LTP in WT (n = 7), p75+/– (n = 8), KI (n = 8), and KI:p75+/– (n = 9) mice at 4 months of age. For each slice, data were normalized to the average slope recorded during baseline. Data represent the mean ± SEM. Statistical differences, compared with pre-tetanus baseline amplitude values, were established using Student’s 2-tailed t test. ***P < 0.001. (D) Western blot for DARPP-32 and actin as loading control in total striatal extracts from WT, p75+/–, KI, and KI:p75+/– mice at 6 months of age (n = 6 per genotype). Right: Representative immunoblots. Plot represents mean ± SEM. Student’s 2-tailed t test was performed; *P < 0.05.
Intrahippocampal infusion of AAV-GFP-p75 in HdhQ7/Q7 WT mice reproduces HD memory deficits.
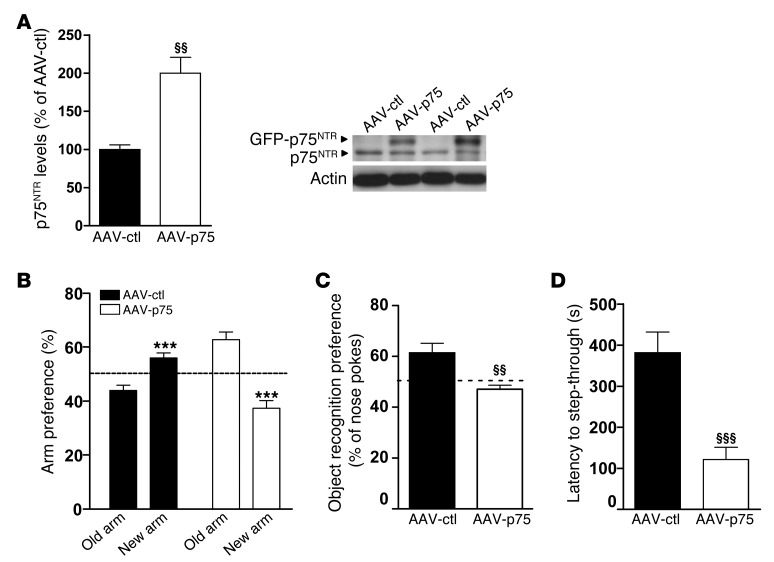
To further evaluate whether the aberrant increase in hippocampal p75NTR levels could contribute to memory impairments in HD mutant mice, we next tested whether overexpression of p75NTR only in the hippocampus of WT mice was able to mimic HD memory deficits. First, we analyzed whether the virus serotype AAV2/8, used to overexpress p75NTR, was able to infect glia and/or neuronal cells (Supplemental Figure 6B). Fluorescence microscopy analysis demonstrated that AAV2/8 efficiently transduces hippocampal neurons but not astrocytes, indicating that the 2/8 serotype is greatly specific for neurons. Then, AAV2/8 serotype specificity was tested in vivo. Slices from WT animals injected with AAV2/8-p75NTR viruses were stained with antibodies against GFAP, and colocalization with GFP was analyzed by confocal microscopy (Supplemental Figure 6C). Consistent with our in vitro data, no overlap in staining between GFAP and GFP was found, indicating that AAV2/8 viruses primarily infect neuronal cells within the hippocampus. First, we confirmed by Western blot analysis higher p75NTR expression in the hippocampus of AAV-p75 mice compared with AAV-GFP (AAV-ctl) animals (Figure 8A). When memory function was evaluated, WT mice overexpressing p75NTR showed impaired spatial and nonspatial long-term memories manifested as reduced preference for either the novel arm or the novel object in the T-SAT and NORT paradigms, respectively (Figure 8, B and C) and by a significant retention deficit in the passive avoidance task (Figure 8D). These experiments demonstrate that overexpression of p75NTR in the WT hippocampus reproduces HD-like memory deficits and support an important role for p75NTR in HD cognitive dysfunction.
Figure 8. Intrahippocampal infusion of AAV-GFP-p75 in WT mice recapitulates the cognitive deficits observed in KI mice.
(A) Western blot for p75NTR and actin as loading control in total hippocampus extracts from AAV-GFP– (AAV-ctl–) and AAV-p75–transduced WT mice. A significant increase in p75NTR levels was found in AAV-p75 mice. Histogram represents mean ± SEM (n = 8 per genotype at 6 months of age). Student’s 2-tailed t test was performed. §§P < 0.01 compared with AAV-ctl mice. (B) Percentage of time spent in arms (old versus novel) from AAV-ctl and AAV-p75 mice at 6 months of age (n = 8–12 per genotype). AAV-p75 mice exhibit no preference for a previously unexposed (novel) arm of a T-maze. (n = 8 per genotype.) Student’s 2-tailed t test was performed. ***P < 0.001 compared with old arm. (C) Percentage of nose pokes to the new object from AAV-ctl and AAV-p75 mice at 6 months of age in the novel object recognition task (n = 8 per genotype). AAV-p75 mice display no preference for the novel object. Student’s 2-tailed t test was performed. §§P < 0.01 compared with AAV-ctl mice. (D) Latency to step-through from light to dark compartment from AAV-ctl and AAV-p75 at 6 months of age (n = 8 per genotype). AAV-p75 mice exhibit worse (shorter latency to crossover) retention performance than AAV-ctl mice. Student’s 2-tailed t test was performed. §§§P < 0.001 compared with AAV-ctl mice. All histograms represent mean ± SEM.
Knockdown of p75NTR expression in the hippocampus of KI mice prevents spatial, object recognition, and associative memories.
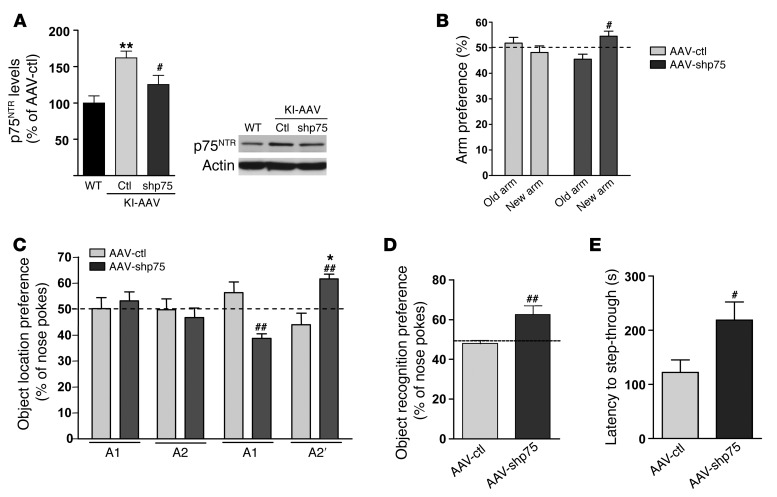
Since hippocampal p75NTR overexpression mirrored HD memory impairments, we next tested whether the specific reduction of p75NTR in the hippocampus of KI mice was sufficient to improve such memory deficits. To this aim, adenoviruses expressing shRNAp75 (AAV-shp75) were infused bilaterally in the dorsal hippocampus of KI mice at 4 months of age, and 1 month later mice were assessed by means of spatial, recognition, and associative memories (Figure 9). First, we tested the efficiency of shRNAp75 in knocking down p75NTR by testing p75NTR hippocampal levels. Western blot analysis revealed a significant decrease of 30% in p75NTR levels in KI mice infused with AAV-shp75 compared with those infused with AAV-ctl (Figure 9A). Then, memory function was assessed. Importantly, reduction of hippocampal p75NTR levels in KI mice to levels of WT completely reversed spatial, recognition, and associative memory deficits (Figure 9, B–E), suggesting that increased expression of p75NTR in the hippocampus of HD mice is a key contributor of cognitive dysfunction in HD.
Figure 9. Intrahippocampal infusion of AAV-shp75 in KI mice at 6 months restores spatial, recognition, and associative memories.
(A) Western blot showing a significant reduction in p75NTR levels in total hippocampus extracts from AAV-shp75–injected KI mice (n = 8 per genotype). **P < 0.01 compared with WT mice, #P < 0.05 compared with AAV-ctl mice. (B) Percentage of time spent in arms from AAV-ctl and AAV-shp75 mice (n = 8–12 per genotype). AAV-shp75 mice exhibit preference for a previously unexposed arm of a T-maze. (n = 8 per genotype.) #P < 0.05 compared with AAV-ctl mice. (C) Percentage of time exploring the new object location (A2) from AAV-ctl and AAV-shp75 mice in the novel-object location task (n = 7–8 per genotype). AAV-shp75 mice showed significantly greater preference for the novel-object location. ##P < 0.01 compared with AAV-ctl mice, *P < 0.05 compared with familiar object location. (D) Percentage of nose pokes to the new object from AAV-ctl and AAV-shp75 mice in the NORT (n = 8 per genotype). AAV-shp75 mice display preference for the novel object. ##P < 0.01 compared with AAV-ctl mice. (E) Latency to step-through from light to dark compartment from AAV-ctl and AAV-shp75 (n = 8 per genotype). AAV-shp75 mice exhibit more latency to crossover than AAV-ctl. #P < 0.05 compared with AAV-ctl mice. All histograms represent mean ± SEM. Student’s 2-tailed t test was performed in A, D, and E and 1-way ANOVA with Bonferroni post hoc comparisons was performed in B and C.
Increased RhoA activity contributes to loss of dendritic spines in KI mutant mice.
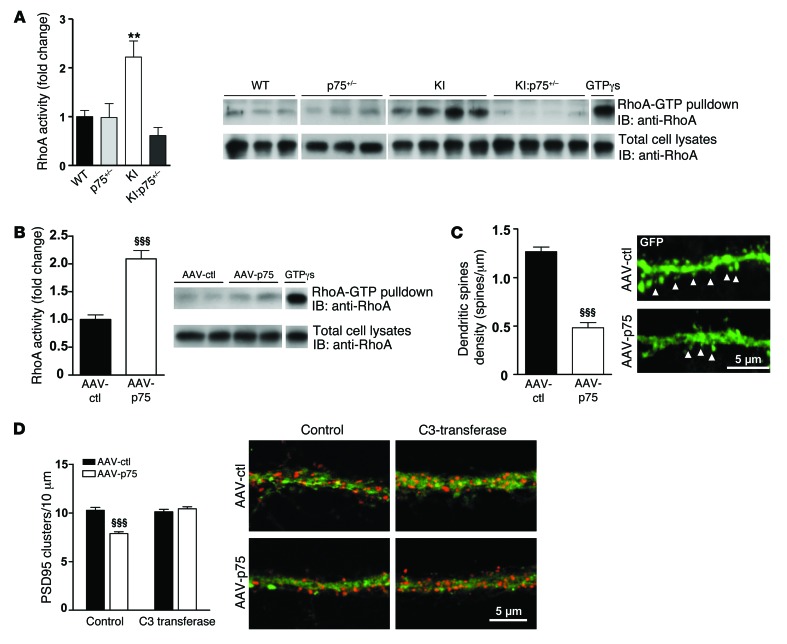
The small GTPase RhoA functions as a negative modulator of dendritic spine formation and maintenance (59, 60), and RhoA activity is modulated by p75NTR signaling (25). Therefore, we hypothesized that aberrant p75NTR expression could contribute to synaptic and memory dysfunction in HD by altering RhoA activity and therefore affecting dendritic spine density. First, we tested RhoA activity in hippocampal extracts from WT, p75+/–, KI, and KI:p75+/– mice by pull-down analysis. Consistent with our hypothesis, KI mutant mice showed a significant increase in hippocampal RhoA activity compared with either WT, p75+/–, or KI:p75+/– mice (Figure 10A). Next, we tested whether p75NTR-mediated RhoA activation could be responsible for hippocampal dendritic spine pathology in HD mice. To this aim, we analyzed whether exogenous overexpression of p75NTR was associated with increased RhoA activity and whether this increase caused dendritic spine alterations. Notably, we found that RhoA activity in the hippocampus of WT mice overexpressing p75NTR was significantly higher (twofold increase) than that in control mice (Figure 10B). Accordingly, the analysis of hippocampal GFP-labeled dendritic spines revealed a significant reduction (~50%) in dendritic spine density in AAV-p75 mice compared with AAV-GFP mice (Figure 10C). We verified that the reduction in spines was not merely due to a decrease in GFP-p75NTR staining by labeling hippocampal slices with an anti-phalloidin antibody (Supplemental Figure 5). Finally, to further validate the role of RhoA in p75NTR-induced dendritic abnormalities, we overexpressed p75NTR in hippocampal primary cultures, and dendritic spine-like structures were analyzed in the presence of the RhoA inhibitor C3 transferase. A significant reduction (~20%; Figure 10D) in the number of spine-like structures counted as PSD95-positive clusters (31) was found in AAV-p75 hippocampal neurons, which agrees with our previous data showing a decrease in dendritic spine density in AAV-p75–transduced mice (Figure 10C). Importantly, by blocking of RhoA activity, the reduction of PSD95-positive clusters was prevented. These experiments suggest that the pathological increase of p75NTR in the hippocampus of HD mice leads to aberrant RhoA activity and consequently dendritic spine alterations.
Figure 10. Increased RhoA activity contributes to p75NTR-mediated dendritic spine loss in mutant KI mice.
(A) Hippocampal lysates from WT, KI, p75+/–, and KI:p75+/– mice at 6 months of age were subjected to RhoA activity assays. Quantification of the RhoA-GTP levels adjusted to the total RhoA is shown. One-way ANOVA with Tukey post hoc comparisons was performed; n = 5 animals per genotype. **P < 0.01 compared with WT mice. (B) Hippocampal lysates from AAV-ctl– and AAV-p75–transduced WT mice were subjected to RhoA activity. Quantification of the RhoA-GTP levels adjusted to the total RhoA is shown. Student’s 2-tailed t test was performed; n = 5–6 per genotype. §§§P < 0.001 compared with AAV-ctl mice. (C) Left: Quantitative analysis showing dendritic spine density. AAV-p75 mice exhibit a significant reduction in dendritic spines compared with AAV-ctl mice. Student’s 2-tailed t test was performed; 32 dendrites (AAV-ctl) and 26 dendrites (AAV-p75) from 3 animals per condition were analyzed. §§§P < 0.001 compared with AAV-ctl mice. Right: Representative confocal microscopy images of CA1 dendrites from AAV-ctl and AAV-p75 mice. Arrowheads indicate dendritic spines. (D) Hippocampal neurons were transduced at DIV14 with AAV-ctl or AAV-p75 and at DIV20 treated with the RhoA inhibitor C3 transferase. Left: Quantitative analysis showing the density of PSD95 clusters. Two-way ANOVA comparing groups × treatment with Bonferroni post hoc comparisons was performed. §§§P < 0.001 compared with control AAV-ctl–treated neurons. Right: Representative confocal microscopy images showing PSD95 clusters (red) in GFP-labeled neurons. All histograms represent mean ± SEM.
Discussion
In the present study we identify p75NTR as an essential contributor to synaptic dysfunction and memory decline in HD and extend previous studies pointing to p75NTR as a negative modulator of hippocampal function (23, 24, 61). We demonstrate an inverse correlation between p75NTR expression in the hippocampus of mutant HD mice and synaptic function manifested as reduced dendritic spine number, altered dendritic morphology, impaired LTP, and memory deficits. In accordance, genetic normalization of p75NTR in HD knock-in mutant mice rescues synaptic morphology, plasticity, and memory deficits, likely by normalization of GTPase RhoA activity. Moreover, overexpression of p75NTR in the hippocampus of WT mice reproduces HD-like memory deficits, while specific hippocampal p75NTR knockdown in HD mutant mice prevents such cognitive impairments.
Decades of research have long established that cognitive function declines in the premanifest and early stages of HD (62–64). Although several studies point to the contribution of other brain structures such as the hippocampus to learning and memory deficits in HD (7–9, 62, 63, 65), data from clinical trials are far from being conclusive on the role of the hippocampus in HD memory deficits. To shed new light on the contribution of the hippocampal circuitry to synaptic and memory decline in HD, we have evaluated spatial and nonspatial memories in a precise genetic mouse model of HD that expresses endogenous levels of mutant huntingtin, HdhQ7/Q111 mice (30), by using a battery of different behavioral tests. We demonstrated impairments in spatial, recognition, and associative memories in HdhQ7/Q111 mutant mice at early and mild disease stages, suggesting hippocampal dysfunction in our HD mouse models. However, we would like to emphasize that even though the contribution of the hippocampus in spatial tasks is known to be critical (66–68), there is no consensus about which brain regions are the most important for nonspatial memory. Then, our behavioral data in HD mice do not completely rule out the contribution of other areas of the limbic system or the prefrontal cortex in such memory impairments.
To get insights into the molecular mechanisms contributing to this memory decline, we focused on p75NTR. The role of p75NTR in the adult brain has been mainly associated with apoptosis, whereas its involvement in synaptic plasticity and memory is poorly understood. Null p75NTR mice exhibit enhanced LTP (21) and impaired LTD (23), while both null and heterozygous p75NTR mice display improved spatial memory (21, 22, 69). In contrast, overexpression of p75NTR negatively modulates dendritic complexity and spine density in hippocampal neurons (24). In this scenario, we show that p75NTR is upregulated in the hippocampus but not in the cortex of HD mice and HD human brain, extending our previous studies showing upregulation of p75NTR in the HD striatum (30). In view of these data we wondered whether normalization of p75NTR levels in KI mice would recover HD memory deficits. In agreement with our hypothesis and supporting a role of aberrant hippocampal p75NTR expression in HD memory impairments, new double-mutant mice expressing mutant huntingtin but “normal” p75NTR levels (KI:p75+/– mice) showed preserved spatial, recognition, and associative memories. Moreover, and according with memory decline, our electrophysiological studies also revealed severe hippocampal LTP deficiencies in HD knock-in mutant mice, in agreement with previous studies showing significant LTP reduction in distinct HD mouse models (14, 70). Importantly, normalization of p75NTR levels in KI:p75+/– mice restored induction and maintenance of LTP, further supporting a critical role for p75NTR in synaptic deficits in HD.
Our data, however, do not directly support the hypothesis that the increase of p75NTR in the hippocampus of HD mice is responsible for synaptic and memory dysfunction. One could argue that downregulation of p75NTR expression in other brain areas in these double-mutant mice could also contribute to the amelioration of HD memory decline. In fact, the striatal p75NTR upregulation previously reported in HD mice (30, 57) raises the question of whether normalization of striatal p75NTR would affect the activity of the corticostriatal circuitries by preventing or improving memory impairments. To answer this question, corticostriatal pathology was also evaluated in KI:p75+/– mutant mice. Interestingly, normalization of striatal p75NTR levels in KI mice did not prevent motor learning deficits or corticostriatal LTP abnormalities, although a recovery of DARPP-32 levels was found. These data suggest that prevention of memory deficits in KI:p75+/– mice is associated with the normalization of p75NTR expression in the hippocampus of KI mice, pointing to p75NTR as a key contributor of synaptic and memory decline in our HD mice. This conclusion is further supported by our findings on overexpression of p75NTR in the hippocampus of WT mice and the selective knocking down of p75NTR in the HD mouse hippocampus. Notably, we found that (a) by increasing p75NTR levels only in the hippocampus of WT mice, it was possible to reproduce those memory deficits observed in HD mice; and (b) the specific reduction of p75NTR in the hippocampus of KI mutant mice completely prevented spatial, recognition, and associative memory deficits. Altogether, these findings suggest a negative effect of hippocampal p75NTR upregulation in synaptic and memory processes in HD mice. These data are in agreement with previous findings demonstrating a critical role of p75NTR in cognitive and synaptic dysfunction in AD. Increased p75NTR expression has been reported in the hippocampus of AD patients (26), while inhibition of p75NTR aberrant signaling by small molecules has been demonstrated to prevent memory deficits and neuritic dystrophy in AD mouse models (27, 28).
Although the precise molecular pathways by which mutant huntingtin alters p75NTR expression in the hippocampus of HD mice remain unclear, our results demonstrate that increased p75NTR protein levels were accompanied by increased p75NTR transcripts. The mechanism of p75NTR transcriptional deregulation under disease conditions is poorly understood. However, it has been proposed that the transcription factor Sp1 drives the expression of p75NTR upon neuronal injury or distinct stress inducers (32, 33). Moreover, Sp1 has been described to be increased in cellular and transgenic models of HD (34). In accordance with these findings, we found a significant increase in Sp1 protein levels in the hippocampus of HD mice and HD human brain, supporting the idea that deregulation of Sp1 activity by mutant huntingtin could underlie the increase in p75NTR mRNA observed in HD hippocampus. However, we cannot rule out that other transcription factors known to regulate p75NTR expression might also contribute to p75NTR deregulation. For instance, it was recently shown that p75NTR is a direct transcriptional target of the transcriptional activator p73 (Tap73) (71), which has also been demonstrated to be hyperactive in HD mouse models and to play a relevant role in HD pathology (72). Interestingly, hippocampal p75NTR and Tap73 expression is increased in AD (73, 74), suggesting that the p73/p75NTR axis may also play an important role in the pathology of neurodegenerative diseases (71).
Several mechanisms can explain how aberrant p75NTR levels mediate synaptic and memory deficits in HD. First, our results indicate that p75NTR directly or indirectly regulates different synaptic-related proteins previously implicated in HD synaptic and/or cognitive deficits, such as CBP, GluA1, CaMKII, or BDNF (14, 15, 54, 55). Indeed, memory improvements in double-mutant KI:p75+/– mice correlated with a recovery of the expression and/or phosphorylation of these molecules, leading us to propose p75NTR as a potential regulator of these pathways in HD synaptic pathology. In fact, altered expression of AMPA receptor subunits is observed in the hippocampus of p75NTR knockout mice (61). Second, p75NTR acts as a negative regulator of dendritic spine density and morphology (24, 75). Accordingly, KI mutant mice show a significant loss of dendritic spines in CA1 pyramidal neurons, a phenotype recovered by normalization of p75NTR levels, suggesting that synaptic and memory deficits in HD could be related to a reduction in the number and complexity of hippocampal dendritic spines. Interestingly, spine loss in HD mice is specific to thin spines, and the remaining ones exhibit shorter neck lengths than the thin spines found in the other 3 genotypes. It has been suggested that spine necks regulate biochemical and electrical signals through compartmentalization of Ca2+ and diffusion of synaptic-related proteins, which could modulate synapse strength (53, 76, 77). Therefore, changes in neck lengths together with altered number and proportion of dendritic spines could mediate the reduced synaptic plasticity observed in KI mice. It is noteworthy that despite the fact that p75+/– and KI:p75+/– mice do not exhibit a significant reduction in the total number of dendritic spines, there was a significant decrease in the proportion of thin spines accompanied by a significant increase in the proportion of mushroom spines, thought to be highly stable with larger postsynaptic densities and to be the locus of long-term memories (78, 79). Actually, an increase in the number of mushroom spines might represent a homeostatic mechanism to compensate for the reduction of thin/learning spines (80, 81). These data suggest that adequate p75NTR levels are required for normal forms of synaptic structural plasticity and cognitive processes and that any aberrant p75NTR expression can be deleterious. We hypothesize that altered p75NTR expression above or beyond an optimal level will be critical for hippocampal function. Thus, levels beyond this threshold will not be sufficient to activate certain transduction pathways important for some forms of synaptic plasticity (impaired performance of p75+/– mice in the passive avoidance), while levels above this optimal expression, as in our HD mice, will induce overactivation of p75NTR signaling and deregulation of p75NTR-modulated molecular pathways essential for memory processes (increased RhoA activity and subsequent altered dendritic spines and cognition in KI mice).
Our results also suggest that the observed loss of spines found in mutant KI mice could be due to altered p75NTR synaptic localization. p75NTR expression in the hippocampus has been classically restricted to afferents coming from basal forebrain cholinergic neurons (82–84). However, recent evidence in vivo and in vitro also demonstrates dendritic spine localization of p75NTR in CA1 and dentate gyrus of hippocampal neurons (19, 23, 85, 86). In accordance with these studies, we found colocalization of p75NTR with MAP2 and PSD95 in hippocampal pyramidal neurons of CA1, revealing a postsynaptic localization of p75NTR. Interestingly, p75NTR was increased in synapses in KI mutant mice, suggesting that postsynaptic p75NTR could have negative effects on synaptic function in HD. Besides p75NTR expression in neurons, it has been reported that astrocytes induce p75NTR expression after different types of injury (36, 86, 87). Increased levels of p75NTR have been detected in both neurons and astrocytes in the hippocampal CA3 region after seizures (37). However, our results showed no colocalization between GFAP and p75NTR in the hippocampus of KI mutant mice, suggesting that the increase in p75NTR levels in these HD mutant mice is predominantly neuronal. Accordingly, specific neuronal overexpression of p75NTR in the hippocampus of WT mice mimics HD-like memory deficits, further supporting the idea that the increase in p75NTR levels in the hippocampal neurons of KI mutant mice is responsible, at least partially, for the synaptic plasticity and memory impairments in HD mice.
How might increased p75NTR levels lead to dendritic spine changes? We built on work showing that p75NTR receptor modulates the activity of RhoA, a Rho family of small GTPases important for the maintenance of the dendritic spines (59, 88). RhoA has complex effects on spines. While sustained elevation of RhoA impairs maturation and maintenance of dendritic spines and reduces dendritic tree and neck length, brief RhoA activation after neuronal activity promotes actin reorganization needed for LTP consolidation (60, 89, 90). Given that p75NTR acts as a constitutive RhoA activator (25, 91), we speculate that prolonged RhoA stimulation due to high p75NTR levels in the hippocampus of KI mutant mice could destabilize actin spine cytoskeleton and compromise the integrity of spines. In this context, we found increased RhoA activation in the hippocampus of mutant KI but not in double-mutant KI:p75+/– mice compared with WT mice, suggesting a direct link between p75NTR-induced dendritic loss and aberrant RhoA activity. In favor of this hypothesis, we found recovery of PSD95-positive clusters in AAV-p75–transduced hippocampal cultures treated with the RhoA inhibitor C3 transferase.
In conclusion, our findings demonstrate a detrimental role of hippocampal p75NTR upregulation in synaptic and memory function in HD. These results, along with previous studies in HD mouse and monkey models reporting hippocampal pathology, underline the need to address the pathogenic significance of the hippocampus in memory and synaptic decline in HD patients. By working on the hypothesis that the hippocampus can play a role in HD memory impairments, new and exciting therapeutic alternatives can be opened. In this view, our data uncovering a new role of p75NTR in memory decline by regulation of structural and functional synaptic plasticity support the possibility of treating synaptic and memory impairments in HD by pharmacological or genetic modulation of p75NTR function.
Methods
Animals.
Heterozygous HdhQ7/Q111 knock-in mice (KI) (92) and p75+/– heterozygous mice (p75NTR/ExonIII) from the Jackson Laboratory were bred. KI and p75+/– were on a C57BL/6 genetic background. Only males from each genotype were used for all experiments. Male R6/1 transgenic mice (B6CBA background) expressing exon 1 of mutant huntingtin with 145 repeats were used (93).
Postmortem brain tissues.
Hippocampal (6 controls and 7 HD patients) and cortical (7 controls and 6 HD patients) brain tissues from patients with HD grades 3 and 4 (mean ± SEM; age 54.5 ± 6.5 years; post-mortem intervals of 4–17 hours) and control cases (mean ± SEM; age 53.5 ± 6.8 years; postmortem intervals of 4–18 hours) were supplied by the Banc de Teixits Neurològics (Biobanc-HC-IDIBAPS).
Behavioral assessment.
The T-maze spontaneous alternation task (T-SAT), novel object recognition task (NORT) (13), object location task (OLT) (94), and passive avoidance paradigm (38) were used to analyze hippocampal-dependent memory. Plus maze and light-dark tests (38) were used to analyze anxiety-like behaviors, while accelerating rotarod (95) was used to analyze motor learning.
T-SAT.
The T-maze apparatus used was a wooden maze consisting of 3 arms, 2 of them situated at 180° from each other, and the third representing the stem arm of the T, situated at 90° with respect to the other 2. All arms were 45 cm long, 8 cm wide, and enclosed by a 20-cm wall. The maze was thoroughly painted with waterproof gray paint. Light intensity was 5 lux throughout the maze. A starting area (10 cm) was located at the end of the stem arm and closed by a wooden guillotine door. Two identical guillotine doors were placed in the entry of the arms situated at 180°. The maze was elevated 60 cm above the floor. In the training trial, 1 arm was closed (novel arm) and mice were placed in the stem arm of the T (home arm) and allowed to explore this arm and the other available arm (familiar arm) for 10 minutes, after which they were returned to the home cage. After 5 hours (long-term memory), mice were placed in the stem arm of the T-maze and allowed to freely explore all 3 arms for 5 minutes. The arm preference was determined by calculation of the time spent in each arm × 100/time spent in both arms (familiar and novel). Animals were tracked and recorded with SMART Junior software (Panlab).
OLT.
The object location memory task evaluates spatial memory and is based on the ability of mice to recognize when a familiar object has been relocated. Exploration took place in an open-top arena with quadrangular form (45 × 45 cm). The light intensity was 40 lux throughout the arena, and the room temperature was kept at 19°C–22°C and 40%–60% humidity. Mice were first habituated to the arena in the absence of objects (2 days, 10 min/d). On the third day during the acquisition phase mice were allowed to explore 2 duplicate objects (A1 and A2), which were placed in the far corners of the arena for 10 minutes. After a delay of 24 hours, 1 object was placed in the corner diagonally opposite. Thus, both objects in the phase were equally familiar, but 1 was in a new location. The position of the new object was counterbalanced between mice. Animals were tracked and recorded with SMART Junior software.
NORT.
The device consisted of a white circular arena with 40 cm diameter and 40 cm high. The light intensity was 40 lux throughout the arena, and the room temperature was kept at 19°C–22°C and 40%–60% humidity. Mice were first habituated to the arena in the absence of objects (2 days, 10 min/d). On the third day, 2 similar objects were presented to each mouse during 10 minutes (A1 and A2 condition), after which they were returned to their home cage. Twenty-four hours later (long-term memory), the same animals were retested for 5 minutes in the arena with a familiar and a new object (A2 and B condition). The object preference was measured as the time exploring each object × 100/time exploring both objects. The arena was rigorously cleaned between animal trials in order to avoid odors. Animals were tracked and recorded with SMART Junior software.
Passive avoidance.
The passive avoidance (light-dark) paradigm was performed as described elsewhere (96, 97) with slight modifications. Experiments are conducted in a 2-compartment box, where 1 compartment is dimly lit (20 lux) and preferable to a rodent, and the other compartment is brightly lit (200 lux); both chambers are connected by a door (5 cm × 5 cm). During training, mice were placed into the aversive brightly lit compartment, and upon the entry into the preferred dimly lit compartment (with all 4 paws inside the dark chamber), mice were exposed to a mild foot shock (2-second foot shock, 1 mA intensity). The latency of mice to enter into the dark chamber was recorded. Twenty seconds after receiving the foot shock, mice were returned to the home cage until testing. After 24 hours (long-term memory) the animal undergoes a retention test (testing). In the retention test, mice were returned to the brightly lit compartment again, and the latency to enter the shock-paired compartment (dark chamber) was measured (retention or recall latency). Ten minutes was used as a time cutoff in the retention test. The animal that learned the task would avoid the location previously paired with the aversive stimulus, and would show greater latency to enter it.
Plus maze.
To analyze mouse anxiety-like behaviors, the plus maze paradigm was performed as previously described (38). Briefly, the raised plus maze was made of wood and consisted of 2 opposing open arms of 30 × 8 cm, and 2 opposing arms of 30 × 8 cm enclosed by 15-cm-high walls. The maze was raised 50 cm from the floor and lit by dim light. Each mouse was placed in the central square of the raised plus maze, facing an open arm, and the behavior was scored for 5 minutes. At the end of each trial, any feces were removed and the apparatus was wiped with 70% alcohol. We recorded the time spent in the open arms, which normally correlates with levels of anxiety. Animals were tracked and recorded with SMART Junior software.
Light-dark paradigm.
The light-dark paradigm was performed as described elsewhere (98) with slight modifications. The light-dark box consisted of 2 compartments, which were connected by a door (5 cm × 5 cm) located at floor level in the center of the partition. The light compartment was white and directly illuminated by a dim light (200 lux), while the dark compartment was black and completely enclosed (20 lux). The test was performed in a dimly lighted room. Each animal was initially confined in the dark compartment and allowed to freely explore the apparatus for 5 minutes. The parameters recorded were time spent by each mouse in the light compartment and latency time for the first crossing to the light compartment. Between animals, the apparatus was carefully cleaned with 70% ethanol solution and allowed to dry.
Accelerating rotarod training procedure.
As previously described (99), animals were placed on a motorized rod (30 mm diameter). The rotation speed gradually increased from 4 to 40 rpm over the course of 5 minutes. The time latency was recorded when the animal was unable to keep up with the increasing speed and fell. Rotarod training and testing was performed 4 times per day during 3 consecutive days.
Viral constructs and stereotaxic injection.
For p75NTR overexpression the plasmid pEGFP-N3-p75 obtained from E. Formaggio (Department of Medicine and Public Health, Pharmacology Section, University of Verona, Verona, Italy) was used to clone into a rAAV2/8-GFP adenoviral vector (BamHI site at 5′ and AgeI at 3′). For knockdown p75NTR expression we designed a siRNA oligomer targeting the mouse p75NTR:Ngfr-1 to clone into a rAAV2/8-GFP adenoviral vector as previously described. Infectious AAV viral particles containing GFP expression cassette with scrambled shRNA (AAV-ctl), shRNAp75 (AAV-shp75), GFP (AAV-GFP), or p75NTR expression cassette (AAV-p75) were generated by the Unitat de Producció de Vectors from the Center of Animal Biotechnology and Gene Therapy at the Universitat Autònoma de Barcelona. Following anesthesia with pentobarbital (30 mg/kg), 5-month-old mice were injected with AAV-GFP, AAV-p75, and AAV-shp75 (viral concentration ranging from 1.53 × 109 to 3.06 × 109 GCs per injection, n = 8 per group) at 2 sites of the hippocampus according to the following coordinates from the bregma (millimeters); anteroposterior, –1.6 and –1.9; lateral, +2.0 and +3.0; and dorsoventral, –0.8 and –1.2. AAVs were injected over 2 minutes, leaving the cannula in place for 5 additional minutes to ensure complete diffusion of the viruses, and then slowly retracted from the brain. The animals were monitored for 2 hours after administration and then returned to the housing facility for 30 days. After this period, animals from experimental groups were subjected to behavioral assessment.
Total protein extraction and subcellular fractionation.
Total protein extraction was performed as described elsewhere (30). For cytosolic and nuclear fractions, tissue was homogenized in lysis buffer (4 mM HEPES, 0.32 M sucrose, 2 mM PMSF, 10 mg/ml aprotinin, 1 mg/ml leupeptin, 2 mM Na3VO4, and 0.1 mg/ml benzamidine) with a Teflon-glass potter and centrifuged at 800 g for 10 minutes to obtain the cytosolic (supernatant) and the nuclear (pellet) fractions. The pellet was resuspended in lysis buffer and centrifuged at 800 g for 10 minutes. The resulting pellet, containing washed nuclear fraction, was then resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 5 mM ZnCl2, 10 mM EGTA, 2 mM PMSF, 10 mg/ml aprotinin, 1 mg/ml leupeptin, 0.1 mg/ml benzamidine, 2 mM Na3VO4, and 100 mM NaF) and incubated for 30 minutes at 4°C in a tube rotator. Finally, after centrifugation for 20 minutes at 15,700 g, the supernatant was collected and stored at –80°C until use. Protein concentration was measured using the DC protein assay kit (Bio-Rad).
Western blotting.
Western blot experiments were performed as previously described (15, 29). Antibodies used for Western blot analysis were anti-p75NTR (1:1,000; Promega); anti-CaMKII, anti–phospho-CaMKIIThr286, anti-BDNF, and anti-CBP (1:1,000; Santa Cruz Biotechnology); anti-GluA1 and anti-GluA2/3 (1:1,000; Upstate Biotechnology); anti–phospho-GluA1Ser831 (1:1,000; Millipore); anti-actin (1:10,000; MP Biochemicals); anti-PSD95 (1:2,000; Affinity Bioreagents); anti-PSD93 (1:1,000; Alomone); anti-SAP102 (1:1,000; Synaptic Systems); anti-spinophilin (1:1,000; Upstate Biotechnology); anti-GluN2B (1:1,000; Chemicon); anti-GluN1 (1:1,000; Chemicon); anti-synaptophysin (1:1,000; Abcam); anti-RhoA (1:1,000; Cell Signaling); anti-SP1 (1:500; Santa Cruz Biotechnology); anti-NeuN (1:1,000; Millipore); anti-DARPP-32 (1:1,000; BD Bioscience); and anti–α-tubulin (1:20,000; Sigma-Aldrich).
RhoA pull-down assay.
The Rho-GTP pull-down assay was performed using a RhoA activation assay kit according to manufacturer instructions (Cell Biolabs). Hippocampal tissue was homogenized in ice-cold 1× assay/lysis buffer. Samples were centrifuged at 14,000 g for 10 minutes at 4°C, and the supernatant (1 mg of protein) was incubated with Rhotekin RBD-agarose beads at 4°C for 1 hour on a rotator. The beads were washed 3 times with 0.5 ml of 1× assay buffer. Protein samples were eluted with 1× SDS-PAGE sample buffer and processed for Western blotting using RhoA antibody.
Real-time quantitative RT-PCR.
Total RNA was isolated from the hippocampus and cortex of HD mutant and WT mice using the Total RNA Isolation Nucleospin RNA II Kit (Macherey-Nagel). Purified RNA (500 ng) was reverse transcribed using the PrimeScript RT Reagent Kit (Perfect Real Time, Takara Biotechnology Inc.). The cDNA synthesis was performed at 37°C for 15 minutes and a final step at 85°C for 5 seconds in a final volume of 20 μl according to the manufacturer’s instructions. The cDNA was then analyzed by quantitative RT-PCR using the following TaqMan Gene Expression Assays (Applied Biosystems): 18S (Hs99999901_s1) and p75NTR (Mm00446294_m1). RT-PCR was performed in 12.5 μl of final volume on 96-well plates using the Premix Ex Taq. Reactions included 40 cycles of a 2-step PCR: 95°C for 5 seconds and 60°C for 20 seconds, after initial denaturation at 95°C for 30 seconds. All quantitative PCR assays were performed in duplicate. To provide negative controls and exclude contamination by genomic DNA, the RT was omitted in the cDNA synthesis step. The quantitative PCR data were quantified using the comparative quantitation analysis program of MxProTM quantitative PCR analysis software version 3.0 (Stratagene) with the 18S gene expression as loading control.
Histology and stereology.
Hippocampal volume estimations were performed following the Cavalieri method. The number of Nissl-positive neurons in the CA1 pyramidal cell layer was established using unbiased design-based stereology blinded to genotype and group and was performed using Computer-Assisted Stereology Toolbox (CAST) software (Olympus Danmark A/S). Specifically, we used the dissector counting procedure in 12 hippocampal coronal sections (30 μm thick) per animal, spaced 240 μm and covering the entire rostrocaudal extent of this brain region.
Primary culture of astrocytes.
Primary astrocyte cultures were obtained from P1 to P3 WT mouse pups by cortical dissection and removal of the meninges. Extracted tissue was mechanically dissociated and resuspended in MEM 1× conditioned media NM-15 (20% FBS, Gibco-BRL; 90 mM d-glucose, Sigma-Aldrich) with l-glutamine and Earle’s salts (Gibco-BRL), plated in 25-cm2 flasks, and cultured at 37°C with 5% CO2. After 2 passages, cultures were purified by shaker agitation during 10 minutes at 400 rpm, and undesired floating cells were discarded. Medium was replaced and seeded astrocytes maintained 2 hours at 37°C in agitation for 16–18 hours at 250 rpm, floating cells removed, and new medium replaced. Twenty-four hours later, astrocytes were trypsinized, seeded at 1 × 106 cells in P100 plates, and allowed to reach 100% confluence. Astrocytes were transduced with AAV-GFP or AAV-p75 (40,000 GCs per cell).
Primary culture of mouse hippocampal neurons and adenovirus-mediated gene transduction.
Primary hippocampal neurons were prepared from E18.5 mouse embryos as previously described (100). The neuronal cell suspension was seeded (70,000 cells/cm2) on coverslips precoated with poly-d-lysine (0.1 mg/ml; Sigma-Aldrich) in 24-well plates. Neurobasal medium containing 1 ml of 1× B27 (50) supplement (Gibco-BRL) and 500 ml of GlutaMAX (100×) (Gibco-BRL) was used to grow the cells in serum-free medium conditions and maintained at 37°C in 5% CO2. Two weeks after plating (DIV14), neurons were transduced with AAV-GFP or AAV-p75 (40,000 GCs per cell) in Neurobasal maintenance medium for up to 1 week. At DIV20, cells were treated with vehicle or C3 ADP ribosyl transferase, a cell-permeable Rho inhibitor (100 ng/ml; Cytoskeleton Inc.), for 24 hours. At DIV21, neurons were fixed for 10 minutes in 4% PFA for immunostaining and confocal analysis.
Immunocytochemistry and image analysis.
Fixed cultures were permeabilized for 10 minutes at room temperature with 0.5% saponin in PBS and blocked with 15% horse serum in PBS. The cells were incubated with anti-PSD95 (1:1,000; Affinity Bioreagents) and secondary antibodies (Cy3, 1:200; Jackson ImmunoResearch). Single images were acquired digitally using Leica Confocal SP5 with a ×63 oil-immersion objective and a digital zoom of 3. Conditions such as pinhole size (1 AU) and frame averaging (4 frames per z-step) were held constant throughout the study. PSD95 clusters in dendrites from GFP-labeled neurons were quantified using the freeware NIH ImageJ version 1.33 by Wayne Rasband (NIH). At least 60 neurites (1–2 neurites per neuron) for each condition in 3 independent experiments were analyzed.
Immunofluorescence and SL confocal analysis.
Immunofluorescence experiments and analysis were carried out as previously described (30, 101). Antibodies against p75NTR (1:100; Promega), GFP (1:1,000; Upstate Biotechnology), PSD95 (1:1,000; Affinity Bioreagents), and MAP2 (1:500; Sigma-Aldrich) and secondary antibodies (Alexa Fluor 488 and 555, Jackson ImmunoResearch) were used. Stained coronal sections were examined blinded to genotype by confocal microscopy, using a Leica TCS SL laser scanning confocal spectral microscope with argon and helium-neon lasers. Images were taken with a 63× numerical aperture lens with 4× digital zoom and standard (1 Airy disc) pinhole. For each mouse, at least 3 slices of 30 μm containing hippocampal tissue were analyzed. Up to 3 representative images, from CA1-stratum radiatum layer, were obtained from each slice. For each image, the entire 3D stack of images was obtained by the use of the Z drive present in the Leica TCS SL microscope, and the size of the optical image was 0.5 μm, with a separation of 2 μm between each. The number of double-labeled p75NTR/PSD95-positive clusters was counted by the freeware NIH ImageJ version 1.33 by Wayne Rasband (NIH).
Dendritic spine dying and SP5 confocal analysis.
Neurons were labeled using the Helios Gene Gun System (Bio-Rad) as previously described (102, 103). Briefly, a suspension containing 3 mg of DiI (Molecular Probes, Invitrogen) dissolved in 100 μl of methylene chloride (Sigma-Aldrich) and mixed with 50 mg of tungsten particles (1.7 mm diameter; Bio-Rad) was spread on a glass slide and air-dried. The mixture was resuspended in 3.5 ml distilled water and sonicated. Subsequently, the mixture was drawn into Tefzel tubing (Bio-Rad), and then removed to allow tube drying during 5 minutes under a nitrogen flow gas. Then, the tube was cut into 13-mm pieces to be used as gene gun cartridges. Dye-coated particles were delivered in the hippocampus using the following protocol. Shooting was performed over 200-μm coronal sections at 80 psi through a membrane filter of 3 μm pore size and 8 × 10 pores/cm2 (Millipore). Sections were stored at room temperature in PBS for 3 hours protected from light and then incubated with DAPI, and mounted in Mowiol to be analyzed. DiI-labeled pyramidal neurons from CA1 of the dorsal hippocampus were imaged using a Leica Confocal SP5 with a ×63 oil-immersion objective. Conditions such as pinhole size (1 AU) and frame averaging (4 frames per z-step) were held constant throughout the study. Confocal z-stacks were taken with a digital zoom of 5, a z-step of 0.2 μm, and at 1,024 × 1,024 pixel resolution, yielding an image with pixel dimensions of 49.25 × 49.25 μm. Z-stacks were deconvolved using the Acoloma plug-ins from ImageJ, to improve voxel resolution and reduce optical aberration along the z-axis. Segments of basal dendrites and proximal apical dendrites were selected for the analysis of spine density and spine morphology according to the following criteria: (a) segments with no overlap with other branches that would obscure visualization of spines and (b) segments either “parallel” to or “at acute angles” relative to the coronal surface of the section to avoid ambiguous identification of spines. Only spines arising from the lateral surfaces of the dendrites were included in the study; spines located on the top or bottom of the dendrite surface were ignored. Given that spine density increases as a function of the distance from the soma, reaching a plateau 45 μm away from the soma, we selected dendritic segments of basal dendrites 45 μm away from the cell body.
For a more precise description of the dendritic shape changes, the spine head diameter was analyzed as a continuous distribution. To this end, we proceeded as follows: first all the identified spines were categorized as spines without neck (stubby spines) or with a clear neck. From the latter, blind measurements of the head diameter were performed manually using ImageJ for all the spines in control mice. Then, a distribution analysis of head diameter was performed. The Gaussian adjustment resulted in a normal distribution with a mean value of 0.5009 μm (Gaussian fit P = 0.90) (Supplemental Figure 4). From the estimated mean value, spine populations were divided into spines with diameters below the mean average (head diameters <0.5009 ± 0.006 μm, thin spines), spines with head diameters above the mean value (head diameters >0.5009 ± 0.006 μm, mushrooms), and a third category including all the non-neck spines (stubby spines). Spine neck was measured as the distance from the dendritic shaft to the head of the spine.
Electrophysiology.
Coronal brain slices (400 μm thickness) were prepared from WT, p75+/–, KI, and KI:p75+/– mice (6 months old) as described previously (104) and incubated for more than 1 hour at room temperature (21°C–24°C) in ACSF (in mM: NaCl 124, KCl 2.69, KH2PO4 1.25, MgSO4 2, NaHCO3 26, CaCl2 2, and glucose 10) and were gassed with 95% O2 and 5% CO2. Slices containing hippocampus or striatum and cortex were transferred to an immersion recording chamber and superfused (2.5 ml/min) with equilibrated ACSF maintained at constant temperature (34°C). To evaluate hippocampal LTP, extracellular field excitatory postsynaptic potentials (fEPSPs) were recorded with a glass microelectrode (impedances 2–3 MΩ; filled with 1 M NaCl) positioned in stratum radiatum area CA1 of the hippocampus. Evoked fEPSPs were elicited by stimulation of the Schaeffer collateral fibers with an extracellular bipolar tungsten electrode via a 2100 isolated pulse stimulator (A-M Systems Inc.). LTP was induced by application of 4 trains (1 second at 100 Hz) spaced 20 seconds, and potentiation was measured for 1 hour after LTP induction at 0.1 Hz. Corticostriatal LTP was induced by application of tetanic stimulation twice (4 trains 1 second at 100 Hz) spaced 10 seconds, and potentiation was measured for 1 hour after LTP induction at 0.1 Hz. For each experiment, population spike (PS) amplitude was expressed as a percentage of average pre-tetanus baseline amplitude. Data were stored on a Pentium-based PC using a PowerLab 4/26 acquisition system (AD Instruments); Scope software (AD Instruments) was used to display PS and measurements of the amplitude of PSs. Statistical differences, compared with pre-tetanus baseline amplitude values, were established using the 2-tailed Student’s t test.
Statistics.
All data are expressed as mean ± SEM. Statistical analysis was performed using the unpaired Student’s t test (2-tailed, 95% confidence), the Kolmogorov-Smirnov test to compare cumulative distributions, 1-way ANOVA, 2-way ANOVA, and the appropriate post hoc tests as indicated in the figure legends. Values of P < 0.05 were considered statistically significant.
Study approval.
Human samples were provided by Banc de Teixits Neurològics (Biobanc-HC-IDIBAPS). The Banc de Teixits Neurològics was established to provide human postmortem brains to researchers in Spain, taking into consideration all the ethical guidelines of the latest Declaration of Helsinki. Informed consent was obtained from all subjects under study. Experimental animal procedures were approved by the Local Ethical Committee of the University of Barcelona (99/01) and the Generalitat de Catalunya (00/1094), following European (2010/63/UE) and Spanish (RD 1201/2005) regulations for the care and use of laboratory animals.
Supplementary Material
Acknowledgments
We thank Ana López and Maria Teresa Muñoz for technical assistance, and Teresa Rodrigo Calduch and the staff of the animal care facility (Facultat de Psicologia, Universitat de Barcelona) for their help. We thank E. Formaggio for providing human p75NTR neurotrophin receptor tagged with GFP at the C-terminus. We are grateful to the Banc de Teixits Neurològics (Biobanc-HC-IDIBAPS) for providing brain samples from control subjects and HD patients. We thank C.A. Saura and members of our laboratory for helpful discussion. This work was supported by grants from the Ministerio de Economía y Competitividad (SAF2012-39142 to S. Ginés, SAF2011-29507 to J. Alberch); the Cure Huntington’s Disease Initiative (CDHI), Centro de Investigaciones Biomédicas en Red sobre Enfermedades Neurodegenerativas (CIBERNED CB06/05/0054 and CB06/05/0042); and the Fondo de Investigaciones Sanitarias Instituto de Salud Carlos III (RETICS: RD06/0010/0006).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(10):4411–4428. doi:10.1172/JCI74809.
Nuria Dominguez-Iturza’s present address is: VIB Center for Biology of Disease and Center for Human Genetics and Leuven Institute for Neuroscience and Disease, KU Leuven, Leuven, Belgium.
References
- 1.Foroud T, et al. Cognitive scores in carriers of Huntington’s disease gene compared to noncarriers. Ann Neurol. 1995;37(5):657–664. doi: 10.1002/ana.410370516. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence AD, Watkins LH, Sahakian BJ, Hodges JR, Robbins TW. Visual object and visuospatial cognition in Huntington’s disease: implications for information processing in corticostriatal circuits. Brain. 2000;123(pt 7):1349–1364. doi: 10.1093/brain/123.7.1349. [DOI] [PubMed] [Google Scholar]
- 3.Lemiere J, Decruyenaere M, Evers-Kiebooms G, Vandenbussche E, Dom R. Cognitive changes in patients with Huntington’s disease (HD) and asymptomatic carriers of the HD mutation — a longitudinal follow-up study. J Neurol. 2004;251(8):935–942. doi: 10.1007/s00415-004-0461-9. [DOI] [PubMed] [Google Scholar]
- 4.Bäckman L, Robins-Wahlin TB, Lundin A, Ginovart N, Farde L. Cognitive deficits in Huntington’s disease are predicted by dopaminergic PET markers brain volumes. Brain. 1997;120(pt 12):2207–2217. doi: 10.1093/brain/120.12.2207. [DOI] [PubMed] [Google Scholar]
- 5.Watkins LH, Rogers RD, Lawrence AD, Sahakian BJ, Rosser AE, Robbins TW. Impaired planning but intact decision making in early Huntington’s disease: implications for specific fronto-striatal pathology. Neuropsychologia. 2000;38(8):1112–1125. doi: 10.1016/S0028-3932(00)00028-2. [DOI] [PubMed] [Google Scholar]
- 6.Giralt A, Saavedra A, Alberch J, Pérez-Navarro E. Cognitive dysfunction in Huntington’s disease: humans, mouse models and molecular mechanisms. J Huntingtons Dis. 2012;1(2):155–173. doi: 10.3233/JHD-120023. [DOI] [PubMed] [Google Scholar]
- 7.Ille R, et al. Emotion recognition and experience in Huntington disease: a voxel-based morphometry study. J Psychiatry Neurosci. 2011;36(3):383–390. doi: 10.1503/jpn.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulsen JS, Smith MM, Long JD, PREDICT HD investigators Coordinators of the Huntington Study Group. Cognitive decline in prodromal Huntington disease: implications for clinical trials. J Neurol Neurosurg Psychiatry. 2013;84(11):1233–1239. doi: 10.1136/jnnp-2013-305114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Bogaard SJ, et al. Shape analysis of subcortical nuclei in Huntington’s disease, global versus local atrophy--results from the TRACK-HD study. J Neurol Sci. 2011;307(1–2):60–68. doi: 10.1016/j.jns.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Berrios GE, Wagle AC, Markova IS, Wagle SA, Rosser A, Hodges JR. Psychiatric symptomsin neurologically asymptomatic Huntington’s disease gene carriers: a comparison with gene negative at risk subjects. Acta Psychiatr Scand. 2002;105(3):224–230. doi: 10.1034/j.1600-0447.2002.0o456.x. [DOI] [PubMed] [Google Scholar]
- 11.Majerová V, et al. Disturbance of real space navigation in moderately advanced but not in early Huntington’s disease. J Neurol Sci. 2012;312(1–2):86–91. doi: 10.1016/j.jns.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Herndon ES, Hladik CL, Shang P, Burns DK, Raisanen J, White CL., 3rd Neuroanatomic profile of polyglutamine immunoreactivity in Huntington disease brains. J Neuropathol Exp Neurol. 2009;68(3):250–261. doi: 10.1097/NEN.0b013e318198d320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giralt A, Saavedra A, Carretón O, Xifró X, Alberch J, Pérez-Navarro E. Increased PKA signaling disrupts recognition memory and spatial memory: role in Huntington’s disease. Hum Mol Genet. 2011;20(21):4232–4247. doi: 10.1093/hmg/ddr351. [DOI] [PubMed] [Google Scholar]
- 14.Lynch G, et al. Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington’s disease. J Neurosci. 2007;27(16):4424–4434. doi: 10.1523/JNEUROSCI.5113-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giralt A, et al. Long-term memory deficits in Huntington’s disease are associated with reduced CBP histone acetylase activity. Hum Mol Genet. 2012;21(6):1203–1216. doi: 10.1093/hmg/ddr552. [DOI] [PubMed] [Google Scholar]
- 16.Simmons DA, et al. Up-regulating BDNF with an ampakine rescues synaptic plasticity and memory in Huntington’s disease knockin mice. Proc Natl Acad Sci U S A. 2009;106(12):4906–4911. doi: 10.1073/pnas.0811228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spires TL, et al. Dendritic spine pathology and deficits in experience-dependent dendritic plasticity in R6/1 Huntington’s disease transgenic mice. Eur J Neurosci. 2004;19(10):2799–2807. doi: 10.1111/j.0953-816X.2004.03374.x. [DOI] [PubMed] [Google Scholar]
- 18.Lu B, Pang PT, Woo NH. The yin and yang of neurotrophin action. Nat Rev Neurosci. 2005;6(8):603–614. doi: 10.1038/nrn1726. [DOI] [PubMed] [Google Scholar]
- 19. Woo NH, Lu B. BDNF in synaptic plasticity and memory. In: Malenka R, ed. Intracellular Communication In The Nervous System. Bethesda, Maryland, USA: NIH; 2009:135–143. [Google Scholar]
- 20.Yoshii A, Constantine-Paton M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol. 2010;70(5):304–322. doi: 10.1002/dneu.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett GL, et al. Enhanced spatial memory and hippocampal long-term potentiation in p75 neurotrophin receptor knockout mice. Hippocampus. 2010;20(1):145–152. doi: 10.1002/hipo.20598. [DOI] [PubMed] [Google Scholar]
- 22.Greferath U, Bennie A, Kourakis A, Bartlett PF, Murphy M, Barrett GL. Enlarged cholinergic forebrain neurons and improved spatial learning in p75 knockout mice. Eur J Neurosci. 2000;12(3):885–893. doi: 10.1046/j.1460-9568.2000.00976.x. [DOI] [PubMed] [Google Scholar]
- 23.Woo NH, et al. Activation of p75NTR by proBDNF facilitates hippocampal long-term depression. Nat Neurosci. 2005;8(8):1069–1077. doi: 10.1038/nn1510. [DOI] [PubMed] [Google Scholar]
- 24.Zagrebelsky M, Holz A, Dechant G, Barde YA, Bonhoeffer T, Korte M. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25(43):9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24(3):585–593. doi: 10.1016/S0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 26.Hu XY, Zhang HY, Qin S, Xu H, Swaab DF, Zhou JN. Increased p75(NTR) expression in hippocampal neurons containing hyperphosphorylated tau in Alzheimer patients. Exp Neurol. 2002;178(1):104–111. doi: 10.1006/exnr.2002.8018. [DOI] [PubMed] [Google Scholar]
- 27.Knowles JK, et al. The p75 neurotrophin receptor promotes amyloid-beta(1-42)-induced neuritic dystrophy in vitro and in vivo. J Neurosci. 2009;29(34):10627–10637. doi: 10.1523/JNEUROSCI.0620-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowles JK, et al. Small molecule p75NTR ligand prevents cognitive deficits and neurite degeneration in an Alzheimer’s mouse model. Neurobiol Aging. 2013;34(8):2052–2063. doi: 10.1016/j.neurobiolaging.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T, et al. Small molecule, non-peptide p75 ligands inhibit Aβ-induced neurodegeneration and synaptic impairment. PLoS One. 2008;3(11):e3604. doi: 10.1371/journal.pone.0003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brito V, Puigdellívol M, Giralt A, del Toro D, Alberch J, Ginés S. Imbalance of p75(NTR)/TrkB protein expression in Huntington’s disease: implication for neuroprotective therapies. Cell Death Dis. 2013;4:e595. doi: 10.1038/cddis.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Bordiuk OL, Ferrante RJ. Experimental models of HD and reflection on therapeutic strategies. Int Rev Neurobiol. 2011;98:419–481. doi: 10.1016/B978-0-12-381328-2.00016-X. [DOI] [PubMed] [Google Scholar]
- 32.Kommaddi RP, Dickson KM, Barker PA. Stress-induced expression of the p75 neurotrophin receptor is regulated by O-GlcNAcylation of the Sp1 transcription factor. J Neurochem. 2011;116(3):396–405. doi: 10.1111/j.1471-4159.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- 33.Ramos A, et al. Hypo-osmolar stress induces p75NTR expression by activating Sp1-dependent transcription. J Neurosci. 2007;27(6):1498–1506. doi: 10.1523/JNEUROSCI.4806-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Z, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J Biol Chem. 2006;281(24):16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- 35.Michaelsen K, et al. Neurotrophin receptors TrkB.T1 and p75NTR cooperate in modulating both functional and structural plasticity in mature hippocampal neurons. Eur J Neurosci. 2010;32(11):1854–1865. doi: 10.1111/j.1460-9568.2010.07460.x. [DOI] [PubMed] [Google Scholar]
- 36.Cragnolini AB, Friedman WJ. The function of p75NTR in glia. Trends Neurosci. 2008;31(2):99–104. doi: 10.1016/j.tins.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Cragnolini AB, Huang Y, Gokina P, Friedman WJ. Nerve growth factor attenuates proliferation of astrocytes via the p75 neurotrophin receptor. Glia. 2009;57(13):1386–1392. doi: 10.1002/glia.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giralt A, et al. Neurobehavioral characterization of Endonuclease G knockout mice reveals a new putative molecular player in the regulation of anxiety. Exp Neurol. 2013;247:122–129. doi: 10.1016/j.expneurol.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 39.Tekinay AB, et al. A role for LYNX2 in anxiety-related behavior. Proc Natl Acad Sci U S A. 2009;106(11):4477–4482. doi: 10.1073/pnas.0813109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assini FL, Duzzioni M, Takahashi RN. Object location memory in mice: pharmacological validation and further evidence of hippocampal CA1 participation. Behav Brain Res. 2009;204(1):206–211. doi: 10.1016/j.bbr.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Cunningham JI, Raudensky J, Tonkiss J, Yamamoto BK. MDMA pretreatment leads to mild chronic unpredictable stress-induced impairments in spatial learning. Behav Neurosci. 2009;123(5):1076–1084. doi: 10.1037/a0016716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deacon RM, Rawlins JN. T-maze alternation in the rodent. Nat Protoc. 2006;1(1):7–12. doi: 10.1038/nprot.2006.2. [DOI] [PubMed] [Google Scholar]
- 43.Sharma S, Rakoczy S, Brown-Borg H. Assessment of spatial memory in mice. Life Sci. 2010;87(17–18):521–536. doi: 10.1016/j.lfs.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachevalier J, Nemanic S. Memory for spatial location and object-place associations are differently processed by the hippocampal formation, parahippocampal areas TH/TF and perirhinal cortex. Hippocampus. 2008;18(1):64–80. doi: 10.1002/hipo.20369. [DOI] [PubMed] [Google Scholar]
- 45.Albasser MM, Davies M, Futter JE, Aggleton JP. Magnitude of the object recognition deficit associated with perirhinal cortex damage in rats: Effects of varying the lesion extent and the duration of the sample period. Behav Neurosci. 2009;123(1):115–124. doi: 10.1037/a0013829. [DOI] [PubMed] [Google Scholar]
- 46.Winters BD, Saksida LM, Bussey TJ. Object recognition memory: neurobiological mechanisms of encoding, consolidation and retrieval. Neurosci Biobehav Rev. 2008;32(5):1055–1070. doi: 10.1016/j.neubiorev.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13(2):93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baarendse PJ, et al. Differential involvement of the dorsal hippocampus in passive avoidance in C57bl/6J and DBA/2J mice. Hippocampus. 2008;18(1):11–19. doi: 10.1002/hipo.20356. [DOI] [PubMed] [Google Scholar]
- 49.Baldi E, Ambrogi LC, Sacchetti B, Tassoni G, Bucherelli C. Effects of coupled perirhinal cortex and medial septal area, fimbria-fornix, entorhinal cortex tetrodotoxin inactivations on passive avoidance consolidation in the rat. Neurosci Lett. 2000;280(2):91–94. doi: 10.1016/S0304-3940(00)00783-7. [DOI] [PubMed] [Google Scholar]
- 50.Burwell RD, Bucci DJ, Sanborn MR, Jutras MJ. Perirhinal and postrhinal contributions to remote memory for context. J Neurosci. 2004;24(49):11023–11028. doi: 10.1523/JNEUROSCI.3781-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milnerwood AJ, et al. Early development of aberrant synaptic plasticity in a mouse model of Huntington’s disease. Hum Mol Genet. 2006;15(10):1690–1703. doi: 10.1093/hmg/ddl092. [DOI] [PubMed] [Google Scholar]
- 52.Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9(8):2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noguchi J, Matsuzaki M, Ellis-Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46(4):609–622. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giralt A, et al. Brain-derived neurotrophic factor modulates the severity of cognitive alterations induced by mutant huntingtin: involvement of phospholipaseCγ activity and glutamate receptor expression. Neuroscience. 2009;158(4):1234–1250. doi: 10.1016/j.neuroscience.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 55.Nithianantharajah J, Barkus C, Murphy M, Hannan AJ. Gene-environment interactions modulating cognitive function and molecular correlates of synaptic plasticity in Huntington’s disease transgenic mice. Neurobiol Dis. 2008;29(3):490–504. doi: 10.1016/j.nbd.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Colbran RJ, Brown AM. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr Opin Neurobiol. 2004;14(3):318–327. doi: 10.1016/j.conb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 2008;18(2):225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bibb JA, et al. Severe deficiencies in dopamine signaling in presymptomatic Huntington’s disease mice. Proc Natl Acad Sci U S A. 2000;97(12):6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20(14):5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tashiro A, Yuste R. Role of Rho GTPases in the morphogenesis and motility of dendritic spines. Methods Enzymol. 2008;439:285–302. doi: 10.1016/S0076-6879(07)00421-1. [DOI] [PubMed] [Google Scholar]
- 61.Rosch H, Schweigreiter R, Bonhoeffer T, Barde YA, Korte M. The neurotrophin receptor p75NTR modulates long-term depression and regulates the expression of AMPA receptor subunits in the hippocampus. Proc Natl Acad Sci U S A. 2005;102(20):7362–7367. doi: 10.1073/pnas.0502460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulsen JS, et al. Clinical and biomarker changes in premanifest Huntington disease show trial feasibility: a decade of the PREDICT-HD study. Front Aging Neurosci. 2014;6:78. doi: 10.3389/fnagi.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tabrizi SJ, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12(7):637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 64.You SC, et al. Executive functions in premanifest Huntington’s disease. Mov Disord. 2014;29(3):405–409. doi: 10.1002/mds.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frodl T, et al. Reduced hippocampal volume correlates with executive dysfunctioning in major depression. J Psychiatry Neurosci. 2006;31(5):316–323. [PMC free article] [PubMed] [Google Scholar]
- 66.Eichenbaum H. How does the hippocampus contribute to memory? Trends Cogn Sci. 2003;7(10):427–429. doi: 10.1016/j.tics.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 67.Hampton RR, Hampstead BM, Murray EA. Selective hippocampal damage in rhesus monkeys impairs spatial memory in an open-field test. Hippocampus. 2004;14(7):808–818. doi: 10.1002/hipo.10217. [DOI] [PubMed] [Google Scholar]
- 68.Nadel L. The hippocampus and space revisited. Hippocampus. 1991;1(3):221–229. doi: 10.1002/hipo.450010302. [DOI] [PubMed] [Google Scholar]
- 69.Wright JW, Alt JA, Turner GD, Krueger JM. Differences in spatial learning comparing transgenic p75 knockout, New Zealand Black, C57BL/6, and Swiss Webster mice. Behav Brain Res. 2004;153(2):453–458. doi: 10.1016/j.bbr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Murphy KP, et al. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J Neurosci. 2000;20(13):5115–5123. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niklison-Chirou MV, et al. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc Natl Acad Sci U S A. 2013;110(47):18952–18957. doi: 10.1073/pnas.1221172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoshino M, et al. Transcriptional repression induces a slowly progressive atypical neuronal death associated with changes of YAP isoforms and p73. J Cell Biol. 2006;172(4):589–604. doi: 10.1083/jcb.200509132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agostini M, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A. 2011;108(52):21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chakravarthy B, et al. Hippocampal membrane-associated p75NTR levels are increased in Alzheimer’s disease. J Alzheimers Dis. 2012;30(3):675–684. doi: 10.3233/JAD-2012-120115. [DOI] [PubMed] [Google Scholar]
- 75.Egashira Y, Tanaka T, Soni P, Sakuragi S, Tominaga-Yoshino K, Ogura A. Involvement of the p75(NTR) signaling pathway in persistent synaptic suppression coupled with synapse elimination following repeated long-term depression induction. J Neurosci Res. 2010;88(16):3433–3446. doi: 10.1002/jnr.22505. [DOI] [PubMed] [Google Scholar]
- 76.Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310(5749):866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- 77.Santamaria F, Wils S, De Schutter E, Augustine GJ. Anomalous diffusion in Purkinje cell dendrites caused by spines. Neuron. 2006;52(4):635–648. doi: 10.1016/j.neuron.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26(7):360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- 79.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33(3):121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 80.Chakraborti A, Allen A, Allen B, Rosi S, Fike JR. Cranial irradiation alters dendritic spine density and morphology in the hippocampus. PLoS One. 2012;7(7):e40844. doi: 10.1371/journal.pone.0040844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu B, Buck CR, Dreyfus CF, Black IB. Expression of NGF and NGF receptor mRNAs in the developing brain: evidence for local delivery and action of NGF. Exp Neurol. 1989;104(3):191–199. doi: 10.1016/0014-4886(89)90029-0. [DOI] [PubMed] [Google Scholar]
- 83.Richardson PM, Issa VM, Riopelle RJ. Distribution of neuronal receptors for nerve growth factor in the rat. J Neurosci. 1986;6(8):2312–2321. doi: 10.1523/JNEUROSCI.06-08-02312.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roux PP, Barker PA. Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol. 2002;67(3):203–233. doi: 10.1016/S0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 85.Bernabeu RO, Longo FM. The p75 neurotrophin receptor is expressed by adult mouse dentate progenitor cells and regulates neuronal and non-neuronal cell genesis. BMC Neurosci. 2010;11:136. doi: 10.1186/1471-2202-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dougherty KD, Milner TA. p75NTR immunoreactivity in the rat dentate gyrus is mostly within presynaptic profiles but is also found in some astrocytic and postsynaptic profiles. J Comp Neurol. 1999;407(1):77–91. doi: 10.1002/(SICI)1096-9861(19990428)407:1<77::AID-CNE6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 87.Rudge JS, et al. Neurotrophic factor receptors and their signal transduction capabilities in rat astrocytes. Eur J Neurosci. 1994;6(5):693–705. doi: 10.1111/j.1460-9568.1994.tb00981.x. [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3(1):217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- 89.Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10(10):927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- 90.Rex CS, et al. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J Cell Biol. 2009;186(1):85–97. doi: 10.1083/jcb.200901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamashita T, Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat Neurosci. 2003;6(5):461–467. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]
- 92.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999;8(1):115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 93.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 94.Murai T, Okuda S, Tanaka T, Ohta H. Characteristics of object location memory in mice: Behavioral and pharmacological studies. Physiol Behav. 2007;90(1):116–124. doi: 10.1016/j.physbeh.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 95.Marco S, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med. 2013;19(8):1030–1038. doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ambrogi Lorenzini CG, Baldi E, Bucherelli C, Sacchetti B, Tassoni G. Role of ventral hippocampus in acquisition, consolidation and retrieval of rat’s passive avoidance response memory trace. Brain Res. 1997;768(1):242–248. doi: 10.1016/s0006-8993(97)00651-3. [DOI] [PubMed] [Google Scholar]
- 97.Lorenzini CA, Baldi E, Bucherelli C, Sacchetti B, Tassoni G. Role of dorsal hippocampus in acquisition, consolidation and retrieval of rat’s passive avoidance response: a tetrodotoxin functional inactivation study. Brain Res. 1996;730(1–2):32–39. doi: 10.1016/0006-8993(96)00427-1. [DOI] [PubMed] [Google Scholar]
- 98.Bourin M, Hascoët M. The mouse light/dark box test. Eur J Pharmacol. 2003;463(1–3):55–65. doi: 10.1016/s0014-2999(03)01274-3. [DOI] [PubMed] [Google Scholar]
- 99.Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462(7275):920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gratacos E, Checa N, Pérez-Navarro E, Alberch J. Brain-derived neurotrophic factor (BDNF) mediates bone morphogenetic protein-2 (BMP-2) effects on cultured striatal neurones. J Neurochem. 2001;79(4):747–755. doi: 10.1046/j.1471-4159.2001.00570.x. [DOI] [PubMed] [Google Scholar]
- 101.Carretón O, et al. Age-dependent decline of motor neocortex but not hippocampal performance in heterozygous BDNF mice correlates with a decrease of cortical PSD-95 but an increase of hippocampal TrkB levels. Exp Neurol. 2012;237(2):335–345. doi: 10.1016/j.expneurol.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 102.Grutzendler J, Tsai J, Gan WB. Rapid labeling of neuronal populations by ballistic delivery of fluorescent dyes. Methods. 2003;30(1):79–85. doi: 10.1016/S1046-2023(03)00009-4. [DOI] [PubMed] [Google Scholar]
- 103.Enriquez-Barreto L, et al. Learning improvement after PI3K activation correlates with de novo formation of functional small spines. Front Mol Neurosci. 2014;6:54. doi: 10.3389/fnmol.2013.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martín ED, Buno W. Stabilization effects of extracellular ATP on synaptic efficiency and plasticity in hippocampal pyramidal neurons. Eur J Neurosci. 2005;21(4):936–944. doi: 10.1111/j.1460-9568.2005.03925.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.