Abstract
Dynamin is a large GTPase that mediates plasma membrane fission during clathrin-mediated endocytosis. Dynamin assembles into polymers on the necks of budding membranes in cells and has been shown to undergo GTP-dependent conformational changes that lead to membrane fission in vitro. Recent efforts have shed new light on the mechanisms of dynamin-mediated fission, yet exactly how dynamin performs this function in vivo is still not fully understood. Dynamin interacts with a number of proteins during the endocytic process. These interactions are mediated by the C-terminal proline-rich domain (PRD) of dynamin binding to SH3 domain-containing proteins. Three of these dynamin-binding partners (intersectin, amphiphysin and endophilin) have been shown to play important roles in the clathrin-mediated endocytosis process. They promote dynamin-mediated plasma membrane fission by regulating three important sequential steps in the process: recruitment of dynamin to sites of endocytosis; assembly of dynamin into a functional fission complex at the necks of clathrin-coated pits (CCPs); and regulation of dynamin-stimulated GTPase activity, a key requirement for fission.
Introduction
The large GTPase dynamin belongs to a family of proteins that mediate various membrane-remodeling events in the cell. Dynamin is best characterized by its role in plasma membrane fission during clathrin-mediated endocytosis [1]. Other members of the dynamin family mediate additional membrane fission and fusion events in the cell, including the fission and fusion of mitochondrial membranes (dynamin-related protein 1 [Drp1], optical atrophy 1 [Opa1] and mitofusin), peroxisome fission (Drp1), and endoplasmic reticulum fusion (atlastin) [2]. All members of the dynamin family contain a G domain that binds and hydrolyses GTP and a stalk domain that promotes self-assembly [3,4]. Dynamin also contains a pleckstrin homology (PH) domain and a PRD. These unique domains almost certainly convey the specific function of dynamin in the cell. The PH domain preferentially binds phosphatidylinositol 4,5-bisphosphate (PIP2) [5], a lipid enriched in the plasma membrane [6], which is believed to function as a key signaling molecule for the recruitment and assembly of the clathrin machinery [7–9]. The PRD provides a platform for dynamin partners to bind via SH3-binding motifs [10–14]. In recent years, a concerted effort has been made to identify the molecular mechanisms that govern dynamin‘s role in membrane fission. This review will discuss current models of how the GTP hydrolysis cycle of dynamin drives fission during clathrin-mediated endocytosis, and how dynamin-binding partners may regulate this process.
Dynamin recruitment to sites of endocytosis
SH3 domain-containing endocytic accessory proteins
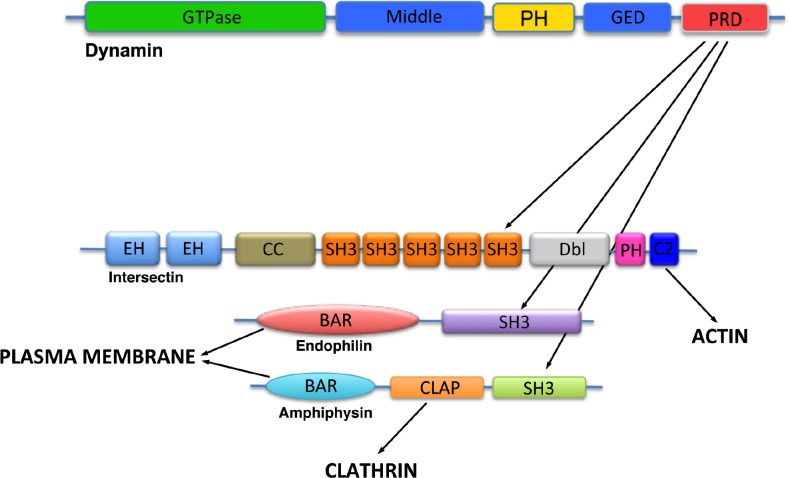
The recruitment of dynamin to sites of endocytosis is dependent on its PRD [12]. Dynamin interacts with a number of endocytic accessory proteins through several SH3-binding motifs located in the PRD [10–14]. Three SH3 domain-containing binding partners of dynamin have been shown to play a role in recruiting dynamin to CCPs on the plasma membrane, intersectin [15–19], amphiphysin [20–25] and endophilin [24,26] (Figure 1).
Figure 1. SH3 domain-containing binding partners of dynamin.
Dynamin interacts with the SH3 domain-containing proteins intersectin, amphiphysin and endophilin via its proline-rich domain (PRD). These three binding partners of dynamin are involved in various aspects of endocytosis. Intersectin functions as a protein scaffold, recruiting dynamin and other endocytic proteins to sites of clathrin-mediated endocytosis. Amphiphysin and endophilin contain N-BIN/amphiphysin/Rvs (BAR) domains and are involved in mediated high membrane curvature during endocytosis, like the formation of the constricted clathrin-coated pit neck. Amphiphysin also binds to clathrin, suggesting it acts as a link between dynamin and the clathrin coat.
PH, pleckstrin homology.
SH3 domain-containing proteins are the best-characterized members of a growing family of protein-protein interaction modules [27]. They recognize proline-rich sequences in a large number of otherwise structurally and functionally diverse proteins. SH3 domains contain a negatively charged pocket that binds proline-rich sequences. These interactions have fairly low affinity and moderately low specificity [28]. It is not uncommon for proteins to have several SH3 domains linked in tandem, suggesting that SH3 domain-containing proteins are capable of mediating the formation of large protein complexes with high rates of assembly and disassembly, like the endocytic machinery. For example, the dynamin-binding partner intersectin contains several SH3 domains that interact with dynamin [15,19,29–32], synaptojanin [33], and the actin network [29,31,34,35].
Intersectin, an endocytic scaffolding protein
In neuronal cells, clathrin-mediated endocytosis is required for synaptic membrane recycling. Upon stimulation of nerve terminals, the Ca2+-dependent phosphatase calcineurin dephosphorylates dynamin and other endocytic proteins (amphiphysin, epsin, eps15 and synaptojanin) [36–38], which leads to their recruitment to sites of vesicle recycling [15,19,26,39]. In neuromuscular junctions of Drosophila, the intersectin homologue Dap160 has been shown to regulate this recruitment of dynamin and other endocytic proteins (endophilin, synaptojanin and AP180) [16–19]. Intersectin also interacts with components of the actin network [29,31,34,35], which suggests that it functions as a scaffold to regulate the recruitment of both the endocytic machinery and the actin network to sites of endocytosis. It has also been shown that actin may be directly involved in regulating the endocytic process [40–45], and recent studies show that its function is tightly coupled to that of dynamin [46,47].
Assembly of dynamin at the necks of coated pits
BIN/amphiphysin/Rvs domain-containing proteins and dynamin assembly
Dynamin co-localizes with intersectin, endophilin, and amphiphysin on clathrin-coated budding vesicles [16,19,45,48–51]. In addition to SH3 domains, endophilin and amphiphysin also contain BIN/amphiphysin/Rvs (BAR) domains. BAR domain-containing proteins mediate membrane sensing and curvature via homodimerization [52–54]. Depending on the shape of the BAR domain dimer, these proteins sense and promote either positive (N- and F [FCH]-BAR proteins) [55–60], or negative curvature (I[inverse]-BAR proteins) [61,62]. BAR domains bind the membrane via electrostatic interactions with lipid head groups. N-BAR proteins contain an amphiphatic helix in their N-terminus (hence the name N-BAR). This helix inserts into the lipid bilayer and may play a role in sensing membrane curvature [55,57]. N-BAR proteins promote high curvatures [55,57,59,60], while proteins containing F-BAR domains have been shown to stabilize more shallow membrane deformations [56,63,64]. It has been shown that the F-BAR proteins FCHo (F-BAR domain-containing Fer/Cip4 homology domain-only protein) [65] and FBP17 (formin binding protein 17) [66] are recruited at early stages of CCP formation along with the clathrin adaptors epsin [67,68] and AP180 [68,69], suggesting that they mediate initial membrane curvature at the sites of clathrin assembly. The membrane neck may then be subsequently narrowed and stabilized by the higher curvature-generating N-BAR proteins endophilin and amphiphysin. This suggests that, following the recruitment to the sites of endocytosis, SH3/BAR domain-containing binding partners of dynamin are continuously involved in promoting dynamin-mediated fission.
BAR domain proteins as membrane templates for dynamin
Once recruited to sites of endocytosis, dynamin must assemble on the necks of CCPs to mediate the release of vesicles. Proper assembly is crucial for dynamin to perform its function. It has been shown that dynamin preferentially binds to high membrane curvature, such as lipid nanotubes [70], and the necks of CCPs [15,26,39,71,72]. In nerve terminals, endophilin and amphiphysin both co-localize with dynamin on constricted CCPs [26]. Injections of antibodies against the BAR domain of endophilin into nerve terminals caused an accumulation of shallow CCPs upon stimulation, and subsequent inhibition of synaptic vesicle recycling, supporting the idea that BAR proteins mediate curvature during CCP formation [73]. Furthermore, when interactions between dynamin and endophilin were selectively perturbed, dynamin failed to localize to CCP necks, despite recruitment to sites of endocytosis [26]. These nerve terminals further failed to maintain synaptic vesicle recycling as evidenced by the accumulation of CCPs, an indication of an inhibition of membrane fission. In cells, the knockdown of both amphiphysin and endophilin decreased the recruitment of dynamin to the plasma membrane and inhibited transferrin uptake [74]. It was further shown in vitro that endophilin facilitated the binding of dynamin to liposomes [26] and clustering of dynamin to giant unilamellar vesicles (GUVs) [74]. Both endophilin and amphiphysin also promoted dynamin-mediated vesiculation from large lipid reservoirs on silica beads [75]. In addition, endophilin promoted recruitment of the dynamin mutant I533A, which fails to bind lipid bilayers, suggesting SH3/BAR protein-mediated curvature can overcome the lipid-binding defects of this mutant, and rescue dynamin assembly [75]. Therefore, it is likely that SH3/BAR proteins function as templates for dynamin at the necks of CCPs, generating the perfect curvature for the efficient recruitment of dynamin and subsequent assembly at the necks of CCPs.
PH domain and PIP2 binding
The PH domain of dynamin binds to lipids, although dynamin recruitment to CCPs is independent of this interaction [24]. The PH domain inserts into the outer leaflet of the lipid bilayer via its variable loop 1, a property that enhances membrane fission [76,77]. Interestingly, the PH domains from different dynamin isoforms convey different lipid-binding properties [78]. Dynamin 2 (ubiquitous isoform) fails to bind larger liposomes unlike the neuronal isoform, dynamin 1 [75,78]. This difference in lipid binding is directly related to their PH domains [78]. Dynamin 1 and 2 share the same amino acid sequence in variable loop 1 but differ in one amino acid in variable loop 3 of the PH domain (Y600 versus L600). Switching these amino acids results in a reverse in the fission activity of the proteins. Generation of a dynamin 2 construct expressing the dynamin 1 PH domain increases transferrin uptake, further indicating that lipid sensing by the PH domain is beneficial in the fission reaction [78]. These data suggest that membrane insertion and sensing are conveyed by the variable loop 1 and 3 respectively, and that both of these components are crucial for fission [76–78]. Also, the addition of endophilin to dynamin 2 “rescues” lipid binding and vesiculation from GUVs, further supporting a role for the SH3/BAR proteins as curvature-generating templates for dynamin [75].
The exact role of the PH domain is still elusive. It has been proposed that insertion of the PH domain into the membrane may act to modify the composition of the neck by sequestering PIP2 [79]. There is emerging evidence that BAR proteins also sequester PIP2 [80,81]. Endophilin clusters PIP2 in GUVs [81] and binding to PIP2 has been shown to facilitate N-BAR protein-mediated curvature [82], suggesting that modulation of lipid composition (as well as curvature) plays a role in fission. In yeast, variation in the local concentration of PIP2 along the length of the CCP necks has been implicated in facilitating the actual fission reaction [43], but yeast lacks dynamin and, as of yet, local variation in the concentration of PIP2 has not been implicated in dynamin-mediated plasma membrane fission.
Taken together, the evidence suggests that amphiphysin and endophilin act as templates for dynamin recruitment and assembly at the necks of CCPs by providing optimal curvature and that they may be integral parts of a pre-fission complex to regulate the actual fission reaction. A potential role of such a complex may be to sequester PIP2 at the neck. Formation of a dynamin-SH3/BAR pre-fission complex may also serve to regulate the GTPase activity of dynamin.
Dynamin-mediated membrane fission
GTP cycling and conformational changes
Membrane fission requires dynamin-stimulated GTPase activity [71] that is promoted by G domain dimerization [83]. G domain dimerization stabilizes the transition state of the GTP hydrolysis reaction, a proposed key determinant for dynamin-mediated fission [84]. G domain dimerization only occurs between rungs of the dynamin helix and, therefore, stimulated hydrolysis only takes place in the assembled polymer when there is close opposition between rungs [84]. Therefore, the architecture of the dynamin polymer ensures that assembly and stimulated GTPase activity are tightly coupled. Stimulated GTP hydrolysis contributes to a conformation change in the dynamin polymer, the dynamin powerstroke, in which the three-helix bundle signaling element (BSE) swings down from the G domain core as a result of GTP hydrolysis [84]. It has been suggested that this powerstroke generates the force necessary to sever the membrane and may be responsible for the super-coiling and twisting of dynamin-decorated tubes observed in vitro as a result of GTP hydrolysis [85–87].
The effect of GTP hydrolysis on the assembled dynamin polymer has been studied for many years. Dynamin assembles on liposomes and forms well-decorated tubes that constrict upon the addition of GTP [87]. These observations led to the postulation that dynamin acts as a mechanochemical enzyme that severs the membrane by force generated from GTP hydrolysis. Three-dimensional reconstructions of dynamin revealed that constriction is mediated by a kink in the stalk domain and assembling into a two-start helix [84,88]. In the presence of a non-hyrolyzable GTP analogue (GMPPCP) a dynamin mutant, lacking the C-terminal PRD (ΔPRD), decreases the inner luminal diameter of the lipid tube from 20 nm to 7 nm [84,85,89–91]. More recently, it has been shown that a transition-state defective mutant, K44A, trapped in a pre-fission state, constricts the inner lumen to 3.7 nm, reaching the theoretical limit required for spontaneous membrane fission [88]. K44A dynamin mediates this “super-constriction” by adopting a two-start helical symmetry that allows more efficient packing of the dynamin subunits in the polymer. This super-constricted K44A dynamin is trapped in a ground state configuration, suggesting that GTP-binding promotes an initial conformational change in the polymer, prior to hydrolysis. In addition, half of the PH domains are tilted out of the membrane in the super-constricted state, compared to the non-constricted dynamin polymer. This suggests that, upon super-constriction, the PH domain re-arranges its orientation in the outer leaflet of the bilayer that may contribute to the destabilization of the lipid and act to promote fission. Also, in the super-constricted structure, the packing of the dynamin subunits is such that the number of G domain dimer interfaces is significantly higher than in an assembled one-start helix. G domain dimerization is required for stimulated GTPase activity and thus, it’s reasonable to assume that a two-start helical symmetry is more favorable for fission. In support of this, wildtype dynamin also assembles into a two-start helix in the presence of GTP, and constricts the membrane to an inner luminal diameter of ~4 nm [88].
In vitro, short scaffolds of dynamin polymers mediate fragmentation of lipid nanotubes in the presence of GTP [92–94]. Furthermore, it has been suggested that two rungs of a dynamin assembly act as a minimal fission machinery [95]. A recent TIRF (total internal reflection fluorescence) microscopy study shows that, in a majority of scission events observed in cells, the number of recruited dynamin molecules is ~26 [46]. Interestingly, this fits well with the number of dynamin molecules that are required for dynamin assembly around a lipid tube.
Exactly how the stimulated GTPase activity of dynamin aims to evoke fission is still unknown. However, it is reasonable to assume that dynamin assembly on the necks of CCPs is followed by a series of conformational changes, super-constriction, the BSE powerstroke, super-coiling and twisting, and re-arrangement of the PH domain, all regulated by the cycling of GTP. Despite recent insights into the structural features of dynamin at various nucleotide states, exactly how these conformational changes contribute to the fission reaction remains to be elucidated.
Three mechanisms for dynamin-mediated plasma membrane fission
Currently, three potential mechanisms have been put forward to describe the final dynamin-mediated fission event. Firstly, GTP-driven constriction and relaxation of the underlying membrane (as a result of assembly and disassembly of short dynamin scaffolds) causes the underlying membrane to be severed [92,94]. Secondly, the GTP-driven constriction of dynamin scaffolds generates torque that acts on areas of the membrane where the difference in curvature is greater (such as the edge of the dynamin scaffold) and the membrane is destabilized to promote fission [93]. Thirdly, PH domain re-arrangement or tilting in the assembled dynamin polymer results in deformation of the lipids, which generates enough energy to overcome the hemi-fission barrier [96]. Although these models are based on single-method studies, and thus each needs to be confirmed by additional techniques, they provide an excellent foundation for future studies. Also these models are most likely not mutually exclusive, and it is plausible that all of these factors work in combination with the GTP-dependent conformational changes described to promote fission. Although these in vitro assays show that dynamin alone can mediate fission, the element of regulation is removed, and hence further studies are necessary to mimic the in vivo environment containing accessory protein and local variations in nucleotide concentration and lipid composition.
Regulation of the fission reaction by SH3/BAR proteins
Dynamin co-assembles with both amphiphysin and endophilin in vitro and at late stages of endocytosis, indicating that these proteins may be involved in regulating the actual fission reaction [26,97]. The GTPase activity of dynamin is influenced by many factors, namely lipid binding, self-assembly and the binding of SH3 proteins to the PRD [5,98–101]. The localization of the PRD within the assembled dynamin polymer is unclear. However, recent evidence suggests that the PRD localizes at the surface of the assembled dynamin polymer, in close proximity to the G domain [88]. Intriguingly, a conserved region of the G domain most proximal to the putative PRD location is only present in dynamin family members containing a PRD. The close proximity of this region of the G domain to the PRD suggests a possible physical link between the two domains. This indicates that SH3/BAR proteins may influence the GTPase activity of the assembled dynamin polymer at late stages of endocytosis. Both amphiphysin and endophilin have been shown to influence the fission activity of dynamin in vitro [74,75]. Therefore, SH3/BAR proteins may act as curvature-inducing templates for dynamin at the necks of CCPs and, hence, either promote fission indirectly by generating optimal conditions for dynamin assembly and stimulated GTPase activity, or directly by interacting with the dynamin polymer during the fission reaction.
Conclusions
The dynamin-binding partners intersectin, endophilin and amphiphysin are crucial in regulating the function of dynamin during clathrin-mediated endocytosis (Figure 2). These proteins act throughout the process to facilitate dynamin-mediated fission and to specifically control the sequential steps of recruitment, assembly and stimulated GTPase activity of dynamin. These SH3-containing proteins act as scaffolds to bring dynamin to the sites of endocytosis in cells, and function as membrane templates at CCP necks to provide the optimal curvature for dynamin recruitment and assembly. They may also promote fission reaction by modulating the local lipid environment. Clues to the mechanisms of dynamin-mediated fission have evolved in recent years, suggesting that assembly and subsequent conformational changes of short scaffolds, differential membrane curvatures, and tilting of the PH domain all contribute to membrane fission. Based on our current understanding as discussed in this review, future studies in vivo will help further elucidate the function of dynamin during the endocytic process in the cell. There are some questions remaining that will push our understanding of how dynamin mediates fission of the plasma membrane, and also how other members of the family operate. Exactly what is the composition of the minimal fission machinery in vivo? What is the role of the powerstroke and other potential conformational changes brought on by GTP hydrolysis? What is the role of the PH domain? Is fission an active or passive reaction?
Figure 2. The sequential steps of dynamin-mediated fission.
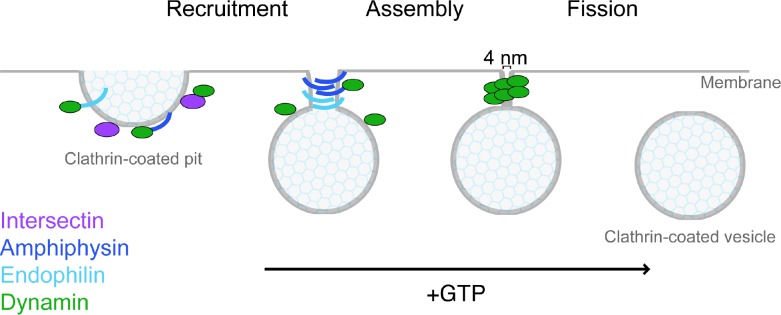
Dynamin‘s role in clathrin-mediated endocytosis migrates through the sequential steps of recruitment, assembly and fission. The SH3 domain-containing binding partners intersectin, amphiphysin and endophilin control these steps and thus promote dynamin-mediated fission. Intersectin acts as a scaffold for dynamin and other endocytic proteins, ensuring its recruitment to sites of endocytosis. Amphiphysin and endophilin are BIN/amphiphysin/Rvs (BAR) proteins and generate constriction of the clathrin-coated pit neck that promotes the dynamin assembly into a polymer. The assembled dynamin polymer undergoes GTP-dependent conformational changes that lead to super-constriction of the neck, the bundle signaling element (BSE) powerstroke, re-arrangement of the pleckstrin homology (PH) domain and polymer disassembly all leading to plasma membrane fission.
Currently, the dynamin field is enjoying an influx of structural data, due to recent advances in the field of cryo-electron microscopy (cryo-EM) and major efforts in crystallography. A new generation of cameras that allow cryo-EM density maps to reach atomic resolution will surely result in a surge of structural information in the coming years and this will ultimately elucidate the intra- and inter-molecular organization of the dynamin fission complex. Still, EM and crystallography alone are insufficient to track the dynamic process of fission. Continuous efforts to elucidate the structural characteristics of the dynamin fission machinery are key but must proceed in collaboration with functional studies. Several “live” fission assays have been published recently that shed light on the transient nature of the fission reaction. However groundbreaking, these are limited by the poor resolution of light and fluorescence microscopy, and thus fail to provide high-resolution information about the organization of the fission complex. Efforts to understand the role of dynamin in cells are relying heavily on protein overexpression, which in itself is problematic. A caveat to the multitude of techniques applied to study the fission reaction is the wide variety in lipid templates used. Fission requires the interplay between proteins and lipids, and the use of lipids must influence the setup and thus the interpretations of the data. Consensus must be reached in the efforts to infer mechanisms of fission in vivo from in vitro results. Hopefully, as we embark on a new and exciting era for structural biology, efforts to reconcile observations in different systems and to adopt multidisciplinary approaches will help us to finally unravel the enigmatic role of dynamin in plasma membrane fission.
Abbreviations
- BAR
BIN/amphiphysin/Rvs
- BSE
bundle signaling element
- CCP
clathrin-coated pit
- cryo-EM
cryo-electron microscopy
- Drp1
dynamin-related protein 1
- GUV
giant unilamellar vesicle
- PIP2
phosphatidylinositol 4, 5-bisphosphate
- PH
pleckstrin homology
- PRD
proline-rich domain
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/6/85
References
- 1.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 2.Praefcke Gerrit JK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–47. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- 5.Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 6.Kwiatkowska K. One lipid, multiple functions: how various pools of PI(4,5)P(2) are created in the plasma membrane. Cell Mol Life Sci. 2010;67:3927–46. doi: 10.1007/s00018-010-0432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 8.Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–9. doi: 10.1016/S0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–67. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McPherson PS. Regulatory role of SH3 domain-mediated protein-protein interactions in synaptic vesicle endocytosis. Cell Signal. 1999;11:229–38. doi: 10.1016/S0898-6568(98)00059-X. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto PM, Herskovits JS, Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–35. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 12.Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–6. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 13.Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS, Schmid SL. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat Cell Biol. 1999;1:119–24. doi: 10.1038/10091. [DOI] [PubMed] [Google Scholar]
- 14.Solomaha E, Szeto FL, Yousef MA, Palfrey HC. Kinetics of Src homology 3 domain association with the proline-rich domain of dynamins: specificity, occlusion, and the effects of phosphorylation. J Biol Chem. 2005;280:23147–56. doi: 10.1074/jbc.M501745200. [DOI] [PubMed] [Google Scholar]
- 15.Evergren E, Gad H, Walther K, Sundborger A, Tomilin N, Shupliakov O. Intersectin is a negative regulator of dynamin recruitment to the synaptic endocytic zone in the central synapse. J Neurosci. 2007;27:379–90. doi: 10.1523/JNEUROSCI.4683-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh T, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Koh T, Korolchuk VI, Wairkar YP, Jiao W, Evergren E, Pan H, Zhou Y, Venken Koen JT, Shupliakov O, Robinson IM, O’Kane CJ, Bellen HJ. Eps15 and Dap160 control synaptic vesicle membrane retrieval and synapse development. J Cell Biol. 2007;178:309–22. doi: 10.1083/jcb.200701030. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1089035
- 18.Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–19. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Winther Åsa ME, Jiao W, Vorontsova O, Rees KA, Koh T, Sopova E, Schulze KL, Bellen HJ, Shupliakov O. The dynamin-binding domains of Dap160/intersectin affect bulk membrane retrieval in synapses. J Cell Sci. 2013;126:1021–31. doi: 10.1242/jcs.118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shupliakov O, Löw P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science. 1997;276:259–63. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- 21.Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell. 1997;8:2003–15. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wigge P, Vallis Y, McMahon HT. Inhibition of receptor-mediated endocytosis by the amphiphysin SH3 domain. Curr Biol. 1997;7:554–60. doi: 10.1016/S0960-9822(06)00254-5. [DOI] [PubMed] [Google Scholar]
- 23.Wigge P, McMahon HT. The amphiphysin family of proteins and their role in endocytosis at the synapse. Trends Neurosci. 1998;21:339–44. doi: 10.1016/S0166-2236(98)01264-8. [DOI] [PubMed] [Google Scholar]
- 24.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–60. doi: 10.1016/S0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida Y, Takei K. Stimulation of dynamin GTPase activity by amphiphysin. Meth Enzymol. 2005;404:528–37. doi: 10.1016/S0076-6879(05)04046-2. [DOI] [PubMed] [Google Scholar]
- 26.Sundborger A, Soderblom C, Vorontsova O, Evergren E, Hinshaw JE, Shupliakov O. An endophilin-dynamin complex promotes budding of clathrin-coated vesicles during synaptic vesicle recycling. J Cell Sci. 2011;124:133–43. doi: 10.1242/jcs.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–92. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 29.Pechstein A, Shupliakov O, Haucke V. Intersectin 1: a versatile actor in the synaptic vesicle cycle. Biochem Soc Trans. 2010;38:181–6. doi: 10.1042/BST0380181. [DOI] [PubMed] [Google Scholar]
- 30.Sakaba T, Kononenko NL, Bacetic J, Pechstein A, Schmoranzer J, Yao L, Barth H, Shupliakov O, Kobler O, Aktories K, Haucke V. Fast neurotransmitter release regulated by the endocytic scaffold intersectin. Proc Natl Acad Sci USA. 2013;110: 8266–71. doi: 10.1073/pnas.1219234110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718006052
- 31.Tsyba L, Nikolaienko O, Dergai O, Dergai M, Novokhatska O, Skrypkina I, Rynditch A. Intersectin multidomain adaptor proteins: regulation of functional diversity. Gene. 2011;473:67–75. doi: 10.1016/j.gene.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–7. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 33.Pechstein A, Bacetic J, Vahedi-Faridi A, Gromova K, Sundborger A, Tomlin N, Krainer G, Vorontsova O, Schäfer JG, Owe SG, Cousin MA, Saenger W, Shupliakov O, Haucke V. Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2. Proc Natl Acad Sci USA. 2010;107:4206–11. doi: 10.1073/pnas.0911073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphries AC, Donnelly SK, Way M. Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J Cell Sci. 2014;127:673–85. doi: 10.1242/jcs.141366. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718190270
- 35.Klein IK, Predescu DN, Sharma T, Knezevic I, Malik AB, Predescu S. Intersectin-2L regulates caveola endocytosis secondary to Cdc42-mediated actin polymerization. J Biol Chem. 2009;284:25953–61. doi: 10.1074/jbc.M109.035071. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877836
- 36.Cousin MA, Tan TC, Robinson PJ. Protein phosphorylation is required for endocytosis in nerve terminals: potential role for the dephosphins dynamin I and synaptojanin, but not AP180 or amphiphysin. J Neurochem. 2001;76:105–16. doi: 10.1046/j.1471-4159.2001.00049.x. [DOI] [PubMed] [Google Scholar]
- 37.Marks B, McMahon HT. Calcium triggers calcineurin-dependent synaptic vesicle recycling in mammalian nerve terminals. Curr Biol. 1998;8:740–9. doi: 10.1016/S0960-9822(98)70297-0. [DOI] [PubMed] [Google Scholar]
- 38.Slepnev VI, Ochoa GC, Butler MH, Grabs D, De Camilli P. Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science. 1998;281:821–4. doi: 10.1126/science.281.5378.821. [DOI] [PubMed] [Google Scholar]
- 39.Evergren E, Tomilin N, Vasylieva E, Sergeeva V, Bloom O, Gad H, Capani F, Shupliakov O. A pre-embedding immunogold approach for detection of synaptic endocytic proteins in situ. J Neurosci Methods. 2004;135:169–74. doi: 10.1016/j.jneumeth.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–42. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1163271
- 41.Ehrlich M, Boll W, van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1021600
- 42.Ferguson SM, Ferguson S, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, O’Toole E, De Camilli P. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–22. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/2104958
- 43.Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877837
- 44.Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1026177
- 45.Cestra G, Toomre D, Chang S, Camilli P de. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:1731–6. doi: 10.1073/pnas.0409376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grassart A, Cheng AT, Hong SH, Zhang F, Zenzer N, Feng Y, Briner DM, Davis GD, Malkov D, Drubin DG. Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J Cell Biol. 2014;205:721–35. doi: 10.1083/jcb.201403041. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718431197
- 47.Taylor MJ, Lampe M, Merrifield CJ. A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol. 2012;10:e1001302. doi: 10.1371/journal.pbio.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/715898015
- 48.Bauerfeind R, Takei K, Camilli P de. Amphphysin I is associated with coated endocytic intermediates and undergoes stimulation-dependent dephosphorylation in nerve terminals. J Biol Chem. 1997;272:30984–92. doi: 10.1074/jbc.272.49.30984. [DOI] [PubMed] [Google Scholar]
- 49.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–9. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 51.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/9749956
- 52.Daumke O, Roux A, Haucke V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell. 2014;156:882–92. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718294065
- 53.Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–6. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718686225
- 54.Gallop JL, McMahon HT. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp. 2005:223–31. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718877838
- 55.Bhatia VK, Madsen KL, Bolinger P, Kunding A, Hedegård P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28:3303–14. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877839
- 56.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–17. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1103374
- 57.Gallop JL, Jao CC, Kent HM, Butler P, Jonathan G, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877840
- 58.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761: 897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N. Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 2006;25:2889–97. doi: 10.1038/sj.emboj.7601176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler P, Jonathan G, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1016996
- 61.Suetsugu S. The proposed functions of membrane curvatures mediated by the BAR domain superfamily proteins. J Biochem. 2010;148:1–12. doi: 10.1093/jb/mvq049. [DOI] [PubMed] [Google Scholar]
- 62.Zhao H, Pykäläinen A, Lappalainen P. I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr Opin Cell Biol. 2011;23:14–21. doi: 10.1016/j.ceb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1031188
- 64.Wang Q, Navarro Marcos VAS, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–5. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–4. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/3501956
- 66.Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem. 2004;279:40091–9. doi: 10.1074/jbc.M404899200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718877841
- 67.Ford Marijn GJ, Mills IG, Peter BJ, Vallis Y, Praefcke Gerrit JK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–6. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1009665
- 68.Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 69.McMahon HT. Endocytosis: an assembly protein for clathrin cages. Curr Biol. 1999;9:R332–5. doi: 10.1016/S0960-9822(99)80206-1. [DOI] [PubMed] [Google Scholar]
- 70.Roux A, Koster G, Lenz M, Sorre B, Manneville J, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–6. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–34. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–90. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 73.Andersson F, Löw P, Brodin L. Selective perturbation of the BAR domain of endophilin impairs synaptic vesicle endocytosis. Synapse. 2010;64:556–60. doi: 10.1002/syn.20772. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718877842
- 74.Meinecke M, Boucrot E, Camdere G, Hon W, Mittal R, McMahon HT. Cooperative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. J Biol Chem. 2013;288:6651–61. doi: 10.1074/jbc.M112.444869. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717970343
- 75.Neumann S, Schmid SL. Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J Biol Chem. 2013;288:25119–28. doi: 10.1074/jbc.M113.490474. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718040325
- 76.Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin’s GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877843
- 77.Ramachandran R, Pucadyil TJ, Liu Y, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20:4630–9. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877844
- 78.Liu Y, Neumann S, Ramachandran R, Ferguson SM, Pucadyil TJ, Schmid SL. Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation. Proc Natl Acad Sci USA. 2011;108:E234–42. doi: 10.1073/pnas.1102710108. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13388956
- 79.Bethoney KA, King MC, Hinshaw JE, Ostap EM, Lemmon MA. A possible effector role for the pleckstrin homology (PH) domain of dynamin. Proc Natl Acad Sci USA. 2009;106:13359–64. doi: 10.1073/pnas.0906945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen Paavo KJ, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1166911
- 81.Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4:1213–23. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718116583
- 82.Yoon Y, Zhang X, Cho W. Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains. J Biol Chem. 2012;287:34078–90. doi: 10.1074/jbc.M112.372789. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718877845
- 83.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–40. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/3811956
- 84.Chappie JS, Mears JA, Fang S, Leonard M, Schmid SL, Milligan RA, Hinshaw JE, Dyda F. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 2011;147:209–22. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13335984
- 85.Danino D, Moon K, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J Struct Biol. 2004;147:259–67. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 86.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–31. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1007616
- 87.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–9. doi: 10.1016/S0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 88.Sundborger AC, Fang S, Heymann JA, Ray P, Chappie JS, Hinshaw JE. A Dynamin Mutant Defines a Superconstricted Prefission State. Cell Reports. 2014;8:734–742. doi: 10.1016/j.celrep.2014.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718519033
- 89.Chen Y, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol. 2004;11:574–5. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1019116
- 90.Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–6. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 92.Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–86. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1138901
- 93.Morlot S, Galli V, Klein M, Chiaruttini N, Manzi J, Humbert F, Dinis L, Lenz M, Cappello G, Roux A. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–29. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717961011
- 94.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–75. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1138900
- 95.Liu Y, Mattila J, Schmid SL. Dynamin-catalyzed membrane fission requires coordinated GTP hydrolysis. PLoS ONE. 2013;8:e55691. doi: 10.1371/journal.pone.0055691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shnyrova AV, Bashkirov PV, Akimov SA, Pucadyil TJ, Zimmerberg J, Schmid SL, Frolov VA. Geometric catalysis of membrane fission driven by flexible dynamin rings. Science. 2013;339:1433–6. doi: 10.1126/science.1233920. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717991294
- 97.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barylko B, Binns D, Lin KM, Atkinson MA, Jameson DM, Yin HL, Albanesi JP. Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J Biol Chem. 1998;273:3791–7. doi: 10.1074/jbc.273.6.3791. [DOI] [PubMed] [Google Scholar]
- 99.Barylko B, Wang L, Binns DD, Ross JA, Tassin TC, Collins KA, Jameson DM, Albanesi JP. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry. 2010;49:10592–4. doi: 10.1021/bi101343p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Herskovits JS, Shpetner HS, Burgess CC, Vallee RB. Microtubules and Src homology 3 domains stimulate the dynamin GTPase via its C-terminal domain. Proc Natl Acad Sci USA. 1993;90:11468–72. doi: 10.1073/pnas.90.24.11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem. 1996;271:22310–4. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]