Abstract
Motor neurons (MNs) are neuronal cells located in the central nervous system (CNS) controlling a variety of downstream targets. This function infers the existence of MN subtypes matching the identity of the targets they innervate. To illustrate the mechanism involved in the generation of cellular diversity and the acquisition of specific identity, this review will focus on spinal MNs (SpMNs) that have been the core of significant work and discoveries during the last decades. SpMNs are responsible for the contraction of effector muscles in the periphery. Humans possess more than 500 different skeletal muscles capable to work in a precise time and space coordination to generate complex movements such as walking or grasping. To ensure such refined coordination, SpMNs must retain the identity of the muscle they innervate. Within the last two decades, scientists around the world have produced considerable efforts to elucidate several critical steps of SpMNs differentiation. During development, SpMNs emerge from dividing progenitor cells located in the medial portion of the ventral neural tube. MN identities are established by patterning cues working in cooperation with intrinsic sets of transcription factors. As the embryo develop, MNs further differentiate in a stepwise manner to form compact anatomical groups termed pools connecting to a unique muscle target. MN pools are not homogeneous and comprise subtypes according to the muscle fibers they innervate. This article aims to provide a global view of MN classification as well as an up-to-date review of the molecular mechanisms involved in the generation of SpMN diversity. Remaining conundrums will be discussed since a complete understanding of those mechanisms constitutes the foundation required for the elaboration of prospective MN regeneration therapies.
Keywords: motor neurons, development, central nervous system, spinal cord, transcription factors, spinal motor neuron, lower motor neuron
Introduction
Motor neurons (MNs) are neuronal cells located in the central nervous system (CNS) controlling a variety of downstream targets. There are two main types of MNs, (i) upper MNs that originate from the cerebral cortex and (ii) lower MNs that are located in the brainstem and spinal cord. Among the latest, spinal MNs (SpMNs) have been intensively studied during the last decades and therefore provide an interesting framework for further molecular characterization. SpMNs are located in the ventral horn of the spinal cord and control effector muscles in the periphery. They form the ultimate and irreplaceable component of the neuronal circuitry since there is no alternative route to convey the commands from the processing centers located in the CNS to the effector muscles in the periphery. Their axon extending through several meters in mammals constitute an exceptional and unique anatomical feature. SpMNs are therefore the longest known cell type.
Complex movements such as walking or grasping require the cooperation of several dozens of muscles. Additionally, sensory-motor feedback loops are essential for the real-time tuning of gestures. To ensure such refined coordination, SpMNs must acquire and retain the identity of muscles they innervate as well as be integrated in a coherent and functional neuronal circuitry. Hollyday et al. (1977) and Landmesser (1978) initially described the anatomical organization of SpMNs with respect to their muscle targets. Authors acknowledged an association between SpMNs' positions and their respective muscle target in the periphery. Ultimately these findings led to the concept of MN pool, which is defined as a compact anatomical group of MNs sharing similar intrinsic characteristics and connecting to a single target in the periphery. Because of their unique and irreplaceable function, diseases that involve loss of MNs such as progressive muscular atrophy, spinal muscular atrophy, primary lateral sclerosis, and amyotrophic lateral sclerosis, are rapidly debilitating, as only symptomatic treatments are available. Understanding the molecular mechanisms underlying SpMN diversity is among the fundamental steps required to elaborate successful regenerative therapies in the future. Here, we provide a complete description of MN classification to then review in depth the organization as well as the molecular mechanisms involved in the generation of SpMNs.
Motor neuron classification
MNs are exceptional cell types that can be divided into two main categories according to the location of their cell body: (i) upper and (ii) lower MNs. Upper and lower MNs must be considered as distinct entities despite of their shared nomenclature. Table 1 summarizes the differences between the two in terms of cell body location, neurotransmitter, targeting, and symptoms upon lesion and emphasizes the inappropriateness of a similar appellation to name both entities.
Table 1.
Comparison between upper and lower MNs.
| Upper MNs | Lower MNs | |
|---|---|---|
| Location | Cortex | Brainstem and SC |
| Neurotransmitter | Glutamate | Acetylcholine |
| Targeting | Within the CNS | Outside the CNS |
| Symptoms upon lesion | Spasticity | Paralysis |
Upper and lower MNs diverge in their cell body location, neurotransmitter, targeting and symptoms upon lesion.
Upper motor neurons
Upper MN cell bodies are located in the pre-motor and primary motor region of the cerebral cortex also known as the “motor strip.” Since upper MNs make glutamatergic connections with lower MNs located in the CNS, they are exclusively confined to the latter. Typical clinical symptoms of upper MN lesion include uncontrolled movement, decreased sensitivity to superficial reflex stimulation and spasticity (Ivanhoe and Reistetter, 2004). The organization of upper MNs is complex and can't be completely and accurately described in this review that primarily focuses on molecular mechanisms that generate SpMN diversity. Readers are invited to refer to the chapter 16 entitled “Upper Motor Neuron Control of the Brainstem and Spinal Cord” from Purves and Williams (2004) for more information.
Lower motor neurons
Lower MN cell bodies are located in specific nuclei in the brainstem as well as in the ventral horn of the spinal cord and therefore, alike upper MNs, are settling within the CNS. The remarkable characteristic of lower MNs is their axonal extension and connection outside of the CNS. Lower MNs are cholinergic and receive inputs from upper MNs, sensory neurons (SNs) as well as from interneurons (INs). Paralysis is a typical clinical symptom of lower MN lesions since once damaged there is no alternative route to convey the information to the muscle targets in the periphery. Lower MNs are classified into three groups according to the type of target they innervate: (i) branchial, (ii) visceral, and (iii) somatic MNs.
Branchial motor neurons
Branchial MNs are located in the brainstem and form, together with SNs, the cranial nuclei. They innervate branchial arch derived muscles of the face and neck through 5 cranial nuclei: the trigeminal (V), facial (VII), glossopharyngeal (IX), vagus (X) and accessory (XI) nerves. Despite their similar function, muscles of the neck and the face differ from other skeletal muscles in their embryological origin since they do not derive from the somites, but instead from the branchial arches. Such developmental difference is mirrored by specific characteristics reviewed in depth by Chandrasekhar (2004).
Visceral motor neurons
Visceral MNs belong to the autonomic nervous system (ANS) responsible for the control of smooth muscles (i.e., heart and arteries) and glands. The ANS can be described as the association of two components: (i) preganglionic MNs located in the CNS connected to ganglionic neurons belonging to the peripheral nervous system (PNS). In turn, peripheral ganglionic neurons target to the final effector organ. Additionally, the ANS is anatomically and functionally divided into two structures: (i) the sympathetic system and (ii) the parasympathetic system.
Motor neurons of the sympathetic system. The sympathetic nervous system is involved in the traditional “fight or flight” responses, recruiting energy storage, increasing awareness, and leading to a global activation of the body metabolism. Central MNs of the sympathetic system are located in the spinal cord from the thoracic segment 1 (T1) to the lumbar segment 2 (L2). These MNs have an intermedio-lateral position and constitute the preganglionic column (PGC) that will be described below. They connect to 3 different targets: two chains of ganglia adjacent to the spinal cord named (i) paravertebral and (ii) prevertebral as well as directly to (iii) the chromaffin cells of the adrenal medulla responsible for the release of the catecholamines (i.e., adrenaline and noradrenaline) in the circulation, in response to stress stimuli. On the other hand, paravertebral and prevertebral ganglia connect to a wide variety of targets including the heart, lungs, kidneys, intestines and the colon.
Motor neurons of the parasympathetic system. The parasympathetic system controls glands secretion and activates the gastrointestinal tract as well as sexual behavior, which are summarized as “rest and digest” functions. Central MNs of the parasympathetic system are located in the brainstem and contribute to the formation of the cranial nerves (III, VII, IX, and X). Parasympathetic MNs are also found in sacral segments 2 to 4 (S2–S4) of the spinal cord. They innervate ganglia located in the proximity of the peripheral targets such as the heart, bladder, lungs, kidneys, and pancreas.
In summary, visceral central MNs from the sympathetic and parasympathetic systems relay information from the CNS to ganglionic neurons of the PNS. In turn those ganglia antagonistically control a large number of various visceral targets. In contrast to branchial mentioned previously and somatic MNs described below, visceral MNs do not directly connect to the final effector. As a result, they constitute an anatomical and functional exception among lower MNs.
Somatic motor neurons
Somatic MNs are located in the Rexed lamina IX in the brainstem and the spinal cord and innervate skeletal muscles responsible for movements (Rexed, 1954). MNs form coherent groups connecting to a unique muscle target defined as MN pools. Somatic MNs can be divided into 3 groups: (i) alpha, (ii) beta, and (iii) gamma according to the muscle fiber type they innervate to within a specific muscle target (Figure 1). A motor unit defines a single MN together with all the muscle fibers it innervates. Interestingly, motor units are homogeneous: a MN innervates muscle fibers of a single type. This observation suggests selectivity in the establishment of neuromuscular connectivity and/or a coordinated maturation between a MN and its targeted fibers. Intuitively, the diversity of MNs mirrors the diversity of targets they innervate. Therefore, to better describe somatic MN diversity, a brief description of skeletal muscle physiology will be provided.
Figure 1.

Muscle innervation. Schematic of muscle fibers on the longitudinal section (adapted from Purves and Williams, 2004). Alpha MN (red) innervates (incoming arrow) extrafusal muscle fibers (EF, brown) whereas gamma MN (purple) connects to intrafusal fibers (IF, blue) within the muscle spindle (MS, light gray) surrounded by the outer capsule (OC, dark gray). Sensory neurons (green) carry information from the intrafusal fibers to the central nervous system (outgoing arrow).
Three classes of muscles can be anatomically and functionally distinguished: (i) cardiac muscles, (ii) smooth muscles and (iii) skeletal muscles. Cardiac muscles are responsible for the rhythmic contraction of the heart while smooth muscles control the diameter of blood vessels and the internal digestive and secretion organs. Both smooth and cardiac muscles are innervated by the ANS (described above). In contrast, somatic MNs exclusively innervate skeletal muscles that are the most abundant muscle class, with around 639 different muscles in the human body (Stone and Stone, 2009). Skeletal muscles are firmly attached to the skeleton by the tendons and are responsible for both posture and movement. Developmentally, skeletal muscles derive from the paraxial mesoderm that produces the somites, which in turn generate muscle precursor cells called myoblasts. Those cells migrate toward the periphery and fuse to form the body of the muscle. Physiologically, skeletal muscles are composed of 2 structures: (i) extrafusal fibers, generating the force and (ii) muscle spindles providing proprioceptive information on the position and extension status of the muscle. Muscle spindles are composed of several intrafusal fibers enveloped by a collagen sheath named the outer capsule. There are three kinds of intrafusal fibers with specific characteristics: (i) dynamic nuclear bag fibers (B1), (ii) static nuclear bag fibers (B2 fibers) and (iii) nuclear chain fibers. Analogously, extrafusal fibers are divided into 3 types according to their physiological and molecular properties: (i) slow-twitch fatigue-resistant (SFR), (ii) fast-twitch fatigue-resistant (FFR) and (iii) fast-twitch fatigable (FF). Table 2 summarizes the principal characteristics of the three extrafusal muscle fibers.
Table 2.
Characteristics of extrafusal muscle fiber types.
| SFR | FFR | FF | ||
|---|---|---|---|---|
| Anatomical properties | Fiber Diameter | Small | Intermediate | Large |
| Color | Red | Light red | White | |
| Capillaries | Many | Many | Few | |
| Myoglobin level | High | High | Low | |
| Mitochondria | Many | Many | Few | |
| Physical properties | Duration use | Hours | Minutes | < Minute |
| Power produce | Low | High | Very high | |
| Recruitment order | First | Second | Third | |
| Fatigue sensitivity | Slow | Intermediate | Fast | |
| Contraction velocity | Slow | Fast | Fast | |
| Function activity | Posture | Normal movements | Intense movements | |
| Metabolic properties | Myosin ATPase activity | Slow | Fast | Fast |
| ATP synthesis | Aerobic | Intermediate | Anaerobic | |
| Glycogen stores | Low | Intermediate | High | |
| Oxidative capacity | High | Intermediate | Low | |
| Glycolytic capacity | Low | Intermediate | High | |
| Energy storage | Triglycerides | Creatine Phosphate | Creatine Phosphate | |
| Glycogen | Glycogen |
Slow-twitch fatigue-resistant (SFR), Fast-twitch fatigue-resistant (FFR), and Fast-twitch fatigable (FF) fibers differ in term of anatomical, physical, and metabolic properties.
Mirroring the diversity of both intra- and extrafusal fiber types in a muscle, somatic MNs are further sub-divided into 3 types: (i) alpha, (ii) beta and (iii) gamma that will be further described below.
Alpha motor neurons. Alpha MNs exclusively innervate extrafusal muscle fibers and are the key of muscle contraction (Figure 1). Anatomically, alpha MNs are characterized by a large cell body and a well-characterized neuromuscular ending. They have an important role in the spinal reflex circuitry by receiving monosynaptic innervation directly from SNs thus minimizing the delay of the response (Eccles et al., 1960). Alpha MNs can be further divided into 3 different subtypes depending on the extrafusal fiber type they innervate: (i) SFR, (ii) FFR, and (iii) FF (Burke et al., 1973) (Figure 2). There is no universal criteria distinguishing alpha MNs subtypes; however, some trends are observed in term of size, excitability, and firing pattern. SFR MNs tend to have a smaller cell body diameter and thus a higher input resistance making them responsive to a lower stimulation threshold. As a result, SFR MNs are recruited first during muscle contraction. They also have the capacity of maintaining a persistent activity even after the stimulation ceased (Lee and Heckman, 1998). On the other hand, FF MNs have often a larger cell body and are firing after the initial recruitment of SFR neurons giving extra strength to the activated muscle. In terms of conduction velocity, MNs innervating fast fibers are substantially faster (100 m/s) than SFR MNs (85 m/s) (Burke et al., 1973). Lastly, little is known about FFR MNs physiology; yet, they are considered to have intermediate characteristics between FF and SFR MNs (Figure 2).
Figure 2.

Characteristics of alpha and gamma MNs. Schematic showing the principal characteristics of alpha and gamma MNs (adapted from Kanning et al., 2010). Within the ventral spinal cord (SC light gray), MN pools (dashed lines) are composed of gamma MNs (blue) as well as three type of alpha MNs: αFF (light brown), αFFR (dark brown), αSFR (green). Alpha MNs have a larger diameter than gamma MNs. Beta MNs are not represented for simplicity. The proportion of alpha MN subtypes varies between MN pools. In the periphery, a muscle is composed of three types of extrafusal fibers: fast-twitch fatigable muscle fibers (light brown, IIb) are innervated by αFF MNs, fast-twitch fatigue-resistant muscle fibers (dark brown, IIa) are innervated by alpha αFFR MNs and slow-twitch fatigue-resistant muscle fibers (green, I) are innervated by αSFR MNs. Intrafusal muscle fibers (blue) reside within a muscle spindle (gray) and are exclusively innervated by gamma MNs. A single MN innervate multiple fibers all of the same type; however, for the schematic simplicity only one fiber is represented.
Beta motor neurons. Beta MNs are smaller and less abundant than other somatic MN subtypes. As a result beta MNs are poorly characterized. They innervate both intrafusal and extrafusal muscle fibers (Bessou et al., 1965) (Figure 3). Therefore, beta MNs constitute an exception to the homogeneity observed in motor-units and control both muscle contraction and responsiveness of the sensory feedback from muscle spindles. They are further subdivided into two subtypes depending on the type of intrafusal fibers they innervate: (i) static, innervating nuclear chain fibers and (ii) dynamic, innervating the nuclear bag fibers of muscle spindles. Static beta MNs increase the firing rate of type Ia and type II sensory fibers at a given muscle length whereas dynamic beta MNs increase the stretch-sensitivity of the type Ia sensory fibers by stiffening the nuclear bag fibers. Beta MNs are mainly characterized anatomically and functionally, further molecular and electrical properties remain to be identified.
Figure 3.
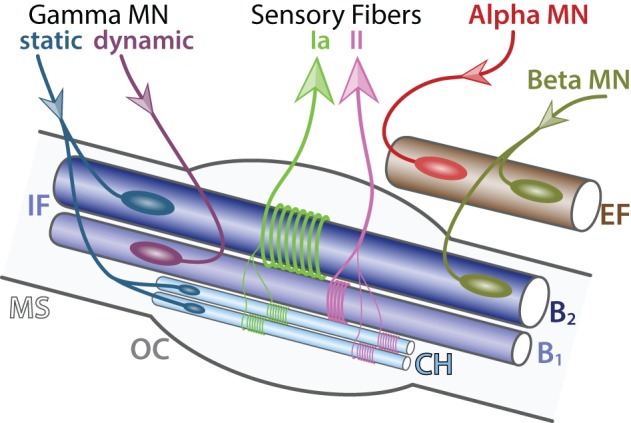
Detailed innervation of a muscle spindle. Schematic of an adult muscle spindle (MS, light gray) on the longitudinal section (adapted from Maier, 1997). Alpha MN (red) exclusively innervates (incoming arrow) extrafusal fibers (EF, brown). Beta MNs (green-brown) innervate both EF and intrafusal fibers (IF, blue). Gamma MNs are divided into two subtypes: static (blue) connecting to nuclear chain (CH, light blue) and nuclear bag 2 (B2, dark blue) fibers and dynamic (purple) connecting to nuclear bag 1 fibers (B1, intermediate blue). Sensory afferent axons Ia (light green) and II (pink) convey information (outgoing arrows) to sensory neurons located in the dorsal root ganglia. The outer capsule (OC) is a dedicated membrane isolating the muscle spindle from the extrafusal fibers. A single MN innervate multiple fibers all of the same type; however, for the schematic simplicity only one fiber is represented.
Gamma motor neurons. Gamma MNs control exclusively the sensitivity of muscle spindles. Their firing increases the tension of intrafusal muscle fibers and therefore mimics the stretch of the muscle. Like beta MNs, gamma MNs are functionally divided into two subtypes: (i) static, innervating nuclear chain fibers and static nuclear bag fibers and (ii) dynamic, innervating the dynamic nuclear bag fibers (Figure 3). Gamma MNs receive only indirect sensory inputs and do not possess any motor function. Therefore, gamma MNs do not directly participate to spinal reflexes (Eccles et al., 1960) but instead contribute to the modulation of muscle contraction.
Summary of motor neuron classification
As seen above, the term “motor neuron” groups a significant diversity of cell types and does not ideally reflect biological reality. Upper and lower MNs are fundamentally different and their shared nomenclature can easily be misleading. For instance, if we define a MN by being a “neuronal cells settling within but projecting outside of the CNS,” upper MNs would be excluded. In fact, upper MNs would be more accurately defined by the terminology “imbuo-neurons” derived from Latin imbuo that signifies “give initial instruction” or by the terminology “didactic-neurons” derived from the Greek didaktikós for instructive. In contrast, lower MNs, with the exception of visceral MNs, connect directly to their muscle targets and constitute the last step of the neuronal circuitry. SpMNs are divided into functional groups, termed pools, mirroring the diversity of muscle targets in the periphery. In addition, a single muscle is composed of several fiber types that are innervated by specific classes of MNs. Therefore MN pools should not be considered as a set of identical cells but instead as a mosaic of MN cell types covering a broad range of functions. The generation of this complex architecture must rely on precise mechanisms ensuring the establishment of the correct connections between matching MN - target pairs. We will review the functional organization of SpMNs as well as the molecular mechanisms leading to their generation.
Generation of spinal motor neurons
The spinal cord offers a relatively simple, yet, powerful experimental model to study neuronal development. It can be schematized as a circuitry formed by three different neuron types. Sensory neurons located in the dorsal root ganglia (DRG) receive input information from the periphery and transmit it either directly to alpha MNs located in the ventral horn (monosynaptic connections) or to association neurons (commissural and interneurons) that, in turn, process and convey the information toward the MNs. MNs then stimulate their respective effector that will generate the appropriate output response (Eccles et al., 1957) (Figure 4). Over the last three decades, many studies have shaded light on important mechanisms governing MN differentiation in the spinal cord. A comprehensive and up-to-date review of those studies will be presented below.
Figure 4.
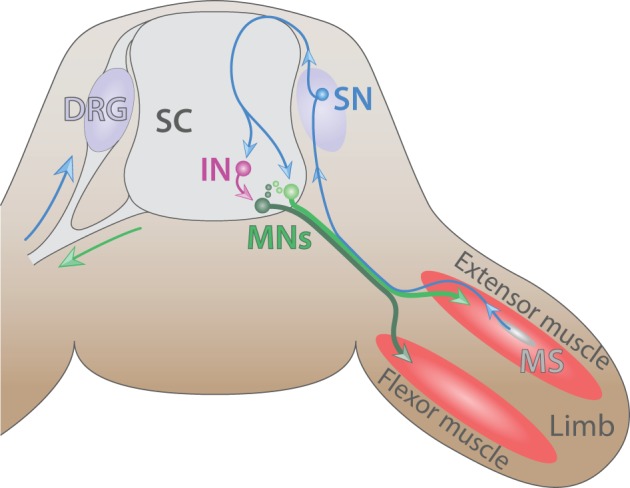
The spinal cord reflex circuitry. Schematic of a myotatic reflex illustrating the spinal cord (SC) circuitry (adapted from Purves and Williams, 2004). Sensory neuron (SN, blue) located in the dorsal root ganglia (DRG) transmits a stretch stimulus sensed by the muscle spindle (MS, gray) to an interneuron (IN, purple) as well as directly to motor neurons (MNs, dark and light green). In turn, MNs stimulate the contraction of extensor muscle (red) and ensure the concomitant relaxation of the antagonist flexor muscle located in the limb.
Developmental origin
During the early phase of embryogenesis, the egg cell undergoes a series of divisions until forming a sphere made of a single layer of cells called the blastula. Subsequently, during a process called gastrulation, a group of cells will enter the blastula cavity leading in triploblastic animals to the formation of the three primary germ layers: (i) the endoderm, (ii) the mesoderm, and (iii) the ectoderm. Individual layers generate progenies restricted to a limited number of distinct fates. The ectoderm undergoes a process called neurulation in which it folds inward and leads to the formation of three ectodermic masses: (i) the neural tube, (ii) the neural crest cells, and (iii) the external ectoderm. The external ectoderm generates the epidermis whereas the neural crest cells form the peripheral ganglion, the pigments of the skin as well as the dorsal root ganglia. Finally, the neural tube gives rise to the CNS, composed of the brain and the spinal cord (Purves and Williams, 2004) (Figure 5A).
Figure 5.
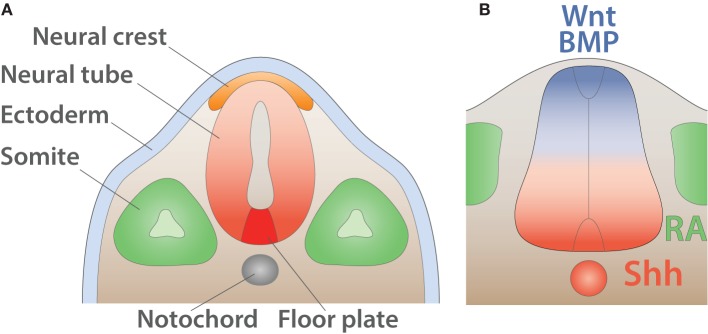
Early anatomy and inductive signals in the neural tube. (A) Schematic of the anatomy of the neural tube after neurulation (adapted from Purves and Williams, 2004). The ectoderm (light blue) is positioned on the external side whereas neural crest (orange) resides underneath. The notochord (gray) induces the differentiation of the floor plate (red). The somites (green) give rise to muscles and bones. (B) Schematic summarizing signals involved in the dorso-ventral pattering of the mouse neural tube shown in transverse section (adapted from Dessaud et al., 2008). Wnt and BMP secreted by the roof plate (blue) as well as retinoic acid (RA) produced by the somites (green) cooperate with Shh expressed by the floor plate and the notochord (red) to pattern the neural tube.
Generation of dedicated spinal cord progenitor domains
Soon after neurulation, the neural tube is surrounded by several inductive signals stimulating the subsequent differentiation process. Members of the wingless-type MMTV integration site family (WNT) (Alvarez-Medina et al., 2008) and of the bone morphogenetic protein family (BMPs) (Mehler et al., 1997) and their regulators Noggin (NOG), Chordin (CHRD), and Follistatin (FST) (Zimmerman et al., 1996; Streit et al., 1998) are expressed in a decreasing dorsal to ventral gradient. Additionally, the surrounding paraxial mesoderm expresses the aldehyde dehydrogenase 1 A2 (ALDH1A2 or RALDH2) (Niederreither et al., 1997), which converts retinaldehyde into retinoic acid (RA) a well-characterized regulator of neuronal differentiation (Pierani et al., 1999; Novitch et al., 2003; Lee et al., 2009). Together those signals collaborate with an increasing ventral to dorsal gradient of sonic hedgehog morphogen (SHH) secreted by the underlying notochord as well as the floor plate (Yamada et al., 1991, 1993; Roelink et al., 1994; Marti et al., 1995a,b; Ericson et al., 1996) (Figure 5B).
Molecularly, SHH binds to the patched homolog 1 receptor (PTCH1) (Stone et al., 1996) and releases its constitutive inhibition of the smoothened homolog (SMO) (Quirk et al., 1997) thereby, preventing the degradation of the GLI-Kruppel family (GLI) proteins (Chen et al., 2011b; Niewiadomski et al., 2014). Hence, SHH signaling correlates directly with GLI activity (GliA) (Figure 6). Conversely, signals from the roof plate induce the expression of GLI repressors (GliR). Together, ventral and dorsal signals lead to a net decreasing gradient of GLI activity from the ventral to the dorsal. In turn, GLI proteins promote or repress in a concentration dependent manner homeodomain transcription factors that can be sorted into two classes: (i) Class-I; paired box 3/6/7 (PAX3/6/7), developing brain homeobox 1 and 2 (DBX1/2), and Iroquois related homeobox 3 (IRX3) are repressed by GliA and thus expressed dorsally whereas (ii) Class-II NK2 homeobox 2 and 9 (NKX2.2/2.9), NK6 homeobox 1 and 2 (NKX6.1/6.2), and oligodendrocyte transcription factor 2 (OLIG2) are induced by GliA and therefore, located ventrally (Shirasaki and Pfaff, 2002). This initial patterning is subsequently refined by cross-repression between pairs of class-I and class-II proteins. Studies using systematic gain or loss of function approaches have identified the specific pairs of class-I and class-II proteins. For example, inactivation of OLIG2 leads to a ventral expansion of IRX3 (Zhou and Anderson, 2002) whereas ectopic expression of NKX6.1 restrains the expression of DBX2 to the most dorsal domain (Briscoe et al., 2000). Thus, cross-repressive interactions between pairs of class-I and class-II proteins guarantee the formation of sharp boundaries between adjacent domains and ensure that they remain mutually exclusive. Ultimately, this process leads to the emergence of five ventral progenitor domains (p0, p1, p2, pMN, and p3) defined by the expression of a unique combination of transcription factors (Ericson et al., 1997; Briscoe et al., 2000; Vallstedt et al., 2001). This simple mechanism is in fact more complex as additional molecules ensure the integrity of each individual progenitor domains. For example both WNT signaling pathway (Lei et al., 2006; Alvarez-Medina et al., 2008; Yu et al., 2008) and the transducin-like enhancer of split 1 (TLE1) (Todd et al., 2012) contribute to reinforce the ventral boundary of the pMN domain. Another level of complexity arises from the interpretation of SHH gradient that is modulated by the downstream network of transcription factors. Hence, tis mechanism creates a feedback loop during the developmental period allowing the modulation of progenitor domain formation (Balaskas et al., 2012).
Figure 6.
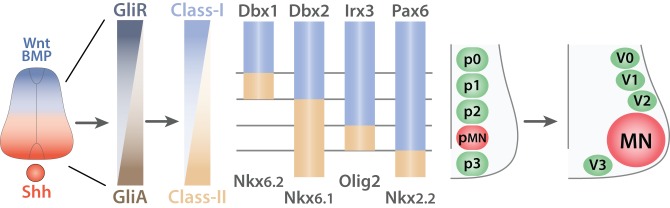
Generation of ventral spinal progenitor domains. Schematic summarizing the mechanisms of progenitor domain formation in the ventral spinal cord (adapted from Ulloa and Marti, 2010). Opposing gradients of Shh (red) and Wnt/BMP proteins (blue) are transduced into Gli protein activity. Gli activators (GliA, brown) in the most ventral region induce the expression of Class-II proteins (light brown) whereas Gli repressors (GliR, dark gray-blue) induce Class-I proteins (light blue) in the dorsal portion of the ventral spinal cord. This initial expression pattern is subsequently refined by cross-repressive interactions between pairs of Class-I and Class-II proteins to generate five exclusive progenitor domains (p0, p1, p2, p3, and pMN). V0, V1, V2, V3, interneurons arise from the p0, p1, p2, and p3 respectively whereas all MNs derive from the pMN progenitors.
Ultimately, the five ventral progenitor domains will generate neuronal cells restricted to a specific lineage (V0, V1, V2, V3, INs, and MNs) (Alaynick et al., 2011). Conceptually, the strategy used for the establishment of the progenitor domains involves inductive gradients interpreted into the expression of specific combinations of transcription factors. Cross-repressive interactions between pairs of transcription factors ensure the creation of mutually exclusive domains. Each progenitor domain then generates progenies restricted to a specific lineage.
Acquisition of motor neuron fate
All SpMNs arise from the unique pMN progenitor domain that expresses the unique combination of the homeodomain proteins NKX6.1, PAX6, and the basic helix-loop-helix (bHLH) protein OLIG2 (Tanabe et al., 1998; Novitch et al., 2001; Vallstedt et al., 2001). To become a mature MN, progenitors need to exit the cell cycle and enter the differentiation process. These events must be tightly regulated in order to generate an appropriate number of differentiated cells at a particular time during neurogenesis. Several mechanisms involved these transitions have been characterized and will be described here.
First, RA described previously as a regulator of progenitor domain formation, is also involved in the acquisition of the MN fate (Novitch et al., 2003). This process illustrates a principle commonly seen in developmental biology and in biology in general, namely, the use of a single cue at multiple steps during development as a mean to reduce the biological cost in energy. RA induces in MN progenitors the expression of glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5 or GDE2) (Jacobson and Rao, 2005; Rao and Sockanathan, 2005). In turn, GDPD5 complexes with the peroxiredoxin 1 (PRDX1) (Yan et al., 2009) and with the GDP form of the G protein alpha subunit i2 (GNAI2) (Hammerle and Tejedor, 2007; Periz et al., 2010; Sabharwal et al., 2011; Park et al., 2013) to promote the MN differentiation program. Similarly, the cut-like homeobox 2 (CUX2) is involved in progenitors' cell cycle progression and cell cycle exit (Iulianella et al., 2008).
In parallel, OLIG1 and 2 contribute to the expression of another bHLH protein named neurogenin 2 (NEUROG2) (Sommer et al., 1996; Novitch et al., 2001, 2003; Scardigli et al., 2001; Lu et al., 2002; Zhou and Anderson, 2002; Lee and Pfaff, 2003; Lee et al., 2004). NEUROG2 interacts with the RA receptor (RAR) and recruits the histone acetyl transferases CREB binding protein (CREBBP) and E1A binding protein p300 (EP300) (Lee et al., 2009) to promote the transcription of downstream MN genes (Mizuguchi et al., 2001; Novitch et al., 2001; Scardigli et al., 2001; Lu et al., 2002; Zhou and Anderson, 2002; Lee and Pfaff, 2003). Interestingly, during the early stage of MN generation OLIG2 and NEUROG2 collaborate to promote MN fate (Mizuguchi et al., 2001). At later stages, the persistence of OLIG1/2 expression and the concomitant down-regulation of NEUROG2 allow the emergence of oligodendrocyte progenitors from the pMN domain (Richardson et al., 2000; Lu et al., 2002; Zhou and Anderson, 2002). Therefore, the dynamic regulation of OLIG2 and NEUROG2 during neurodevelopment allows the sequential generation of MNs and oligodendrocytes at different time from a common progenitor domain (Lee et al., 2005). An important downstream target of OLIG2 and NEUROG2 signaling is the motor neuron and pancreas homeobox 1 (MNX1 or HB9) (Tanabe et al., 1998; Lee et al., 2005, 2009). Remarkably, MNX1 stimulates its own expression (Tanabe et al., 1998) providing to developing MNs their independence from SHH and RA signaling. Therefore, MNX1 has been used as a reliable and specific marker of post-mitotic SpMNs.
Although cell fates seem to be established early in development, some evidences suggest that additional mechanisms that ensure their maintenance are required. For example, MNs and V2 INs are generated by two adjacent progenitor domains (Figure 6). Inactivation of MNX1 in developing MNs induces a switch toward V2 IN fate (Arber et al., 1999; Thaler et al., 1999). Comparably, the runt-related transcription factor 1 (RUNX1), whose expression is restricted to selected post-mitotic cervical MNs (Theriault et al., 2004; Stifani et al., 2008; Guizard et al., 2010) is important for the consolidation of MN phenotype by ensuring the persistent suppression of the IN program (Stifani et al., 2008). The molecular mechanism underlying the divergence between V2 INs and MNs have been remarkably revealed by Pfaff and colleagues and involves the transient expression of the LIM homeobox 3 (LHX3) in developing MNs and V2 INs (Thaler et al., 2002). In prospective V2 INs, LHX3 forms a complex with the LIM domain binding 1 (LDB1 or NLI) and promotes the IN fate via the LIM domain only 4 (LMO4) (Thaler et al., 2002; Lee et al., 2008). In prospective MNs, the ISL1 transcription LIM homeodomain (ISL1) is induced by SHH secreted by the notochord and floor plate (Yamada et al., 1991; Ericson et al., 1992) and inserts into the LHX3-LDB1 complex to induce a switch toward MN specification (Thaler et al., 2002; Song et al., 2009). Most importantly ISL1-LHX3 complex directly binds and induce the expression genes involved in cholinergic neurotransmission, a fundamental characteristic of SpMNs (Cho et al., 2014; Kania, 2014a). Despite these important findings the acquisition and the maintenance of MN fate remain to be fully understood. A recent study has shed light on the mechanisms by which MN precursors detach from neuroepithelium and migrate laterally as they exit the cell cycle (Rousso et al., 2012). What are the molecular mechanisms controlling such migration? Rousso et al. (2012) remarkably identify the role of the forkhead box P2 and 4 (FOXP2/4) in promoting the detachment of newly born MNs from the ventricular zone. Additionally, the authors elegantly linked nuclear gene regulation to effector protein at the membrane. Namely, FOXP2/4 repress the expression cadherin 2 (CDH2) responsible for the attachment of developing progenitors to the neuroepithelium.
In conclusion, the emergence of newborn MNs from the pMN progenitor domain relies on the precise control of the balance between proliferation and differentiation. Although OLIG2 and NEUROG2 have prominent roles into MN fate commitment, additional mechanisms are required to ensure the consolidation of this phenotype. Following the acquisition of their general identity, MNs need to differentiate and acquire features required for their respective function. This process, termed patterning, will be described hereafter.
Patterning in spinal motor neuron development
Following the initial acquisition of their general fate, newborn SpMNs are required to further differentiate to adopt an identity reminiscent of their respective muscle targets. The general strategy is, at least in part, comparable to the mechanisms leading to the emergence of spinal progenitor domains. Globally, SpMN specification follows a temporal gradient along the ventro-dorsal and rostro-caudal axes (Nornes and Carry, 1978). MNs located more ventrally and more rostrally are generated earlier. This temporal regulation reflects two mechanisms: (i) the progressive expansion of the total volume of the neural tissue and (ii) the generation of specific cell types along the rostro-caudal axis.
Several proteins including the fibroblast growth factor (FGF), the growth differentiation factor 11 (GDF11 or BMP-11), members of transforming growth factor beta (TGFB) family as well as RA (Durston et al., 1989; Muhr et al., 1999; Liu et al., 2001; Bel-Vialar et al., 2002; Sockanathan et al., 2003; Liu, 2006) form a gradient along the rostro-caudal axis and induce, in a concentration-dependent manner, the expression of protein of the homeobox (HOX) family (Ensini et al., 1998; Lance-Jones et al., 2001). Hox genes are arranged into genomic clusters and their response to FGF and RA concentration is correlated to their position within a cluster (Liu et al., 2001; Bel-Vialar et al., 2002). Genes located at the 5′ end are induced by high concentration of FGF and thus expressed in more caudal regions. Conversely, genes at the 3′ end are induced by low concentrations of FGF and therefore expressed in more rostral regions (Liu et al., 2001; Bel-Vialar et al., 2002; Dasen et al., 2003; Mazzoni et al., 2013b). After the initial activation of HOX protein expression, further refinement is achieved at the rostral boundary by histone modifications performed by the Bmi1 polycomb ring finger oncogene (BMI1) part of the polycomb repressive complex (Golden and Dasen, 2012). At the caudal edge, cross-repressive interactions between pairs of HOX proteins (Dasen et al., 2003, 2005) lead to non-overlapping domains. For instance, HOX6, 9, and 10 expression correlates with the brachial, thoracic, and lumbar segments, respectively.
Subsequently, HOX patterning induces the formation of anatomical columns termed motor columns along the rostro-caudal axis (Shah et al., 2004; Dasen et al., 2005; Jung et al., 2010). The underlying mechanisms have been partially defined since HOX patterning converges toward the expression of FOXP1 (Arber, 2008; Dasen et al., 2008; Pfaff, 2008; Rousso et al., 2008; Palmesino et al., 2010). Mechanistically, HOX6 and 10 at brachial and lumbar segments respectively direct the expression of FOXP1, which in turn cooperate with HOX proteins to induce the formation of limb specific MNs at the expense of thoracic MNs (Dasen et al., 2008; Rousso et al., 2008). Additionally, HOXC9 has a critical role in restricting appendage specific MNs to the limb innervating segments by selectively excluding them from thoracic segments (Jung et al., 2010, 2014). This effect is at least partially mediated by direct and indirect repression of FOXP1 in thoracic segments.
In summary, after the formation of dedicated progenitor domains, intrinsic and extrinsic molecules cooperate to promote a general MN fate. Inductive signals along the rostro-caudal axis profile developing MNs to adjust to specific local needs. Together these mechanisms lead to the formation of anatomically defined motor columns. We will describe hereafter each column by providing information on their molecular specificity as well as mechanisms of their formation.
Columnar organization of spinal motor neurons
SpMNs are organized into distinct anatomical columns extending along the rostro-caudal axis and called motor columns (Figure 7). Previous studies have described four main columns: the median motor column (MMC), the lateral motor column (LMC), the hypaxial motor column (HMC), and the preganglionic column (PGC) (Prasad and Hollyday, 1991; Tsuchida et al., 1994; Jessell, 2000; Alaynick et al., 2011; Francius and Clotman, 2014) Each column possesses a coherent gene expression profile as well as a uniform axonal projection pattern (Figure 8). We will hereafter describe their molecular identity as well as the developmental mechanisms required for their formation. Moreover, we will complete this picture by describing the less well-characterized spinal accessory column (SAC) and the phrenic motor column (PMC).
Figure 7.

Segmental organization of spinal motor columns. Schematic summarizing the segmental distribution of spinal motor columns (adapted from Dasen and Jessell, 2009). While the medial motor column (MMC, brown) is present all along the rostro-caudal axis, the spinal accessory column (SAC, purple) is restricted to the five first cervical segments (C1–C5). The phrenic motor column (PMC, red) is confined between C3 and C5. The preganglionic column (PGC, orange) extends through the thoracic segments until the second lumbar segments (L2) as well as well as between sacral segments 2 and 4 (S2–S4). The hypaxial motor column (HMC, light blue) is exclusive of the thoracic segment where as the lateral motor column (LMC, dark and light green) is located at limb levels: brachial (C5-T1) and lumbar segments (L1–L5).
Figure 8.
Organization of SpMNs at cervical, brachial/lumbar and thoracic levels. Schematic summarizing the characteristics of spinal motor columns at cervical (A), brachial/lumbar (B) and thoracic (C) levels (adapted from Dasen and Jessell, 2009). MMC MNs (brown) are located medially and connect to the axial musculature (Epaxial). PMC MNs (red) have an inter-medio-lateral position and connect to the diaphragm. SAC MNs (purple) exit the CNS via the lateral exit point (LEP) and connect to mastoid and neck muscles. LMC MNs (green) are divided into two divisions medial (m, dark green) and lateral (l, light green). LMCm MNs connect to the ventral (v) part of the limb whereas LMCl MNs innervate the dorsal (d) region. HMC MNs (light blue) are located in the medio-lateral region and connect to the body wall and intercostal muscles (Hypaxial). PGC MNs (orange) are positioned dorso-laterally and innervate to the sympathetic chain ganglia (SCG) and chromaffin cells of the adrenal gland (AdrG). Proteins expressed by each column are depicted with their respective color code.
The median motor column
MMC MNs are located in the medial region of the ventral spinal cord and target to the dermomyotome (Gutman et al., 1993; Tsuchida et al., 1994), which gives rise to the axial musculature later in development (Fetcho, 1987; Gutman et al., 1993). Axial muscles are mainly involved in the maintenance of the body posture and are found all along the body axis. Therefore, MMC MNs are not segmentally restricted and are found all along the spinal cord (Figure 7). MMC MNs are characterized by the co-expression of MNX1, ISL1/2, and LHX3 (Tsuchida et al., 1994) (Figure 8). In mature MNs, LHX3 is unique to MMC MNs and its forced expression is sufficient to impose MMC identity (Sharma et al., 1998, 2000). LHX3 is therefore commonly used as a reliable marker of MMC MNs; however, as mentioned earlier, LHX3 is also transiently expressed developing MN in which it contributes to the establishment of their identity at the expense of the V2 INs. Interestingly, MMC MNs present an exception to the rostro-caudal patterning of HOX proteins. How do MMC MNs escape HOX rostro-caudal patterning? Molecularly, proteins from the WNT family (WNT4/5A/5B) are expressed in a ventral to dorsal decreasing gradient (Agalliu et al., 2009) and permit the persistence of LHX3/4 expression (Shirasaki and Pfaff, 2002; Agalliu et al., 2009) in the most ventral region. In turn, LHX3/4 make MMC MNs unresponsive to HOX patterning (Dasen et al., 2005, 2008). As suggested by Dasen and colleagues, this unique feature likely reflects the ancestral properties of the MMC from which other motor column have derived during evolution (Dasen et al., 2008; Dasen, 2009; Dasen and Jessell, 2009; Jung et al., 2010; Philippidou and Dasen, 2013).
The spinal accessory column
SAC MNs are located in the intermedio-lateral region of the spinal cord and expand from the end of the medulla until the 5th cervical segment (C1–C5) (Jacobson and Marcus, 2007; Ullah et al., 2007) (Figure 7). SAC MNs innervate mastoid muscles as well as four muscles of the neck (Sternomastoid, Cleidomastoid, Cleidotrapezius, and Acromiotrapezius) (Brichta et al., 1987; Watanabe and Ohmori, 1988). While SAC MNs innervating the mastoid muscles are located in the rostral portion, MNs innervating the trapezius muscles are located in the most caudal segments of the C1–C5 segment of the spinal cord (Ullah et al., 2007; Stifani et al., 2008). SAC MNs are different from other SpMNs because they innervate muscles that derive from branchial arches (Pabst et al., 1998; Aldskogius et al., 2009) and because their axons penetrate the periphery by exiting through the lateral exit point (LEP) located midway along the dorso-ventral axis of the spinal cord (Sharma et al., 1998; Schubert and Kaprielian, 2001; Pabst et al., 2003; Dillon et al., 2005; Bravo-Ambrosio and Kaprielian, 2011) (Figure 8). Therefore, SAC MNs are also referred as dorsal MNs (dMNs) as opposed to ventral MNs (vMNs) exiting classically via the ventral root. Molecularly, SAC MNs have been successfully distinguished from other MNs by the use of different markers such as activated leukocyte cell adhesion molecule (ALCAM or BEN) (Schubert and Kaprielian, 2001; Dillon et al., 2005) as well as ISL1, RUNX1 and the paired-like homeobox 2b (PHOX2B) (Pattyn et al., 2000; Amiel et al., 2009; Dubreuil et al., 2009; Stifani and Ma, 2009; Kobayashi et al., 2013; Mazzoni et al., 2013a; Laumonnerie et al., 2014).
Developmentally, the NK2 homeobox 9 (NKX2.9) has been shown to be required for proper SAC MN generation (Pabst et al., 2003) as well as for SAC axonal projection (Dillon et al., 2005). Conversely, LHX3/4 inactivation leads to an increase number of SAC MNs (Sharma et al., 1998) whereas LHX3 is sufficient to promote vMNs at the expense of dMNs (Lieberam et al., 2005; Hirsch et al., 2007; Kobayashi et al., 2013). Together these results suggest that the expression of LHX3/4 allows vMNs generation at the expense SAC MNs.
SAC MNs are a very peculiar population of SpMNs as they are the exclusive cells of branchial type in the spinal cord. Several characteristics described above are reminiscent of hindbrain branchiomotor and visceromotor MN populations and largely differ from other SpMNs. SAC MNs may appear therefore as a transitional population between the hindbrain and cervical MNs.
The phrenic motor column
Phrenic MNs are located in the cervical segments C2–C6 at embryonical stages and become progressively confined between cervical levels C3–C5 by birth (Webber and Pleschka, 1976; Allan and Greer, 1997a,b; Song et al., 2000) (Figure 7). They connect to a particular muscle: the diaphragm. This muscle is essential for respiration and therefore is under constant rhythmic activity. This characteristic also applicable to the cardiac muscle differs diametrically from skeletal muscles required to generate unsystematic contraction. The diaphragm is involved in both inspiration and expiration, both conscious and unconscious. Because of its unique function, phrenic MNs are required to produce a perpetual rhythmic firing as early as the first instant after birth and throughout life. Although phrenic MNs have been well characterized in terms of cell body position and anatomical properties, until recently little was known about their molecular characteristics. In a well-detailed study, Philippidou and colleagues identified for the first time the molecular profile of phrenic MNs (Philippidou et al., 2012). Namely, phrenic MNs are under the control of HOX5 patterning and are expressing high levels of the POU domain class 3 transcription factor 1 (POU3F1 or SCIP) as well as ISL1/2, and MNX1 (Thaler et al., 1999; Rousso et al., 2008; Castellani and Kania, 2012; Philippidou et al., 2012; Philippidou and Dasen, 2013) (Figure 8). Interestingly, soon after birth, phrenic MNs as well as the diaphragm muscle itself undergo significant anatomical and functional transformations including dendritic arborization and electrical activity properties (Cameron et al., 1991; Prakash et al., 2000). These changes reflect the passage from the aquatic intrauterine gestation to the aerobic life. Therefore, although phrenic MNs are established early in development to ensure their functionality for the time of birth, additional mechanisms, yet to be characterized, are likely occurring after birth to guarantee further post-natal maturation. The vital role of the PMC coupled to the recent molecular findings cited above encouraged Machado and colleagues to induce the differentiation of embryonic stem cells into phrenic MNs in vitro (Machado et al., 2014). This prodigious achievement carries great hope for stem cell based regenerative therapies as patients suffering from SpMN diseases ultimately face respiratory impairments. Nevertheless, additional efforts are still required to establish a viable therapy from this initial breakthrough.
The preganglionic motor column
PGC MNs also known as spinal visceral MNs constitute the CNS component of the ANS. They are located in the thoracic and upper lumbar spinal segments (T1–L2) (Figure 7) where they occupy an intermedio-lateral location (Figure 8). They do not innervate skeletal muscles as other somatic SpMNs do but instead connect to the sympathetic ganglia. Thus, PGC MNs are involved in stimulation of smooth muscles as well as in control of glands secretions. They can be molecularly identified by the expression of the SMAD family member 1 (SMAD1 or pSMAD1) (Dasen et al., 2008), nitric oxide synthase 1 neuronal (NOS1 or nNOS) (Saito et al., 1994; Wetts and Vaughn, 1994; Dasen et al., 2003), zinc finger E-box binding homeobox 2 (ZEB2 or SIP1) (Roy et al., 2012), as well as low level of FOXP1 (Dasen et al., 2008; Rousso et al., 2008; Morikawa et al., 2009).
Despite extreme functional differences, somatic and visceral SpMNs arise from common precursors expressing ISL1/2 (Prasad and Hollyday, 1991; Thaler et al., 2004). The maintenance of ISL2 in maturing MNs leads to the generation of somatic MNs whereas its down-regulation together with the persistence of ISL1 guides maturing MNs toward the visceral fate (Thaler et al., 2004). Recently, the one cut domain family members (ONECUT1/2/3), expressed in newly born MNs (Francius and Clotman, 2010, 2014; Audouard et al., 2012), have been found to bind directly to a specific enhancer region of Isl1 gene to maintain its expression (Roy et al., 2012) resulting in a limitation of PGC MN formation. This consequence is challenged by the effect of ZEB2 promoting PGC formation. Therefore, opposing and cooperating mechanisms ensure the proper divergence between somatic and visceral SpMNs.
As mentioned earlier, spinal visceral MNs are also found in the sacral segments (S2–S4). However, these cells belong to the parasympathetic system (rest and digest) while thoracic PGC MNs belong to the sympathetic autonomic system (fight or flight). In addition to these functional differences, thoracic and sacral PGC MNs also differ in terms of axonal projections. While thoracic PGC MNs connect to the sympathetic chain ganglia located in the proximity of the spine, sacral PGC MNs connect to ganglia in the vicinity of the effector targets (kidney, bladder, gonads). Therefore, the molecular properties of sacral PGC MNs that remain largely unknown are presumably substantially different from thoracic PGC MNs.
The hypaxial motor column
Initially the MMC had been separated in two divisions: (i) a medial MMC (MMCm), described above as MMC, targeting to axial musculature and present all along the rostro-caudal axis and (ii) a lateral MMC (MMCl) targeting to the body wall and present only in the thoracic segments (Gutman et al., 1993; Jessell, 2000). However, recent molecular findings have associated MMCl MNs with PGC and LMC MNs rather than with MMC MNs (Dasen et al., 2008; Rousso et al., 2008). Therefore, the MMCl has been referred to as the hypaxial motor column (HMC) (Dasen et al., 2008; Agalliu et al., 2009). This new nomenclature better reflects HMC MN molecular nature and avoids confusion with MMC MNs. HMC MNs are located in the ventro-lateral spinal cord and innervate muscles derived from the ventral mesenchyme (Smith and Hollyday, 1983). The ventral mesenchyme gives rise to the body wall musculature composed of the intercostal and abdominal muscles present only at thoracic level (Prasad and Hollyday, 1991). Therefore, HMC MNs are only present at thoracic level (Tsuchida et al., 1994; Sharma et al., 2000) (Figure 7). Molecularly, HMC MNs are characterized by the expression of MNX1, ISL1, ETS variant 1 (ETV1 or ER81) and low levels of ISL2 (Dasen et al., 2008; Rousso et al., 2008) (Figure 8). Interestingly, FOXP1 inactivation converts both PGC and LMC MNs to a HMC phenotype (Dasen et al., 2008). As suggested by Dasen and Jessell (2009), HMC and MMC MNs likely reflect the vestige of an ancestral spinal motor column organization from which other motor columns derived (Jung et al., 2014). Finally, because intercostal and abdominal muscles are involved in respiration, HMC MNs could presumably be somehow related to PMC MNs described previously. To our knowledge no experiment has been reported to address this suggestion that remains to be tested.
The lateral motor column
LMC MNs are located in the most lateral portion of the ventral spinal cord (Bueker, 1944). They connect to muscles of the appendages and therefore are present only at limb levels also defined as brachial (C5 to T1) and lumbar levels (L1–L5) (Hollyday and Hamburger, 1977; Hollyday and Jacobson, 1990) (Figure 7). This segmentation reflects the rostro-caudal patterning of HOX proteins (Kessel and Gruss, 1991; Liu et al., 2001; Dasen et al., 2003) controlled by local inductive signals (Ensini et al., 1998).
LMC MNs are further separated into two divisions: medial and lateral (Tosney et al., 1995). These divisions retain a topographic correspondence with the localization of their target in the periphery. Medial LMC (LMCm) MNs target to the ventral part of the limb whereas lateral LMC (LMCl) MNs innervate the dorsal limb musculature (Landmesser, 1978; Tosney and Landmesser, 1985a,b; Kania et al., 2000) (Figure 8). Molecularly, LMC MNs are characterized by the expression of ISL2, FOXP1, and ALDH1A2 and do not sustain LHX3 expression (Tsuchida et al., 1994; Sockanathan and Jessell, 1998; Dasen et al., 2008; Rousso et al., 2008). Sockanathan and Jessell (1998) have remarkably revealed the molecular mechanism leading to the emergence of LMC divisions. At limb levels, the paraxial mesoderm secretes RA that induces the generation of LMC MNs (Ensini et al., 1998; Sockanathan et al., 2003; Vermot et al., 2005). Early born LMC MNs co-express ISL1/2 as well as ALDH1A2 and in turn secrete RA. This additional signal induces the down-regulation of ISL1 to the profit of the Lim homeobox 1 (LHX1) in later born LMC. Furthermore, cross-repressive interactions allow both divisions to remain mutual exclusive (Kania and Jessell, 2003). ISL1 and LHX1 also control the differential segregation of the cell body position of LMC divisions (Sockanathan and Jessell, 1998; Kania and Jessell, 2003; Rousso et al., 2008). Interestingly, matured LMCm MNs down-regulate MNX1 expression (Kania and Jessell, 2003; Rousso et al., 2008); a unique characteristic among SpMNs. Yet, the functional relevance of this distinct observation remains to be elucidated. Further information about LMC will be provided in the section dedicated to axonal targeting.
To date, 6 different motor columns have been identified in mouse the spinal cord. The SAC located in the rostral cervical segments is the only representative of the branchial category whereas the PGC in the thoracic and sacral segments is the only visceral motor column. In contrast, MMC, HMC, PMC, and LMC are somatic and innervate skeletal muscles belonging to different groups. However, to date SpMNs haven't been mapped at the single cell resolution levels (Wichterle et al., 2013). Therefore, the possibility of having uncharacterized discrete SpMN populations can't be excluded. Furthermore, SpMN diversity expands beyond the columnar organization described above. In fact, SpMNs form muscle specific groups termed pools. We will review hereafter the mechanisms driving motor pool formation.
Specification of motor neuron pools
A remarkable event in SpMN development is the acquisition of MN pool identity, assigning to a given group a specific muscle target. The coordination between more than 50 different muscles in the typical amniotes' limb required to perform complex movements implies a precise mechanism to assign to each muscle a corresponding MN pool (Romanes, 1941; Sullivan, 1962). Previous studies have described the localization of individual MN pools according to specific targets (Landmesser, 1978; Hollyday and Jacobson, 1990; Choi and Hoover, 1996; Ryan et al., 1998) and suggest that MN pools respect a topographic organization (reviewed by Kania, 2014b). The more rostral a MN pool is positioned, the more anterior and proximal the target is located. Interestingly, MNs possess predetermined intrinsic features independent of the presence of peripheral targets that control at least partially pool specification (Phelan and Hollyday, 1990). Therefore, MN pool determination can be divided in two phases (i) purely intrinsic and (ii) extrinsically induced (Dasen, 2009).
The intrinsic molecular mechanisms of MN pool specification are not yet fully understood, however it appears to rely on the combinatorial expression of HOX proteins. Dasen et al. (2005) have performed an extensive screen of the expression of 39 Hox genes as well as HOX cofactors. Their results demonstrate that within a specific rostro-caudal segment, cross-repressive interactions between HOX members produce a unique combinatorial code that directs MN pool identity (Dasen et al., 2005; Lacombe et al., 2013). This identity is revealed by the activation of pool specific proteins such as the ETV1 and ETV4 (or PEA3) (Lin et al., 1998; Ladle and Frank, 2002; Livet et al., 2002), RUNX1 (Theriault et al., 2004; Dasen et al., 2005; Stifani et al., 2008; Zagami et al., 2009; Lamballe et al., 2011) and POU3F1 (Dasen et al., 2005; Rousso et al., 2008). By doing so, Dasen et al. (2005) have remarkably linked the intrinsic HOX combinatorial network to extrinsically induced factors whose expressions are dependent on a signal from the periphery (Lin et al., 1998; Haase et al., 2002) described in more detail below. However, to date the entire mapping of HOX proteins in SpMN pools remains unpublished. Furthermore, molecular effectors of pool specificity downstream of the HOX combinatorial network remain elusive.
In parallel to intra-segmental HOX combinatorial network, NKX6.1 contributes to the intrinsic mechanisms of MN pool specification (De Marco Garcia and Jessell, 2008). First, NKX6.1 is expressed in subsets of LMC MNs independently of the presence of its muscle target. Second, NKX6.1 inactivation leads to persistent muscle targeting errors. These results strongly suggest that NKX6.1 participates in controlling MN pool specificity and uncover two sequential roles of NKX6.1 in MN development. In the early phase, it takes part in the specification of progenitor domains in response to SHH gradient whereas in the late phase, it contributes to the specification of discrete MN pools. Strategically, intrinsic cues allow the development and the maturation of MNs independently of their location. This approach provides plasticity and tolerance to adapt to changes in the peripheral environment.
Unlike NKX6.1 and the HOX combinatorial intra-segmental network described above, extrinsically induced players are expressed in developing MNs upon reception of a specific signal. This mechanism can be considered as a checkpoint ensuring further developmental refinements only after the completion of prerequisite steps. What are the extrinsic signals allowing further MN differentiation? So far, only one factor has been unambiguously identified. Namely, the glial cell derived neurotrophic factor (GDNF) is secreted by both Cutaneous maximus (CM) and Latissimus dorsi (LD) muscles and induces the expression of ETV4 in the corresponding MN pools (Lin et al., 1998; Haase et al., 2002; Livet et al., 2002). The analysis of ETV4 mutant animals revealed that even though some aspects of MN development are pre-established by intrinsic cues, later signals are further required for the maintenance of MN pool characteristics such as cell body position, axonal arborization and dendritic patterning ensuring the establishment of correct input connections (Ladle and Frank, 2002; Livet et al., 2002; Vrieseling and Arber, 2006). Additionally, after the initial expression of ETV4 induced by GDNF, CM MNs recruits adjacent MNs and induce in a non-cell autonomous manner the expression of ETV4 (Helmbacher et al., 2003). Therefore, one of the strategy initial differentiation followed by the recruitment in situ of neighboring MNs.
Together these results illustrate the coordination between intrinsic and extrinsically-induced cues. While the first group allows MN development independently of the environment, the second ensures the completion of essential steps. Together these mechanisms create a flexible process allowing MNs to adapt to environment variability. By definition, MN pool specification is intimately linked to axonal targeting. Intensive works have identified various molecules involves in SpMN axonal targeting. We will dedicate the next section to the review the known molecular mechanisms controlling SpMN axonal targeting.
Motor neuron axonal targeting
Axonal targeting is a critical process of MN development. MN axons emerge within the CNS and transit through different tissues to reach and connect to their specific muscle target in the periphery. Axonal targeting not only provides to MNs their unique anatomical characteristic and therefore their irreplaceable function but also ensures their persistence through the action of trophic signals. In order to complete such critical process, MNs combine several mechanisms in a stepwise manner (Figure 9). Several “checkpoints” are established along the axonal route, each one requiring a choice to orientate toward a particular direction. While the initial steps rely on intrinsic mechanisms, the late aspects of MN axonal targeting rely on signals received at the growth cone, and inducing molecular and anatomical modifications.
Figure 9.

Steps of MN axonal targeting. Schematic summarizing the steps of MN axonal targeting (adapted from Dasen and Jessell, 2009). The first step termed “CNS exit” reflect the choice of developing MNs to exit the CNS via the ventral root (vMNs, blue) or through the lateral exit point (LEP) (dMNs, red). The second choice labeled “Columns” return to the motor column: MMC MNs (brown) target to epaxial musculature whereas LMC MNs (green) project to the limb. The third step named “Divisions” refers to the choice made by the medial and lateral divisions of the LMC. LMCl MNs (light green) invade the dorsal part of the limb (d) whereas LMCm (dark green) MNs target to the ventral region (v). The fourth step termed “Pool intrinsic” refers to the selection of a specific muscle target (red) and is controlled by intrinsic cues. The last step named “Pool extrinsic” illustrates the induction of specific protein expression upon a signal from the muscle target, which coordinates the terminal arborization of MN axons. Proteins involved in each step are indicated.
The very first choice SpMN axons make occurs within the spinal cord (termed “CNS exit” in Figure 9). vMNs leave the CNS via the ventral root whereas dMNs exit more dorsally via the LEP. This decision is at least partially controlled by LHX3 and 4 (Sharma et al., 1998; Bravo-Ambrosio and Kaprielian, 2011). Yet, LHX3/4 are transcription factors and therefore are unlikely the effectors of this axonal targeting decision made at the growth cone. Instead, the chemokine (C-X-C motif) receptor 4 (CXCR4) is expressed by vMN axons and its ligand CXCL12 localizes in the ventral mesenchyme surrounding the spinal cord. This molecular signal attracts vMN axons toward the ventral root (Lieberam et al., 2005). Conversely, dMNs express the netrin receptor deleted in colorectal carcinoma (DCC) and are repelled away from the midline expressing netrin 1 (NTN1) (Dillon et al., 2005) (Figure 10A). The complete molecular mechanisms allowing dMNs to escape the classical ventral root exit are yet to be characterized. As dMNs are absent outside of the cervical regions, novel molecules involved in SAC MNs axonal targeting could presumably be restricted to the first cervical segments. Unbiased differential screenings of genes downstream of transcription factors exclusive of dMNs (PHOX2B) or vMNs (LHX3/4) may identify new effector molecules involved in their divergence.
Figure 10.

Guiding cues of SpMN axonal targeting. Schematic summarizing guiding cues important for MN axonal targeting. (A) Ventral exiting MNs (vMN, blue) express CxcR4 and are attracted (plus signs) by CxcL12 expressed by the mesenchyme (dark green). Dorsal MNs (dMN, purple) express DCC and are repelled (minus sign) away from the midline expressing Ntn1 (light green). (B) MMC MNs (brown) expressing both FgfR1 and EphA3/4 are attracted by Fgf secreted by the dermomyotome but repelled by Ephrin-As expressed by the dorsal root ganglion. LMC MNs (green) target to the limb and pause before further growth. This pause is mediated by Npn1-Sema3A repellent signaling expressed by LMC MNs and the limb respectively. (C) LMCm MN (dark green) axons express EphB1 and Npn2 and are constrained into the ventral limb by Sema3F and Ephrin-Bs expressed by the dorsal limb mesenchyme (dark brown). Conversely, LMCl MN (light green) axons express Ephrin-As and EphA4 and are restricted to the dorsal part of the limb by a combination of Ephrin-As repulsive signal from the ventral limb mesenchyme (light brown) and EphAs (red) attractive signal from the dorsal part of the limb.
The second step in MN axonal growth consists in selecting the orientation toward their forthcoming muscle target (termed “Columns” in Figure 9). Schematically, growing vMN axons can adopt three directions: (i) dorsal, toward the axial musculature (MMC), (ii) lateral, invading the limb (LMC) or (iii) ventral, toward the sympathetic chain or to the body wall musculature (PGC and HMC, respectively). This schematic intentionally omits PMC targeting for simplicity. These decisions are comprised within the identity of a particular motor column and therefore considered as intrinsic. Presumably, the unique combinatorial expression of transcription factors controls downstream effectors and modulators of axonal growth. Although the molecular mechanisms remain largely unknown, MMC MNs express the fibroblast growth factor receptor 1 (FGFR1) and are attracted by the dermomyotome secreting FGF (Shirasaki et al., 2006; Soundararajan et al., 2010). Additionally, MMC axons expressing the Eph receptor A3 and 4 (EPHA3 and 4) are constrained by repellent contact with sensory DRG neurons expressing ephrin-As (EFNA1) (Gallarda et al., 2008). Together these mechanisms lead MMC axons to bypass the DRG and target to the axial musculature (Figure 10B). The molecules leading LMC axons to initially target the limb are unknown, however Huber et al. (2005) revealed the role of Semaphorin-Neuropillin in controlling the timing and the fasciculation of LMC axons. Neuropilin 1 (NRP1) expressed by LMC axons mediates the repulsion from the limb mesenchyme expressing semaphoring 3A (SEMA3A). Inactivation of SEMA3A-NRP1 signaling results in a premature invasion of the limb bud. Interestingly, NRP1 is expressed by both MN and SN axons and contributes to MN axon fasciculation along the sensory axons (Huettl et al., 2011). This example illustrates the use of a single molecule to synchronize sensory and motor development (Wang et al., 2011; Fukuhara et al., 2013). Such strategy ensures the formation of a coherent and functional circuitry. Lastly, PGC and HMC axons specifically turn ventrally toward the sympathetic chain and the body wall musculature, respectively. To date the mechanisms of such decision remain unidentified.
The lateral and medial divisions of the LMC have provided a powerful framework to study MN axonal decisions. After entering the base of the limb LMC axons pause before targeting toward the dorsal or the ventral parts of the limb (Tosney and Landmesser, 1985a; Wang and Scott, 2000). LMCm MNs express ISL1 and target to the ventral part of the limb whereas LMCl MNs express LHX1 and connect to the dorsal part of the limb (termed “Division” in Figure 9). Interestingly, LHX1 inactivation does not perturb LMCl formation but instead impairs the dorsal/ventral axonal projection specificity (Kania et al., 2000). Reciprocally, the LIM homeobox transcription factor 1 beta (LMX1B) expressed in a decreasing dorsal to ventral gradient in the limb mesenchyme is also important for LMC divisions axonal targeting (Kania et al., 2000). The molecular mechanisms of LMC axonal targeting rely prominently on Ephrin-Eph signaling and have been the source of recent exciting discoveries summarized by Bonanomi and Pfaff (2010) and reviewed in depth by Kao et al. (2012). In brief, LMCl MNs express LHX1 that induces the expression of EPHA4. LMCl axons are repelled away from the ventral limb mesenchyme expressing EFNAs (Helmbacher et al., 2000; Kania and Jessell, 2003). Similarly, LMCm MNs express EPHB1 are repulsed from the dorsal limb mesenchyme expressing EFNBs (Luria et al., 2008). Therefore, cross-repulsive Ephrin-Eph signaling mediates the correct segregation of LMCl and LMCm (Figure 10C). However, additional mechanisms contribute as well to LMC MNs axonal targeting. For example, GDNF and GDNF family receptor alpha 1 (GFRA1) cooperate with EFNAs-EPHAs signaling to control LMC MN dorso-ventral choice (Kramer et al., 2006). More recently, new discoveries have enriched Ephrin-Eph signaling with additional levels of complexity. Trans forward and reverse signaling (Dudanova et al., 2012) as well as interaction in cis (Kao and Kania, 2011) regulate LMC MN axonal targeting. Furthermore, the tyrosine kinase receptor Ret proto-oncogene (RET) acts co-receptor for both GDNF and ephrin-As modulating their response and thus adding another layer of complexity in LMC MN axonal targeting (Bonanomi et al., 2012). Together these results demonstrate that LMC targeting is complex and tightly regulated. Further experiments will permit a better understanding of this multifaceted process.
After making their initial decisions MN axons need to select their specific muscle target. This step is closely related to the formation of MN pools discussed above. MNs are programmed to recognize their muscle target (Lance-Jones and Landmesser, 1980). Intrinsic cues are expressed in a pool specific manner to direct MN axons toward their specific muscle target (termed “Pool Intrinsic” in Figure 9). NKX6 (De Marco Garcia and Jessell, 2008) as well as the HOX combinatorial network (Dasen et al., 2005) have been proposed as intrinsic regulators of muscle target selection. Presumably, other molecules, yet to characterize, play a role in the establishment of specific connections between a MN pool and its respective muscle target. Among them, the downstream molecular effectors that regulate axonal path finding remain to be identified.
Finally, after reaching their appropriate muscle, MN axons need to form functional connections with their target. Interestingly, studies focusing on the CM and LD muscles have revealed that this process is initiated upon receiving a signal from the peripheral target and therefore is considered as an extrinsic event (termed “Pool extrinsic” in Figure 9). MN pools innervating these two muscles are characterized by the expression of ETV4 (Ladle and Frank, 2002). It has been remarkably shown that the initial expression of ETV4 is induced by GDNF expressed by the CM and LD muscles (Haase et al., 2002; Helmbacher et al., 2003). In turn, ETV4 is responsible for inducing the terminal axonal arborization (Livet et al., 2002) as well as the dendritic refinement of these specific MN pools (Vrieseling and Arber, 2006). The molecular effectors of terminal arborization are still unknown; however, downstream targets of GDFN and/or ETV4 signaling could be good candidates for further investigation. Recently, Audouard et al. (2012) have identified for the first time a transcriptional regulator of neuromuscular junction formation. The analysis of ONECUT1 inactivated animals demonstrates a peculiar hind limb locomotion pattern resulting from impairments in neuromuscular junction formation. These findings open new opportunities to further characterize downstream molecular effectors important for the formation of functional connections between MNs and their respective muscle targets.
Period of natural cell death
MNs are generated in excess and then progressively decrease in number during a natural cell death period (Oppenheim, 1991). This process ensures the generation of the appropriate number of MNs and guarantees the elimination of aberrant cells. This strategy can also result from the requirement of a temporary function; for example, certain MNs may initially be generated to ensure a particular developmental function and are subsequently eliminated. Regardless of the reasons, natural MN death leads to the removal of around 40% of the initially generated MNs (Hamburger, 1975). This loss can be comprehensively divided into two phases (Yaginuma et al., 1996). The early phase is independent of any peripheral signal and likely reflects a negative selection of unsuitable MNs. The subsequent phase has been described more intensively and is dependent on survival signals from the periphery and thus reflects the refinement of mature MN innervations. Temporally, natural MN cell death in mice starts progressively from embryological day (E) 11.5 in most rostral segments and spreads gradually to the caudal levels with a peak occurring at E14 (Yamamoto and Henderson, 1999). The absence of MN cell death postnatally suggests a necessity to reach completion of MN development before birth (Oppenheim, 1986). Numerous molecules have been involved in MN survival signaling. The initial discoveries of the nerve growth factor (NGF), neurotrophins (NTFs) and brain derived neurotrophic factor (BDNF) (Snider, 1994) led to the characterization of additional molecules involved in neuronal survival, including cytokines (ciliary neurotrophic factor CNTF, leukemia inhibitory factor LIF) (Dechiara et al., 1995; Li et al., 1995), the TGFB family (GDNF, neurturin NRTN, persephin PSPN) (Henderson et al., 1994; Poulsen et al., 1994; Oppenheim et al., 1995, 2000), the hepatocyte growth factor (HGF) as well as FGF1, 2 and 5 (Henderson, 1996; Oppenheim, 1996).
Interestingly, in parallel of the general survival mechanisms introduced above, results suggest the existence of pool specific survival signals (Gould and Oppenheim, 2004). Gu and Kania (2010) undertook the profiling of survival receptors expression in lumbar LMC MN pools as well as survival molecules in the corresponding limb muscles. Although their results did not reveal a general mechanism linking MN pool specific survival and combination of trophic factors expressed in the muscles, they emphasized the complexity of MN survival. Indeed, the authors discuss several indications supporting a plausible convergence between the mechanisms controlling axon guidance and MN survival into a unified and coherent process.
Since this article does not primarily focus on MN cell death and selective survival, the following reviews are recommended to provide a detailed description of this complex and indispensable process (Oppenheim, 1991; Hamburger, 1992; Henderson, 1996; Pettmann and Henderson, 1998; Gould and Enomoto, 2009).
Specification of motor neuron subtypes within a pool
The diversity of SpMNs is not limited to the specification of MN pools but also impinges on muscle fiber structural and functional diversity. Despite the detailed columnar classification of SpMNs, little is known about the mechanisms causing a specific MN to recognize and connect to a unique fiber type within its individual muscle target. We will describe recent studies that shed light on the mechanisms controlling alpha and gamma MN differentiation as well as between fast and slow alpha MNs.
Alpha vs. gamma MNs
The divergence between alpha and gamma MNs is poorly characterized (Eccles et al., 1960; Bryan et al., 1972; Westbury, 1982). Evidence from several studies suggests that alpha and gamma MN identities are fated early during embryonic stages. For example, inactivation of the programmed cell death in MNs leads to an increased number of MNs with gamma characteristics (Buss et al., 2006). This result implies that alpha and gamma MN are differentiated prior to axon outgrowth and trophic support requirement. During the first weeks after birth, alpha and gamma MNs can be molecularly identified by the differential expression of the RNA binding protein, fox-1 homolog 3 (RBFOX3 or NeuN), the estrogen-related receptor gamma (ESRRG) (Friese et al., 2009), the GDNF family receptor alpha 1 (GFRA1) (Shneider et al., 2009), the serotonin receptor 1D (HTR1D) (Enjin et al., 2010) as well as the ATPase, Na+/K+ transporting, alpha 1 (ATP1A1) (Edwards et al., 2013). Alpha MNs maintain high levels of RBFOX3 expression after birth whereas gamma MNs up-regulate ESRRG and GFRA1 and simultaneously down-regulate RBFOX3. These markers are segregated only at post-natal stages and are therefore unlikely participate in the early phase of alpha and gamma MN divergence. A recent study identified the first embryological marker of gamma MNs (Ashrafi et al., 2012). Namely, WNT7A is selectively expressed in gamma MNs at late embryological stages. The authors also revealed that its expression is dependent on a muscle spindle-derived signal that is not GDNF, previously characterized as required for their survival (Gould et al., 2008; Shneider et al., 2009). These results open new perspectives to further characterize the molecular mechanisms controlling alpha vs. gamma MN divergence.
Fast vs. slow MNs
Alpha MNs can be classified according to the type of extrafusal fiber they innervate (FF, FFR, SFR). MNs are intrinsically competent to recognize and connect to either fast or slow muscle fibers (Rafuse et al., 1996; Landmesser, 2001). Studies have proposed that the synaptic vesicle glycoprotein 2a (SV2A) (Chakkalakal et al., 2010) as well as the estrogen-related receptor beta (ESRRB) (Enjin et al., 2010) are restricted to slow MNs soon after birth. Conversely, the calcitonin-related polypeptide alpha (CALCA) and the chondrolectin (CHODL) are restricted to fast MNs (Enjin et al., 2010). More recently, Muller et al. (2014) elegantly identified the non-canonical Notch ligand delta-like homolog 1 (DLK1) as a regulator necessary and sufficient to promote fast MN phenotype. These results identify for the first time a molecular regulator of the fast vs. slow MN divergence. This initial breakthrough will indubitably facilitate further identification of the mechanisms of fiber-type-specific alpha MN differentiation.
In summary, SpMN diversity expends beyond the formation of MN pools. In fact, SpMN identity impregnates into muscle fiber types characteristics. Recent findings lead to the identification of key players as well as molecular markers of MN subtype populations. These discoveries open new avenue for further characterization.
Conclusion and perspectives on the generation of spinal motor neurons
SpMNs are unique and irreplaceable neuronal cells connecting the CNS to targets in the periphery. While visceral SpMNs of the thoracic and sacral regions control autonomic functions, somatic SpMNs regulate movements by controlling the contraction of individual muscles. These crucial roles lead to inexorable impairments when affected by diseases. Thus, intensive research has focused on understanding MN biology and diseases.
Over the years, studies have accumulated data and revealed mechanisms driving MN properties and behaviors. Remarkably, the diversity of SpMNs mirrors the variety of targets they innervate but also impinges within individual muscle fiber types. This exceptional diversity is acquired progressively during development and has been reviewed here. The ventralization of the neural tube has been described as a consequence of surrounding molecules expressed in a gradient fashion and inducing in a concentration dependent manner the expression of sets of homeodomain proteins leading the emergence of exclusive progenitor domains. All SpMNs arise from the pMN domain from which SpMN precursors exit the cell cycle and migrate away from the neuroepithelium while acquiring post-mitotic MN features. Concomitantly, patterning molecules along the rostro-caudal axis induce in a concentration-dependant manner the expression of several transcription factors notably members of the HOX family. In turn, these proteins define exclusive rostro-caudal segments (brachial, thoracic, lumbar). Subsequently, while SpMNs strengthen their motor identity, they segregate into anatomical columns termed motor columns. Combinations of LIM homeodomain proteins provide a unique molecular profile for each motor column. In parallel, the LIM code induces the initial steps of a crucial process: MN axonal targeting. SpMN axonal targeting and further differentiation occurs in a step-wise manner. Checkpoints are established along the route to ensure the completion of critical steps. Furthermore, these checkpoints are informative and instruct developing SpMNs of the environment at the growth cone. The SpMN target can be seen as the last checkpoint of the chain. Upon reaching their final destination, SpMNs are required to complete their differentiation process and form functional connection with their target. SpMN identity echoes muscle fiber type properties. Finally, as a mechanism controlling the integrity of SpMN development, naturally programmed cell death induces the elimination of inadequate MNs and ensure the formation of a coherent circuitry.
Although, the overall strategy as well as the intrinsic transcription factors governing the generation of SpMN diversity have been, at least partially characterized and summarized here, our review emphasizes the poor knowledge about the downstream molecular effectors of MN development. In fact, the more differentiated SpMNs become the more fragmentary our understanding is. This is particularly important in regard to prospective MN regeneration therapies for which understanding MN general identity will not be sufficient. Instead, tweaking subtype-specific effector molecules may be a powerful strategy to regenerate functional MNs in fully developed adults. The identification of additional effectors can be achieved in two ways: (i) oriented investigation of downstream targets of known intrinsic regulators such as LIM and/or HOX proteins for example and (ii) unbiased screenings combining, viral retrograde tracing (Stepien et al., 2010; Tripodi et al., 2011), laser capture micro-dissection (Bandyopadhyay et al., 2014) and RNA sequencing (Enjin et al., 2010). Such approaches would indubitably unveil new regulators and effectors of SpMN subtype specification. Furthermore, several studies have already shed light on the role of non-coding micro RNAs (miRNAs) in MN development (Cao et al., 2007; Visvanathan et al., 2007; Otaegi et al., 2011a,b). For example, Chen and Wichterle (2012) demonstrate that the inactivation of the Endoribonuclease Dicer (DICER1), an important player of double strand RNA post-transcription gene silencing, perturbs the formation of PGC and LMC MNs. Similarly, OLIG2 repression initiated at the p2-pMN border relies on mir-17–3 p miRNA-mediated silencing of Olig2 mRNA (Chen et al., 2011a). The implication of non-coding miRNAs is likely more complex and numerous findings will likely arise from this recent and mostly unexplored field of research. The unbiased screenings mentioned above could identify novel regulatory mechanisms of SpMN diversity involving non-coding RNAs.
SpMNs are anatomically well organized. This morphological arrangement correlates with the position of their respective target in the periphery as reviewed by Kania (2014b). Thus, SpMN settling position and axonal targeting must be somehow molecularly connected. An ingenious strategy to further understand the mechanisms driving SpMN specification consists in uncoupling MN differentiation processes such as column formation, cell body positioning, and axonal targeting. One naturally occurring opportunity to study MN differentiation processes independently from one another could lie on the analysis of rhomboideus MN pool. These neurons constitute, in fact, the only known exception to the MN columnar organization described earlier. Although innervating an axial muscle, this MN pool is located in the lateral component of the ventral horn at caudal brachial segments; a position typical of LMC MNs (Straznicky and Tay, 1983; Hollyday and Jacobson, 1990; Tsuchida et al., 1994; Rousso et al., 2008). Therefore, molecular profiling of this particular MN pool may be interesting to identify new effectors and regulators of SpMN organization.
Finally, this review deliberately focused on SpMN development from a motor perspective. However, SpMNs are “only” one constituent of a larger coherent circuitry. Complex movements require the control of individual muscles in a collaborating manner. This coordination relies on a highly organized circuitry between SNs, association neurons, and SpMNs as reviewed by Ladle et al. (2007). In a perspective of regeneration therapies, SpMNs with the correct identity should insert in a pre-existing neuronal circuitry. Such possibility infers that (i) regenerated SpMNs settle at their appropriate location, (ii) that SpMNs' inputs are plastic to form new functional connections and (iii) that regenerated SpMNs project to their appropriate target across a fully developed living organism. These are the challenges the scientific MN community will have to resolve in the coming future.
Author contributions
Nicolas Stifani reviewed the relevant literature, wrote the manuscript and generated the figures.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to acknowledge Dr. Stefano Stifani, Dr. Artur Kania, Dr. Ed Ruthazer, Dr. Tim Kennedy, Dr. Donald Von Meyel and Dr. Christine Vande Velde for comments and corrections. Gene and protein names and symbols follow HUGO Gene Nomenclature Committee at the European Bioinformatics Institute freely available at http://www.genenames.org.
References
- Agalliu D., Takada S., Agalliu I., Mcmahon A. P., Jessell T. M. (2009). Motor neurons with axial muscle projections specified by Wnt4/5 signaling. Neuron 61, 708–720 10.1016/j.neuron.2008.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaynick W. A., Jessell T. M., Pfaff S. L. (2011). SnapShot: spinal cord development. Cell 146, 178–178 e1. 10.1016/j.cell.2011.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldskogius H., Berens C., Kanaykina N., Liakhovitskaia A., Medvinsky A., Sandelin M., et al. (2009). Regulation of boundary cap neural crest stem cell differentiation after transplantation. Stem Cells 27, 1592–1603 10.1002/stem.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D. W., Greer J. J. (1997a). Development of phrenic motoneuron morphology in the fetal rat. J. Comp. Neurol. 382, 469–479 [DOI] [PubMed] [Google Scholar]
- Allan D. W., Greer J. J. (1997b). Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J. Comp. Neurol. 382, 459–468 [DOI] [PubMed] [Google Scholar]
- Alvarez-Medina R., Cayuso J., Okubo T., Takada S., Marti E. (2008). Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development 135, 237–247 10.1242/dev.012054 [DOI] [PubMed] [Google Scholar]
- Amiel J., Dubreuil V., Ramanantsoa N., Fortin G., Gallego J., Brunet J. F., et al. (2009). PHOX2B in respiratory control: lessons from congenital central hypoventilation syndrome and its mouse models. Respir. Physiol. Neurobiol. 168, 125–132 10.1016/j.resp.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Arber S. (2008). FoxP1: conducting the Hox symphony in spinal motor neurons. Nat. Neurosci. 11, 1122–1124 10.1038/nn1008-1122 [DOI] [PubMed] [Google Scholar]
- Arber S., Han B., Mendelsohn M., Smith M., Jessell T. M., Sockanathan S. (1999). Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23, 659–674 10.1016/S0896-6273(01)80026-X [DOI] [PubMed] [Google Scholar]
- Ashrafi S., Lalancette-Hebert M., Friese A., Sigrist M., Arber S., Shneider N. A., et al. (2012). Wnt7A identifies embryonic gamma-motor neurons and reveals early postnatal dependence of gamma-motor neurons on a muscle spindle-derived signal. J. Neurosci. 32, 8725–8731 10.1523/JNEUROSCI.1160-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouard E., Schakman O., Rene F., Huettl R. E., Huber A. B., Loeffler J. P., et al. (2012). The Onecut transcription factor HNF-6 regulates in motor neurons the formation of the neuromuscular junctions. PLoS ONE 7:e50509 10.1371/journal.pone.0050509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaskas N., Ribeiro A., Panovska J., Dessaud E., Sasai N., Page K. M., et al. (2012). Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284 10.1016/j.cell.2011.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U., Fenton W. A., Horwich A. L., Nagy M. (2014). Production of RNA for transcriptomic analysis from mouse spinal cord motor neuron cell bodies by laser capture microdissection. J. Vis. Exp. 13:e51168 10.3791/51168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S., Itasaki N., Krumlauf R. (2002). Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 129, 5103–5115 [DOI] [PubMed] [Google Scholar]
- Bessou P., Emonet-Denand F., Laporte Y. (1965). Motor fibres innervating extrafusal and intrafusal muscle fibres in the cat. J. Physiol. 180, 649–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D., Chivatakarn O., Bai G., Abdesselem H., Lettieri K., Marquardt T., et al. (2012). Ret is a multifunctional coreceptor that integrates diffusible- and contact-axon guidance signals. Cell 148, 568–582 10.1016/j.cell.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D., Pfaff S. L. (2010). Motor axon pathfinding. Cold Spring Harb. Perspect. Biol. 2:a001735 10.1101/cshperspect.a001735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Ambrosio A., Kaprielian Z. (2011). Crossing the border: molecular control of motor axon exit. Int. J. Mol. Sci. 12, 8539–8561 10.3390/ijms12128539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta A. M., Callister R. J., Peterson E. H. (1987). Quantitative analysis of cervical musculature in rats: histochemical composition and motor pool organization. I. Muscles of the spinal accessory complex. J. Comp. Neurol. 255, 351–368 10.1002/cne.902550304 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M., Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 10.1016/S0092-8674(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Bryan R. N., Trevino D. L., Willis W. D. (1972). Evidence for a common location of alpha and gamma motoneurons. Brain Res. 38, 193–196 10.1016/0006-8993(72)90602-6 [DOI] [PubMed] [Google Scholar]
- Bueker E. D. (1944). Differentiation of the lateral motor column in the avian spinal cord. Science 100:169 10.1126/science.100.2591.169 [DOI] [PubMed] [Google Scholar]
- Burke R. E., Levine D. N., Tsairis P., Zajac F. E. 3rd. (1973). Physiological types and histochemical profiles in motor units of the cat gastrocnemius. J. Physiol. 234, 723–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss R. R., Gould T. W., Ma J., Vinsant S., Prevette D., Winseck A., et al. (2006). Neuromuscular development in the absence of programmed cell death: phenotypic alteration of motoneurons and muscle. J. Neurosci. 26, 13413–13427 10.1523/JNEUROSCI.3528-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron W. E., He F., Kalipatnapu P., Jodkowski J. S., Guthrie R. D. (1991). Morphometric analysis of phrenic motoneurons in the cat during postnatal development. J. Comp. Neurol. 314, 763–776 10.1002/cne.903140409 [DOI] [PubMed] [Google Scholar]
- Cao X., Pfaff S. L., Gage F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536 10.1101/gad.1519207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani V., Kania A. (2012). Breathless without Hox. Nat. Neurosci. 15, 1607–1609 10.1038/nn.3272 [DOI] [PubMed] [Google Scholar]
- Chakkalakal J. V., Nishimune H., Ruas J. L., Spiegelman B. M., Sanes J. R. (2010). Retrograde influence of muscle fibers on their innervation revealed by a novel marker for slow motoneurons. Development 137, 3489–3499 10.1242/dev.053348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar A. (2004). Turning heads: development of vertebrate branchiomotor neurons. Dev. Dyn. 229, 143–161 10.1002/dvdy.10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. A., Huang Y. P., Mazzoni E. O., Tan G. C., Zavadil J., Wichterle H. (2011a). Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 69, 721–735 10.1016/j.neuron.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. A., Wichterle H. (2012). Apoptosis of limb innervating motor neurons and erosion of motor pool identity upon lineage specific dicer inactivation. Front. Neurosci. 6:69 10.3389/fnins.2012.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sasai N., Ma G., Yue T., Jia J., Briscoe J., et al. (2011b). Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 9:e1001083 10.1371/journal.pbio.1001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. H., Cargnin F., Kim Y., Lee B., Kwon R. J., Nam H., et al. (2014). Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 10:e1004280 10.1371/journal.pgen.1004280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Hoover J. E. (1996). The organization of acromiodeltoid and spinodeltoid motor nuclei in rat spinal cord. Brain Res. 738, 146–149 10.1016/0006-8993(96)00958-4 [DOI] [PubMed] [Google Scholar]
- Dasen J. S. (2009). Transcriptional networks in the early development of sensory-motor circuits. Curr. Top. Dev. Biol. 87, 119–148 10.1016/S0070-2153(09)01204-6 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., De Camilli A., Wang B., Tucker P. W., Jessell T. M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316 10.1016/j.cell.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., Jessell T. M. (2009). Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 88, 169–200 10.1016/S0070-2153(09)88006-X [DOI] [PubMed] [Google Scholar]
- Dasen J. S., Liu J. P., Jessell T. M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933 10.1038/nature02051 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., Tice B. C., Brenner-Morton S., Jessell T. M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491 10.1016/j.cell.2005.09.009 [DOI] [PubMed] [Google Scholar]
- Dechiara T. M., Vejsada R., Poueymirou W. T., Acheson A., Suri C., Conover J. C., et al. (1995). Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell 83, 313–322 10.1016/0092-8674(95)90172-8 [DOI] [PubMed] [Google Scholar]
- De Marco Garcia N. V., Jessell T. M. (2008). Early motor neuron pool identity and muscle nerve trajectory defined by postmitotic restrictions in Nkx6.1 activity. Neuron 57, 217–231 10.1016/j.neuron.2007.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E., Mcmahon A. P., Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489–2503 10.1242/dev.009324 [DOI] [PubMed] [Google Scholar]
- Dillon A. K., Fujita S. C., Matise M. P., Jarjour A. A., Kennedy T. E., Kollmus H., et al. (2005). Molecular control of spinal accessory motor neuron/axon development in the mouse spinal cord. J. Neurosci. 25, 10119–10130 10.1523/JNEUROSCI.3455-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V., Barhanin J., Goridis C., Brunet J. F. (2009). Breathing with phox2b. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2477–2483 10.1098/rstb.2009.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudanova I., Kao T. J., Herrmann J. E., Zheng B., Kania A., Klein R. (2012). Genetic evidence for a contribution of EphA:ephrinA reverse signaling to motor axon guidance. J. Neurosci. 32, 5209–5215 10.1523/JNEUROSCI.5707-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston A. J., Timmermans J. P., Hage W. J., Hendriks H. F., De Vries N. J., Heideveld M., et al. (1989). Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature 340, 140–144 10.1038/340140a0 [DOI] [PubMed] [Google Scholar]
- Eccles J. C., Eccles R. M., Iggo A., Lundberg A. (1960). Electrophysiological studies on gamma motoneurones. Acta Physiol. Scand. 50, 32–40 10.1111/j.1748-1716.1960.tb02070.x [DOI] [PubMed] [Google Scholar]
- Eccles J. C., Eccles R. M., Lundberg A. (1957). The convergence of monosynaptic excitatory afferents on to many different species of alpha motoneurones. J. Physiol. 137, 22–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards I. J., Bruce G., Lawrenson C., Howe L., Clapcote S. J., Deuchars S. A., et al. (2013). Na+/K+ ATPase alpha1 and alpha3 isoforms are differentially expressed in alpha- and gamma-motoneurons. J. Neurosci. 33, 9913–9919 10.1523/JNEUROSCI.5584-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjin A., Rabe N., Nakanishi S. T., Vallstedt A., Gezelius H., Memic F., et al. (2010). Identification of novel spinal cholinergic genetic subtypes disclose Chodl and Pitx2 as markers for fast motor neurons and partition cells. J. Comp. Neurol. 518, 2284–2304 10.1002/cne.22332 [DOI] [PubMed] [Google Scholar]
- Ensini M., Tsuchida T. N., Belting H. G., Jessell T. M. (1998). The control of rostrocaudal pattern in the developing spinal cord: specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development 125, 969–982 [DOI] [PubMed] [Google Scholar]
- Ericson J., Briscoe J., Rashbass P., Van Heyningen V., Jessell T. M. (1997). Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harb. Symp. Quant. Biol. 62, 451–466 10.1101/SQB.1997.062.01.053 [DOI] [PubMed] [Google Scholar]
- Ericson J., Morton S., Kawakami A., Roelink H., Jessell T. M. (1996). Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 87, 661–673 10.1016/S0092-8674(00)81386-0 [DOI] [PubMed] [Google Scholar]
- Ericson J., Thor S., Edlund T., Jessell T. M., Yamada T. (1992). Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science 256, 1555–1560 10.1126/science.1350865 [DOI] [PubMed] [Google Scholar]
- Fetcho J. R. (1987). A review of the organization and evolution of motoneurons innervating the axial musculature of vertebrates. Brain Res. Rev. 12, 243–280 10.1016/0165-0173(87)90001-4 [DOI] [PubMed] [Google Scholar]
- Francius C., Clotman F. (2010). Dynamic expression of the onecut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience 165, 116–129 10.1016/j.neuroscience.2009.09.076 [DOI] [PubMed] [Google Scholar]
- Francius C., Clotman F. (2014). Generating spinal motor neuron diversity: a long quest for neuronal identity. Cell. Mol. Life Sci. 71, 813–829 10.1007/s00018-013-1398-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friese A., Kaltschmidt J. A., Ladle D. R., Sigrist M., Jessell T. M., Arber S. (2009). Gamma and alpha motor neurons distinguished by expression of transcription factor Err3. Proc. Natl. Acad. Sci. U.S.A. 106, 13588–13593 10.1073/pnas.0906809106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara K., Imai F., Ladle D. R., Katayama K., Leslie J. R., Arber S., et al. (2013). Specificity of monosynaptic sensory-motor connections imposed by repellent Sema3E-PlexinD1 signaling. Cell Rep. 5, 748–758 10.1016/j.celrep.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallarda B. W., Bonanomi D., Muller D., Brown A., Alaynick W. A., Andrews S. E., et al. (2008). Segregation of axial motor and sensory pathways via heterotypic trans-axonal signaling. Science 320, 233–236 10.1126/science.1153758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden M. G., Dasen J. S. (2012). Polycomb repressive complex 1 activities determine the columnar organization of motor neurons. Genes Dev. 26, 2236–2250 10.1101/gad.199133.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould T. W., Enomoto H. (2009). Neurotrophic modulation of motor neuron development. Neuroscientist 15, 105–116 10.1177/1073858408324787 [DOI] [PubMed] [Google Scholar]
- Gould T. W., Oppenheim R. W. (2004). The function of neurotrophic factor receptors expressed by the developing adductor motor pool in vivo. J. Neurosci. 24, 4668–4682 10.1523/JNEUROSCI.0580-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould T. W., Yonemura S., Oppenheim R. W., Ohmori S., Enomoto H. (2008). The neurotrophic effects of glial cell line-derived neurotrophic factor on spinal motoneurons are restricted to fusimotor subtypes. J. Neurosci. 28, 2131–2146 10.1523/JNEUROSCI.5185-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. X., Kania A. (2010). Examining the combinatorial model of motor neuron survival by expression profiling of trophic factors and their receptors in the embryonic Gallus gallus. Dev. Dyn. 239, 965–979 10.1002/dvdy.22215 [DOI] [PubMed] [Google Scholar]
- Guizard N., Coupe P., Stifani N., Stifani S., Collins D. L. (2010). Robust 3D reconstruction and mean-shift clustering of motoneurons from serial histological images. Med. Imag. Augment. Real. 6326, 191–199 10.1007/978-3-642-15699-1_20 [DOI] [Google Scholar]
- Gutman C. R., Ajmera M. K., Hollyday M. (1993). Organization of motor pools supplying axial muscles in the chicken. Brain Res. 609, 129–136 10.1016/0006-8993(93)90865-K [DOI] [PubMed] [Google Scholar]
- Haase G., Dessaud E., Garces A., De Bovis B., Birling M., Filippi P., et al. (2002). GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron 35, 893–905 10.1016/S0896-6273(02)00864-4 [DOI] [PubMed] [Google Scholar]
- Hamburger V. (1975). Cell death in the development of the lateral motor column of the chick embryo. J. Comp. Neurol. 160, 535–546 10.1002/cne.901600408 [DOI] [PubMed] [Google Scholar]
- Hamburger V. (1992). History of the discovery of neuronal death in embryos. J. Neurobiol. 23, 1116–1123 10.1002/neu.480230904 [DOI] [PubMed] [Google Scholar]
- Hammerle B., Tejedor F. J. (2007). A novel function of DELTA-NOTCH signalling mediates the transition from proliferation to neurogenesis in neural progenitor cells. PLoS ONE 2:e1169 10.1371/journal.pone.0001169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbacher F., Dessaud E., Arber S., Delapeyriere O., Henderson C. E., Klein R., et al. (2003). Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron 39, 767–777 10.1016/S0896-6273(03)00493-8 [DOI] [PubMed] [Google Scholar]
- Helmbacher F., Schneider-Maunoury S., Topilko P., Tiret L., Charnay P. (2000). Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development 127, 3313–3324 [DOI] [PubMed] [Google Scholar]
- Henderson C. E. (1996). Role of neurotrophic factors in neuronal development. Curr. Opin. Neurobiol. 6, 64–70 10.1016/S0959-4388(96)80010-9 [DOI] [PubMed] [Google Scholar]
- Henderson C. E., Phillips H. S., Pollock R. A., Davies A. M., Lemeulle C., Armanini M., et al. (1994). GDNF: a potent survival factor for motoneurons present in peripheral nerve and muscle. Science 266, 1062–1064 10.1126/science.7973664 [DOI] [PubMed] [Google Scholar]
- Hirsch M. R., Glover J. C., Dufour H. D., Brunet J. F., Goridis C. (2007). Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev. Biol. 303, 687–702 10.1016/j.ydbio.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Hollyday M., Hamburger V. (1977). An autoradiographic study of the formation of the lateral motor column in the chick embryo. Brain Res. 132, 197–208 10.1016/0006-8993(77)90416-4 [DOI] [PubMed] [Google Scholar]
- Hollyday M., Hamburger V., Farris J. M. (1977). Localization of motor neuron pools supplying identified muscles in normal and supernumerary legs of chick embryo. Proc. Natl. Acad. Sci. U.S.A. 74, 3582–3586 10.1073/pnas.74.8.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyday M., Jacobson R. D. (1990). Location of motor pools innervating chick wing. J. Comp. Neurol. 302, 575–588 10.1002/cne.903020313 [DOI] [PubMed] [Google Scholar]
- Huber A. B., Kania A., Tran T. S., Gu C., De Marco Garcia N., Lieberam I., et al. (2005). Distinct roles for secreted semaphorin signaling in spinal motor axon guidance. Neuron 48, 949–964 10.1016/j.neuron.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Huettl R. E., Soellner H., Bianchi E., Novitch B. G., Huber A. B. (2011). Npn-1 contributes to axon-axon interactions that differentially control sensory and motor innervation of the limb. PLoS Biol. 9:e1001020 10.1371/journal.pbio.1001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A., Sharma M., Durnin M., Vanden Heuvel G. B., Trainor P. A. (2008). Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development 135, 729–741 10.1242/dev.013276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanhoe C. B., Reistetter T. A. (2004). Spasticity: the misunderstood part of the upper motor neuron syndrome. Am. J. Phys. Med. Rehabil. 83, S3–S9 10.1097/01.PHM.0000141125.28611.3E [DOI] [PubMed] [Google Scholar]
- Jacobson M., Rao M. S. (2005). Developmental Neurobiology. New York, NY: Kluwer Academic/Plenum [Google Scholar]
- Jacobson S., Marcus E. M. (2007). Neuroanatomy for the Neuroscientist. New York, NY; London: Springer [Google Scholar]
- Jessell T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29 10.1038/35049541 [DOI] [PubMed] [Google Scholar]
- Jung H., Lacombe J., Mazzoni E. O., Liem K. F., Jr., Grinstein J., Mahony S., et al. (2010). Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67, 781–796 10.1016/j.neuron.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Mazzoni E. O., Soshnikova N., Hanley O., Venkatesh B., Duboule D., et al. (2014). Evolving hox activity profiles govern diversity in locomotor systems. Dev. Cell 29, 171–187 10.1016/j.devcel.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A. (2014a). Concocting cholinergy. PLoS Genet. 10:e1004313 10.1371/journal.pgen.1004313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A. (2014b). Spinal motor neuron migration and the significance of topographic organization in the nervous system. Adv. Exp. Med. Biol. 800, 133–148 10.1007/978-94-007-7687-6_8 [DOI] [PubMed] [Google Scholar]
- Kania A., Jessell T. M. (2003). Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron 38, 581–596 10.1016/S0896-6273(03)00292-7 [DOI] [PubMed] [Google Scholar]
- Kania A., Johnson R. L., Jessell T. M. (2000). Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell 102, 161–173 10.1016/S0092-8674(00)00022-2 [DOI] [PubMed] [Google Scholar]
- Kanning K. C., Kaplan A., Henderson C. E. (2010). Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440 10.1146/annurev.neuro.051508.135722 [DOI] [PubMed] [Google Scholar]
- Kao T. J., Kania A. (2011). Ephrin-mediated cis-attenuation of Eph receptor signaling is essential for spinal motor axon guidance. Neuron 71, 76–91 10.1016/j.neuron.2011.05.031 [DOI] [PubMed] [Google Scholar]
- Kao T. J., Law C., Kania A. (2012). Eph and ephrin signaling: lessons learned from spinal motor neurons. Semin. Cell Dev. Biol. 23, 83–91 10.1016/j.semcdb.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Kessel M., Gruss P. (1991). Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 67, 89–104 10.1016/0092-8674(91)90574-I [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Homma S., Okada T., Masuda T., Sato N., Nishiyama K., et al. (2013). Elucidation of target muscle and detailed development of dorsal motor neurons in chick embryo spinal cord. J. Comp. Neurol. 521, 2987–3002 10.1002/cne.23326 [DOI] [PubMed] [Google Scholar]
- Kramer E. R., Knott L., Su F., Dessaud E., Krull C. E., Helmbacher F., et al. (2006). Cooperation between GDNF/Ret and ephrinA/EphA4 signals for motor-axon pathway selection in the limb. Neuron 50, 35–47 10.1016/j.neuron.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Lacombe J., Hanley O., Jung H., Philippidou P., Surmeli G., Grinstein J., et al. (2013). Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons. PLoS Genet. 9:e1003184 10.1371/journal.pgen.1003184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladle D. R., Frank E. (2002). The role of the ETS gene PEA3 in the development of motor and sensory neurons. Physiol. Behav. 77, 571–576 10.1016/S0031-9384(02)00907-1 [DOI] [PubMed] [Google Scholar]
- Ladle D. R., Pecho-Vrieseling E., Arber S. (2007). Assembly of motor circuits in the spinal cord: driven to function by genetic and experience-dependent mechanisms. Neuron 56, 270–283 10.1016/j.neuron.2007.09.026 [DOI] [PubMed] [Google Scholar]
- Lamballe F., Genestine M., Caruso N., Arce V., Richelme S., Helmbacher F., et al. (2011). Pool-specific regulation of motor neuron survival by neurotrophic support. J. Neurosci. 31, 11144–11158 10.1523/JNEUROSCI.2198-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance-Jones C., Landmesser L. (1980). Motoneurone projection patterns in the chick hind limb following early partial reversals of the spinal cord. J. Physiol. 302, 581–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance-Jones C., Omelchenko N., Bailis A., Lynch S., Sharma K. (2001). Hoxd10 induction and regionalization in the developing lumbosacral spinal cord. Development 128, 2255–2268 [DOI] [PubMed] [Google Scholar]
- Landmesser L. (1978). The distribution of motoneurones supplying chick hind limb muscles. J. Physiol. 284, 371–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L. T. (2001). The acquisition of motoneuron subtype identity and motor circuit formation. Int. J. Dev. Neurosci. 19, 175–182 10.1016/S0736-5748(00)00090-3 [DOI] [PubMed] [Google Scholar]
- Laumonnerie C., Da Silva R. V., Kania A., Wilson S. I. (2014). Netrin 1 and Dcc signalling are required for confinement of central axons within the central nervous system. Development 141, 594–603 10.1242/dev.099606 [DOI] [PubMed] [Google Scholar]
- Lee R. H., Heckman C. J. (1998). Bistability in spinal motoneurons in vivo: systematic variations in persistent inward currents. J. Neurophysiol. 80, 583–593 [DOI] [PubMed] [Google Scholar]
- Lee S. K., Jurata L. W., Funahashi J., Ruiz E. C., Pfaff S. L. (2004). Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development 131, 3295–3306 10.1242/dev.01179 [DOI] [PubMed] [Google Scholar]
- Lee S. K., Lee B., Ruiz E. C., Pfaff S. L. (2005). Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 19, 282–294 10.1101/gad.1257105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Pfaff S. L. (2003). Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731–745 10.1016/S0896-6273(03)00296-4 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee B., Joshi K., Pfaff S. L., Lee J. W., Lee S. K. (2008). A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 14, 877–889 10.1016/j.devcel.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee B., Lee J. W., Lee S. K. (2009). Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron 62, 641–654 10.1016/j.neuron.2009.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q., Jeong Y., Misra K., Li S., Zelman A. K., Epstein D. J., et al. (2006). Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev. Cell 11, 325–337 10.1016/j.devcel.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Li M., Sendtner M., Smith A. (1995). Essential function of LIF receptor in motor neurons. Nature 378, 724–727 10.1038/378724a0 [DOI] [PubMed] [Google Scholar]
- Lieberam I., Agalliu D., Nagasawa T., Ericson J., Jessell T. M. (2005). A Cxcl12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron 47, 667–679 10.1016/j.neuron.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Lin J. H., Saito T., Anderson D. J., Lance-Jones C., Jessell T. M., Arber S. (1998). Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell 95, 393–407 10.1016/S0092-8674(00)81770-5 [DOI] [PubMed] [Google Scholar]
- Liu J. P. (2006). The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development 133, 2865–2874 10.1242/dev.02478 [DOI] [PubMed] [Google Scholar]
- Liu J. P., Laufer E., Jessell T. M. (2001). Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 32, 997–1012 10.1016/S0896-6273(01)00544-X [DOI] [PubMed] [Google Scholar]
- Livet J., Sigrist M., Stroebel S., De Paola V., Price S. R., Henderson C. E., et al. (2002). ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35, 877–892 10.1016/S0896-6273(02)00863-2 [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D., et al. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Luria V., Krawchuk D., Jessell T. M., Laufer E., Kania A. (2008). Specification of motor axon trajectory by ephrin-B:EphB signaling: symmetrical control of axonal patterning in the developing limb. Neuron 60, 1039–1053 10.1016/j.neuron.2008.11.011 [DOI] [PubMed] [Google Scholar]
- Machado C. B., Kanning K. C., Kreis P., Stevenson D., Crossley M., Nowak M., et al. (2014). Reconstruction of phrenic neuron identity in embryonic stem cell-derived motor neurons. Development 141, 784–794 10.1242/dev.097188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A. (1997). Development and regeneration of muscle spindles in mammals and birds. Int. J. Dev. Biol. 41, 1–17 [PubMed] [Google Scholar]
- Marti E., Bumcrot D. A., Takada R., Mcmahon A. P. (1995a). Requirement of 19K form of Sonic hedgehog for induction of distinct ventral cell types in CNS explants. Nature 375, 322–325 10.1038/375322a0 [DOI] [PubMed] [Google Scholar]
- Marti E., Takada R., Bumcrot D. A., Sasaki H., Mcmahon A. P. (1995b). Distribution of Sonic hedgehog peptides in the developing chick and mouse embryo. Development 121, 2537–2547 [DOI] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Closser M., Morrison C. A., Nedelec S., Williams D. J., et al. (2013a). Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16, 1219–1227 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E. O., Mahony S., Peljto M., Patel T., Thornton S. R., Mccuine S., et al. (2013b). Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 16, 1191–1198 10.1038/nn.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler M. F., Mabie P. C., Zhang D., Kessler J. A. (1997). Bone morphogenetic proteins in the nervous system. Trends Neurosci. 20, 309–317 10.1016/S0166-2236(96)01046-6 [DOI] [PubMed] [Google Scholar]
- Mizuguchi R., Sugimori M., Takebayashi H., Kosako H., Nagao M., Yoshida S., et al. (2001). Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757–771 10.1016/S0896-6273(01)00413-5 [DOI] [PubMed] [Google Scholar]
- Morikawa Y., Komori T., Hisaoka T., Senba E. (2009). Detailed expression pattern of Foxp1 and its possible roles in neurons of the spinal cord during embryogenesis. Dev. Neurosci. 31, 511–522 10.1159/000243715 [DOI] [PubMed] [Google Scholar]
- Muhr J., Graziano E., Wilson S., Jessell T. M., Edlund T. (1999). Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron 23, 689–702 10.1016/S0896-6273(01)80028-3 [DOI] [PubMed] [Google Scholar]
- Muller D., Cherukuri P., Henningfeld K., Poh C. H., Wittler L., Grote P., et al. (2014). Dlk1 promotes a fast motor neuron biophysical signature required for peak force execution. Science 343, 1264–1266 10.1126/science.1246448 [DOI] [PubMed] [Google Scholar]
- Niederreither K., Mccaffery P., Drager U. C., Chambon P., Dolle P. (1997). Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 62, 67–78 10.1016/S0925-4773(96)00653-3 [DOI] [PubMed] [Google Scholar]
- Niewiadomski P., Kong J. H., Ahrends R., Ma Y., Humke E. W., Khan S., et al. (2014). Gli protein activity is controlled by multisite phosphorylation in vertebrate hedgehog signaling. Cell Rep. 6, 168–181 10.1016/j.celrep.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nornes H. O., Carry M. (1978). Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 159, 1–6 10.1016/0006-8993(78)90105-1 [DOI] [PubMed] [Google Scholar]
- Novitch B. G., Chen A. I., Jessell T. M. (2001). Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789 10.1016/S0896-6273(01)00407-X [DOI] [PubMed] [Google Scholar]
- Novitch B. G., Wichterle H., Jessell T. M., Sockanathan S. (2003). A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 40, 81–95 10.1016/j.neuron.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Oppenheim R. W. (1986). The absence of significant postnatal motoneuron death in the brachial and lumbar spinal cord of the rat. J. Comp. Neurol. 246, 281–286 10.1002/cne.902460211 [DOI] [PubMed] [Google Scholar]
- Oppenheim R. W. (1991). Cell death during development of the nervous system. Annu. Rev. Neurosci. 14, 453–501 10.1146/annurev.ne.14.030191.002321 [DOI] [PubMed] [Google Scholar]
- Oppenheim R. W. (1996). Neurotrophic survival molecules for motoneurons: an embarrassment of riches. Neuron 17, 195–197 10.1016/S0896-6273(00)80151-8 [DOI] [PubMed] [Google Scholar]
- Oppenheim R. W., Houenou L. J., Johnson J. E., Lin L. F., Li L., Lo A. C., et al. (1995). Developing motor neurons rescued from programmed and axotomy-induced cell death by GDNF. Nature 373, 344–346 10.1038/373344a0 [DOI] [PubMed] [Google Scholar]
- Oppenheim R. W., Houenou L. J., Parsadanian A. S., Prevette D., Snider W. D., Shen L. (2000). Glial cell line-derived neurotrophic factor and developing mammalian motoneurons: regulation of programmed cell death among motoneuron subtypes. J. Neurosci. 20, 5001–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G., Pollock A., Hong J., Sun T. (2011a). MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 31, 809–818 10.1523/JNEUROSCI.4330-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G., Pollock A., Sun T. (2011b). An optimized sponge for microrNA miR-9 affects spinal motor neuron development in vivo. Front. Neurosci. 5:146 10.3389/fnins.2011.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O., Herbrand H., Arnold H. H. (1998). Nkx2-9 is a novel homeobox transcription factor which demarcates ventral domains in the developing mouse CNS. Mech. Dev. 73, 85–93 10.1016/S0925-4773(98)00035-5 [DOI] [PubMed] [Google Scholar]
- Pabst O., Rummelies J., Winter B., Arnold H. H. (2003). Targeted disruption of the homeobox gene Nkx2.9 reveals a role in development of the spinal accessory nerve. Development 130, 1193–1202 10.1242/dev.00346 [DOI] [PubMed] [Google Scholar]
- Palmesino E., Rousso D. L., Kao T. J., Klar A., Laufer E., Uemura O., et al. (2010). Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating reelin signalling. PLoS Biol. 8:446 10.1371/journal.pbio.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C., Sabharwal P., Zhang M., Meyers C. L., Sockanathan S. (2013). GDE2 promotes neurogenesis by glycosylphosphatidylinositol-anchor cleavage of RECK. Science 339, 324–328 10.1126/science.1231921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Hirsch M., Goridis C., Brunet J. F. (2000). Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development 127, 1349–1358 [DOI] [PubMed] [Google Scholar]
- Periz G., Yan Y., Bitzer Z. T., Sockanathan S. (2010). GDP-bound Galphai2 regulates spinal motor neuron differentiation through interaction with GDE2. Dev. Biol. 341, 213–221 10.1016/j.ydbio.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettmann B., Henderson C. E. (1998). Neuronal cell death. Neuron 20, 633–647 10.1016/S0896-6273(00)81004-1 [DOI] [PubMed] [Google Scholar]
- Pfaff S. L. (2008). Developmental neuroscience: hox and fox. Nature 455, 295–297 10.1038/455295a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K. A., Hollyday M. (1990). Axon guidance in muscleless chick wings: the role of muscle cells in motoneuronal pathway selection and muscle nerve formation. J. Neurosci. 10, 2699–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Dasen J. S. (2013). Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80, 12–34 10.1016/j.neuron.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Walsh C. M., Aubin J., Jeannotte L., Dasen J. S. (2012). Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 15, 1636–1644 10.1038/nn.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierani A., Brenner-Morton S., Chiang C., Jessell T. M. (1999). A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell 97, 903–915 10.1016/S0092-8674(00)80802-8 [DOI] [PubMed] [Google Scholar]
- Poulsen K. T., Armanini M. P., Klein R. D., Hynes M. A., Phillips H. S., Rosenthal A. (1994). TGF beta 2 and TGF beta 3 are potent survival factors for midbrain dopaminergic neurons. Neuron 13, 1245–1252 10.1016/0896-6273(94)90062-0 [DOI] [PubMed] [Google Scholar]
- Prakash Y. S., Mantilla C. B., Zhan W. Z., Smithson K. G., Sieck G. C. (2000). Phrenic motoneuron morphology during rapid diaphragm muscle growth. J. Appl. Physiol. 89, 563–572 [DOI] [PubMed] [Google Scholar]
- Prasad A., Hollyday M. (1991). Development and migration of avian sympathetic preganglionic neurons. J. Comp. Neurol. 307, 237–258 10.1002/cne.903070207 [DOI] [PubMed] [Google Scholar]
- Purves D., Williams S. M. (2004). Neuroscience. Sunderland, MA: Sinauer Associates [Google Scholar]
- Quirk J., Van Den Heuvel M., Henrique D., Marigo V., Jones T. A., Tabin C., et al. (1997). The smoothened gene and hedgehog signal transduction in Drosophila and vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 62, 217–226 10.1101/SQB.1997.062.01.027 [DOI] [PubMed] [Google Scholar]
- Rafuse V. F., Milner L. D., Landmesser L. T. (1996). Selective innervation of fast and slow muscle regions during early chick neuromuscular development. J. Neurosci. 16, 6864–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M., Sockanathan S. (2005). Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science 309, 2212–2215 10.1126/science.1117156 [DOI] [PubMed] [Google Scholar]
- Rexed B. (1954). A cytoarchitectonic atlas of the spinal cord in the cat. J. Comp. Neurol. 100, 297–379 10.1002/cne.901000205 [DOI] [PubMed] [Google Scholar]
- Richardson W. D., Smith H. K., Sun T., Pringle N. P., Hall A., Woodruff R. (2000). Oligodendrocyte lineage and the motor neuron connection. Glia 29, 136–142 10.1002/(SICI)1098-1136(20000115)29:2%3C136::AID-GLIA6%3E3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- Roelink H., Augsburger A., Heemskerk J., Korzh V., Norlin S., Ruiz i Altaba A., et al. (1994). Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 76, 761–775 10.1016/0092-8674(94)90514-2 [DOI] [PubMed] [Google Scholar]
- Romanes G. J. (1941). The development and significance of the cell columns in the ventral horn of the cervical and upper thoracic spinal cord of the rabbit. J. Anat. 76, 112–130 [PMC free article] [PubMed] [Google Scholar]
- Rousso D. L., Gaber Z. B., Wellik D., Morrisey E. E., Novitch B. G. (2008). Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron 59, 226–240 10.1016/j.neuron.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso D. L., Pearson C. A., Gaber Z. B., Miquelajauregui A., Li S., Portera-Cailliau C., et al. (2012). Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron 74, 314–330 10.1016/j.neuron.2012.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Francius C., Rousso D. L., Seuntjens E., Debruyn J., Luxenhofer G., et al. (2012). Onecut transcription factors act upstream of Isl1 to regulate spinal motoneuron diversification. Development 139, 3109–3119 10.1242/dev.078501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. M., Cushman J., Jordan B., Samuels A., Frazer H., Baier C. (1998). Topographic position of forelimb motoneuron pools is conserved in vertebrate evolution. Brain Behav. Evol. 51, 90–99 10.1159/000006531 [DOI] [PubMed] [Google Scholar]
- Sabharwal P., Lee C., Park S., Rao M., Sockanathan S. (2011). GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron 71, 1058–1070 10.1016/j.neuron.2011.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Kidd G. J., Trapp B. D., Dawson T. M., Bredt D. S., Wilson D. A., et al. (1994). Rat spinal cord neurons contain nitric oxide synthase. Neuroscience 59, 447–456 10.1016/0306-4522(94)90608-4 [DOI] [PubMed] [Google Scholar]
- Scardigli R., Schuurmans C., Gradwohl G., Guillemot F. (2001). Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31, 203–217 10.1016/S0896-6273(01)00358-0 [DOI] [PubMed] [Google Scholar]
- Schubert W., Kaprielian Z. (2001). Identification and characterization of a cell surface marker for embryonic rat spinal accessory motor neurons. J. Comp. Neurol. 439, 368–383 10.1002/cne.1356 [DOI] [PubMed] [Google Scholar]
- Shah V., Drill E., Lance-Jones C. (2004). Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev. Dyn. 231, 43–56 10.1002/dvdy.20103 [DOI] [PubMed] [Google Scholar]
- Sharma K., Leonard A. E., Lettieri K., Pfaff S. L. (2000). Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature 406, 515–519 10.1038/35020078 [DOI] [PubMed] [Google Scholar]
- Sharma K., Sheng H. Z., Lettieri K., Li H., Karavanov A., Potter S., et al. (1998). LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95, 817–828 10.1016/S0092-8674(00)81704-3 [DOI] [PubMed] [Google Scholar]
- Shirasaki R., Lewcock J. W., Lettieri K., Pfaff S. L. (2006). FGF as a target-derived chemoattractant for developing motor axons genetically programmed by the LIM code. Neuron 50, 841–853 10.1016/j.neuron.2006.04.030 [DOI] [PubMed] [Google Scholar]
- Shirasaki R., Pfaff S. L. (2002). Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 25, 251–281 10.1146/annurev.neuro.25.112701.142916 [DOI] [PubMed] [Google Scholar]
- Shneider N. A., Brown M. N., Smith C. A., Pickel J., Alvarez F. J. (2009). Gamma motor neurons express distinct genetic markers at birth and require muscle spindle-derived GDNF for postnatal survival. Neural Dev. 4:42 10.1186/1749-8104-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Hollyday M. (1983). The development and postnatal organization of motor nuclei in the rat thoracic spinal cord. J. Comp. Neurol. 220, 16–28 10.1002/cne.902200104 [DOI] [PubMed] [Google Scholar]
- Snider W. D. (1994). Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell 77, 627–638 10.1016/0092-8674(94)90048-5 [DOI] [PubMed] [Google Scholar]
- Sockanathan S., Jessell T. M. (1998). Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell 94, 503–514 10.1016/S0092-8674(00)81591-3 [DOI] [PubMed] [Google Scholar]
- Sockanathan S., Perlmann T., Jessell T. M. (2003). Retinoid receptor signaling in postmitotic motor neurons regulates rostrocaudal positional identity and axonal projection pattern. Neuron 40, 97–111 10.1016/S0896-6273(03)00532-4 [DOI] [PubMed] [Google Scholar]
- Sommer L., Ma Q., Anderson D. J. (1996). neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8, 221–241 10.1006/mcne.1996.0060 [DOI] [PubMed] [Google Scholar]
- Song A., Ashwell K. W., Tracey D. J. (2000). Development of the rat phrenic nucleus and its connections with brainstem respiratory nuclei. Anat. Embryol. 202, 159–177 10.1007/s004290000096 [DOI] [PubMed] [Google Scholar]
- Song M. R., Sun Y., Bryson A., Gill G. N., Evans S. M., Pfaff S. L. (2009). Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development 136, 2923–2932 10.1242/dev.037986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan P., Fawcett J. P., Rafuse V. F. (2010). Guidance of postural motoneurons requires MAPK/ERK signaling downstream of fibroblast growth factor receptor 1. J. Neurosci. 30, 6595–6606 10.1523/JNEUROSCI.4932-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien A. E., Tripodi M., Arber S. (2010). Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron 68, 456–472 10.1016/j.neuron.2010.10.019 [DOI] [PubMed] [Google Scholar]
- Stifani N., Freitas A. R., Liakhovitskaia A., Medvinsky A., Kania A., Stifani S. (2008). Suppression of interneuron programs and maintenance of selected spinal motor neuron fates by the transcription factor AML1/Runx1. Proc. Natl. Acad. Sci. U.S.A. 105, 6451–6456 10.1073/pnas.0711299105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani S., Ma Q. (2009). ‘Runxs and regulations’ of sensory and motor neuron subtype differentiation: implications for hematopoietic development. Blood Cells Mol. Dis. 43, 20–26 10.1016/j.bcmd.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134 10.1038/384129a0 [DOI] [PubMed] [Google Scholar]
- Stone R. J., Stone J. A. (2009). Atlas of Skeletal Muscles. Boston, MA: McGraw-Hill Higher Education [Google Scholar]
- Straznicky C., Tay D. (1983). The localization of motoneuron pools innervating wing muscles in the chick. Anat. Embryol. (Berl.) 166, 209–218 10.1007/BF00305083 [DOI] [PubMed] [Google Scholar]
- Streit A., Lee K. J., Woo I., Roberts C., Jessell T. M., Stern C. D. (1998). Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development 125, 507–519 [DOI] [PubMed] [Google Scholar]
- Sullivan G. E. (1962). Anatomy and embryology of the wing musculature of the domestic fowl (gallus). Aust. J. Zool. 10, 458–518 10.1071/ZO9620458 [DOI] [Google Scholar]
- Tanabe Y., William C., Jessell T. M. (1998). Specification of motor neuron identity by the MNR2 homeodomain protein. Cell 95, 67–80 10.1016/S0092-8674(00)81783-3 [DOI] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S. L. (1999). Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 23, 675–687 10.1016/S0896-6273(01)80027-1 [DOI] [PubMed] [Google Scholar]
- Thaler J. P., Koo S. J., Kania A., Lettieri K., Andrews S., Cox C., et al. (2004). A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron 41, 337–350 10.1016/S0896-6273(04)00011-X [DOI] [PubMed] [Google Scholar]
- Thaler J. P., Lee S. K., Jurata L. W., Gill G. N., Pfaff S. L. (2002). LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110, 237–249 10.1016/S0092-8674(02)00823-1 [DOI] [PubMed] [Google Scholar]
- Theriault F. M., Roy P., Stifani S. (2004). AML1/Runx1 is important for the development of hindbrain cholinergic branchiovisceral motor neurons and selected cranial sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 10343–10348 10.1073/pnas.0400768101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd K. J., Lan-Chow-Wing N., Salin-Cantegrel A., Cotter A., Zagami C. J., Lo R., et al. (2012). Establishment of motor neuron-V3 interneuron progenitor domain boundary in ventral spinal cord requires Groucho-mediated transcriptional corepression. PLoS ONE 7:e31176 10.1371/journal.pone.0031176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosney K. W., Hotary K. B., Lance-Jones C. (1995). Specifying the target identity of motoneurons. BioEssays 17, 379–382 10.1002/bies.950170503 [DOI] [PubMed] [Google Scholar]
- Tosney K. W., Landmesser L. T. (1985a). Development of the major pathways for neurite outgrowth in the chick hindlimb. Dev. Biol. 109, 193–214 10.1016/0012-1606(85)90360-4 [DOI] [PubMed] [Google Scholar]
- Tosney K. W., Landmesser L. T. (1985b). Specificity of early motoneuron growth cone outgrowth in the chick embryo. J. Neurosci. 5, 2336–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi M., Stepien A. E., Arber S. (2011). Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature 479, 61–66 10.1038/nature10538 [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Ensini M., Morton S. B., Baldassare M., Edlund T., Jessell T. M., et al. (1994). Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79, 957–970 10.1016/0092-8674(94)90027-2 [DOI] [PubMed] [Google Scholar]
- Ullah M., Mansor O., Ismail Z. I., Kapitonova M. Y., Sirajudeen K. N. (2007). Localization of the spinal nucleus of accessory nerve in rat: a horseradish peroxidase study. J. Anat. 210, 428–438 10.1111/j.1469-7580.2007.00709.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa F., Marti E. (2010). Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Developmental Dyn. 239, 69–76 10.1002/dvdy.22058 [DOI] [PubMed] [Google Scholar]
- Vallstedt A., Muhr J., Pattyn A., Pierani A., Mendelsohn M., Sander M., et al. (2001). Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 31, 743–755 10.1016/S0896-6273(01)00412-3 [DOI] [PubMed] [Google Scholar]
- Vermot J., Schuhbaur B., Le Mouellic H., Mccaffery P., Garnier J. M., Hentsch D., et al. (2005). Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development 132, 1611–1621 10.1242/dev.01718 [DOI] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J. W., Lee S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749 10.1101/gad.1519107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieseling E., Arber S. (2006). Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell 127, 1439–1452 10.1016/j.cell.2006.10.042 [DOI] [PubMed] [Google Scholar]
- Wang G., Scott S. A. (2000). The “waiting period” of sensory and motor axons in early chick hindlimb: its role in axon pathfinding and neuronal maturation. J. Neurosci. 20, 5358–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Klein R., Zheng B., Marquardt T. (2011). Anatomical coupling of sensory and motor nerve trajectory via axon tracking. Neuron 71, 263–277 10.1016/j.neuron.2011.06.021 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Ohmori Y. (1988). Location of motoneurons supplying upper neck muscles in the chicken studied by means of horseradish peroxidase. J. Comp. Neurol. 270, 271–278 10.1002/cne.902700207 [DOI] [PubMed] [Google Scholar]
- Webber C. L., Jr., Pleschka K. (1976). Structural and functional characteristics of individual phrenic motoneurons. Pflugers Arch. 364, 113–121 10.1007/BF00585178 [DOI] [PubMed] [Google Scholar]
- Westbury D. R. (1982). A comparison of the structures of alpha and gamma-spinal motoneurones of the cat. J. Physiol. 325, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetts R., Vaughn J. E. (1994). Choline acetyltransferase and NADPH diaphorase are co-expressed in rat spinal cord neurons. Neuroscience 63, 1117–1124 10.1016/0306-4522(94)90577-0 [DOI] [PubMed] [Google Scholar]
- Wichterle H., Gifford D., Mazzoni E. (2013). Neuroscience. Mapping neuronal diversity one cell at a time. Science 341, 726–727 10.1126/science.1235884 [DOI] [PubMed] [Google Scholar]
- Yaginuma H., Tomita M., Takashita N., Mckay S. E., Cardwell C., Yin Q. W., et al. (1996). A novel type of programmed neuronal death in the cervical spinal cord of the chick embryo. J. Neurosci. 16, 3685–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Pfaff S. L., Edlund T., Jessell T. M. (1993). Control of cell pattern in the neural tube: motor neuron induction by diffusible factors from notochord and floor plate. Cell 73, 673–686 10.1016/0092-8674(93)90248-O [DOI] [PubMed] [Google Scholar]
- Yamada T., Placzek M., Tanaka H., Dodd J., Jessell T. M. (1991). Control of cell pattern in the developing nervous system: polarizing activity of the floor plate and notochord. Cell 64, 635–647 10.1016/0092-8674(91)90247-V [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Henderson C. E. (1999). Patterns of programmed cell death in populations of developing spinal motoneurons in chicken, mouse, and rat. Dev. Biol. 214, 60–71 10.1006/dbio.1999.9413 [DOI] [PubMed] [Google Scholar]
- Yan Y., Sabharwal P., Rao M., Sockanathan S. (2009). The antioxidant enzyme Prdx1 controls neuronal differentiation by thiol-redox-dependent activation of GDE2. Cell 138, 1209–1221 10.1016/j.cell.2009.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Mcdonnell K., Taketo M. M., Bai C. B. (2008). Wnt signaling determines ventral spinal cord cell fates in a time-dependent manner. Development 135, 3687–3696 10.1242/dev.021899 [DOI] [PubMed] [Google Scholar]
- Zagami C. J., Zusso M., Stifani S. (2009). Runx transcription factors: lineage-specific regulators of neuronal precursor cell proliferation and post-mitotic neuron subtype development. J. Cell. Biochem. 107, 1063–1072 10.1002/jcb.22221 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Anderson D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73 10.1016/S0092-8674(02)00677-3 [DOI] [PubMed] [Google Scholar]
- Zimmerman L. B., De Jesus-Escobar J. M., Harland R. M. (1996). The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86, 599–606 10.1016/S0092-8674(00)80133-6 [DOI] [PubMed] [Google Scholar]



