Significance
Vibrio cholerae causes the life-threatening diarrheal disease cholera. These bacteria live in aquatic environments and are introduced into humans through contaminated water sources. The bacteria express virulence factors upon entry into the human gastrointestinal tract. The present study shows that virulence factor expression is controlled by the shift in temperature to 37 °C, through an RNA thermometer that controls translation of the major virulence regulator protein ToxT. This simple mechanism allows maximal virulence factor expression within the mammalian host and may be more widespread among human pathogens than currently known.
Abstract
Vibrio cholerae is the bacterium that causes the diarrheal disease cholera. The bacteria experience a temperature shift as V. cholerae transition from contaminated water at lower temperatures into the 37 °C human intestine. Within the intestine, V. cholerae express cholera toxin (CT) and toxin-coregulated pilus (TCP), two main virulence factors required for disease. CT and TCP expression is controlled by the transcriptional activator protein ToxT. We identified an RNA thermometer motif in the 5′ UTR of toxT, with a fourU anti-Shine-Dalgarno (SD) element that base pairs with the SD sequence to regulate ribosome access to the mRNA. RNA probing experiments demonstrated that the fourU element allowed access to the SD sequence at 37 °C but not at 20 °C. Moreover, mutations within the fourU element (U5C, U7C) that strengthened base-pairing between the anti-SD and SD sequences prevented access to the SD sequence even at 37 °C. Translation of ToxT-FLAG from the native toxT UTR was enhanced at 37 °C, compared with 25 °C in both Escherichia coli and V. cholerae. In contrast, the U5C, U7C UTR prevented translation of ToxT-FLAG even at 37 °C. V. cholerae mutants containing the U5C, U7C UTR variant were unable to colonize the infant mouse small intestine. Our results reveal a previously unknown regulatory mechanism consisting of an RNA thermometer that controls temperature-dependent translation of toxT, facilitating V. cholerae virulence at a relevant environmental condition found in the human intestine.
The aquatic, Gram-negative bacterium Vibrio cholerae is responsible for the diarrheal disease cholera. Cholera has swept through global human populations in large pandemics, with the first six pandemics being caused by the O1 classical biotype, and the seventh current pandemic caused by the O1 El Tor biotype. The organisms infect humans through the consumption of contaminated water sources. Once V. cholerae is inside the human small intestine, the transcription factor protein ToxT directly activates the ctx and tcp genes, which encode the essential virulence factors cholera toxin (CT) and toxin-coregulated pilus (TCP), respectively (1). TCP is required for intestinal colonization, and CT induces the profuse watery diarrhea that is the hallmark of this disease (2, 3). V. cholerae strains that lack toxT are unable to express CT or TCP and are unable to cause disease (4). A regulatory cascade that incorporates input from various environmental signals, the ToxR regulon, controls the transcription of toxT to ensure maximal expression within the intestine (1, 5). Additionally, ToxT-dependent transcriptional activity is controlled by environmental signals found within the intestine (6, 7). We report here a third mechanism that controls virulence factor expression in V. cholerae via temperature-dependent modulation of the translation of ToxT.
RNA thermometers are temperature-sensing RNA sequences found in the 5′ UTR of mRNA (8). The RNA thermometer folds into a structure to prevent access of the ribosome to the Shine-Dalgarno (SD) sequence within the mRNA at lower temperatures. As temperature increases the structure unfolds and facilitates mRNA translation at elevated temperatures. One type of RNA thermometer is the fourU element, which is characterized by a UUUU motif that base pairs with the SD sequence to form a stem-loop structure that unfolds by a zipper mechanism as temperature increases (9). The stem-loop can be stabilized by strengthening the base pairing between the fourU element and SD sequence through the substitution of G-C base pairs, and this results in an elevated melting temperature (10). The best-characterized fourU thermometer controls temperature-dependent translation of a small heat shock protein, AgsA, in Salmonella enterica (11). FourU thermometers also control temperature-dependent translation of the virulence factor LcrF in Yersinia spp. (12) and iron-acquisition genes in Shigella dysenteriae and Escherichia coli (13), ensuring appropriate expression of factors important for virulence within host tissues. Other types of RNA thermometers control virulence gene expression in Listeria monocytogenes (14) and Neisseria meningitidis (15).
Here we show that toxT translation in V. cholerae is controlled in a temperature-dependent manner by a fourU element within the 5′ UTR. At human body temperature, the thermometer structure within the mRNA opens to allow ToxT translation and subsequent virulence factor expression. Mutations within the fourU element that strengthen the secondary structure and prevent melting at 37 °C also prevent virulence of V. cholerae within an animal model for cholera, indicating that the RNA thermometer is an important modulator of V. cholerae virulence.
Results
The toxT 5′ UTR Contains a FourU RNA Thermometer Element.
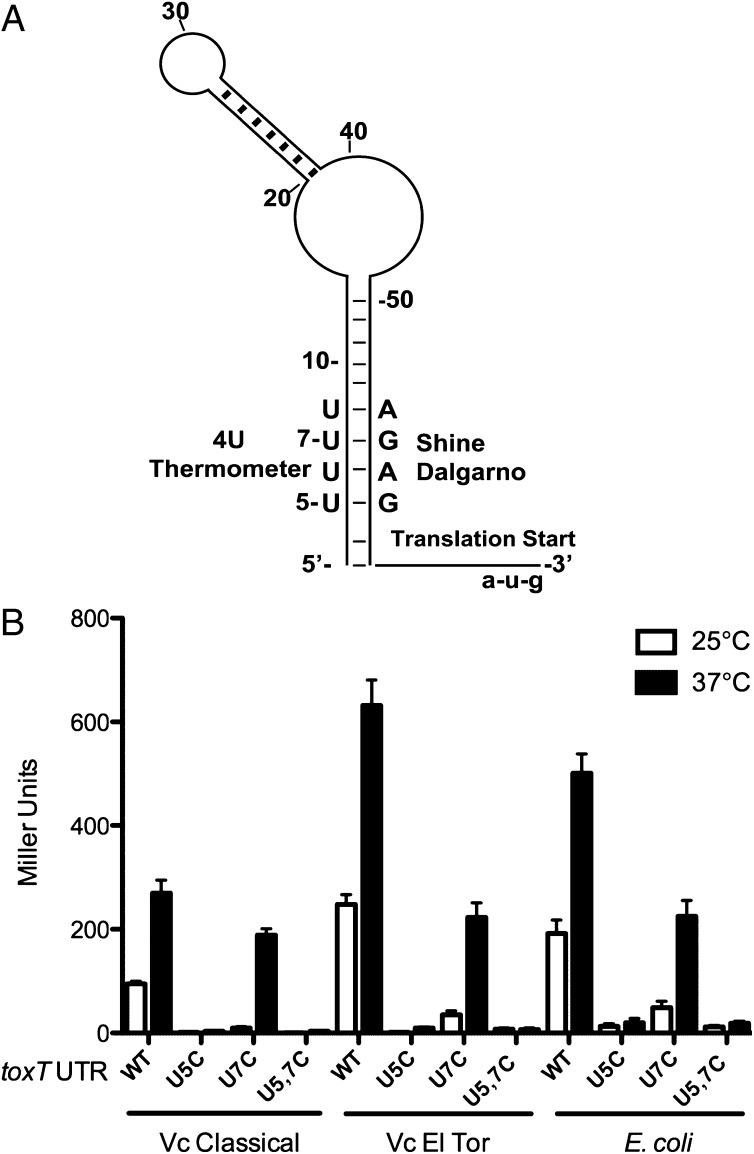
An increase of temperature to 37 °C signifies entry into a mammalian host to a pathogenic bacterium, and temperature-dependent virulence factor expression has been shown to be regulated by RNA thermometers in Yersinia spp. (12) and Listeria monocytogenes (14). We identified a potential RNA thermometer in the 5′ UTR of V. cholerae toxT that has characteristics of a fourU element (Fig. 1A). The Mfold RNA structure prediction program (16) predicted that the four uridine residues in the mRNA at positions +5 to +8 with respect to the transcription start site base pair with the SD ribosome binding site at residues +56 to +59. This forms a stem-loop structure containing canonical and noncanonical base-pairing between the fourU element and the SD sequence that is characteristic of an RNA thermometer (11).
Fig. 1.
The toxT 5′ UTR modulates temperature-dependent translation of LacZ. (A) Schematic of the predicted structure of the toxT 5′ UTR. The secondary structure was derived from Mfold (16). Shown are the relevant nucleotides within the putative fourU thermometer and the SD sequence; the complete nucleotide sequence and structure is shown in Fig. S1. The start codon and the U5 and U7 residues are indicated. (B) Effect of temperature on LacZ translation from toxT 5′ UTR. The toxT 5′UTR-lacZ (WT) construct, and the U5C, U7C, and U5C U7C (U5, 7C) mutant toxT 5′ UTR-lacZ constructs were expressed from the pBAD promoter in plasmids pBO1296, pBO1826, pBO1829, and pKEK1807, respectively. Plasmids were transformed into V. cholerae O1 strains O395 (Classical), C6706 (El Tor), and E. coli strain DH5α, and strains assayed for β-galactosidase activity during midlog growth (OD600 of 0.4–0.5) at the temperatures indicated in the presence of 0.1% arabinose. Translation from the WT UTR was significantly greater at 37 °C than at 25 °C (P < 0.01), and translation from WT UTR was significantly greater than from U5C and U5,7C UTRs at 37 °C (P < 0.001); Student’s two-tailed t test.
To determine whether this UTR could impart temperature-dependent control over translation, we created a translational fusion of the toxT 5′ UTR to lacZ in pBAD18-lacZ481 (12). High-level transcription was induced by the addition of arabinose, and we measured translation at 25 °C and 37 °C by determining β-galactosidase levels in two different V. cholerae O1 biovars, classical (strain O395) and El Tor (strain C6706), as well as in E. coli (strain DH5α) (Fig. 1B). In both V. cholerae strains as well as in E. coli, translation from the native toxT UTR (WT) was ∼150% greater at 37 °C compared with 25 °C, indicating the presence of an RNA thermometer in the UTR.
The noncanonical U-G base pairs in the fourU RNA thermometer can be strengthened by substituting a C residue in place of the U residue(s), which raises the melting temperature of the fourU-SD sequence double-stranded stem (10). Using site-directed mutagenesis we created toxT 5′ UTRs with U5C, U7C, and U5C U7C (U5, 7C). These mutant UTRs were fused to lacZ in pBAD18 as above and measured for β-galactosidase activity at 25 °C and 37 °C in V. cholerae and E. coli in the presence of arabinose (Fig. 1B). The U5C substitution caused a dramatic reduction in LacZ translation at 37 °C in both V. cholerae strains and E. coli (95–98%) compared with translation from the native UTR. In contrast, the U7C substitution led to only modest reductions (25–60%) in LacZ translation at 37 °C compared with the native UTR. These results are consistent with the zipper-type unfolding model of fourU thermometer melting (9), because the U5C mutation would be predicted to stabilize the entire stem-loop structure, whereas the U7C mutation would not. The U5, 7C double mutation led to a near absence (96–98% reduction) of detectable LacZ translation at 37 °C in all three bacterial strains.
RNA Structure Probing Reveals Melting of toxT UTR Thermometer at Elevated Temperature.
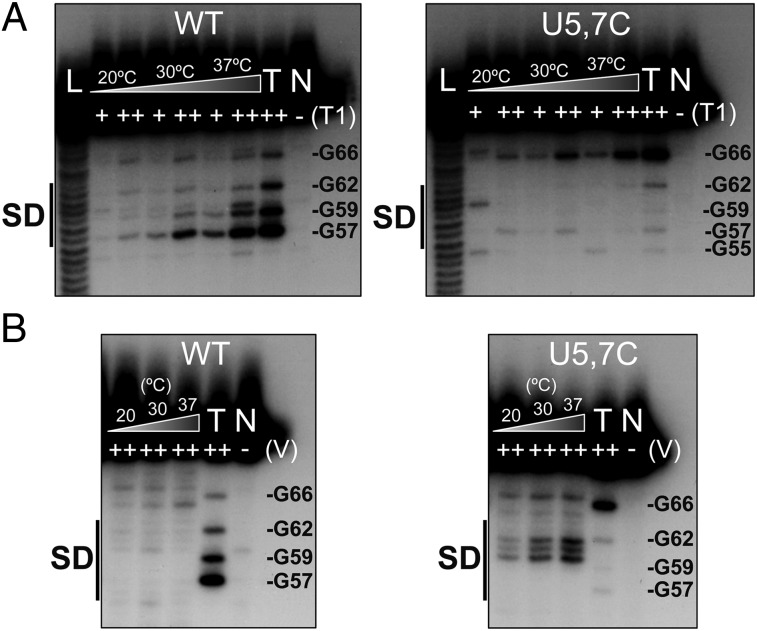
The secondary structures of 5′-labeled toxT UTRs, both the native UTR and the U5, 7C mutant UTR, were probed at increasing temperatures using RNase T1 (cuts 3′ of single-stranded guanines) and RNase V1 (specific for double-stranded regions). The cleavage products were separated on a denaturing 15% polyacrylamide gel. Focusing on the SD region of the native toxT UTR, T1 cleavage revealed that the SD region (G57, G59) becomes single-stranded as the temperature increases from 20 °C to 37 °C (Fig. 2A; the entire UTR structure can be found in Fig. S2). In contrast, no melting of the SD region of the U5, 7C toxT UTR occurs even at the elevated temperature of 37 °C, as evidenced by lack of cleavage of G57 and G59 by RNase T1. These results are consistent with the lack of translation from the U5, 7C UTR at 37 °C as shown above (Fig. 1B), being due to lack of melting of the RNA thermometer.
Fig. 2.
The toxT 5′ UTR fourU RNA thermometer facilitates temperature-dependent access to the SD sequence. Enzymatic cleavage of 5′ 32P-end-labeled toxT RNA was carried out at 20 °C, 30 °C, and 37 °C. RNA fragments were separated on 15% polyacrylamide. (A) RNase T1 (cuts 3′ of single-stranded guanines) was used at the indicated concentrations [0.01 U (+) and 0.1 U (++)]. (B) RNase V1 (cuts double-stranded, stacked regions) was used at 0.1 U. Lane L is the alkaline ladder, lane N contains no RNase, and RNA was cleaved with 0.1 U T1 at 42 °C in lane T. WT indicates the native toxT 5′ UTR, and U5, 7C indicates the U5C U7C toxT 5′ UTR. The SD sequence is labeled, and select nucleotides are noted.
The toxT native and U5, 7C UTRs were also probed with RNase V1 to detect double-stranded regions (Fig. 2B). Conditions were chosen that highlighted differences between these RNA structures. RNase V1 probing revealed that the region between G59 and G62, which includes the lower part of the SD sequence and stem-loop structure, became more obviously double-stranded in the U5, 7C mutant UTR at increasing temperature up to 37 °C. In contrast, under identical conditions, no enhanced double-stranded base pairs could be detected in this region with increasing temperature in the native toxT UTR. These results complement the RNase T1 cleavage patterns and indicate that the native UTR thermometer melts at 37 °C, whereas the U5, 7C thermometer remains clamped in a double-stranded structure at higher temperatures.
The FourU Thermometer Mediates Temperature-Dependent Translation of ToxT.
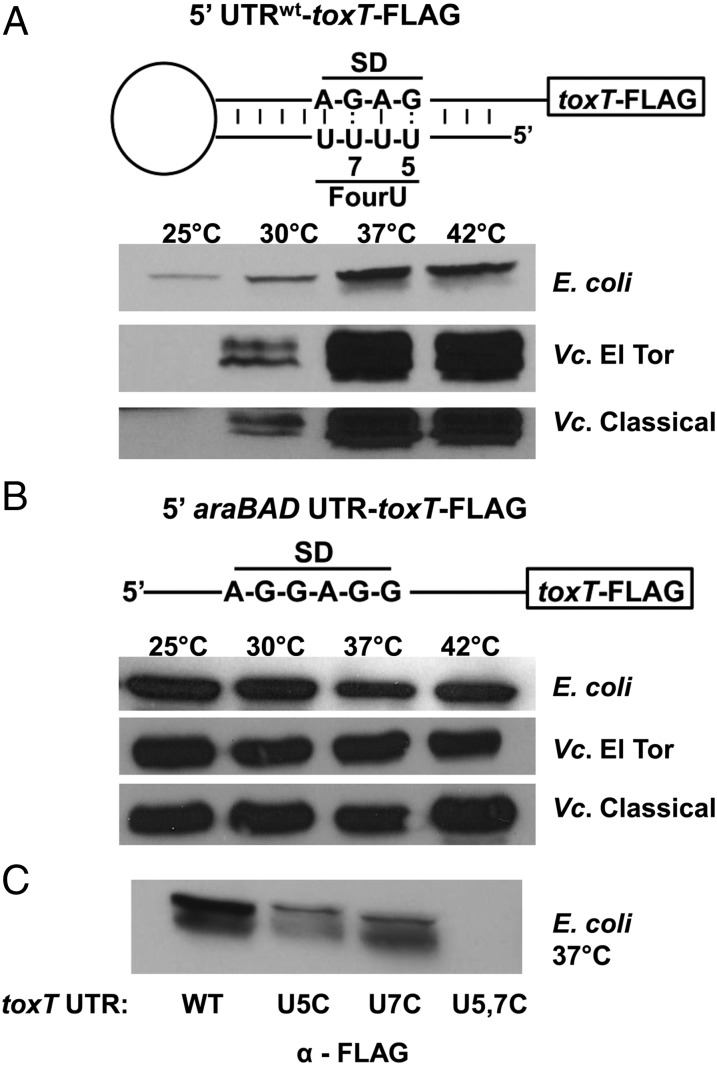
Transcription of toxT within V. cholerae is complex and is subject not only to many environmental cues feeding into the ToxR- and TcpP-dependent transcription of toxT (5) but also to an autoregulatory loop whereby ToxT activates its own transcription from the tcpA promoter (17). To avoid these overlapping regulatory mechanisms that converge on toxT transcription and focus instead on temperature-dependent translation of ToxT, we expressed toxT with its native 5′ UTR from the arabinose-inducible pBAD promoter within plasmid pBAD18 (18). A FLAG tag was added to ToxT to facilitate detection by Western immunoblot with anti-FLAG antibody. As a control for temperature-independent translation, we also created a plasmid that expresses toxT-FLAG from the arabinose-inducible pBAD promoter but with the 5′ UTR from araBAD [present in plasmid pBAD24 (18)] replacing the native toxT 5′ UTR.
When toxT is transcribed at high levels in E. coli from the pBAD promoter in the presence of arabinose, translation of ToxT-FLAG from the native toxT UTR is temperature-dependent (Fig. 3A). Low levels of ToxT-FLAG could be detected at 25 °C, with more ToxT-FLAG detectable at 30 °C and maximum amounts of ToxT-FLAG detectable at 37 °C and 42 °C. When toxT-FLAG is transcribed at high levels in V. cholerae from the pBAD promoter in the presence of arabinose, translation of ToxT-FLAG from the native toxT UTR was even more strongly temperature-dependent (Fig. 3A). The toxT-FLAG transcript with the native toxT UTR was expressed in both V. cholerae biotypes, classical and El Tor, and these strains additionally carried chromosomal deletions in toxT, to avoid any potential complications due to the presence of the chromosomally encoded ToxT. In both classical and El Tor biotypes, no ToxT-FLAG was detectable at 25 °C, modest amounts of ToxT-FLAG could be detected at 30 °C, and maximum amounts of ToxT-FLAG were detectable at 37 °C and 42 °C. [ToxT is known to undergo proteolytic degradation, and a smaller ToxT degradation product could also be detected in these assays (19).]
Fig. 3.
The toxT 5′ UTR fourU RNA thermometer facilitates temperature-dependent translation of ToxT. (A) Schematic representation of the 5′-toxT UTR-toxT-FLAG transcript of pKEK1733. E. coli DH5α (E. coli), V. cholerae O1 ΔtoxT strains KKV2426 (Vc El Tor) and KKV2288 (Vc Classical) carrying plasmid pKEK1733 were grown at the indicated temperatures in the presence of 0.1% arabinose. (B) Schematic representation of the 5′-araBAD-UTR-toxT-FLAG transcript of pKEK1789. E. coli DH5α (E. coli), V. cholerae O1 strain C6706 (Vc El Tor), and V. cholerae O1 strain O395 (Vc Classical) carrying plasmid pKEK1789 (5′-araBAD-UTR) were grown at the indicated temperatures in the presence of 0.1% arabinose. Western immunoblot using anti-FLAG monoclonal antibody was performed in A and B with whole-cell lysates (OD600 = 0.6). (C) Mutations in the fourU thermometer disrupt temperature-dependent translation of ToxT. E. coli DH5α carrying plasmids pKEK1733 (5′-toxT UTR), pKEK1734 (5′-U5C toxT UTR), pKEK1735 (5′-U7C toxT UTR), or pKEK1736 (5′-U5C U7C toxT UTR) were grown at 37 °C in the presence of 0.1% arabinose. Western immunoblot using anti-FLAG monoclonal antibody was performed with whole-cell lysates (OD600 = 0.6).
Temperature-dependent translation is specifically due to the presence of the toxT 5′ UTR, because when toxT-FLAG was instead transcribed from the same promoter with the araBAD 5′ UTR replacing the native toxT UTR, similar levels of ToxT-FLAG were translated at all temperatures in E. coli and both V. cholerae biotypes (Fig. 3B). These results demonstrate that the native toxT 5′ UTR acts as an RNA thermometer to control temperature-dependent translation of ToxT.
Strengthening Base-Pairing Within the FourU Thermometer Inhibits ToxT Translation at Elevated Temperatures.
The mutant toxT UTRs with strengthened stem-loop binding (U5C, U7C, and U5, 7C) were also fused to toxT-FLAG, and these transcripts were expressed from the pBAD promoter in E. coli at 37 °C (Fig. 3C). As was seen above, the native toxT UTR allows translation of ToxT-FLAG at 37 °C. The introduction of the U5C or the U7C mutation into the toxT UTR results in less ToxT-FLAG translation, whereas the introduction of both U5C and U7C mutations (U5, 7C) results in no detectable ToxT-FLAG translation. These results are consistent with the RNA probing experiments above, in which the U5, 7C UTR remained double-stranded at 37 °C, which would sequester the SD sequence and prevent ToxT translation.
The FourU Thermometer Controls Virulence Factor Expression in V. cholerae in Vitro.
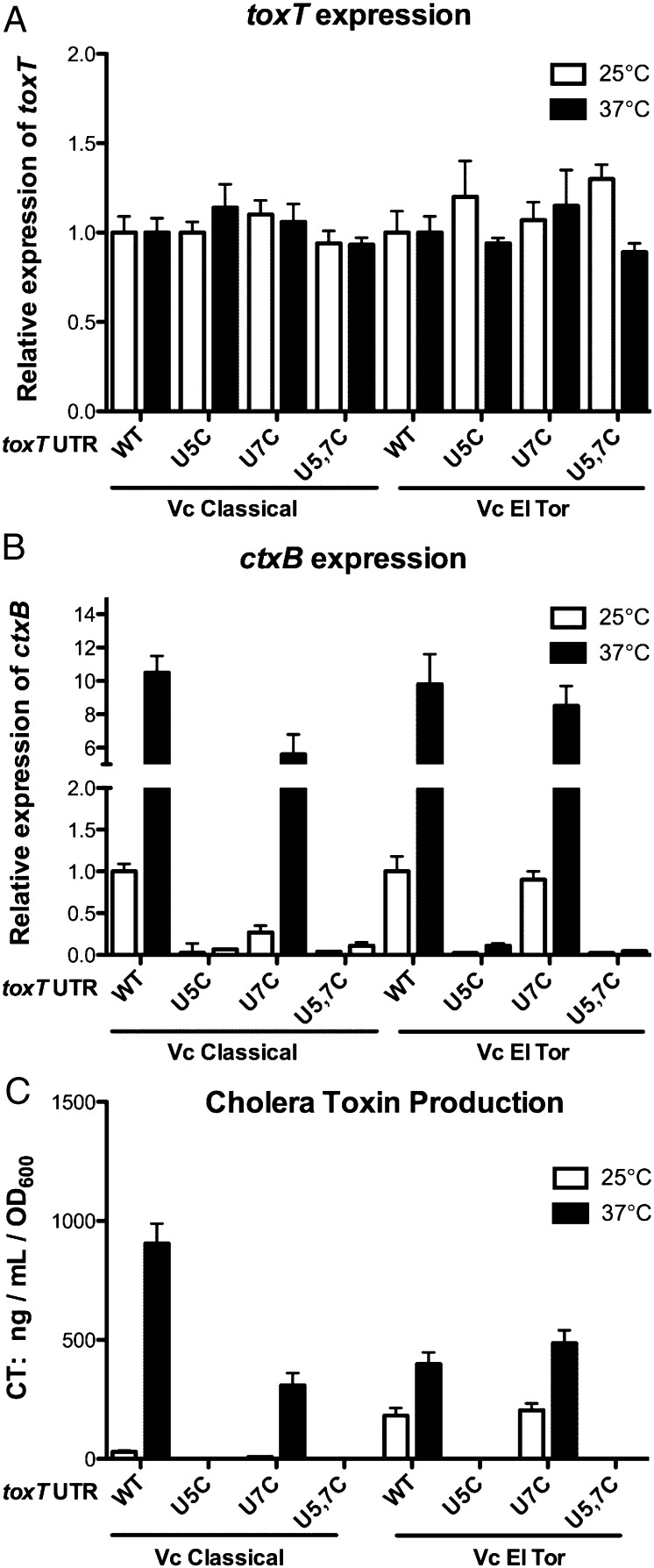
As mentioned above, a complicated regulatory cascade controls transcription of toxT that could mask the effects of temperature-dependent translation of ToxT. To determine whether the fourU thermometer controls temperature-dependent virulence factor expression, we transcribed toxT with either native or mutant 5′ UTR from the pBAD promoter in V. cholerae O1 classical and El Tor strains that contained deletions of the chromosomal toxT gene, during growth at 25 °C and 37 °C. All cultures were assayed at the same stage of midlog growth (OD600 of 0.6). Transcript levels of toxT were measured by quantitative RT-PCR, which demonstrated that the amount of toxT transcripts was similar at both temperatures for the wild-type and mutant UTR-toxT transcripts in both classical and El Tor V. cholerae (Fig. 4A; no significant difference in expression in any sample; Student t test).
Fig. 4.
The toxT 5′ UTR fourU RNA thermometer facilitates temperature-dependent expression of CT. V. cholerae O1 ΔtoxT strains KKV2288 (Classical) and KKV2426 (El Tor) carrying plasmids pKEK1682 (5′-toxT UTR-toxT), pKEK1683 (5′-U5C toxT UTR-toxT), pKEK1685 (5′-U7C toxT UTR- toxT), or pKEK1686 (5′-U5C U7C toxT UTR- toxT) were grown at 25 °C and 37 °C in the presence of 0.1% arabinose and harvested at identical cell densities (OD600 = 0.6). mRNA abundance for (A) toxT and (B) ctxB were determined by quantitative RT-PCR (Materials and Methods). (C) Supernatants were assayed for CT.
We also measured transcript levels of the ToxT-dependent virulence gene ctxB in the same strains grown at 25 °C and 37 °C (Fig. 4B). When normalized to transcript levels of these genes at 25 °C in the same strain expressing the native UTR-toxT gene, it could be confirmed that (i) ctx transcription was higher at 37 °C than at 25 °C in strains expressing the native UTR-toxT, (P < 0.001 classical and P < 0.01 El Tor), (ii) the U5C and U5C U7C UTRs caused significant decreases in ctx transcription (P < 0.001 at 37 °C for both classical and El Tor), and (iii) the U7C UTR had little effect on ctx transcription (P < 0.05 classical 37 °C, no significant difference El Tor). Similar results were seen when transcript levels of the ToxT-dependent virulence gene tcpB were measured in the same strains (Fig. S3). These results are consistent with the function of the RNA thermometer in the toxT UTR controlling virulence gene transcription by modulating ToxT translation.
Finally, we measured CT expression in these same strains during growth at 25 °C and 37 °C (Fig. 4C). As anticipated, CT expression mirrored ctxB transcription in these strains. Under these conditions, CT expression was strongly temperature-dependent in the O1 classical V. cholerae strain translating ToxT from the native UTR, with little CT expression at 25 °C, and 30-fold higher CT expressed at 37 °C. CT expression was also temperature-dependent in the O1 El Tor V. cholerae strain translating ToxT from the native UTR, with twofold higher CT expressed at 37 °C than at 25 °C. The introduction of the U7C mutation into the UTR caused a modest reduction in CT expression at 37 °C in the classical strain and had no effect on CT expression in the El Tor strain. In contrast, introduction of the U5C mutation into the UTR resulted in no detectable CT expression at either temperature in both the classical and El Tor strains. Likewise, introduction of both U5C and U7C mutations (U5, 7C) into the UTR resulted in no detectable CT expression at either temperature in both classical and El Tor strains. These results confirm that the toxT UTR controls temperature-dependent expression of CT. Moreover, the U5C UTR mutation has a stronger negative effect on virulence factor expression than the U7C mutation, consistent with the results seen with LacZ translation from the various toxT UTRs (Fig. 1). These results also suggest there may be additional mechanism(s) controlling ToxT translation that vary between V. cholerae biotypes.
The FourU Thermometer Controls Virulence of V. cholerae in Vivo.
The ability of V. cholerae to colonize the infant mouse small intestine is correlated with its ability to cause disease in humans (20). We reasoned that strengthening the fourU thermometer by the introduction of the U5C and/or U7C mutations into the genome of V. cholerae would reveal whether the fourU thermometer controlled temperature-dependent virulence in vivo. V. cholerae classical O1 strains were constructed with U5C, U7C, and U5C U7C toxT UTR mutations, and a V. cholerae El Tor O1 strain was constructed with the U5C U7C mutations. We used a competition assay in the infant mouse to determine the relative ability of these V. cholerae strains to colonize the infant mouse small intestine.
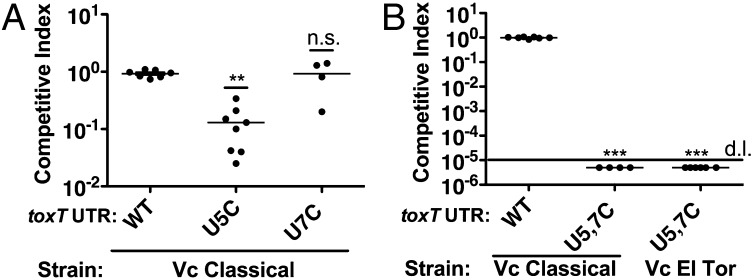
The U5C toxT UTR mutant showed an approximate 10-fold defect in its ability to colonize the infant mouse small intestine [competitive index (C.I.) = 0.13, P < 0.01] (Fig. 5A). In contrast, the U7C toxT UTR mutant colonized the infant mouse intestine similar to the wild-type strain. These results are consistent with the relative effect these single UTR mutations had upon LacZ/ToxT translation and CT expression at 37 °C, with the U5C mutation having a stronger negative effect than the U7C mutation.
Fig. 5.
The toxT 5′ UTR fourU RNA thermometer controls virulence of V. cholerae. (A) V. cholerae O1 classical strains KKV598 (WT), KKV2333 (U5C 5′toxT UTR), and KKV2368 (U7C 5′toxT UTR) were coinoculated with the wild-type strain O395 perorally into infant mice. **P < 0.01, Student t test; n.s., not significant. (B) V. cholerae O1 classical strains KKV598 (WT) and KKV2425 (U5C, U7C 5′toxT UTR), and V. cholerae O1 El Tor strain KKV2382 (U5C, U7C 5′toxT UTR) were coinoculated with the wild-type strains O395 or C6706 perorally into infant mice. No U5C U7C mutant bacteria were recovered from the intestine, detection limit (d.l.) is 5 × 10−5. ***P < 0.001, Student t test. Inocula in A and B contained a ratio of ∼1:1 mutant/wild-type; intestinal homogenates were recovered at 24 h after inoculation and cfu of wild-type and mutant strains determined. The competitive index is given as the output ratio of mutant/wild-type divided by the input ratio of mutant/wild-type; each value shown is from an individual mouse.
The U5C U7C toxT UTR mutations in both classical and El Tor O1 strains rendered the bacteria completely incapable of colonizing the infant mouse intestine (Fig. 5B); no mutant V. cholerae strains were recovered from any mice (C.I. < 5 × 10−5; P < 0.001). These results are consistent with the inhibitory effect of the U5C U7C mutations on ToxT translation at 37 °C (Fig. 3C), which is due to the lack of melting of the RNA thermometer (Fig. 2) and sequestration of the SD sequence. Our results demonstrate the importance of the RNA thermometer in regulating the virulence of V. cholerae.
Discussion
Controlled access to the ribosome binding site (RBS) within mRNA is a mechanism that modulates the rate of protein synthesis in bacteria. RBS access can be controlled by RNA–RNA interactions, including trans-acting small RNAs that bind to the SD sequence and prevent 30S ribosome binding (21). RBS access can also be controlled by cis-acting RNA sequences that base pair with the SD sequence and form a secondary structure that prevents ribosome binding. These cis-acting sequences include riboswitches that bind diverse ligands and primarily control expression of biosynthetic pathways (22). Another type of cis-acting sequence is RNA thermometers, which control temperature-dependent translation of proteins (8). RNA thermometers such as the ROSE element are typically found within the 5′ UTR of heat shock protein genes; ROSE prevents translation until heat shock temperature, by sequestration of the SD sequence through basepairing (23).
Another type of RNA thermosensor is the fourU element, which has been best characterized in the 5′ UTR of an S. enterica heat shock gene, agsA (11). The four consecutive uridine residues within this element base pair with the SD sequence and form a stem-loop that prevents ribosome binding at lower temperatures. Critical to the function of this thermometer are two noncanonical base pairs between the uridine residues within the element and G residues in the SD sequence. This thermometer opens upon increased temperature through a zippering mechanism initiated at the base of the stem-loop closest to the translation start (9). Our results have demonstrated that a similar fourU thermometer is present in the 5′ UTR of toxT mRNA, and it functions to regulate temperature-dependent ToxT translation in V. cholerae. This regulatory mechanism allows for high levels of ToxT at the mammalian host temperature of 37 °C, and low to no ToxT protein at the lower temperatures found in the aquatic environmental reservoir where V. cholerae are naturally found. Because ToxT is the direct transcriptional activator of the most important V. cholerae virulence factors, CT and TCP, the fourU thermometer controls temperature-dependent induction of virulence as the bacteria move from the aquatic environment into the human host.
Several lines of evidence demonstrated the function of this thermosensor in the 5′ UTR of toxT, including direct RNA probing, lacZ translational fusions, Western immunoblot for ToxT protein, and measurement of ToxT-dependent virulence factor expression. Moreover, the substitution of C-G base pairs in place of the U-G base pairs (U5C and/or U7C) within the stem-loop structure inhibited or prevented thermal induction of ToxT translation and subsequent virulence, as predicted. On the basis of previous NMR studies (9), the fourU RNA thermometer opens by a zippering mechanism initiating from the base of the stem loop, and thus the U5C mutation would be expected to act like a clamp and have a stronger negative effect on thermal induction of ToxT translation than the U7C mutation. Indeed, the U7C mutation had only modest effects on ToxT translation and no significant effect on intestinal colonization, whereas the U5C mutation caused significant decreases in ToxT translation and an approximately 10-fold reduction in intestinal colonization. Importantly, the 5′ UTR containing both U5C and U7C mutations prevented ToxT translation and subsequent virulence factor expression at 37 °C, and V. cholerae O1 pandemic strains (classical and El Tor) containing these mutations were completely unable to colonize the intestine. RNA probing revealed that the stem-loop structure of the U5C U7C 5′ UTR remained double-stranded at 37 °C, emphasizing the importance of the fourU thermometer in the temperature-dependent virulence of V. cholerae.
As V. cholerae transition from the aquatic environment into the human intestine, the environment encountered within the small intestine leads to the ToxT-dependent induction of TCP and CT expression and subsequent disease. A number of different environmental signals have been identified that modulate the ToxR- and TcpP-dependent induction of toxT transcription, a critical event in this virulence cascade (5), as well as those that modulate the activity of ToxT (6, 7). The increase to 37 °C as the bacteria move into the host seems like an obvious environmental change that signifies optimal disease conditions, and our results demonstrate how the RNA thermometer in the 5′ UTR of toxT modulates virulence by enhancing ToxT translation at temperatures found within the intestine. Our studies reveal yet another layer of control over V. cholerae pathogenesis that ensures the correct temporal and spatial expression of virulence factors. The RNA thermometer mechanism provides temperature-dependent control over translation from a preexisting mRNA, and thus may allow for quicker or more precise control over virulence gene expression when entering or exiting a host. FourU RNA thermometers have been identified that control virulence by temperature-dependent expression of LcrF in Yersinia spp. (12) and iron-acquisition genes in Shigella dysenteriae and enteropathogenic E. coli (13). RNA thermometers represent a simple yet elegant mechanism for controlling temperature-dependent gene expression and may be more widespread among pathogenic bacteria that interact with mammalian hosts than is currently known.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli strains DH5α (24) and Top10 (Invitrogen) were used for cloning, and SM10λpir was used for conjugation. V. cholerae O1 strains O395 [classical biotype (25)] and C6706 [El Tor biotype (26)] were used. V. cholerae strain KKV2426 (C6706 ΔtoxT::Cm) was created via chitin transformation (27) using genomic DNA from strain SY1002 (28). V. cholerae strains KKV2333 (O395 toxT 5′ UTR U5C), KKV2368 (O395 toxT 5′ UTR U7C), KKV2425 (O395 toxT 5′ UTR U5C, U7C), KKV2382 (C6706 toxT 5′ UTR U5C, U7C), as well as KKV598 (O395 native toxT 5′ UTR) and KKV2372 (C6706 native toxT 5′ UTR), were constructed by using plasmids pKEK1445, pKEK1468, pKEK1469, and pKEK1808 to introduce the mutations into V. cholerae strains KKV2288 (29) and KKV2426 by allelic exchange. A complete list of primers and plasmids used is provided in Tables S1 and S2.
Strains were grown in Luria broth (LB) at the indicated temperatures. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/mL; chloramphenicol (Cm), 2 μg/mL; streptomycin (Sm), 100 μg/mL and 1 mg/mL. When required, arabinose was added to the media at a concentration of 0.1%. Plasmids used are listed in Table S2, and details on their construction are included in SI Materials and Methods. All constructs were verified by sequencing.
β-Galactosidase Assays and Western Immunoblot.
For β-galactosidase assays, overnight cultures were diluted 1:100 in LB plus 100 μg/mL ampicillin and 0.1% arabinose, grown at the indicated temperatures to an optical density at 600 nm of 0.3–0.8, permeabilized with chloroform and SDS, and then assayed for enzymatic activity by the method of Miller (30). All assays were performed independently at least twice, with triplicate samples. For Western immunoblots, strains were grown overnight in LB + ampicillin (Amp) at 37 °C. They were then diluted 1:100 in LB + Amp + 0.1% arabinose, grown at 25 °C, 30 °C, 37 °C, or 42 °C and harvested at OD600 of 0.6. Cell cultures were normalized by OD600, resuspended in SDS sample buffer, boiled for 10 min, separated by SDS/PAGE, and transferred to nitrocellulose membranes. Western immunoblot was performed with anti-FLAG M2 monoclonal antibody (Sigma), and detection with ECL reagent (GE Healthcare).
Structure Probing Experiments.
RNAs were synthesized in vitro by runoff transcription with T7 RNA polymerase from linearized plasmid templates. Plasmids pBO1299 and pBO1845 (linearized with MlsI) were used to generate RNA for enzymatic structure probing. RNA structure probing was carried out at 20 °C, 30 °C, and 37 °C. RNA was 5′-end-labeled as previously described (31). The labeled RNA was then digested with ribonucleases T1 and V1 as previously described (32), and the RNA fragments were separated on denaturing 15% polyacrylamide gels. Alkaline ladders were generated as previously described (31).
CT Assays.
V. cholerae ΔtoxT strains KKV2288 and KKV2426 were transformed with plasmids that transcribe toxT from the pBAD promoter and translate ToxT from the native or mutant toxT 5′ UTRs (pKEK1682, pKEK1683, pKEK1684, and pKEK1685). Cultures were used to measure CT production, as well as toxT transcript levels as described below. CT was measured in the supernatant of cultures at OD600 of 0.6 grown in LB + Amp + 0.1% arabinose at 25 °C and 37 °C. The ganglioside M1 ELISA (GM1-ELISA) was used with polyclonal rabbit serum directed against the purified B subunit of CT as previously described (33).
Quantitative RT-PCR.
Total RNA was isolated from V. cholerae strains KKV2288 and KKV2426 transformed with plasmids that transcribe toxT from the pBAD promoter and translate ToxT from the native or mutant toxT 5′ UTRs (pKEK1682, pKEK1683, pKEK1684, pKEK1685) using TRIzol Max Bacterial RNA Isolation Kit (Ambion). Overnight cultures were inoculated at 1:100 into LB + Amp with 0.1% arabinose. They were grown at 25 °C and 37 °C to OD600 of 0.6. The RNA was treated with Turbo DNase (Ambion), and reverse transcription reactions were performed using iScript Reverse Transcription supermix (BioRad); RT-negative controls were included for every RNA sample. cDNA was amplified in the Applied Biosystems ABI 7500 Real-Time PCR System, using SsoAdvanced Universal Syber Green supermix (BioRad), and specific primers for toxT, ctxB, tcpB, and gyrA were used. gyrA was used as the control. Assays were performed in triplicate. The relative expression values (R) were calculated by using the formula R = 2 − dCt target − dCt endogenous control, where Ct is the fractional threshold cycle.
In Vivo Colonization Assay.
V. cholerae mutant strains were each mixed in a 1:1 ratio with the isogenic wild-type strain and then inoculated intragastrically into 5-d-old CD-1 suckling mice (∼106 mutant: 106 wild type). Infected pups were placed in a humidified 30 °C incubator. After 20 h the mice were killed, and their small intestines were isolated and homogenized. The mutant/wild-type ratios were determined by plating dilutions on LB agar containing X-Gal (5-bromo-4-chloro-3-indolyl-d-galactopyranoside). The competitive index is given as the output ratio of mutant:wild-type divided by the input ratio of mutant:wild-type; Prism 5.0b was used for statistical analyses.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant RO1 AI51333 (to K.E.K.), by German Research Foundation Priority Program 1258 (to F.N.), and by the Studienstiftung des Deutschen Volkes (J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.K.W. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411570111/-/DCSupplemental.
References
- 1.DiRita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88(12):5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84(9):2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmgren J, Svennerholm AM. Mechanisms of disease and immunity in cholera: A review. J Infect Dis. 1977;136(Suppl):S105–S112. doi: 10.1093/infdis/136.supplement.s105. [DOI] [PubMed] [Google Scholar]
- 4.Champion GA, Neely MN, Brennan MA, DiRita VJ. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23(2):323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 5.Childers BM, Klose KE. Regulation of virulence in Vibrio cholerae: The ToxR regulon. Future Microbiol. 2007;2(3):335–344. doi: 10.2217/17460913.2.3.335. [DOI] [PubMed] [Google Scholar]
- 6.Schuhmacher DA, Klose KE. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J Bacteriol. 1999;181(5):1508–1514. doi: 10.1128/jb.181.5.1508-1514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abuaita BH, Withey JH. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun. 2009;77(9):4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortmann J, Narberhaus F. Bacterial RNA thermometers: Molecular zippers and switches. Nat Rev Microbiol. 2012;10(4):255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 9.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010;38(11):3834–3847. doi: 10.1093/nar/gkq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 2011;39(18):8258–8270. doi: 10.1093/nar/gkr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: A novel type of RNA thermometer in Salmonella. Mol Microbiol. 2007;65(2):413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 12.Böhme K, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 2012;8(2):e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouse AB, Righetti F, Kortmann J, Narberhaus F, Murphy ER. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One. 2013;8(5):e63781. doi: 10.1371/journal.pone.0063781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson J, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110(5):551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 15.Loh E, et al. Temperature triggers immune evasion by Neisseria meningitidis. Nature. 2013;502(7470):237–240. doi: 10.1038/nature12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown RC, Taylor RK. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16(3):425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 18.Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abuaita BH, Withey JH. Termination of Vibrio cholerae virulence gene expression is mediated by proteolysis of the major virulence activator, ToxT. Mol Microbiol. 2011;81(6):1640–1653. doi: 10.1111/j.1365-2958.2011.07798.x. [DOI] [PubMed] [Google Scholar]
- 20.Klose KE. The suckling mouse model of cholera. Trends Microbiol. 2000;8(4):189–191. doi: 10.1016/s0966-842x(00)01721-2. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12):a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1-2):17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narberhaus F. Translational control of bacterial heat shock and virulence genes by temperature-sensing mRNAs. RNA Biol. 2010;7(1):84–89. doi: 10.4161/rna.7.1.10501. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Mekalanos JJ, Collier RJ, Romig WR. Enzymic activity of cholera toxin. II. Relationships to proteolytic processing, disulfide bond reduction, and subunit composition. J Biol Chem. 1979;254(13):5855–5861. [PubMed] [Google Scholar]
- 26.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64(7):2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marvig RL, Blokesch M. Natural transformation of Vibrio cholerae as a tool—optimizing the procedure. BMC Microbiol. 2010;10:155. doi: 10.1186/1471-2180-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto S, Izumiya H, Morita M, Arakawa E, Watanabe H. Application of lambda Red recombination system to Vibrio cholerae genetics: Simple methods for inactivation and modification of chromosomal genes. Gene. 2009;438(1-2):57–64. doi: 10.1016/j.gene.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Childers BM, et al. N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J Biol Chem. 2011;286(32):28644–28655. doi: 10.1074/jbc.M111.258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. A Short Course in Bacterial Genetics. 2nd Ed. Plainview, NY: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 31.Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: An in vitro study of plasmid pIP501. EMBO J. 1994;13(15):3599–3607. doi: 10.1002/j.1460-2075.1994.tb06667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 2011;39(7):2855–2868. doi: 10.1093/nar/gkq1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ristaino PA, Levine MM, Young CR. Improved GM1-enzyme-linked immunosorbent assay for detection of Escherichia coli heat-labile enterotoxin. J Clin Microbiol. 1983;18(4):808–815. doi: 10.1128/jcm.18.4.808-815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.