Abstract
Large-scale surveys of single-cell gene expression have the potential to reveal rare cell populations and lineage relationships, but require efficient methods for cell capture and mRNA sequencing1–4. Although cellular barcoding strategies allow parallel sequencing of single cells at ultra-low depths5, the limitations of shallow sequencing have not been directly investigated. By capturing 301 single cells from 11 populations using microfluidics and analyzing single-cell transcriptomes across downsampled sequencing depths, we demonstrate that shallow single-cell mRNA sequencing (~50,000 reads per cell) is sufficient for unbiased cell-type classification and biomarker identification. In developing cortex we identify diverse cell types including multiple progenitor and neuronal subtypes, and we identify EGR1 and FOS as previously unreported candidate targets of Notch signaling in human but not mouse radial glia. Our strategy establishes an efficient method for unbiased analysis and comparison of cell populations from heterogeneous tissue by microfluidic single-cell capture and low-coverage sequencing of many cells.
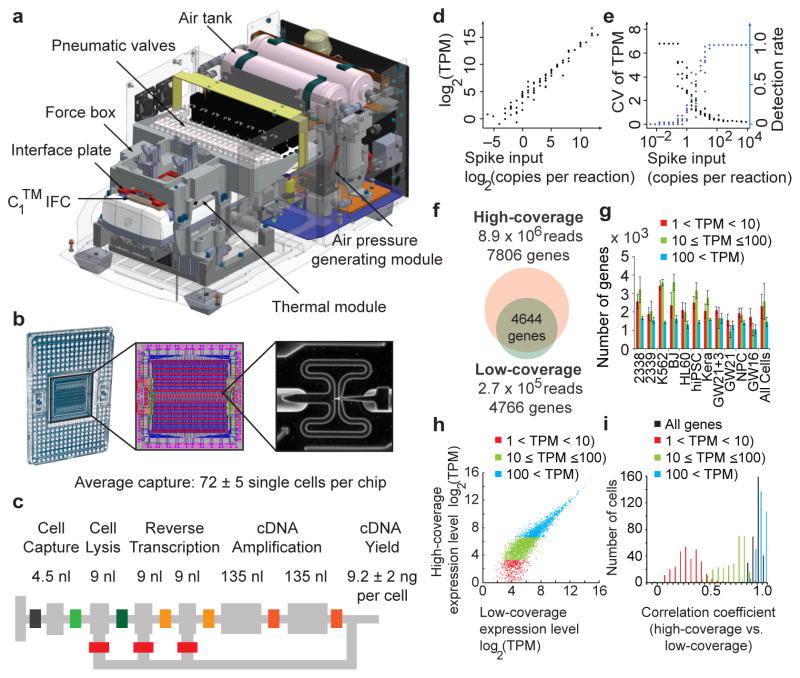
To routinely capture single cells, we designed the C1™ Single-Cell Auto Prep System (Fig. 1a). The microfluidic system performs reverse transcription and cDNA amplification in nanoliter reaction volumes (Fig. 1b–c), which increases the effective concentration of reactants and may improve the accuracy of mRNA Seq6. We sequenced libraries from single cells at high-coverage (~8.9 × 106 reads per cell) and used the results as a reference to explore the consequences of reduced sequencing depth. To explore current practical limits of low-coverage sequencing, we pooled dozens of barcoded single-cell libraries in single MiSeq® System runs (Illumina, ~2.7 × 105 reads per cell) and downsampled high-coverage results to ultra low depths. We prepared sequencing libraries after cDNA amplification with the SMARTer® Ultra™ Low RNA Kit for Illumina® Sequencing (Clontech) and the Nextera® XT kit (Illumina). Genomic alignment rates and other quality metrics were similar across libraries, whereas empty negative control wells showed no appreciable sequence alignment (<1%) (Supplementary Table 1).
Figure 1.
Capturing single cells and quantifying mRNA levels using the C1™ Single-Cell Auto Prep System. (a) Key functional components of the C1™ System are labeled, including the pneumatic components necessary for control of the microfluidic integrated fluidic circuit (IFC) and the thermal components necessary for preparatory chemistry. (b) Left panel- the complete IFC with carrier; reagents and cells are loaded into dedicated carrier wells and reaction products are exported to other dedicated carrier wells. Middle panel- diagram of the IFC: Connections between polydimethylsiloxane microfluidic chip and carrier (pink circles), control lines (red), fluidic lines for preparatory chemistry (blue), and lines connecting control lines (green). Right panel- a single cell captured in a 4.5 nL capture site; there are 96 captures sites per IFC. The average single cell capture rate was 72 ± 5 cells (mean ± s.e.m.) per chip (Supplementary Tables 1, 2). (c) Schematic for a C1™ reaction line is shown with reaction line colored light grey and isolation valves in varied colors. All reagents are delivered through a common central bus line (segment of bus line shown on far left). Each reaction begins in the 4.5 nL capture site. Delivery of the lysis reagent expands the reaction to also include the first 9 nL chamber. The reaction is expanded again upon delivery of the reverse transcription (RT) reagent to include the second and third 9 nL chambers. Finally, the two 135 nL reaction chambers are included to provide the larger volume required for the PCR reagents. After the addition of RT reagent, the contents of the reaction line are pumped in a loop using a bypass line (bottom) for mixing and the IFC is then incubated at 42°C for RT. Mixing is repeated after the addition of PCR reagents and thermal cycling is performed. Following preparatory chemistry, each single-cell reaction product exits the chip using a dedicated fluidic path to the carrier (path shown to the right). (d) Sequencing of reaction products from 46 K562 cells at low-coverage (1.7 × 105 reads per cell) reveals that expression level estimates correlate strongly with known copy numbers of input spikes (Pearson’s r = 0.968) from External RNA Controls Consortium (ERCC) RNA Spike-In Control Mix 1 (2.8 × 104 copies/reaction). (e) The fraction of positive reactions where ERCC transcripts are detected above 1 TPM in single cells and the coefficient of variation for ERCC levels are both plotted versus the spike input amounts. (f–i) Pools of barcoded libraries from 301 cells were sequenced at high coverage by HiSeq® and at low coverage by MiSeq®. (f) In a representative cell, 4644 genes were detected above 1 TPM in both datasets. (g) Graph showing the average number of genes expressed at various levels detected by high coverage sequencing in each cell type (Methods). (h) In a representative cell, expression levels of genes detected in high- and low-coverage datasets were highly correlated (r = 0.91). (i) Histogram of correlation coefficients for all single cells (n = 301). The mean correlation coefficients increased with expression level: 0.25 (1<TPM<10, red), 0.66 (10≤TPM≤100, green), 0.93 (TPM>100, blue) and 0.91 (all genes with TPM>1).
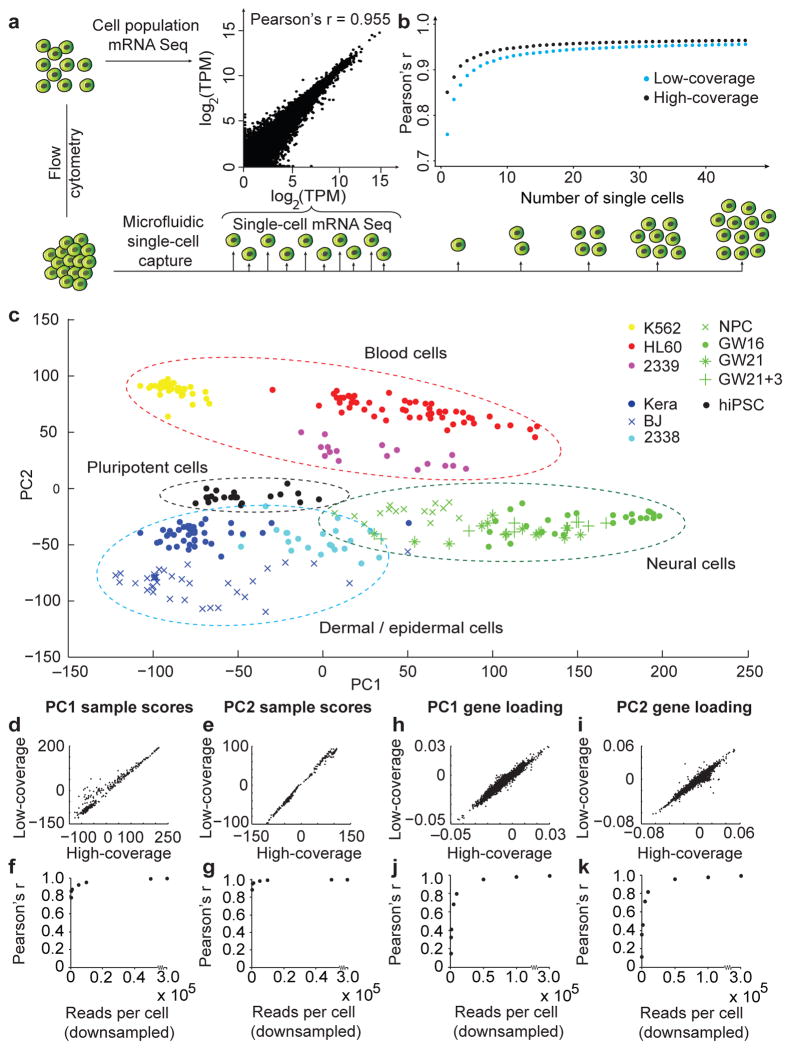
We assessed the accuracy, detection rates and variance of RNA level estimates from low-coverage sequencing of single-cell libraries by comparing the results with known quantities of spike-in RNA transcripts7 and with high-coverage sequencing of the same libraries. Levels of RNA spikes determined by low-coverage mRNA sequencing correlated strongly with known input quantities (r = 0.968). For inputs above 32 copies, all spikes could be detected in all samples with minimal variance (Fig. 1d–e)6,8. In a representative cell, the majority of genes detected by high-coverage sequencing were also detected by low-coverage sequencing (Fig. 1f). Of the genes detected by high- but not low-coverage sequencing, the vast majority (98%) were not expressed at high levels (transcript per million, TPM>100) and most (63%) were expressed at low levels (1<TPM<10, Supplementary Fig. 1). Across 301 cells from a range of sources, the average correlation between estimates of single-cell gene expression from low-and high-coverage sequencing was 0.91 (Fig. 1f–g, Supplementary Fig. 2). However, for transcripts with low expression levels (1<TPM<10), the correlation dropped to 0.25, demonstrating a limitation of quantifying low abundance transcripts in individual cells using shallow sequencing. Despite this limitation, combining low-coverage results from as few as 10 individual K562 cells accurately reflected results from a pooled population of K562 cells captured by flow cytometry (r>0.92) (Fig. 2a–b). We concluded that single-cell capture and low-coverage sequencing can be used to profile gene expression of individual cells and that combined results reflect properties of a given cell population.
Figure 2.
Low-coverage single-cell mRNA sequencing is sufficient to detect genes contributing to cell identity. (a) The average expression levels from single-cell mRNA sequencing of 46 K562 cells correlate strongly with expression levels from a population of 100 K562 cells isolated by flow cytometry. (b) The correlation between individual K562 cells and the population improves with diminishing returns as additional single cell results are combined. (c) Distinct groups of cells corresponding to pluripotent, blood, skin, and neural cells can be identified by PCA of 301 cells sequenced at low coverage. (d–g) Sample scores from low- and high-coverage data were calculated using the eigenvectors from high-coverage data and correlate strongly across all 301 cells for PC1 (d, r = 0.973) and PC2 (e, r = 0.997). The strong sample score correlations (r > 0.92) persist with as few as 5000 reads per cell for PC1 (f) and PC2 (g). (h–k) Similarly, eigenvectors derived from low- and high-coverage datasets correlate strongly for the eigenvectors defining PC1 (h, r = 0.980) and PC2 (i, r = 0.956), but strong correlations of eigenvectors (r>0.95) for PC1 (j) and PC2 (k) require at least 50,000 reads per cell.
To examine whether low-coverage sequencing can distinguish between cell types, we first compared cells from sources expected to show robust differences in gene expression: pluripotent cells, skin cells, blood cells, and neural cells. We performed principal component analysis (PCA) of low-coverage sequencing data to identify genes explaining variation across cells. PCA separated cells into groups corresponding to the source populations (Fig. 2c, Supplementary Figs. 3–5) and genes distinguishing each group reflected biological properties of the cell types (Supplementary Fig. 5, Supplementary Table 3). PCA of low- and high-coverage sequencing data revealed a remarkably similar graphical distribution of analyzed cells, and the majority (78%) of the top 500 PCA genes were shared between PCA performed on low- and high-coverage data (Supplementary Figs. 4, 6 and Supplementary Table 4). We next examined the minimal depth at which low-coverage sequencing could be applied to describe variation across diverse cell types. The positions of cells along PC1 and PC2 were highly correlated between low- and high-coverage sequencing results (Fig. 2d–e) and could be accurately predicted by sequencing cells at ultra-low depths of less than 10,000 reads per cell (Fig 2f-g, Supplementary Fig. 4). Similarly, low-coverage sequencing provided accurate estimates of the contribution of genes to the loading of PC1 and PC2 (Fig. 2h–i), but required at least 50,000 reads per cell (Fig. 2j–k). Thus, even at ultra-low depths where the levels of individual genes are difficult to estimate, the combination of abundant genes that vary across cells permits classification of cells.
To explore whether low-coverage single-cell mRNA Seq is sufficient to distinguish closely-related cell types from heterogeneous populations, we further analyzed single cells derived from developing human cortex during phases of neurogenesis. Excitatory neurons of the cerebral cortex are born from radial glia, which reside in the germinal zones of the dorsal telencephalon: the ventricular zone and the subventricular zone9. Before reaching their terminal positions in the cortical plate, newborn neurons migrate through the germinal and intermediate zones. Defects in specification and migration of newborn neurons underlie the pathogenesis of many neurodevelopmental disorders10, but studying these transient populations of cells in heterogeneous tissue has been challenging. We collected single cells from the germinal zone of gestational week 16 (GW16) human fetal cortex aiming to capture radial glia and newly-generated cortical neurons. To analyze cell diversity in the context of neural differentiation, we also collected primary cells from GW21 cortex, and further cultured a subset of these cells for three weeks (GW21+3). Similarly, to represent more immature neuroepithelial cells, we collected neural progenitors derived from pluripotent stem cells (Fig. 3a).
Figure 3.
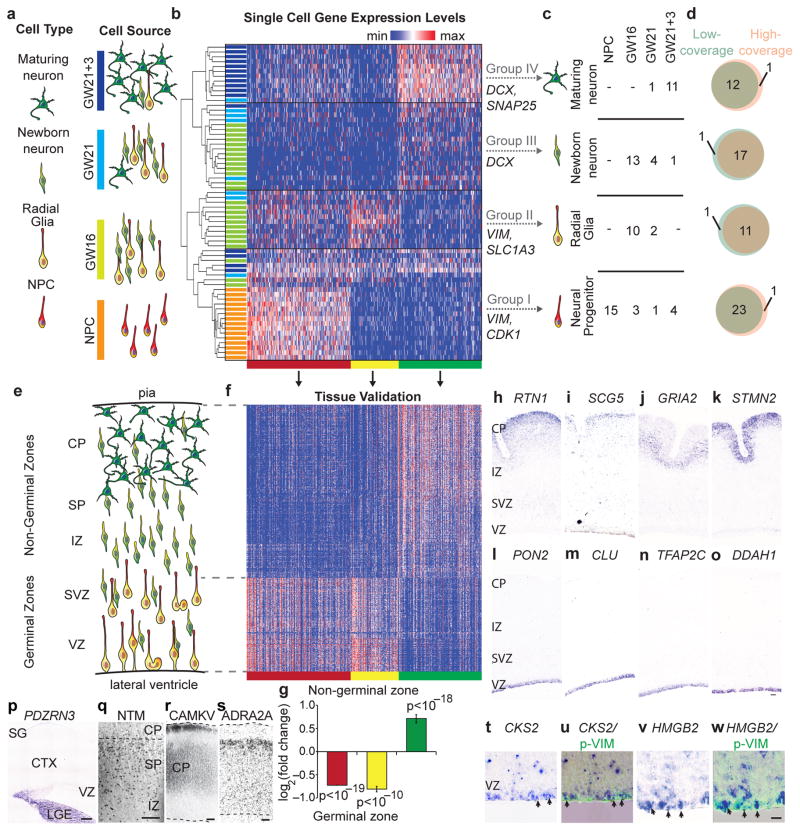
Low-coverage single-cell mRNA sequencing distinguishes diverse neural cell types and identifies biomarkers in heterogeneous tissue. (a) Schematic of cell types and sources selected to represent stages of neuronal differentiation. Cultured neural progenitors represent early undifferentiated stages, while primary cortical samples are expected to contain radial glia, newborn, and maturing neurons. (b) Hierarchical clustering of 65 single cells across 500 genes with the strongest PC1-3 loading scores identifies four major groups of cells (I–IV) and k-means clustering identifies three clusters of genes (red, yellow, green). (c) Major groups can be interpreted based on the expression of known genes. Table shows the number of cells of specific types captured from each source. (d) Cell classification based on low-coverage data largely overlaps with classification based on high-coverage data. (e) Schematic of the distribution of cell types in developing cortex at mid-gestation. (f) Heatmap of gene expression values for PCA genes (columns) in 599 regions of the developing cortex11 (rows). (g) Genes belonging to the red cluster (n = 218) and yellow cluster (n = 98) are enriched in the ventricular (VZ) and subventricular zones (SVZ), while genes belonging to the green cluster (n = 176) are enriched in the intermediate zone (IZ), subplate (SP), and cortical plate (CP); p values were calculated using Wilcoxon signed-rank test. (h–o) In situ hybridization for representative genes belonging to the neuronal (green) cluster including RTN1 (h), SCG5 (i), GRIA2 (j), STMN2 (k), and genes belonging to the radial glia (yellow) cluster including PON2 (l), CLU (m), TFAP2C (n), DDAH1 (o), in GW 14.5 human cortical sections. (p–s) Distinct expression patterns were observed for candidate novel markers of subgroups. (p) In situ hybridization for the candidate immature inhibitory neuron marker PDZRN3 in GW16.5 human cortex (CTX). (q) Immunostaining for the candidate newborn neuron marker NTM in IZ, SP, and CP. (r–s) Immunostaining for markers distinguishing maturing neuronal subgroups CAMKV (r) and ADRA2A (s) in the CP of GW24.5 human cortex. Abbreviations SG - subpial granular layer, LGE – lateral ganglionic eminence. (t–w) In situ hybridization for candidate cell division markers in the progenitor gene cluster (red) showing CKS2 (t) and HMGB2 (v) expression in radial glia undergoing mitosis at the edge of the ventricular surface revealed by immunoreactivity for the phosphorylated (ser82) Vimentin (u, w).
Variation across cells derived from these four neural sources was analyzed using PCA (Supplementary Figs. 7, 8). Hierarchical clustering of cells based on the 500 genes explaining the most variation in PC1, PC2 and PC3 separated the cells into four broad groups with cells from each source contributing to multiple groups (Fig. 3b–c). Group memberships of individual cells largely overlapped between high- and low-coverage sequencing data (Fig. 3d), and could be identified at downsampled depths between 5,000 and 50,000 reads per cell (Supplementary Figs. 9, 10). Cells in groups I and II expressed high levels of neuroepithelial markers, including VIM, SOX2, and PAX6. In addition, cells in group I also expressed high levels of proliferative markers CDK1 and ASPM, while cells in group II expressed high levels of mature radial glial markers SLC1A3 and HES1. In contrast, cells in groups III–IV expressed the pan-neuronal marker DCX, and cells in group IV also expressed many markers of neuronal maturation, including MEF2C, SATB2 and SNAP25 (Supplementary Fig. 10–12). Thus, we interpreted the groups to represent dividing neural progenitors (group I), radial glia (group II), newborn neurons (group III), and maturing neurons (group IV). To independently validate our results, we examined the expression of genes distinguishing each group across 599 tissue samples collected from distinct regions of the developing human cortex11. Genes defining neural progenitors and radial glia in single cell analysis were strongly enriched in the germinal zones, while genes defining newborn and maturing neurons were strongly enriched outside of germinal zones (Fig. 3e–g, Supplementary Table 5). Similarly, in situ hybridization confirmed that novel markers of radial glia, newborn and maturing neurons are expressed in zones where these cell types are abundant (Fig. 3h–o, Supplementary Fig. 13).
In addition to the four broad groups identified using hierarchical clustering, distinct subgroups corresponded to other known and potentially novel cell types (Supplementary Fig. 10–11). For example, cells in group Ib expressed multiple markers of intermediate neural progenitors3. Cells in group IIIb expressed canonical markers of inhibitory interneurons GAD1 and DLX genes, as well as novel markers such as PDZRN3 (Fig. 3p), while the remaining cells in group III expressed proneural genes NEUROD1 and NEUROD6. In addition, group III cells expressed UNC5D, a gene transiently up-regulated in newly-generated mouse excitatory neurons required for the earliest phases of migration12, and other genes such as ROBO2 and NTM (Fig. 3q), whose possible roles in newborn cortical neurons remain to be investigated. Group IV could be further divided into maturing neurons expressing high levels of CAMKV and cells expressing high levels of ADRA2A (Supplementary Fig. 11). Complementary expression patterns of CAMKV and ADRA2A proteins in the cortical plate (Fig 3r–s) indicate these finer subgroups may reflect additional heterogeneity within maturing cortical neurons. Although many of the genes explaining variation across single cells related to cell identity, a subset of genes with strong PCA loading were enriched for mitotic markers and have not been studied in radial glia development (Supplementary Fig. 13). Candidate mitotic markers CKS2 and HMGB2 were detected specifically in a subset of human radial glia undergoing cell division at the edge of the lateral ventricle (Fig. 3t–w). Thus, low-coverage sequencing of single cells collected from primary tissue can be used to identify cell types, states, and candidate biomarkers.
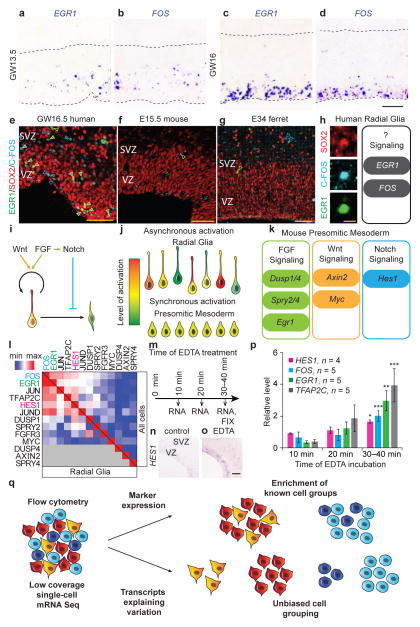
Transcription of immediate early genes has been extensively studied in activated neurons13–15, but the strong PCA loading scores of EGR1 and FOS suggested their expression may also reflect important aspects of cellular diversity in the developing cortex (Supplementary Fig. 13, Supplementary Table 5). Indeed, in situ hybridization revealed mosaic expression of EGR1 and FOS in the ventricular zone (Fig. 4a–d). The levels of EGR1 and FOS were highly correlated across single radial glial cells, and the proteins were co-expressed in a subset of radial glia, suggesting these genes could be transcribed in response to the same signaling pathway (Fig. 4e,i, Supplementary Fig. 14). Multiple signaling pathways including FGF, Notch and Wnt, orchestrate radial glia development, but asynchronous activation of these signaling pathways in neighboring cells makes identifying downstream effector genes challenging16,17. Coordinated patterns of pathway activation in other tissues have facilitated identification of candidate downstream effector genes, but these target genes often depend on cellular context and vary across species16,18.
Figure 4.
EGR1 and FOS are candidate targets of Notch signaling in human radial glia identified using low-coverage single-cell mRNA Seq. (a–b) In situ hybridization images of human VZ at GW13.5 showing cells sparsely labeled for EGR1 (a) and FOS (b). (c–d) At GW16, pronounced mosaic expression of EGR1 (c) and FOS (d) was detected in the apical portion of VZ. Dashed lines indicate apical and basal edges of the VZ. (e–g) EGR1 and FOS proteins are detected in a subset of SOX2-expressing cells in the human ventricular zone (e), but rarely co-label with SOX2-expressing cells in mouse (f) or ferret (g) at similar developmental stages. Filled arrows: triple labeled cells; yellow arrows: EGR1/SOX2-expressing cells; blue arrows: C-FOS/SOX2-expresing cells; scale bar 50 μm. (h) High magnification example of a SOX2 (red) expressing cell in human VZ that is immunoreactive for C-FOS (cyan) and EGR1 (green), scale bar is 10 μm. Schematic represents hypothesis that EGR1 and FOS expression in vivo in human radial glia could be elicited in response to activated signaling pathways. (i) Schematic showing the key developmental signaling pathways regulating radial glia development. (j) Asynchronous activation of signaling pathways makes identification of downstream target genes challenging in heterogeneous tissue. (k) Schematic showing key candidate effector genes of FGF, Wnt and Notch signaling in mouse presomitic mesoderm. (l) Heatmap shows correlation coefficients between mRNA levels for EGR1, FOS, other immediate early genes, and canonical effectors of FGF, Notch and Wnt signaling pathway across all 65 neural cells (above diagonal) and within radial glia (below diagonal). (m) Schematic showing experimental design for stimulating Notch signaling in organotypic slice cultures of human fetal cortex using EDTA. (n-o) In situ hybridization for HES1 in control (n) and experimental (o) slices. (p) Quantification of mRNA levels of HES1, FOS, EGR1 and TFAP2C (n = 4–5 independent samples, 2–3 slices per condition). All qRT-PCR results represent average ± s.e.m calculated using −ΔΔCt method, p values were calculated using paired two-tailed Student t-test, * p < 0.05, ** p < 0.01, *** p < 0.001. (q) Low-coverage mRNA Seq of single cells permits in silico sorting of cells based on cell type or state. Flow cytometry uses established staining characteristics to enrich for known cell types in heterogeneous samples. In contrast, low-coverage single-cell mRNA Seq identifies the major genes explaining variation across single cells allowing for unbiased discovery and further analysis of distinct cell populations and states.
To determine which signaling pathway might be responsible for the coordinated activation of immediate early genes in human radial glia, we examined the correlation of EGR1 and FOS mRNA levels with the levels of canonical signaling pathway effector genes established by studies of other developmental processes. Across single cells, EGR1 and FOS mRNA correlated more strongly with the Notch effector HES1 than with FGF effectors DUSP1/4, SPRY2/4 or WNT effectors AXIN2 or MYC (Fig. 4l). To examine if activation of Notch signaling induces changes in EGR1 and FOS expression, we activated Notch signaling in cultured human cortical slices by removing extracellular calcium19. Incubation of primary human cortical slices with EDTA induced a rapid (30–40 minutes) increase in the levels of HES1 as well as EGR1, FOS, and another highly-correlated gene, TFAP2C (Fig. 4m–p). In other stem cell contexts, EGR1 and FOS play a role in quiescence and retention in the stem cell niche20–22, but the role of these genes as candidate Notch targets in radial glia remains to be examined. Surprisingly, EGR1 and C-FOS were rarely detected in mouse or ferret radial glia (Fig 4e–g, Supplementary Fig. 14), indicating that these factors could contribute to differences in radial glia development across species, which include a dramatically longer G1 phase and increased proliferative capacity in human radial glia23. Together, our findings suggest that low-coverage single-cell analysis can be more generally applied to identify cells in different states of signaling pathway activation and candidate downstream target genes.
Identifying gene expression profiles of cells of the same type or state has numerous applications in modern biology. Here we demonstrate that ultra low-coverage sequencing of single cells (<10,000 reads) is sufficient for unbiased classification of diverse cell types in heterogeneous tissue, but that finer distinctions within categories and resolution of the set of genes explaining variation require moderately higher depths (~50,000 reads). Increased sequencing coverage beyond these depths likely provides diminishing returns because single cells contain a limited number of transcripts (~300,000) and the amplification steps used to generate sequencing libraries sample a subset (~40%) of the transcriptome24–26. Using our shallow sequencing strategy, we identified numerous cell type-specific biomarkers across a range of cell types in midgestation human cortex, including radial glia in different stages of cell cycle progression and signaling pathway activation, and newly-generated neurons in the earliest phases of migration. Few specific markers exist for purification of these distinct cell states and transient developmental intermediates using flow cytometry. In contrast to flow cytometry, low-coverage single-cell sequencing detects thousands of abundant transcripts that can be analyzed to group cells according to cell type or state (Fig. 4q). Although the observed level of a given transcript in a single cell can vary due to transcriptional bursts and technical noise associated with low quantities of input RNA8,27,28, the simultaneous profiling of multiple differentially expressed transcripts enables unbiased discovery of cell groups based on shared signatures of gene expression29–31. By grouping cells of a given identity, the bulk transcriptome for that population may then be accurately reconstructed6. We anticipate that the unbiased classification of cells by efficient low-coverage single-cell sequencing will be applied to large-scale surveys of primary tissue samples to identify cell-type-specific biomarkers, compare gene expression in cells of a given type across samples, and reconstruct developmental lineages of related cell types.
Online Methods
Origin of cell lines and tissue samples
Human induced pluripotent stem cells (hiPSCs) were originally derived from neonatal male human foreskin BJ fibroblasts by Dr. Guangwen Wang at the Department of Genetics at Stanford University, using Sendai virus from Life Technologies (Cat # A16517). Cultured undifferentiated hiPSCs were maintained in Essential 8™ Medium (Life Technologies). StainAlive™ Tra-1-60 Antibody (DyLight™ 488) staining (Stemgent) was used to confirm undifferentiated state. Following a dissociation with StemPro® Accutase® Cell Dissociation Reagent (Life Technologies), single cells were plated onto Matrigel®-coated plates at 2.5 × 105 cells/cm2. Neural progenitor cell (NPC) differentiation was induced using DMEM/F12 media (GIBCO® Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 from Life Technologies), supplemented with B27 (without Vitamin A, Life Technologies, Cat # 12587010), N2, 0.1 mM non-essential amino acids, 0.5% bovine serum albumin, 1 mM BME, 50 nM LDN-193189 (Stemgent), 5 μM SB431542 (Stemgent), 1 μM Stemolecule™ Cyclopamine (Stemgent). After 12 days in culture, >90% of cells were immunopositive for PAX6.
ATCC® PCS-200-010™ cell (foreskin keratinocytes, abbreviated ‘Kera’) culture was maintained in dermal cell basal medium ATCC® PCS-200-030™ supplemented with the keratinocyte growth kit (ATCC® PCS-200-040™). ATCC® CRL-2338™ cells (derived from a primary stage IIA, grade 3 invasive ductal carcinoma with no lymph node metastases, abbreviated ‘CRL-2338’) were cultured in complete growth medium RPMI-1640 (ATCC® 30-2001™) supplemented with 10% Fetal Bovine Serum (FBS, GIBCO® 16000-077™). ATCC® CRL-2339™ cells (Epstein-Barr virus transformed B lymphoblasts, abbreviated ‘CRL-2339’) were cultured in growth medium RPMI-1640 medium (ATCC® 30-2001™) supplemented with 10% FBS. ATCC® CCL-240™ cells (promyeloblastic peripheral blood leukocytes obtained by leukopheresis from a patient with acute promyelocytic leukemia, abbreviated ‘HL60’) were cultured in Iscove’s Modified Dulbecco’s Media (IMDM) (ATCC® 30-2005™) supplemented with 20% FBS. ATCC® CCL-243™ cells (lymphoblastic cells isolated from the pleural effusion of a patient with chronic myelogenous leukemia in terminal blast crises, abbreviated ‘K562’) were cultured in IMDM (ATCC® 30-2005™) media supplemented with 10% FBS (Life Technologies, Cat # 16000-077). Stemgent® BJ human fibroblasts were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FBS. All cultures were passaged using 0.05% Trypsin supplemented with 0.02% EDTA or using 1X TrypLE™ Select (Life Technologies). For systems verification tests of capture efficiency, primary cells were obtained from splenocytes (AllCells, LLC PB003F) and peripheral blood mononuclear cells (AllCells, LLC PB003F)
De-identified fetal cortical tissue samples were collected from elective pregnancy termination specimens at San Francisco General Hospital. Tissue was collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Human Gamete, Embryo and Stem Cell Research Committee (institutional review board) at the University of California, San Francisco. Neocortical tissue sample spanning the thickness of the cortical wall was embedded in 3.5% low melting point agarose (Fisher) and sectioned using a Leica VT1200S vibrating blade microtome to 300 μm slices in Artificial Cerebrospinal Fluid (ACSF) containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1.25 mM NaH2PO4. The germinal region of the GW16 neocortex was microdissected using a microsurgical blade. The samples were centrifuged for 5 minutes at 300g and residual ACSF was replaced with a pre-warmed working solution of Papain/ freshly diluted in Earl’s Balanced Salt Solution according to manufacturer’s instructions (Worthington Biochem. Corp.). The samples were incubated at 37 °C for 20–30 minutes and centrifuged for 5 minutes at 300g. After removing the Papain/DNaseI supernatant, tissue was resuspended in 1 mL of sterile Dulbecco’s Phosphate Buffered Saline (DPBS) containing 3% FBS (Sigma) and manually triturated by pipetting up and down approximately 10 times. The suspension was passed through a 40 μm strainer cap (BD Falcon) to yield a uniform single cell suspension. Cells collected from primary GW21 cortex (ScienCell™, Cat. No. 1520, Lot No. 9298) were thawed and either mixed directly with C1™ Cell Suspension Reagent for cell loading, or cultured in 6-well plate pre-coated with poly-L-Ornithine (Sigma) and Laminin (Sigma) at 10 μg/mL. Complete neuronal medium (ScienCell™, Cat. No. 1521) was replaced every other day for 19 days.
Cell loading, mRNA Seq library preparation and sequencing
Adherent cultures were dissociated using 0.05% Trypsin supplemented with 0.02% EDTA or using TrypLE™ Select (Life Technologies). Following centrifugation and removal of the dissociation medium, cells were resuspended at a concentration of 150–500 cells/μL. This cell suspension was mixed with C1™ Cell Suspension Reagent (Fluidigm, Cat # 634833) at the recommended ratio of 3:2 immediately before loading 5 μL of this final mix on the C1™ IFC along with 20 μL of freshly prepared staining buffer (2.5 μL ethidium homodimer-1 and 0.625 μL Calcein AM from Life Technology’s LIVE/DEAD® Viability/Cytotoxicity Kit added to 1.25 mL C1™Cell Wash Buffer) in their respective input wells. Images of captured cells were collected Leica DMI 4000B microscope in the brightfield, GFP, and CY3 channels using the Surveyor V7.0.0.9 MT software (Objective Imaging).
Single-cell RNA extraction and mRNA amplification were performed on the C1™ Single-Cell Auto Prep Integrated Fluidic Circuit (IFC) following the methods described in the protocol (PN 100–7168, http://www.fluidigm.com/). For experiments where exogenous spike-in controls were used, the spikes were added to the lysis mix at a 20,000-fold dilution. The PCR thermal protocol was adapted from a recent publication that optimized template-switching chemistry for single-cell mRNA Seq32 and is outlined in the C1™ Single-Cell Auto Prep System protocol. For the population control experiment, we used reagent formulations and workflows exactly as described in the SMARTer® Ultra Low RNA Kit user manual (Cat# 634833,1 kit for 10 C1™ IFCs), except that the thermal protocol followed the recommendations outlined in the C1™ Single-Cell Auto Prep System user guide (PN 100–7168).
For the population control experiment, we sorted 100 K562 cells into 3.5 μL of Clontech Reaction Buffer containing exogenous spike-in controls using a BD FACSAria™ III. The 20,000-fold diluted ERCC spike-in controls were further diluted (9:3500) in Clontech Reaction Buffer such that an equal mass (rather than an equal concentration) of the spikes was included in the population control reaction. Following the sort, cells were frozen at −80 °C overnight before continuing the SMARTer® Ultra Low RNA Kit protocol according to manufacturer’s recommendations.
The cDNA reaction products were quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies) and high sensitivity DNA chips (Agilent) and were then diluted to a final concentration of 0.15–0.30 ng/μL using C1™ Harvest Reagent. The diluted cDNA reaction products were then converted into mRNA Seq libraries using the Nextera® XT DNA Sample Preparation Kit (Illumina, FC-131-1096 and FC-131-1002, 1 kit used for 4 C1™ IFCs/384 samples) following manufacturer’s instructions, with minor modifications. Specifically, reactions were run at one quarter of the recommended volume, the tagmentation step was extended to 10 minutes, and the extension time during the PCR step was increased from 30 seconds to 60 seconds. After the PCR step, samples were pooled, cleaned twice with 0.9X Agencourt AMPure XP SPRI beads (Beckman Coulter), eluted in TE buffer and quantified using a high sensitivity DNA chip (Agilent). For high-coverage sequencing, libraries from a subset of captured cells from each source were pooled to reach a target of ten million aligned reads per cell.
Processing mRNA sequencing data
An index for RNA-Seq by Expectation-Maximization (RSEM) was generated based on the hg19 RefSeq transcriptome downloaded from the UCSC Genome Browser database33 (23,637 total genes). Read data was aligned directly to this index using RSEM/bowtie29, 34. FASTQ files from high-coverage sequencing data were downsampled to 11 seq-depths: 100, 500, 1,000, 5,000, 10,000, 50,000, 100,000, 150,000, 200,000, 250,000 and 300,000 reads using a Python script to randomly select reads, and downsampled results were also aligned to the same index using RSEM/bowtie. Quantification of gene expression levels in transcripts per million (TPM) for all genes in all samples was performed using RSEM v1.2.429. Genomic mappings were performed with TopHat v2.0.435, and the resulting alignments were used to calculate genomic mapping percentages. Raw sequencing read data was directly aligned to human rRNA sequences NR_003287.1 (28s), NR_003286.1 (18S), and NR_003285.2 (5.8S) via bowtie v2.0, and the percent of reads aligned to rRNA was then calculated as reads aligned to these sequences divided by total reads. Linear expression data was imported into the Fluidigm® SINGuLAR™ Analysis Toolset 2.0 (R-scripts and user guide can be found at http://www.fluidigm.com) and converted into log-space. Transcripts with TPM values less than one were dropped from further analysis prior to log transformation. To identify outlier cells from each chip, a set of genes detected in at least half of the samples was considered, and samples with median expression values below the 15th percentile for these genes were removed using the identify Outliers function using the SINGuLAR™ package. No additional normalization was performed between individual samples. Sequencing results obtained from capture sites with no detectable Calcein AM staining that were not flagged as sequencing outliers were retained in the dataset. Capture sites containing multiple live cells based on Calcein AM staining or brightfield microscopy were removed from further analysis. To assess technical variation during library preparation, cDNA from a single cell (GW21+3_1) was split and two independent libraries were prepared with the Nextera® XT DNA Sample Preparation Kit (Illumina). The correlation between log2 TPM expression values for technical replicates (0.993) was greater than that between any pair of distinct cells.
Principal Components Analysis and Clustering
PCA was performed in the Fluidigm® SINGuLAR™ Analysis Toolset 2.0 R package, which calls the princomp R package (http://stat.ethz.ch/R-manual/R-patched/library/stats/html/princomp.html). The 500 top-ranked PCA genes were selected based on the maximum absolute value of each gene loading score in the first three eigenvectors (PC1, PC2 and PC3). To compare sample scores between downsampled low-coverage datasets with high-coverage mRNA Seq datasets (Fig. 2, Supplementary Fig. 9), the eigenvectors derived from the high-coverage data were applied to the low-coverage data using the applyPCA function in the SINGuLAR™ package. Hierarchical clustering of the top 500 PCA genes across 301 cells was also performed in the Fluidigm® SINGuLAR™ package. Genes are clustered based on the Pearson correlation. Samples are clustered based on a Euclidian distance matrix with complete linkage. Significance of cluster assignment in Supplementary Figure 9 and 10 was tested using “Pvclust”36, which employs a multiple bootstrap resampling algorithm to calculate the approximately unbiased (AU) probability values for cluster distinctions. We performed the clustering for 50,000 bootstraps.
Comparison of low- and high-coverage gene expression data
Pearson’s correlation coefficients were calculated using the log-transformed TPM values for genes shared in both the low- and high-coverage datasets (TPMlow>1 and TPMhigh>1), and also separately for all genes in Supplementary Figure 2. In addition, gene transcripts were binned, based on the high-coverage data, into low expression (1<TPMhigh<10), medium expression (10≤TPMhigh≤100), and high expression (100<TPMhigh) bins and Pearson’s correlation coefficients were again calculated for each of these subsets. We assessed the number of dropouts (TPM<1) that were excluded from the correlation analysis by counting the number of genes that were detected only in the low-coverage data (1<TPMlow and TPMhigh≤1) and the number of genes that were detected only in the high-coverage data (1 < TPMhigh and TPMlow≤1).
Validation using K562 cell and population mRNA Seq data including spikes
An additional validation dataset was generated using K562 cells with exogenous spike-in controls (Life Technologies, Cat # 4456740) delivered in the lysis reagent at a 20,000-fold dilution as described above. A total of 46 captured single cells, one empty reaction line, and one population sample (100 K562 cells sorted into a standard SMARTer™ Ultra Low RNA Kit reaction) were sequenced at both low- and high-coverage for this validation dataset. The sequences for the 92 External RNA Controls Consortium (ERCC) spike-in controls were added to our RSEM index, and RSEM v1.2.429 was used to quantify gene expression levels in units of TPM. The average expression level, coefficient of variation, and detection frequency (based on a limit of detection of TPM>1) was calculated for each of the 92 spike-in controls across the 46 single-cell capture events and plotted against known inputs (copies per reaction) of each spike-in control (Fig. 1d–e). In addition, these cells were used to determine the correlation between aggregated single-cell and population data as a function of the number of single-cell datasets included in the ensemble. For a range of ensemble sizes shown along the horizontal axis in Fig. 2a, we randomly selected 10 ensembles, measured the Pearson’s correlation of each ensemble with the population, and then averaged the Pearson’s correlation across the 10 ensembles. These 46 average correlation values between various cell groupings and the overall population were then plotted as a function of the number of cells included in the ensembles (Fig. 2b).
Analysis of PCA genes and candidate biomarkers in neural cells
To examine the number of distinct gene clusters among the top 500 PCA genes explaining variation across the neural cells, we performed consensus clustering using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). Heatmaps were visually inspected to identify the optimal number of gene clusters. Based on these results, K-means clustering with three clusters was performed with a Euclidean distance matrix (2000 iterations) and 20 resampling iterations. To identify candidate cell-type-specific biomarkers, we examined the Pearson correlation between each gene with that of an idealized gene with binary expression in only one group of cells as determined by grouping relationships in hierarchical clustering.
Gene expression data for the top 500 PCA genes included in Fig. 3f–g were obtained from BrainSpan11 across all cortical samples for all probes from post-conception week 15 (GW17), post-conception week 16 (GW18), and two post-conception week 21 (GW23) samples. For genes containing multiple probes, the log-transformed gene expression values were averaged. For each gene, expression values across 211 cortical germinal zone samples (ventricular zone and subventricular zone regions) and 388 non-germinal zone samples (intermediate zone, subplate and cortical plate regions) were displayed on a heatmap in Fig. 3f using GENE-E, maintaining the order of genes in Fig. 3b. To evaluate the distribution of genes in the red, yellow, and green gene clusters, we performed a Wilcoxon signed-rank test comparing each gene between the averaged germinal zone and non-germinal zone samples. 8/500 genes were not represented by microarray probes in the BrainSpan11 dataset: HEPN1, SNURF, ZNF286B, LOC100507246, MPC1, ATRAID, TECR, and FRMD6-AS1. The same approach was used to examine the expression of candidate cell type-specific biomarkers in Supplementary Fig. 11, but samples from marginal zone and subpial granular layer were also included in the analysis, and results across samples were further averaged for distinct laminae.
Immunohistochemistry and in situ hybridization
Timed-pregnant Swiss Webster mice were obtained from Simonsen Laboratories and maintained according to protocols approved by the UCSF Institutional Animal Care and Use Committee. Pregnant dams were deeply anesthesized with inhaled isoflurane and euthanized by cervical dislocation and two litters were collected. Embryos were decapitated and dissected brains were fixed in 4% paraformaldehyde overnight. Timed-pregnant ferret (Marshall BioResources) was maintained according to protocols approved by the UCSF Institutional Animal Care and Use Committee. E35 pregnant dam was deeply anesthetized with ketamine prior to the administration of inhaled isoflurane. Ovariohysterectomy for fetus collection was performed for embryo collection. Embryos were perfused transcardially with cold phosphate buffered saline and 4% paraformaldehyde. Dissected brains were fixed in 4% paraformaldehyde overnight.
For immunohistochemistry and in situ hybridization, human fetal cortical samples were fixed overnight in 4% paraformaldehyde, cryoprotected in 30% sucrose and embedded in a 1:1 mixture of 30% sucrose and optimal cutting temperature (Thermo Scientific). Thin 20 μm cryosections were collected on superfrost slides (VWR) using Leica CM3050S cryostat. For immunohistochemistry, heat-induced antigen retrieval was performed in 10 mM sodium citrate buffer, pH 6. For antibodies against CAMKV, NTM and SYNC we did not perform antigen retrieval. Primary antibodies against ADRA2A (1:100, Thermo Scientific PA1-048), CAMKV (1:100, Novus Biologicals NBP1-68097), EGR1 (1:50, Cell Signaling 41535), C-FOS (1:100, Santa Cruz SC-8047), CTIP2 (1:500, Abcam ab18465), NTM (1:100, R&D Systems AF1235), phosphorylated Vimentin ser82 (1:500, MBL International D095-3), SATB2 (1:250, Santa Cruz SC81376), SOX2 (1:200, Santa Cruz SC17320), SYNC, isoform 1 (1:100, a kind gift from Kay Davis, University of Oxford) were diluted in blocking buffer containing 10% Donkey Serum, 0.5 % Triton™-X100 and 0.2% gelatin. Binding was revealed using an appropriate Alexa Fluor™ 488 (A21206), Alexa Fluor™ 546 (A11056), and Alexa Fluor™ 647 (A31471) fluorophore-conjugated secondary antibody (Life Technologies). Cell nuclei were counter-stained using DAPI (Life Technologies). Images were collected using a Leica TCS SP5 X Confocal microscope and processed using ImageJ or Imaris (Bitplane).
Probes complementary to target human mRNA used for RNA in situ hybridization were generated specifically for this study except for EMX2 which was generated against mouse sequence and generously provided by Antonio Simeone (Institute of Genetics and Biophysics, Adriano Buzzati-Traverso). To generate RNA in situ probes, total RNA was extracted from primary human fetal cortical samples age GW14–21 using the RNeasy RNA extraction kit (Qiagen) and reverse transcribed with Superscript III First Strand Synthesis System with random hexamers (Life Technologies). Primers specific to target genes of interest were designed using Primer3 and amplified by PCR using Phusion proofreading DNA polymerase (Thermo Scientific). Specific genes were amplified using the following primers: ANXA2: forward primer – CCA GGA GCT GCA GGA AAT TA, reverse primer – TGT TAG CTG GAA GCA TGG TG (it should be noted that target ANXA2 mRNA sequence is indistinguishable from a related retrotransposed pseudogene, ANXAP2, and our probe would not distinguish between transcripts from these loci); C1ORF61: forward primer – TCC AAG AAG AAG CAG CCT CA, reverse primer – CAG GTA CAG TGG GCT TCC TG; CKS2: forward primer – GCG CTC TCG TTT CAT TTT CT, reverse primer – GCA CTT AAG AGA AAA ACT GAC TGG; CLU: forward primer – CGG AGG CCT CAC TTC TTC TT, reverse primer – GTA TTC CTG CAG CGC TTT CT; DDAH1: forward primer – CCC CTA AGC CTC CCG AAG, reverse primer – TAG CGG TGG TCA CTC ATC TG; EGR1: forward primer – CTG CAC GCT TCT CAG TGT TC, reverse primer – CAT GTC CCT CAC AAT TGC AC; FOS: forward primer – AGC AGT GAC CGT GCT CCT AC, reverse primer – CAG GAA CCC TCT AGG GAA GA; GRIA2: forward primer – TGT TTT ATT GCA AGT GGT CCA A, reverse primer – ATC CAC ACT GGG CAT ATT AAA; HES1: forward primer – TTT AGC ACT CCT TCC CGT TG, reverse primer – AAA CAC CTT AGC CGC CTC TC; HMGB2: forward primer – GCC ATT TTT CAA ACC CTC TTC, reverse primer – CAC CTT TGG GAG GAA CGT AA; NNAT: forward primer – TTT CTC GAC CAC CCA CCT AC, reverse primer – AGG AGC ACC TGA TGA TAC GG; PDZRN3: forward primer – AGC AAC GAG TCT TTC ATT TCG, reverse primer – GCT CTC CGC TCT TTG CTT T; PON2: forward primer – CCG AAG GTA TCT GGG GAA AT, reverse primer – TTG ATC CCA TTT GCT GAA TC; RTN1: forward primer – CCC CTC CCT CCA GTA CCA TA, reverse primer – TGA ATC CAT TAG GAA CTA CAG AGA AA; SCG5: forward primer – GGT ACC CAG ACC CTC CAA AT, reverse primer – CCA AGG GCT GGA TGA ACT AC; SPARC: forward primer – CTT CAG ACT GCC CGG AGA, reverse primer – CAG GCG CTT CTC ATT CTC AT; SRGAP3: forward primer – CCG AGA AGA TGT TCC CCA AC, reverse primer – CGC AGT TAC TAT GGG CCT TT; STMN2: forward primer – AAT GGA TCA TGC GAT ATC AGG, reverse primer – GCC AAA GCA CAT TTG TAG CA; TAGLN3: forward primer – GGG CTT GAT TGA CAC AGG AG, reverse primer – GAA CTG GGA GAT TTG CTC CA; TFAP2C: forward primer – GAC CCC TAC TCG CAT CTG G, reverse primer – AGA GTC ACA TGA GCG GCT TT; TTYH1: forward primer – GGC AAC AGT GAG ACC AGT GA, reverse primer – AAC TGA GGC ACA GCT TCT CG. PCR products of predicted band size were gel extracted and A-tailed using GoTaq® DNA Polymerase (Promega) and ligated into the pGEM®T-Easy Vector System (Promega). Ligation products were transfected into One Shot TOP10 Chemically Competent E.coli (Life Technologies). Cloned probe sequences were confirmed by sequencing. Digoxigenin labeled RNA probes for in situ hybridization were generated by linearizing the pGEM®T-Easy Vector and in vitro transcribing the probe using T7 or SP6 RNA Polymerase (Roche) in the presence of DIG-RNA Labeling Mix (Roche). In situ hybridization was performed blinded to the sense/antisense status for each probe and sense control probes gave no signal (data not shown). The in situ hybridization protocol has been described before37. For subsequent immunolabelling, slides were subjected to antigen retrieval as described above. Images were collected with a Leica DMI 4000B microscope using a Leica DFC295 camera.
Organotypic slice cultures
Human fetal cortical slices were collected as described above. Slices were transferred into slice culture inserts (Millicell) in 6-well culture plates (Corning) and cultured in culture medium containing 66% BME, 25% Hanks, 5% FBS, 1% N-2, 1% penicillin/streptomycin, glutamine (Life Technologies). Slices were cultured in a 37 °C incubator at 5% CO2, 8% O2 for one day. To induce Notch signaling, culture medium was completely replaced with Ca2+-free ACSF containing 126 mM NaCl, 3 mM KCl, 1.2 mM NaH2PO4, 26 mM NaHCO3, 2 mM MgCl2, 1 mM EDTA and 10 mM D-glucose. Control slices were incubated in parallel with ACSF containing 126 mM NaCl, 3 mM KCl, 1.2 mM NaH2PO4, 26 mM NaHCO3, 1.3 mM MgCl2, 2.4 mM CaCl2 and 10 mM D-glucose. All slice cultures were placed in a 37 °C incubator at 5% CO2, 8% O2 for the duration of the treatment. After 10 minutes, 20 minutes, and 30–40 minutes incubation the slices were either frozen on dry ice and stored at −80 °C in RNase-free tubes or fixed with 4% paraformaldehyde for 20 minutes at 4 °C and processed for cryosectioning as described above.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
RNA extraction and cDNA synthesis were performed as described above and qRT-PCR was performed using the QuanTitect SYBR® Green PCR Mix (Qiagen) in a Roche LightCycler 480 II. The following primer pairs were used in this study to detect specific mRNAs, blinded to the treatment status of each sample: GAPDH: forward primer – GAG TCA ACG GAT TTG GTC GT, reverse primer – TTG ATT TTG GAG GGA TCT CG; ACTB: forward primer – GGA CTT CGA GCA AGA GAT GG, reverse primer – AGC ACT GTT GGC GTA CAG; HPGK: forward primer –CTG TGG GGG TAT TTG AAT GG, reverse primer – CTT CCA GGA GCT CCA AAC TG; TFAP2C: forward primer – TCA GTC CCT GGA AGA TTG TCG, reverse primer: - CCA GTA ACG AGG CAT TTA AGC A; EGR1: forward primer – ACC CCT CTG TCT ACT ATT AAG GC, reverse primer – TGG GAC TGG TAG CTG GTA TTG. Quantification and comparisons of gene expression levels were performed using the −ΔΔCt method and statistical analysis of differences between control and EDTA- treated samples was performed using paired two-tailed Student t-test.
Supplementary Material
Acknowledgments
We thank Marina Bershteyn (UCSF), Elizabeth Di Lullo (UCSF), Caitlyn Gertz (UCSF), Cory McLean (23andMe), Florence Ng (Fluidigm Corporation), John Liu (UCSF), Craig Lowe (Stanford University), Michael Oldham (UCSF), Aaron Diaz (UCSF), and Hanna Retallack (UCSF) for reading the manuscript and for useful suggestions, Shaohui Wang and Yingying Wang for technical assistance, Kay Davies (University of Oxford) for providing the syncoilin antibody, Shuyuan Yao and Sherman Ku (Allen Institute for Brain Research) for assistance in the neural progenitor induction protocol, Guangwen Wang (Stanford University) for providing hiPS cells, and the staff at the San Francisco General Hospital for providing access to donated fetal tissue. LS, NL, and MAU are partially supported by CIRM Tools and Technologies II grant RT2-02052 to MAU, and the generation and maintenance of the hiPS and neural progenitor cells was also supported by RT2-02052 to MAU. A.A.P. is supported by a Damon Runyon Cancer Research Foundation postdoctoral fellowship (DRG-2166-13). This research was also supported by NINDS awards R01NS075998 and R01NS072630 to ARK.
Footnotes
Competing Financial Interests
A.A.L., A.P.M., B.A., B.C., B.F., B.N.J., D.K., D.T., G.S., J.A.A.W., J.S., J.W., L.K., L.S., M.A.U., M.N., M.T., M.W., N.L., N.R., P.C., R.C.J., R.L., S.W., S.W., and X.W. have a financial interest in Fluidigm Corporation as employees and/or stockholders.
Accessions
Relevant data sets, including raw reads, have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession number SRP041736.
Author Contribution
A.A.P., A.A.L., J.A.A.W., J.S., T.J.N., and X.W. conceived and designed the study. A.A.P., A.A.L., B.A., J.H.L. J.S., J.W., L.K., L.S., N.L., P.C., and T.J.N. performed experiments. A.A.P., A.A.L., J.S., S.W., T.J.N., and X.W. analyzed the data. A.A.L., A.P.M., B.C., B.F., B.N.J., D.K., D.T., G.S., J.S., J.A.A.W., J.W., M.T., M.N., M.W., N.R., P.C., R.L., L.S.W., and X.W. were involved in system development. A.P.M., A.R.K., J.A.A.W., M.A.U, and R.C.J. supervised the project, helped with design and interpretation, and provided laboratory space and financial support. A.A.P., J.S., and T.J.N. wrote the manuscript with input from all authors.
References
- 1.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature reviews Genetics. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi A, et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 4.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014 doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nature methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome research. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 9.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature reviews Neuroscience. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 10.Ross ME, Walsh CA. Human brain malformations and their lessons for neuronal migration. Annual review of neuroscience. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- 11.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyoshi G, Fishell G. Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron. 2012;74:1045–1058. doi: 10.1016/j.neuron.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarcic G, et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieche I, et al. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:6789–6795. doi: 10.1158/1078-0432.CCR-04-0306. [DOI] [PubMed] [Google Scholar]
- 15.Fischer AJ, Scott MA, Ritchey ER, Sherwood P. Mitogen-activated protein kinase-signaling regulates the ability of Muller glia to proliferate and protect retinal neurons against excitotoxicity. Glia. 2009;57:1538–1552. doi: 10.1002/glia.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krol AJ, et al. Evolutionary plasticity of segmentation clock networks. Development. 2011;138:2783–2792. doi: 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Hansson ML, et al. MAML1 acts cooperatively with EGR1 to activate EGR1-regulated promoters: implications for nephrogenesis and the development of renal cancer. PloS one. 2012;7:e46001. doi: 10.1371/journal.pone.0046001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Housden BE, et al. Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes. PLoS genetics. 2013;9:e1003162. doi: 10.1371/journal.pgen.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min IM, et al. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell stem cell. 2008;2:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Okada S, Fukuda T, Inada K, Tokuhisa T. Prolonged expression of c-fos suppresses cell cycle entry of dormant hematopoietic stem cells. Blood. 1999;93:816–825. [PubMed] [Google Scholar]
- 22.Bonnert TP, et al. Molecular characterization of adult mouse subventricular zone progenitor cells during the onset of differentiation. The European journal of neuroscience. 2006;24:661–675. doi: 10.1111/j.1460-9568.2006.04912.x. [DOI] [PubMed] [Google Scholar]
- 23.Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 26.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nature reviews Genetics. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 27.Faddah DA, et al. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell stem cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature methods. 2013 doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 29.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nature biotechnology. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Developmental cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Fan JB, et al. Highly parallel genome-wide expression analysis of single mammalian cells. PloS one. 2012;7:e30794. doi: 10.1371/journal.pone.0030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic acids research. 2011;39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 37.Wallace VA, Raff MC. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development. 1999;126:2901–2909. doi: 10.1242/dev.126.13.2901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.