Abstract
We used high-resolution mass spectrometry to measure the abundance of more than 9,000 proteins in 19 individually dissected colorectal tumors representing lymph node metastatic (n=10) and non-metastatic (n=9) phenotypes. Statistical analysis identified MX1 and several other proteins as overexpressed in lymph node positive tumors. MX1, IGF1-R and IRF2BP1 showed significantly different expression in IHC validation (Wilcoxon test p=0.007 for IGF1-R, p=0.04 for IRF2BP1, and p=0.02 for MX1 at the invasion front) in the validation cohort. Knockout of MX1 by siRNA in cell cultures and wound healing assays provided additional evidence for the involvement of this protein in tumor invasion. The collection of identified and quantified proteins to our knowledge is the largest tumor proteome dataset available at the present. The identified proteins can give insights in the mechanisms of lymphatic metastasis in CRC and may act as prognostic markers and therapeutic targets after further prospective validation.
Keywords: colorectal cancer, drug targets, biomarkers, metastasis, mass spectrometry, MX1
Introduction
Colorectal cancer is one of the major causes of tumor related death in western countries. The prognosis becomes worse and 5-year survival rates decrease down to ~60% when lymphatic metastasis occurs. In recent years post-genomic biology brought about major shift in the way cancer research is performed. It is expected to eventually lead to mechanistic elucidation of the disease and to the development of new approaches for early diagnosis and targeted treatments. The sequencing of human genome and subsequent resequencing of large number of cancer genomes revealed a complex landscape of driver and passenger mutations that affect as many as 80 genes in each individual tumor examined, but with only a handful of less than 15 mutations that occur at statistically significant frequencies1, 2. To make it more complicated, recent studies suggest that epigenetic alterations might be as important as mutations in the aetiology of the disease, and that cancer might be a systemic type of disease that is defined as much by the specifics of the individual organism as by the properties of the primary tumor and its distant metastases. One of the major challenges facing the modern post-genomic cancer biology is the elucidation of the complex regulatory mechanisms that control protein abundance, which very often shows poor correlation with transcript abundances as comparative studies have demonstrated 3. Genetic mutations and epigenetic alterations in cancer cells exert their effect most likely by affecting the abundance and the properties of specific groups of proteins. However the stochastic nature of transcription and the complex mechanisms that regulate protein synthesis, degradation, and stability downstream of transcription, make it very difficult to predict how mutations and epigenetic changes would affect the abundance and the function of relevant proteins.
This study focuses on the proteome as a more direct approach to establish the molecular hallmarks that distinguish individual tumors and tumors of different stages of the disease, and which may be utilized to develop better approaches to diagnosis and therapy. We used the latest generation high-resolution hybrid mass spectrometry to assess the expression of more than 9000 proteins in a collection of manually dissected colorectal tumors. A subset of the samples was analyzed in parallel with DNA microarrays. This allowed us to perform comparative analysis of protein and transcript abundances on a genomic scale and identify protein candidates that show differential expression in the context of tumor progression from stage UICC II phenotypes without lymph node metastases to stage UICC III phenotype with lymph node metasases. Lymphatic metastasis is an independent strong predictor for outcome in CRC. Therefore for stage UICC III CRC adjuvant chemotherapy is recommended after surgery. Nevertheless ~30% of these tumors develop recurrent disease which has to be treated by further chemotherapy, radiation or surgery. Therefore molecular markers are needed to identify high risk cases and new more effective therapeutic targets. Our findings provide an insight which transcribed genes will occur as translated and functionally relevant proteins, and which expressed proteins tend to be more abundant in the metastatic CRC compared to the non-metastatic tumors.
Materials and Methods
Patients
Nineteen patients with histopathologically verified primary adenocarcinoma of the colorectum were included in the study for proteome analysis. From this cohort immediately after surgery the resected specimens were evaluated by a pathologist and tumor samples were harvested in liquid nitrogen. The samples were stored at -80°C before further work up. In a validation cohort comprizing 40 patients with colon carcinomas (stage UICC II: n=20; stage UICC III: n=20) immunohistochemical (IHC) investigations of the paraffin embedded tumor samples were performed. Patients that have received radiotherapy or suffering from hereditary syndromes (e.g. familial adenomatous polyposis, HNPCC) or inflammatory bowel disease (crohn's disease, colitis ulcerosa) were excluded. After histopathological staging of the whole removed tumor bearing tissue, the samples were divided in groups either belonging to tumors with (stage UICC III) or without (stage UICC II) lymph node metastases. The demographic patient data and detailed histopathological results were selected from the Erlangen Registry for Colorectal Carcinomas (ERCRC) (Supplemental tables 1 and 4).
Tissue workup for proteome analysis
The tissue workup was performed by cryotomy after manual dissection (CMD) 4. The harvested tumor samples were inserted into a cryotube (Roth, Karlsruhe, Germany) and covered with Tissue-Tek (Zakura, Zoeterwoude, Netherlands). The tissue was immediately shock frozen in liquid nitrogen. Initially a control slice was dissected from the block and stained with hematoxylin-eosin (HE) dye. Any identified connective tissue or healthy mucosa was removed from the Tissue-Tek embedded specimen. On a further control slice, the purity of the carcinoma tissue was checked again and the procedure repeated. When the carcinoma portion of the Tissue-Tek embedded specimen was judged to be above 80% continuous series of 10 slices (40 μm) were dissected and collected in a cryotube. The dissected slices were immediately shock frozen in liquid nitrogen and stored at -80°C until proteome analysis.
Reagents
Unless indicated otherwise in the text, chemicals and HPLC solvents were purchased from Sigma-Aldrich. The highest available grades were used.
Protein extraction, separation, digestion, and preparation of samples for mass spectrometry
The proteins were extracted from the frozen tumor samples with 2X SDS sample buffer, reduced, alkylated and separated by gel electrophoresis as previously described 5. The gel lanes were sliced and digested as described previously 5.
Nano-scale LC-/MS/MS analysis
Protein digests analysis was carried out as described in Greenwood et al (4). Briefly, electrospray ionization MS was performed on a hybrid LTQ/Orbitrap Velos instrument (Thermo Fisher, USA) interfaced to a split-less nano-scale HPLC (Ultimate 3000, Dionex, USA). The peptides were desalted at 1 μl.min-1 on a 2 cm long, 0.1 mm i.d. trap column packed with 5 μm C18 particles (Dionex, USA). The peptides were then eluted from the trap column and separated in a 90-min gradient of 2-30% (v/v) acetonitrile in 0.1% (v/v) formic acid at a flow rate of 0.3 μl.min-1. The separation column was a 15 cm long, 0.1 mm i.d. pulled tip packed with 5 μm C18 particles (Nikkyo Technos Co., Tokyo, Japan). The eluting peptides were ionized by applying 1.75 kV via a liquid junction interface. The LTQ/Orbitrap Velos was operated in positive ion mode and the Top20 data-dependent scanning mode was used where the instrument first executes 2 high-resolution scans at a resolution of 30,000 (at 400 m/z) and then 20 MS/MS scans for the 20 most abundant peptide ions having a charge state > 1. During the high-resolution scans the Orbitrap analyzer accumulated 106 ions for the maximum of 0.5 s. During MS/MS scans the LTQ was filled with 5,000 precursor ions for the maximum of 0.1 s. We used normalized collision energy of 30, minimum signal intensity of 500, activation time of 10 ms and activation Q of 0.250. A dynamic exclusion to avoid repetitive analysis of abundant peptide ions was used as follows: after a peptide has been analyzed once its m/z was put in the exclusion list for 30 seconds. The instrument performed an internal mass calibration by a lock mass 6. All samples were analyzed at least three times by LC-MS/MS to allow assessment of reproducibility and statistical analysis.
Data analysis of proteins
MS/MS data were analyzed by CPAS (Computational Proteomics Analysis System) as described in 5. In addition LC-MS/MS data were also analyzed by MaxQuant and the Andromeda search engine and label free quantitation was performed as described in 7-9. Protein abundance was assessed by the spectral counting method and by summing up the peptide ion intensities as determined by a replicate high-resolution scan in the Orbitrap mass analyzer.
Statistical analysis
Protein identification data were assessed for significance using the PeptideProphet and ProteinProphet programs from the Transproteomic pipeline incorporated into CPAS as described previously 5. The MaxQuant searches were performed as described in Cox et al. 7, using a reverse database to calculate false discovery rate (FDR). Results from the Andromeda engine were filtered at both peptide and protein level. In both cases the cutoff was at 1% FDR.
Identification of differentially expressed proteins was performed in R using the packages permax 10 and locfdr 11, 12. First, the rank-transformed spectral counts output by MaxQuant were used to calculate permutation-based test statistics, using permax and the non-parametric two-sample Wilcoxon test. Then the local false discovery rate (fdr) was calculated for each protein using locfdr. Protein with local fdr less than 0.15 were selected as candidates. Similar results were obtained using the R package Significance Analysis of Microarrays (SAM) package 13-15 in its RNA-seq mode (data not shown).
Validation by immunohistochemical staining
Several proteins selected on the basis of SAM q-values and permax p-values were further subjected to validation experiments utilizing immunohistochemical staining of paraffin-embedded formaldehyde-fixed tissue (FFPE) sections from an independent cohort of colon carcinoma samples.
IHC validation of selected markers was performed in 20 colon carcinomas with (stage UICC III) and in 20 cases without (stage UICC II) lymph node metastases. Patient and tumor details are listed in Supplemental Table 4. After formalin fixation paraffin embedded tumor samples were made immediately after surgery from the resected specimens. 4 μm slices were cut, rehydrated with xylol and ethanol. After incubation in target retrival solution endogen peroxidase was blocked. Primary antibodies were added: DNAJA2 (3A1) (ab124017, Abcam, UK), DNAJB11 (ab75107, Abcam, UK), IGF1 Receptor (phospho Y1161) (ab39398, Abcam UK), IRF2BP1 (HPA042164, Sigma-Aldrich, Sweden), M143 (anti-M1), Preparation 11/2005/S417 (Dr. Kochs, University of Freiburg, Germany). Staining was performed as described elsewhere after adding secondary biotinylated antibodies and staining reagents 16-19. Marker expression results were counted in % of IHC positive cells separately for tumor center and invasion front (Table 2).
Table 2.
Expression of markers in the tumor center and invasion front detected by immunohistochemistry, stage UICC II: lymph node negative colon carcinomas, stage UICC III: lymph node positive colon carcinomas, values are presented as percent positive tumor cells. The p value is calculated by the two-sample Wilcoxon test. Significant proteins are shown in bold.
| UICC II Mean (median); % | UICC III Mean (median); % | p | |
|---|---|---|---|
| Tumor center | |||
| IGF1-R | 79 (85) | 82 (80) | 0.42 |
| MX1 | 13 (5) | 17 (17) | 0.19 |
| DNAJA2 | 92(95) | 87 (95) | 0.76 |
| IRF2BP1 | 69 (75) | 17 (0) | 0.008 |
| DNAJB11 | 78 (90) | 69 (80) | 0.47 |
| Invasion front | |||
| IGF1-R | 84 (90) | 89 (90) | 0.007 |
| MX1 | 24 (7.5) | 46 (47.5) | 0.02 |
| DNAJA2 | 93 (95) | 86 (90) | 0.12 |
| IRF2BP1 | 62 (80) | 37 (0) | 0.04 |
| DNAJB11 | 81 (90) | 83 (80) | 0.60 |
siRNA-mediated MX1 knockdown, Western blotting and wound-healing assays
MX1 was knocked down in two colorectal cell lines, DLD1 and SW450, using commercially available siRNA reagent (sc45260, Santa Cruz Biotechnology, CA) with DharmaFECT transfection reagent (Thermo Fisher) following manufacturer's instructions. The wound-healing assays were performed as previously described (18). For each transfection reaction, including controls mock-transfected with reagent only, multiple replicate wells of a 96-well tissue culture plate were seeded with approximately 5000 cells and incubated for 3 hours to allow the cells to attach. Wounds were created after 24h by manually scratching each well with a yellow pipette tip and the plate was then gently washed with pre-warmed medium to remove detached cells and imaged on a Nikon Ti – E wild field inverted microscope using scan large image option at 10x magnification. The plate was then incubated for 24 hours and imaged again. The images were processed by the NIS Elements software, to calculate wound closure rate (LUNDEBERG et al.,1992) and determine statistical significance whenever judged necessary. After being imaged the cells were lysed, separated by SDS-PAGE and transferred to PDVF membrane. Anti- MX1/2/3 (C1) mouse monoclonal antibody (Santa Cruz Biotechnology, sc-166412) was used for detection of MX1. Imaging was done using Li-Cor infrared Odyssey system.
Results
Quantitative proteome profiling
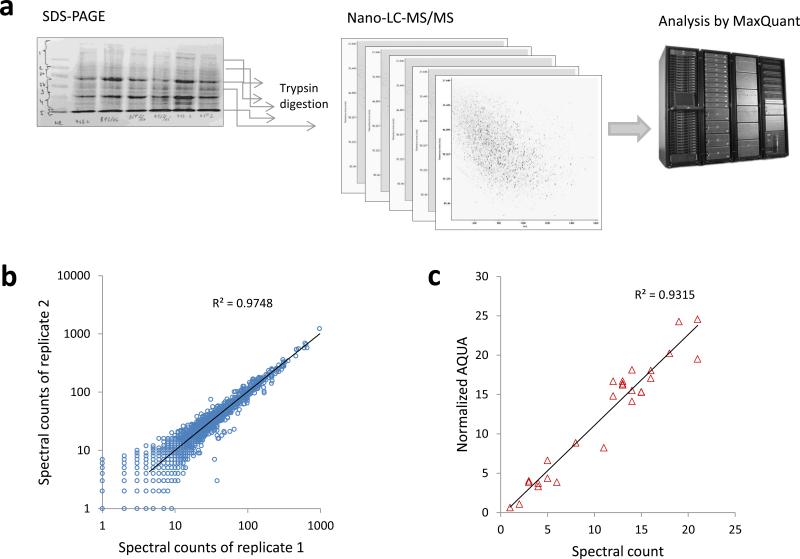
Figure 1 summarizes the procedures undertaken to quantify the colorectal tumor proteomes and illustrates the reproducibility and quantitative precision of the label-free quantitation approach. We analyzed 19 individual tumors to generate the raw dataset for quantitation. The initial intention was to analyze 10 stage UICC III and 10 stage UICC II tumors but one of the stage II tumors did not pass quality control because the total amount of the protein extracted from the tissue sample was too low. Therefore the analyzed cohort consisted of 19 tumor samples. The proteome of each tumor was fractionated into size-resolved fractions, digested and analyzed by nano-scale liquid chromatography and high-resolution tandem mass spectrometry on an LTQ Orbitrap Velos instrument (Fig. 1a). Quantitative accuracy and reproducibility were assessed by comparing technical replicates (Fig. 1b) and by comparing the abundance estimates obtained by spectral counts (SpC) and label-free peptide ion intensities (data not shown). Figure 1b illustrates technical reproducibility of the total analysis: summed spectral counts for each detected protein from one set of LC-MS/MS runs analyzing all fractions obtained by electrophoresis were plotted against another set of LC-MS/MS runs analyzing the same set of samples. In addition, we performed paralleled quantitative analysis by the accurate isotope dilution method 20, 21. We followed a modified AQUA procedure that takes advantage of the high-resolution mass analysis enabled by the LTQ Orbitrap instrument 22. The results for one protein, KCD12, are shown in Fig. 1c. Similar results were obtained for other proteins such as Stat1 (data not shown). As shown in Fig. 1b, the technical reproducibility is excellent with coefficient of correlation exceeding 0.99. The absolute amounts of the protein measured by internal labeled standards correlated very well with the abundance estimates obtained by the spectral counting method (Pearson r= 0.96, Fig. 1c).
Figure 1.
Large-scale analysis of protein abundance in manually-dissected colorectal tumors. a: A workflow diagram illustrating the individual steps of the analysis. Proteins are extracted, fractionated by PAGE, digested and analyzed by nano-scale LC-MS/MS on a hybrid high-resolution LTQ/Orbitrap Velos instrument. The individual proteins are then quantified by label-free techniques. Altogether we performed more than 300 individual LC-MS/MS runs to analyze the 19 tumor specimens. b: Scatter plot comparing data from 2 technical replicate analyses. The R-squared is shown on the plot. c: Validation of the spectral count data by a modified AQUA assay. A labeled peptide derived from KCD12 was spiked into the protein digests and used as an internal standard to quantify the endogenous KCD12.
Supplemental Table 2 gives the average numbers of proteins identified in each tumor and in total for the 19 analyzed tumors. The numbers apply to the processed dataset that was filtered at 1% FDR at both, protein and peptide level. The heterogeneity of the dataset is worth noting; the proteomes of individual tumors overlap but each tumor can be characterized by a unique pattern of protein expression, possibly underscoring the specifics of the individual cell and molecular evolution that enabled its formation. A core CRC proteome comprising about 3,000 proteins is detected in all of the tumors. The most numerous are the proteins involved in metabolic processes (1,909) and biological regulation (1,267). Soluble, nuclear, and membrane proteins are detected at comparable rate indicating that the technique we chose to use does not suffer from the well-known bias toward soluble proteins that affects other approaches relying on multidimensional gel separation.
Stage UICC III vs. stage UICC II comparison and identification of candidate markers for lymphatic metastasis
Stage UICC III and UICC II tumors showed very similar total number of proteins identified per individual tumor, total number of tandem mass spectra acquired, and protein abundance distributions. Statistical analysis performed in R using the packages permax and locfdr, and the RNA-seq implementation of the SAM package identified a number of proteins as significantly overexpressed (local fdr<0.15) in the 10 metastatic tumors compared to the 9 non-metastatic tumors. MX1, an interferon gamma-induced antiviral protein, and several other proteins were further studied by IHC in an independent patients cohort. The statistically significant candidate proteins identified by the proteomics screen are presented in Table 1.
Table 1.
Analysis of protein expression in Stage III against stage II in colorectal tumors by permax and locfdr. The protein spectral counts assigned at 1% false discovery rate (fdr) by MaxQuant at both, peptide and protein level were used to perform the calculations. Local fdr was calculated in R using the package locfdr and the two-sample Wilcoxon test statistics calculated by permax. The individual one-sided p values in the table are based on the two-sample Wilcoxon statistics.
| Gene | IPI | Wilcoxon | p | Local fdr |
|---|---|---|---|---|
| MX1 | IPI00167949 | —4.55 | 0.00003 | 0.0002 |
| DNAJA2 | IPI00032406 | —4.18 | 0.000014 | 0.0012 |
| CASP7 | IPI00216675 | —3.57 | 0.000178 | 0.0180 |
| DNAJB11 | IPI00008454 | —3.57 | 0.000180 | 0.0182 |
| GOLPH3L | IPI00514951 | —3.39 | 0.000350 | 0.0340 |
| AIP | IPI00925804 | —3.07 | 0.001081 | 0.0885 |
| IGF1R | IPI00027232 | —3.07 | 0.001081 | 0.0889 |
| RNF4 0 | IPI00162563 | —3.07 | 0.001081 | 0.0889 |
| CDC27 | IPI00794278 | —3.07 | 0.001081 | 0.0889 |
| CLN5 | IPI00026050 | —2.99 | 0.001389 | 0.1076 |
| TTC1 | IPI00016912 | —2.92 | 0.001776 | 0.1294 |
| HAT1 | IPI00024719 | —2.92 | 0.001776 | 0.1294 |
| UBAC1 | IPI00305442 | —2.92 | 0.001776 | 0.1294 |
| NUDC | IPI00550746 | —2.92 | 0.001776 | 0.1294 |
| OLA1 | IPI00916847 | —2.92 | 0.001776 | 0.1294 |
| COX7A2L | IPI00022421 | 3.22 | 0.000635 | 0.1346 |
| PMPCA | IPI00166749 | 3.22 | 0.000635 | 0.1346 |
| GLS | IPI00289159 | 3.22 | 0.000635 | 0.1346 |
| CASP2 | IPI00291570 | 3.22 | 0.000635 | 0.1346 |
| IRF2BP1 | IPI00645608 | 3.22 | 0.000635 | 0.1346 |
| HSPB6 | IPI00908768 | 3.22 | 0.000635 | 0.1346 |
| FRMD8 | IPI00011090 | 3.39 | 0.000350 | 0.0823 |
| ALDH5A1 | IPI00336008 | 3.39 | 0.000350 | 0.0823 |
| NT5DC2 | IPI00783118 | 3.39 | 0.000350 | 0.0823 |
| RPL14 | IPI00555744 | 3.76 | 0.000086 | 0.0211 |
| MRPS33 | IPI01010059 | 3.96 | 0.000037 | 0.0083 |
| DAG1 | IPI00028911 | 4.18 | 0.000014 | 0.0031 |
| CCDC93 | IPI00154668 | 4.18 | 0.000014 | 0.0031 |
| MESDC2 | IPI00399089 | 4.18 | 0.000014 | 0.0031 |
Comparative analysis of protein abundance and mRNA expression in 6 colorectal carcinomas
In a previously published study the mRNA expression of 14,500 genes was assessed in a cohort of 80 colorectal tumors obtained and dissected by the same protocols we used for proteome analysis. We therefore searched the available samples from the mRNA profiling study and were able to obtain frozen samples from 6 of the tumors. These were included in the proteome analysis along with 13 additional samples. The obtained protein abundance data were correlated with the available gene array data. Supplemental Figure 2 shows scatter plots and the Spearman correlation coefficients of spectral count data (for protein abundance) and the oligonucleotide array data for the 6 colorectal tumor samples. There is a positive but modest correlation with mean r=0.43, recapitulating the now well acknowledged fact that protein abundance is as much, if not more determined by the rate of translation and by post-transcriptional control mechanisms, than by the abundance of the corresponding mRNA 3, 23.
MX1 overexpression in stage UICC III compared to stage UICC II colorectal tumors
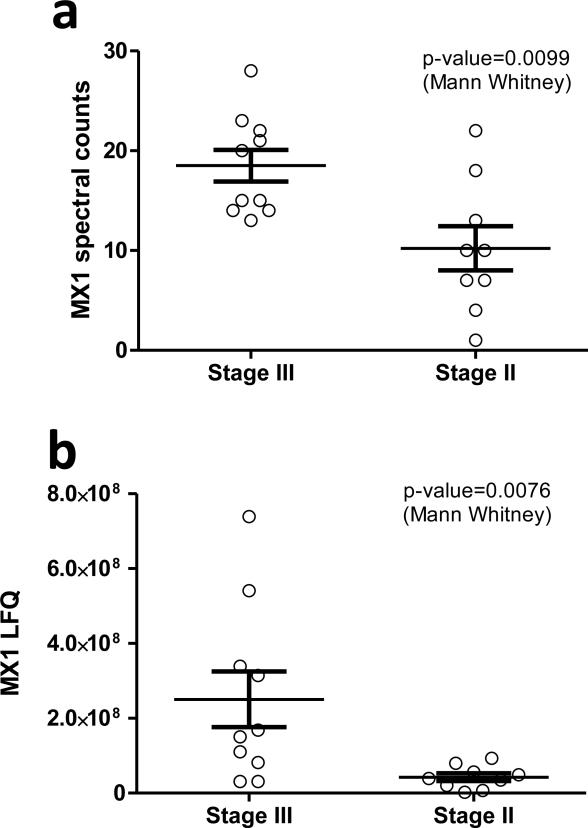
Among the proteins identified to be overexpressed in stage UICC III tumors is the interferon gamma induced protein MX1. This result is intriguing since in a recently published study MX1 was identified to be overexpressed in metastatic triple-negative breast cancer. Therefore we carried out additional statistical analyses to evaluate whether MX1 could be considered as a marker for lymphatic metastasis of colorectal tumors as well. Figure 2 shows analysis of MX1 abundance in all the 19 tumors analyzed in this study. The box plot in Fig3a shows spectral count data indicating overexpression in stage III tumors. An alternative quantitation approach, a measurement based on integrated and summed up peptide ion intensities further corroborates this conclusion and is presented in Fig3b. In this analysis Mx1 abundance was assessed on the basis of the signal intensities generated by the peptide ions corresponding to each of the many identified MX1 peptides.
Figure 2.
Quantitation of MX1 in 19 colorectal tumors by two label-free approaches. A: MX1 was quantified by spectral counting. B: Quantitation was done by label-free peptide intensities integration. In both analyses the non-parametric Mann-Whitney t-test was used to assess if the means are significantly different between stage UICC III and stage UICC II tumors.
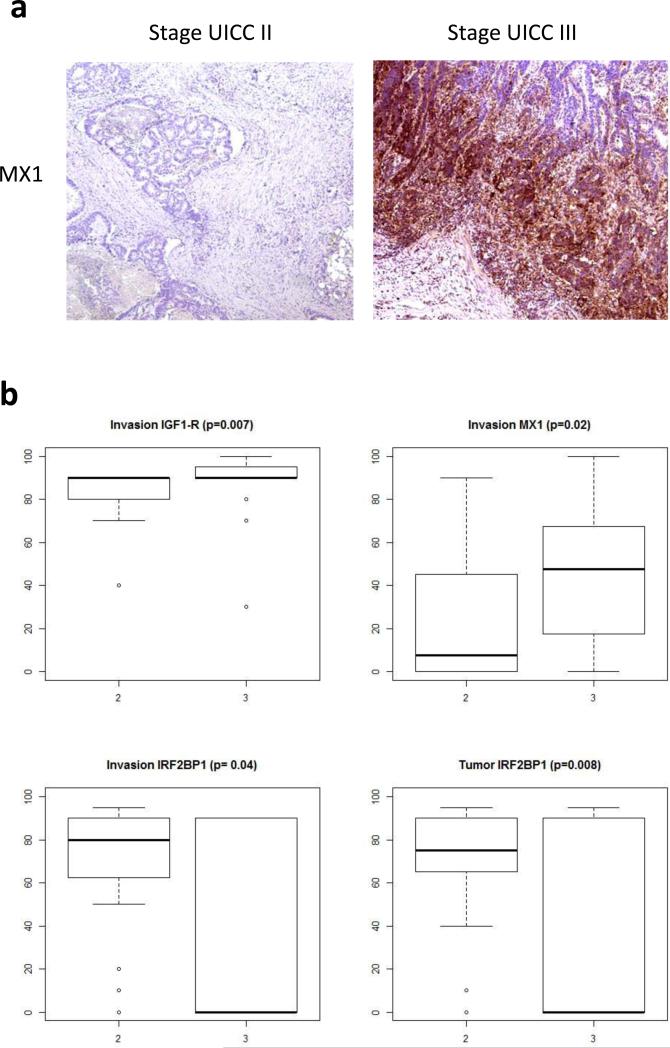
Figure 3.
Validation of candidate proteins by IHC. A: Representative IHC staining of MX1 in stage UICC III and stage UICC II tumors. B: Boxplots for MX1, IGF1-R, and IRF2BP1. The p values calculated by the two-sample Wilcoxon test are indicated for the significant proteins. The validation cohort comprised 20 stage UICC III samples and 20 stage UICC II samples.
Validation by immunohistochemistry and follow-up mechanistic cell culture studies
In the next stage of the study several candidate proteins were subjected to validation by orthogonal experimental approaches. The results from these validation and mechanistic experiments are summarized in Table 2, and Figures 3 and 4. Three proteins were positively validated by immunohistochemistry in an independent cohort of samples from 20 stage UICC III and 20 stage UICC II patients. These were MX1 and IGF1-R, found to be overexpressed particularly at the invasion front of stage UICC III tumors, and IRF2BP1, found to be significantly decreased in stage UICC III in both, tumor centers and invasion fronts compared to UICC stage II tumors.
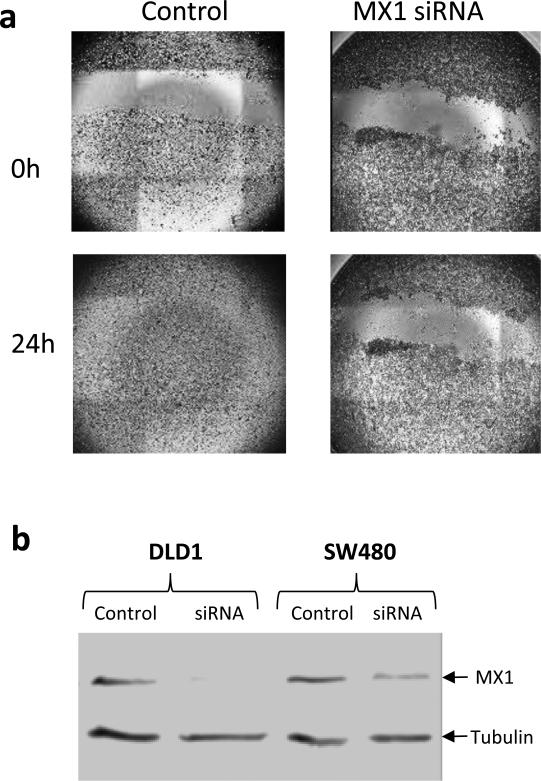
Figure 4.
MX1 Knock-down and wound-healing assays with DLD1 cells. A: Wound-healing assays with cells transfected with MX1-specific siRNA or mock-transfected. B: Western blotting analysis of lysates from control and MX1 knock-down DLD1 and SW480 cells. MX1 and tubulin bands are indicated with arrows.
In addition to the validation experiments utilizing antibody-based staining of tumor tissue, we undertook to evaluate the potential involvement of MX1 in tumor cells' migration and invasion. To this end we knocked-down the expression of MX1 in 2 colorectal cancer cell lines using MX1-specific siRNA and carried out wound-healing assays. The results, shown in Figure 4 clearly demonstrate that MX1 knock-down strongly inhibits wound healing of DLD1 cells. The second cell line, SW480, was also affected but to a lesser extent, although the results were highly reproducible (data not shown). This could possibly be explained by the facts that SW480 were not as migratory as DLD1 and also, the knock-down of MX1 was not as efficient as in DLD1 as shown on the western blot in Figure 4.
Discussion
The sequencing of the human genome and the development of high-throughput technologies that allow the activities of thousands of genes to be assayed simultaneously, and in only a minute amount of clinical sample, enabled a plethora of new approaches that can be used for identification and validation of biomarkers and drug target candidates in oncology. In particular, it is now possible to not only map the entire landscape of genomic mutations in individual tumors, but also, using array technologies or next-generation sequencing, to measure the activity of the tumor genome in a highly quantitative and comprehensive way 1, 24, 25. These new capabilities are expected to enable a new and more efficient personalized approach to treating cancer and other diseases. However, one important shortcoming of clinical genomics cannot be overlooked: many proteins that are key players in cancer biology are known to be regulated at post-transcriptional level. Such proteins will slip through any mutation and gene expression screen and remain undetected as causative agents or biomarkers because our knowledge of the regulation of protein abundance in the cell is far from complete. Thus, if we were to base our attempts to develop personalized cancer treatments solely on mutation and gene expression data, these attempts are destined to fail, or at best, to deliver very modest results. Therefore genomics needs to be complemented with protein level analysis for both, drug target identification and development of novel diagnostic assays.
Here we applied recently developed mass spectrometry-based techniques that can be used to acquire an almost genome-scale quantitative snapshot of protein abundance in tumors. Such data could be extremely useful and complementary to genomics in a number of ways: it can provide validation of candidate genes, it can lead to the identification of likely drug targets that are overexpressed in a subset of tumors due to post-transcriptional mechanisms, it can provide candidate proteins for the development of new types of multiplex diagnostics with increased specificity and sensitivity.
We used a recently developed hybrid high-resolution mass spectrometry technology 26 to analyze 19 colorectal tumors grouped by stage into metastatic (stage UICC III) and non-metastatic (stage UICC II) classes. The tumor tissue was manually dissected to ensure tumor enrichment, homogeneity and to maximize the coverage of the proteome analysis. As a result we achieved an analytical depth of more than 9,000 proteins identified in the 19 tumor samples, to our knowledge the largest tumor proteome data set to date. The proteins abundance was estimated by spectral counting and by label-free intensity methods. In the subsequent analyses we used protein spectral counts to identify differentially expressed proteins because of the robustness and reproducibility of this approach and its applicability to unlabeled clinical samples 5, 22, 27. This led to the identification of several proteins that were significantly overexpressed in the stage UICC III tumors compared to the non-metastatic stage UICC II tumors (Table 1). Among the proteins pinpointed as significant three proteins were selected for further validation. These were MX1, a GTP-binding protein involved in antiviral responses28, 29, IGF1-R a growth factor receptor known to be involved in cancer (reviewed in 30) and IRF2BP1, a protein involved in the regulation of interferon-induced gene expression31. The identification of MX1 as the top proteomic candidate marker for distinguishing between the stage UICC III and stage UICC II tumors in the analyzed cohort is intriguing because of its apparent involvement in antiviral responses and also, because we recently identified this protein to be among the proteins that are overexpressed in metastatic triple-negative breast cancer 22. To further investigate this we performed wound-healing experiments, which confirmed the possible involvement of MX1 in colorectal tumor cells' invasion and metastasis (Fig. 5). Validation by immunohistochemical methods provided further evidences in this direction (Fig. 4 and Table 2).
Concluding remarks
In this study we have achieved 9,000+ proteins coverage of the colorectal tumor proteome, which led to the identification of candidate markers of lymphatic metastasis. Simultaneous measurement of mRNA and proteins abundances in 6 tumors showed that the correlation between protein and message abundances is about 40%, which suggests that tumor genomics should always be complemented with paired proteome analysis. Furthermore, the quantitative atlas of protein abundance in colorectal tumor generated by this study can be explored in the future to identify and/or validate candidate drug targets and diagnostic markers, and to identify molecular pathways that contribute to tumor invasion and metastasis. An example of such candidate marker/target is MX1 which was the top candidate selected by proteomics, and was successfully validated in an independent cohort of samples and in cell-based mechanistic studies utilizing siRNA-mediated knock-down and wound-healing assays.
Supplementary Material
Novelty and impact.
We report very large-scale proteome analysis of 19 colorectal tumors. More than 9,000 proteins were identified, which makes the generated dataset the largest colorectal tumor proteome to date. The study identified candidate biomarkers for metastasis. Three of the proteins, MX1, IGF1-R, and IRF2BP1, were further validated in an independent cohort of 40 tumor samples and MX1 was also studied in knockout experiments using small interfering RNA.
Acknowledgements
The methodology for large-scale tumor proteome analysis and related bioinformatics pipeline were developed with support from the NIH, grant 1RO3CA150131 to MM. We are also grateful to University of Essex for continuing support to the proteomics unit at the School of Biological Sciences, particularly for providing the funding for the acquisition of the Orbitrap Velos instrument. The study was supported by the German Research Foundation (DFG: CR136/2), the German Federal Department for Education and Research (BMBF, Polyprobe) and the ELAN-Foundation of the University Elangen-Nuremberg.
References
- 1.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 4.Croner RS, Guenther K, Foertsch T, Siebenhaar R, Brueckl WM, Stremmel C, Hlubek F, Hohenberger W, Reingruber B. Tissue preparation for gene expression profiling of colorectal carcinoma: three alternatives to laser microdissection with preamplification. J Lab Clin Med. 2004;143:344–51. doi: 10.1016/j.lab.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Alldridge L, Metodieva G, Greenwood C, Al-Janabi K, Thwaites L, Sauven P, Metodiev M. Proteome profiling of breast tumors by gel electrophoresis and nanoscale electrospray ionization mass spectrometry. J Proteome Res. 2008;7:1458–69. doi: 10.1021/pr7007829. [DOI] [PubMed] [Google Scholar]
- 6.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 8.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 32:279–89. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 10.Gray R. Permax. R package version 1.2. 1:2005. [Google Scholar]
- 11.Efron B. Empirical Bayes Estimates for Large-Scale Prediction Problems. Journal of the American Statistical Association. 2009;104:1015–28. doi: 10.1198/jasa.2009.tm08523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genetic epidemiology. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Tibshirani R. Finding consistent patterns: A nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2011 doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012;13:523–38. doi: 10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croner RS, Schellerer V, Demund H, Schildberg C, Papadopulos T, Naschberger E, Sturzl M, Matzel KE, Hohenberger W, Schlabrakowski A. One step nucleic acid amplification (OSNA) - a new method for lymph node staging in colorectal carcinomas. Journal of translational medicine. 8:83. doi: 10.1186/1479-5876-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schellerer VS, Mueller-Bergh L, Merkel S, Zimmermann R, Weiss D, Schlabrakowski A, Naschberger E, Sturzl M, Hohenberger W, Croner RS. The clinical value of von Willebrand factor in colorectal carcinomas. American journal of translational research. 3:445–53. [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer A, Merkel S, Bruckl W, Schellerer V, Schildberg C, Campean V, Hohenberger W, Croner RS. Cdc2 as prognostic marker in stage UICC II colon carcinomas. Eur J Cancer. 2009;45:1466–73. doi: 10.1016/j.ejca.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Naschberger E, Croner RS, Merkel S, Dimmler A, Tripal P, Amann KU, Kremmer E, Brueckl WM, Papadopoulos T, Hohenadl C, Hohenberger W, Sturzl M. Angiostatic immune reaction in colorectal carcinoma: Impact on survival and perspectives for antiangiogenic therapy. Int J Cancer. 2008;123:2120–9. doi: 10.1002/ijc.23764. [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–73. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenwood C, Metodieva G, Al-Janabi K, Lausen B, Alldridge L, Leng L, Bucala R, Fernandez N, Metodiev MV. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J Proteomics. 2012;75:3031–40. doi: 10.1016/j.jprot.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 23.Schwanhausser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–9. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 26.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–69. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Sadygov RG, Yates JR., 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical chemistry. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 28.Horisberger MA. Interferons, Mx genes, and resistance to influenza virus. American journal of respiratory and critical care medicine. 1995;152:S67–71. doi: 10.1164/ajrccm/152.4_Pt_2.S67. [DOI] [PubMed] [Google Scholar]
- 29.Horisberger MA, Hochkeppel HK. IFN-alpha induced human 78 kD protein: purification and homologies with the mouse Mx protein, production of monoclonal antibodies, and potentiation effect of IFN-gamma. Journal of interferon research. 1987;7:331–43. doi: 10.1089/jir.1987.7.331. [DOI] [PubMed] [Google Scholar]
- 30.Hartog H, Wesseling J, Boezen HM, van der Graaf WT. The insulin-like growth factor 1 receptor in cancer: old focus, new future. Eur J Cancer. 2007;43:1895–904. doi: 10.1016/j.ejca.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Childs KS, Goodbourn S. Identification of novel co-repressor molecules for Interferon Regulatory Factor-2. Nucleic acids research. 2003;31:3016–26. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.