Abstract
Adipose tissue macrophage (ATM)-driven inflammation plays a key role in insulin resistance; however, factors activating ATMs are poorly understood. Using a proteomics approach, we show that markers of classical activation are absent on ATMs from obese humans, but readily detectable on airway macrophages of patients with cystic fibrosis, a disease of chronic bacterial infection. Moreover, treating macrophages with glucose, insulin, and palmitate – conditions characteristic of the metabolic syndrome – produces a ‘metabolically-activated’ phenotype distinct from classical activation. Markers of metabolic activation are expressed by pro-inflammatory ATMs in obese humans/mice and are positively correlated with adiposity. Metabolic activation is driven by independent pro- and anti-inflammatory pathways, which regulate balance between cytokine production and lipid metabolism. We identify PPARγ and p62/SQSTM1 as two key proteins that promote lipid metabolism and limit inflammation in metabolically-activated macrophages. Collectively, our data provide important mechanistic insights into pathways that drive the metabolic disease-specific phenotype of macrophages.
INTRODUCTION
Macrophages accumulate in adipose tissue of obese mice and humans (Weisberg et al., 2003; Xu et al., 2003) and are key contributors to inflammation and obesity-induced insulin resistance (Chawla et al., 2011; Lumeng and Saltiel, 2011; Olefsky and Glass, 2010; Wellen and Hotamisligil, 2005). The evidence implicating adipose tissue macrophage (ATM) inflammation in potentiating insulin resistance is substantial. Indeed, ablation of pro-inflammatory (CD11c+) ATMs using a diphtheria toxin system led to a rapid improvements in insulin sensitivity and glucose tolerance, associated with marked decreases in local and systemic inflammation in obese mice (Patsouris et al., 2008). Moreover, targeting pathways that mediate inflammation in the macrophage revealed significant roles for TLR4, JNK, and IKKβ in potentiating ATM inflammation and insulin resistance in mice (Arkan et al., 2005; Han et al., 2013; Saberi et al., 2009). Anti-inflammatory effects may also help explain the insulin-sensitizing action of thiazolidinediones (TZDs). Indeed, myeloid-specific deletion of PPARγ, the molecular target of TZDs, exacerbated macrophage inflammation and insulin resistance (Odegaard et al., 2007).
Macrophages are heterogeneous, and based on patterns of gene expression and function they have been classified as classically (M1) or alternatively (M2) activated (Gordon and Taylor, 2005). The M1 phenotype is promoted by Th1 mediators such as LPS and IFNγ and is characterized by the overproduction of pro-inflammatory cytokines. In contrast, Th2 mediators (eg. IL-4) drive the M2 phenotype, which activates expression of immunosuppressive factors and peroxisome proliferator-activated receptor gamma (PPARγ) that promote tissue remodeling and helps resolve inflammation (Odegaard et al., 2007). It has been proposed that during weight gain macrophages undergo a ‘phenotypic switch’ from an anti-inflammatory M2 phenotype to a pro-inflammatory M1 state, a conversion that has been linked to the emergence of systemic insulin resistance (Lumeng et al., 2007).
Although it is clear that ATM activation is involved in regulating insulin sensitivity, the mechanisms that underlie transition to the pro-inflammatory ATM phenotype and corresponding signature of cell surface markers are poorly understood. For example, CD11c, the most commonly used marker of pro-inflammatory ATMs in mice and humans, is suppressed by the classical activation paradigm of LPS stimulation (Becker et al., 2012). More recently, Ferrante and colleagues provided evidence that ATMs in obese mice exhibit increased lipid metabolism and further suggested that increases in ATM number, rather than ATM activation, may be responsible for the low-grade inflammation observed in obesity (Xu et al., 2013). In addition, several studies have described a ‘mixed’ M1/M2 phenotype for ATMs in obese mice and humans, suggesting that ATMs adopt more complex states in vivo (Sica and Mantovani, 2012; Zeyda et al., 2007). However the etiology of such mixed M1/M2 phenotypes are incompletely understood.
Here we combine proteomic, gene expression, and flow cytometric analyses of human and murine macrophages; studies of cells from obese, non-obese, and cystic fibrosis patients; and murine knockout studies to characterize pro-inflammatory ATMs, and to identify the molecular mechanisms that produce them. Our findings demonstrate that treating macrophages with mixtures of glucose, insulin, and palmitate (i.e., ‘metabolic activation’) produces a complex macrophage phenotype that is mechanistically distinct from classical activation, suggesting that metabolic disease-specific pathways drive macrophage inflammation via mechanisms that are different from those operative during infection.
RESULTS
Plasma membrane proteomics identifies markers of M1 macrophages
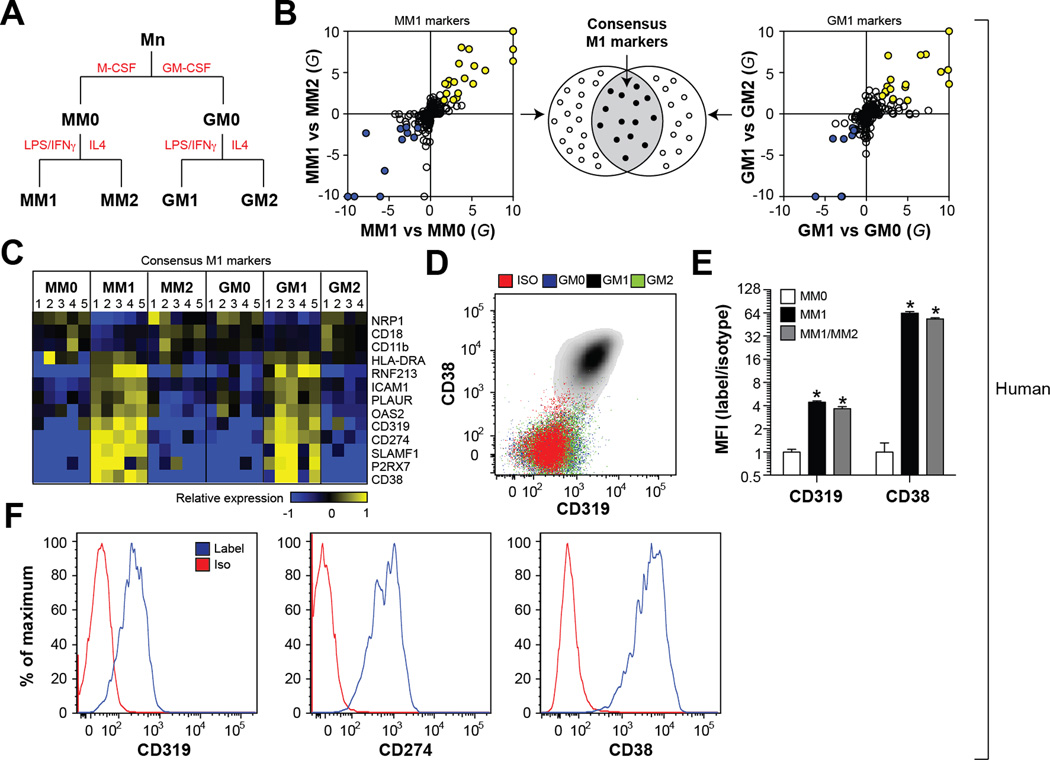
The lack of specific markers for human macrophage subsets is a major hurdle to understanding ATM inflammation in obese and diabetic patients (Geissmann et al., 2010). We therefore used a plasma membrane (PM) proteomics approach to define protein expression patterns diagnostic of pro-inflammatory human macrophages. Initial studies focused on a widely used in vitro system that included classically activated (M1, pro-inflammatory), alternatively activated (M2, anti-inflammatory), and un-stimulated (M0) macrophages generated from human peripheral blood monocytes (Mn) in the presence of M-CSF (MM0, MM1, MM2) or GM-CSF (GM0, GM1, GM2) (Fig. 1A). Classical and alternative activation were confirmed by measuring expression of genes associated with the M1 and M2 phenotypes (Fig. S1).
Fig. 1. Plasma membrane proteomics identifies markers of M1 macrophages.
Panel A: Conditions for differentiating and activating human monocyte-derived macrophages. Panel B: Proteins consistently up- (or down-) regulated in M1 relative to M0 and M2 macrophages were identified using a combination of the G-test and t-test. Panel C: The consensus markers of M1 macrophages; heatmap depicts regulation of protein expression across all cells types (blue = down-regulated, yellow = up-regulated). Panel D: Validation of M1 markers by flow cytometry. Panel E: Cell surface levels of CD319 and CD38 in macrophages exposed to M1 or M1/M2 (LPS, IFNγ, and IL4); results are signal-to-noise ratio in mean fluorescence intensity (MFI). Panel F: Cell surface CD319, CD274, and CD38 levels in airway macrophages (defined as CD14+CD15−) of a CF patient. Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to MM0; N=6. See also Figs. S1, S2; Table S1.
PM-associated proteins were biotinylated, affinity-isolated, and analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry. This approach identified 349 proteins with high confidence (Table S1). Gene ontology analysis of those proteins validated our analytical strategy by revealing strong enrichments in functional annotations consistent with the PM compartment of macrophages (Fig. S1).
Protein levels were quantified by spectral counting (Liu et al., 2004), a measure of relative protein abundance, which were closely correlated with flow cytometry data (Fig. S1). Markers of classically activated macrophages were selected by identifying proteins consistently up- (or down-) regulated by both the t-test and G-test (Becker et al., 2010) in M1 cells relative to M2 and M0 cells. We identified 52 significantly regulated PM proteins in M1 macrophages differentiated by M-CSF or GM-CSF treatment (Fig. 1B, Table S1). We further required that such markers were reproduced in macrophages differentiated in the presence of M-CSF and GM-CSF, as we thought this would increase their utility as markers in diverse environments in vivo. Based on these stringent criteria, we identified 13 membrane proteins as putative cell surface markers of M1 macrophages (Fig. 1B–C, Table S1).
We previously used a similar plasma membrane proteomics approach to define cell surface markers of M1 macrophages in mice (Becker et al., 2012). Interestingly, only 6 of the M1 markers identified in human macrophages overlapped with those found in classically activated bone marrow-derived murine macrophages (BMDMs). To promote continuity between human and mouse studies, our subsequent analyses focused on three such conserved markers (CD274, CD38, and CD319). Flow cytometric measurements validated the ability of CD274, CD38, and CD319 to reliably identify classically activated macrophages ex vivo using both human and murine macrophages (Fig. 1D, Fig. S1). Importantly, the induction of CD38 and CD319 on the cell surface of M1 cells was not attenuated by simultaneously exposing macrophages to M1 and M2 stimuli (Fig. 1E), suggesting that these markers can report exposure to classical stimuli even in the context of a ‘mixed’ macrophage phenotype, which is likely to be present in more complex conditions in vivo (Mosser and Edwards, 2008).
To validate the ability of these markers to identify macrophages inflamed by bacterial stimuli in vivo, we tested the expression of CD38, CD319, and CD274 on airway macrophages from infected patients with cystic fibrosis (CF), a disease characterized by heightened inflammatory responses in the airways due to chronic respiratory tract infection with pathogenic bacteria like Pseudomonas aeruginosa (Rowe et al., 2005). Flow cytometric analysis detected strong expression levels for all 3 markers, in all CF patients examined (Fig. 1F, Fig. S2). Thus, our proteomics approach successfully identified macrophage markers expressed in an in vivo inflammatory condition caused by chronic bacterial infection.
ATMs from obese humans and mice do not express M1 markers
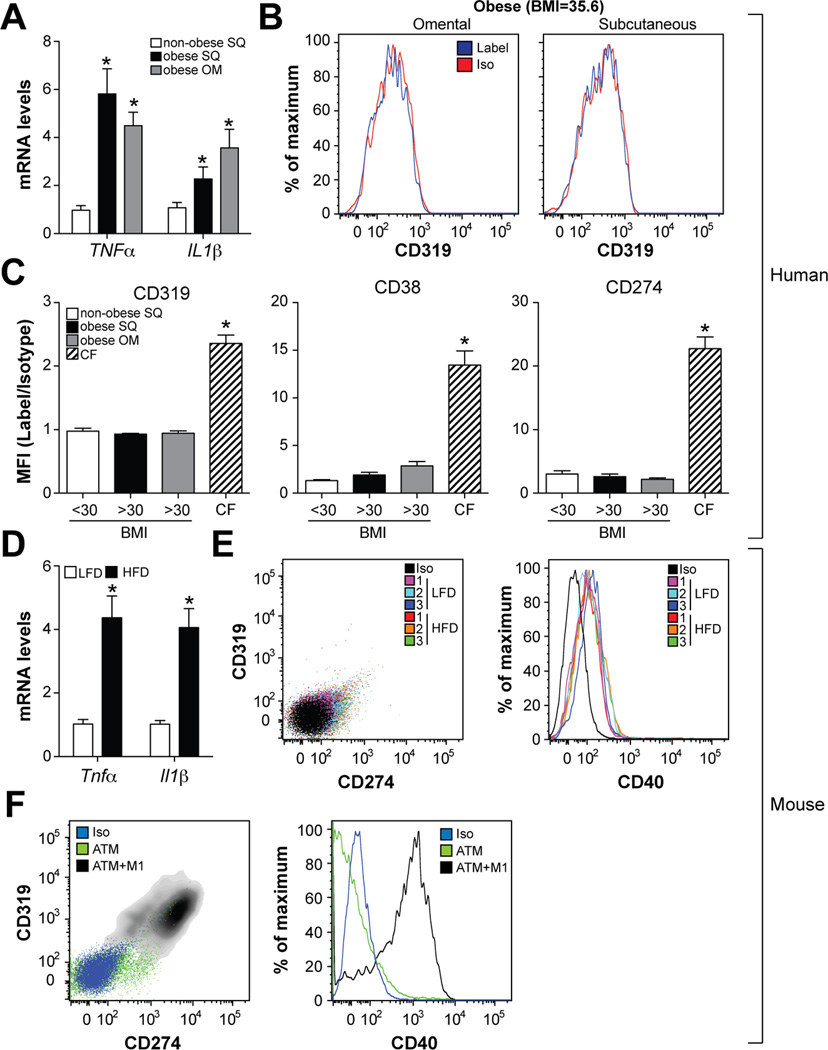
During obesity, macrophages have been proposed to undergo a ‘phenotypic switch’ from an anti-inflammatory M2 phenotype to a pro-inflammatory M1 state (Lumeng et al., 2007). To determine if the M1 markers discovered by proteomics and validated in patients with CF could identify pro-inflammatory ATMs, we studied macrophages from omental and subcutaneous adipose tissue obtained from obese (BMI>30kg/m2) subjects undergoing bariatric surgery. For comparative purposes, we also investigated ATMs in subcutaneous adipose tissue obtained from non-obese (BMI<30kg/m2) subjects undergoing abdominoplasty. Although pro-inflammatory cytokine expression was elevated in both omental and subcutaneous adipose tissue from obese subjects (Fig. 2A), cell surface markers for M1 activation (CD38, CD274, CD319) were either not present or only very weakly expressed by ATMs in either of the adipose tissue depots examined (Fig. 2B–C, Fig. S3).
Fig. 2. ATMs of obese humans and mice are not classically activated.
Panels A-C: Omental (OM) and subcutaneous (SQ) adipose tissue was collected from obese subjects (BMI>30kg/m2) undergoing bariatric surgery (N=7). For comparative purposes, subcutaneous adipose tissue was collected from non-obese subjects (BMI<30kg/m2) undergoing abdominoplasty (N=7). Panel A: Pro-inflammatory cytokine expression. Panel B: Cell surface CD319 levels in ATMs (defined as CD14+CD15−CD206+CD1c−) from a representative obese subject. Panel C: Cell surface levels of CD38, CD274, and CD319 in ATMs from non-obese and obese subjects and airway macrophages from CF patients; results are signal-to-noise ratio in mean fluorescence intensity (MFI). Panels D-F: Male C57BL/6 mice were place on a low-fat diet (LFD) or high-fat diet (HFD) for 16 weeks and ATMs from epididymal fat were isolated. Panel D: Pro-inflammatory cytokine expression. Panel E: ATM cell surface CD319, CD274, and CD40 levels. Panel F: ATM cell surface CD319, CD274, and CD40 levels in the presence and absence of M1 stimuli. Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to non-obese or LFD; N=4–7. See also Fig. S3.
We also tested the ability of M1 markers to identify pro-inflammatory ATMs in mice. To this end, we used a mouse model for diet-induced obesity and insulin resistance (DIO), where male C57BL/6 mice were placed on low-fat or high-fat diets for 16 weeks. Analysis of ATMs from lean and obese mice recapitulated the findings obtained in human subjects. Thus, ATMs from obese mice were inflamed relative to their lean counterparts (Fig. 2D), but did not overexpress CD319, CD274, (Fig. 2E), or CD40 (Fig. 2E), a previously characterized M1 marker in mice (Becker et. al., 2012). Importantly, treating ATMs (purified from obese mice) with LPS/IFNγ in vitro induced expression of CD319, CD274, and CD40, suggesting that ATMs are capable of adopting an M1 phenotype given the appropriate environment (Fig. 2F). Collectively, these findings suggest that distinct surface markers report macrophage inflammation in a setting of bacterial infection and metabolic dysfunction.
‘Metabolically’ and classically activated macrophages express distinct cell surface proteins
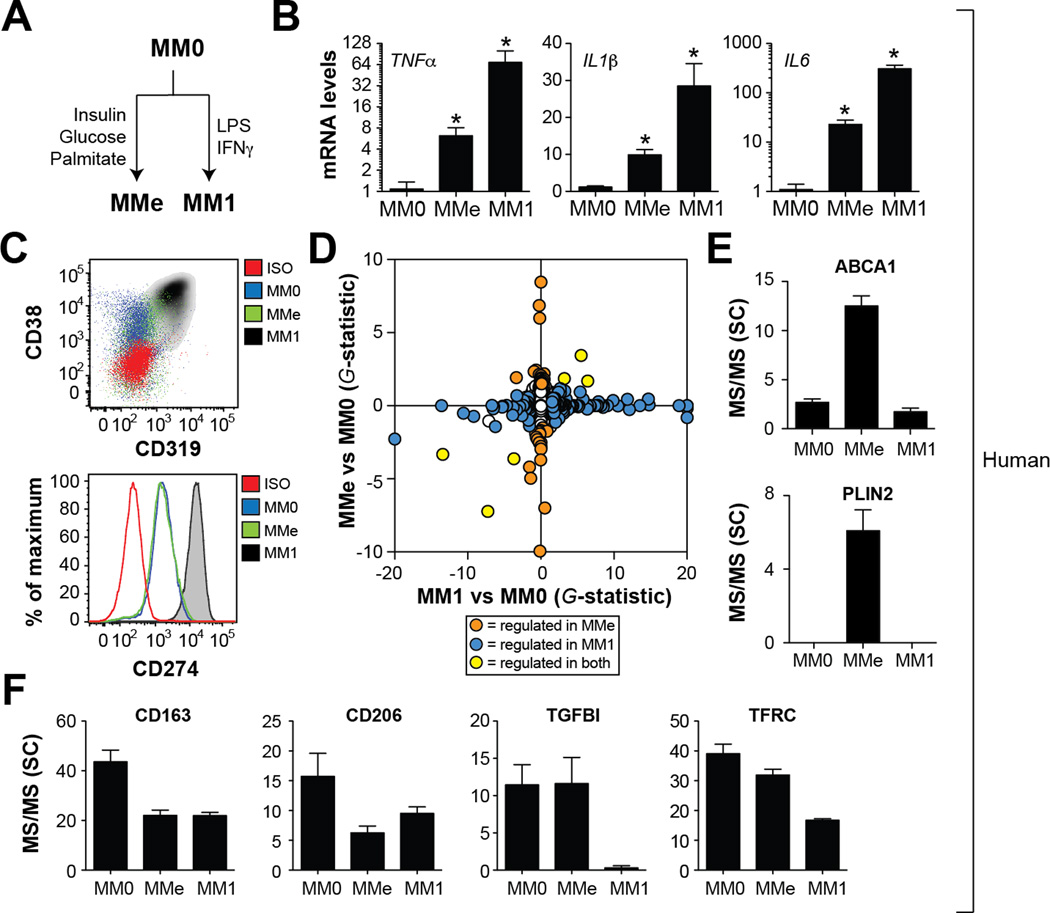
The lack of M1 cell surface marker expression by pro-inflammatory ATMs in obese humans and mice suggested that distinct pathways might promote macrophage inflammation in the context of obesity, a hypothesis that has been proposed by Hotamisligil and colleagues (Calay and Hotamisligil, 2013; Hotamisligil and Erbay, 2008). In the context of metabolic disease, macrophage inflammation may be regulated by molecules such as glucose, insulin, and palmitate, which are elevated in patients with metabolic disease (Despres and Lemieux, 2006). Thus, we hypothesized that exposure to high levels of glucose, insulin, and palmitate (‘metabolic activation’; MMe) may produce a unique pro-inflammatory macrophage phenotype.
Classical and metabolic activation (Fig. 3A) both induced pro-inflammatory cytokine expression (Fig. 3B). However, MMe macrophages failed to express cell surface markers diagnostic of classically activated macrophages including CD38, CD319, and CD274 (Fig. 3C), a finding that resembled our findings for pro-inflammatory ATMs isolated from obese humans and mice (Figs. 2B–C). To identify diagnostic cell surface markers of metabolic activation we performed membrane proteomics of MM0, MM1, and MMe macrophages. Proteomics analysis of metabolically and classically activated macrophages revealed remarkably distinct cell surface protein expression patterns (<5% overlap; Fig. 3D, Table S2). For example, cell surface proteins specifically over-expressed by MMe macrophages included ABCA1, CD36, and PLIN2 (Fig. 3E); proteins involved in lipid metabolism that have been associated with M2 macrophages. However, the M2 markers CD163, CD206, TFRC, and TGFBI (Becker et al., 2012; Martinez et al., 2006) were suppressed or not induced in MMe macrophages (Fig. 3F), indicating that metabolic and alternative activation programs are distinct. In addition, proteomics analysis confirmed that MMe macrophages did not express markers of classical activation (Table S2).
Fig. 3. Metabolically and classically activated macrophages are distinct.
Panel A: Conditions for metabolic (MMe) or classical (MM1) activation of human monocyte-derived macrophages. Panel B: Pro-inflammatory cytokine expression. Panel C: Flow cytometric analysis of cell surface CD38, CD319, and CD274 levels in MM0, MM1, and MMe macrophages. ISO= isotype control. Panel D: Plasma membrane proteomics analysis of MM0, MM1, and MMe macrophages. Panels E-F: Quantification of cell surface proteins in MM0, MM1, and MMe macrophages by mass spectrometry (spectral count, SC). Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to MM0; N=6. See also Table S2.
Adipose tissue media promotes ‘metabolic activation’ of macrophages
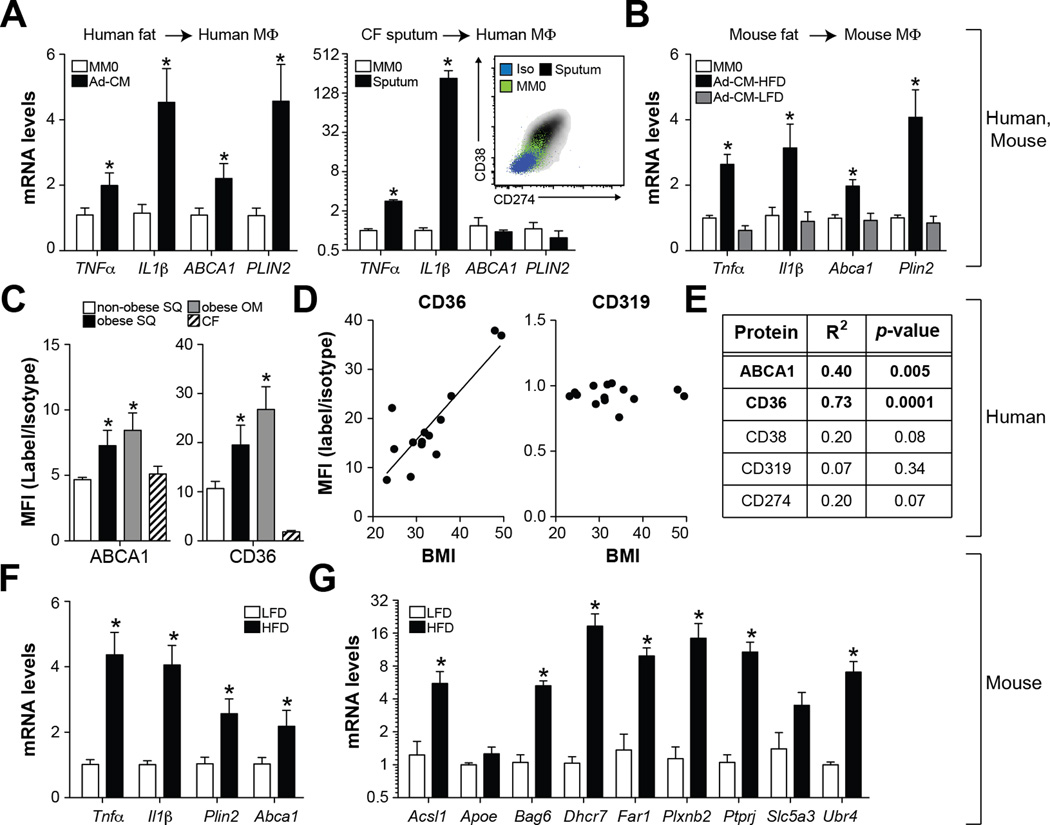
We considered the possibility that experimental exposure to mixtures of glucose, insulin, and palmitate may inadequately model in vivo ATM conditioning. To explore this, we treated human monocyte-derived macrophages with media conditioned by visceral adipose tissue from obese humans or mice, and found that IL1β, TNFα, ABCA1, and CD36 expression were all induced (Fig. 4A–B). In sharp contrast, treatment with sputum from a CF patient enhanced IL1β and TNFα expression, and surface levels of CD274 and CD38, but failed to induce CD36 and PLIN2 in human macrophages (Fig. 4A). Thus, treating naïve macrophages with disease-specific biological fluids (adipose media vs. CF sputum) reproduces the distinct surface marker expression observed in airway macrophages and ATMs in vivo (Fig. 2,4).
Fig. 4. ATMs display evidence of metabolic activation.
Panel A: qRT-PCR analysis of human MM0 macrophages treated with media conditioned by human omental fat (Ad-CM) or sputum collected from a CF patient. Inset: Surface levels of CD38 and CD274. Panel B: qRT-PCR analysis of murine macrophages (MM0) treated with media conditioned by epididymal adipose tissue collected from obese (Ad-CM-HFD) or lean (Ad-CM-LFD) mice. Panels C-E: Omental (OM) and subcutaneous (SUBQ) adipose tissue was collected from obese subjects (BMI>30kg/m2) undergoing bariatric surgery (N=7). For comparative purposes, subcutaneous adipose tissue was collected from non-obese subjects (BMI<30kg/m2) undergoing abdominoplasty (N=7). Panel C: Cell surface levels of CD36 and ABCA1 in ATMs; results are signal-to-noise ratio in mean fluorescence intensity (MFI). Panel D: Relationships between BMI and cell surface CD36 or CD319 levels on ATMs (N=14). Panel E: Summary of linear regression analyses of cell surface protein levels on ATMs and BMI. Panels F-G: Male C57BL/6 mice were place on a low-fat diet (LFD) or high-fat diet (HFD) for 16 weeks and ATMs from epididymal fat were purified and analyzed by qRT-PCR. Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to MM0 or non-obese or LFD; N=4–7. See also Fig. S3.
Because adipocytes undergo lipolysis during culture and preferentially release palmitate into the medium, we hypothesized that palmitate was the major driver of metabolic activation. To test this hypothesis, we compared MMe macrophages with MM0 macrophages treated with glucose, insulin, or palmitate in isolation. Treatment with palmitate was sufficient to drive PLIN2 and IL1β expression, suggesting that it may be the main driver of metabolic activation (Fig. S4). We further confirmed that palmitate levels at the lower limit in type 2 diabetic patients (0.1mM) (Reaven et al., 1988) could stimulate the MMe phenotype as well (Fig. S4).
ATMs of obese humans and mice display evidence of ‘metabolic activation’
We investigated if markers of metabolic activation were present on ATMs in vivo by quantifying ATM cell surface ABCA1 and CD36 levels in omental and subcutaneous adipose tissue from obese subjects undergoing bariatric surgery. As before, subcutaneous adipose tissue obtained from non-obese (BMI<30kg/m2) subjects undergoing abdominoplasty were used for comparative purposes. Cell surface CD36 and ABCA1 levels were elevated on ATMs from omental and subcutaneous adipose tissue in obese relative to non-obese subjects and relative to airway macrophages of CF patients (Fig. 4C, Fig. S3). Moreover, linear regression analysis revealed strong relationships between BMI and ATM CD36 levels (R2 = 0.73, p=0.0001, N=14) or ABCA1 levels (R2 = 0.40, p=0.02, N=14) in subcutaneous adipose tissue (Figs. 4D–E), suggesting that macrophage metabolic activation correlates with adiposity. In sharp contrast, such relationships were absent for markers of M1 macrophages assessed in the same patients (CD38, CD319, CD274; Figs. 4D–E).
Analysis of ATMs from obese mice produced similar findings. Indeed, we observed increases in the expression of pro-inflammatory cytokines (Tnfα and Il1β) and MMe markers (Plin2, Abca1) in ATMs isolated from obese mice (Fig. 4F). Moreover, ATMs isolated from obese mice overexpressed 7 of 9 additional MMe markers identified in our proteomics analysis of human MMe macrophages (Fig. 4G). Collectively, these findings are consistent with the notion that palmitate released from adipocytes promotes macrophage metabolic activation in vivo.
Manuscript re-write begin.
Pro-inflammatory signaling does not drive MMe surface marker expression
The fact that M1 and MMe macrophages differed markedly in surface marker expression was surprising given that palmitate is thought to mimic LPS action by binding to TLRs (Himes and Smith, 2010; Shi et al., 2006). This finding led us to hypothesize that different signaling pathways mediate the M1 and MMe phenotypes. To begin to test this hypothesis, we used bioinformatics to identify specific cell signaling pathways linked to cell surface proteins expressed by MM1 (but not MMe) macrophages.
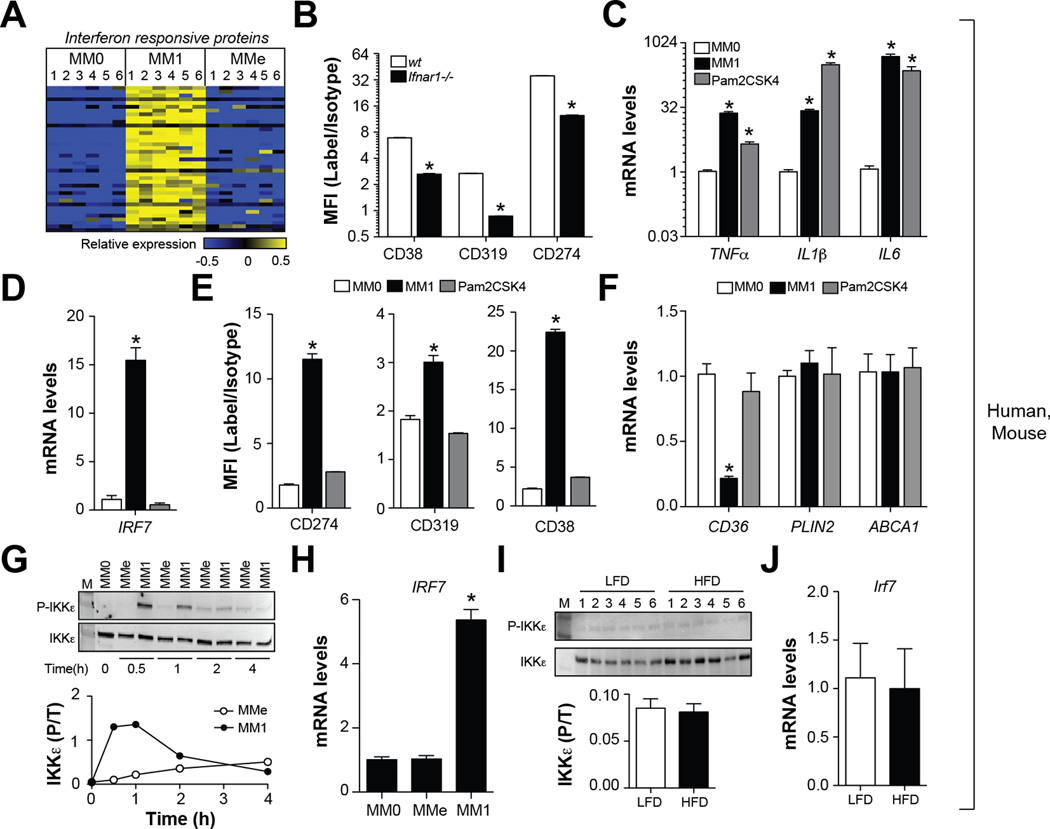
LPS binding to TLR4 stimulates pro-inflammatory cytokine production through the NFκB pathway and induces the type I interferon response via IRF7 (Beutler, 2004). Consistent with this idea, bioinformatics analysis suggested that cell surface proteins selectively induced by MM1 macrophages were associated with the type I interferon response (p=10−30; Fig. 5A). To confirm the role of the type I interferon pathway in regulating M1 surface markers, we performed two experiments that engaged the NFκB pathway without a type I interferon response. First, we compared CD38, CD274, and CD319 expression in MM1 cells made from wild type and type I interferon pathway deficient (Ifnar−/−) mice (Muller et al., 1994). Ablation of Ifnar1 significantly lowered cell surface CD38, CD274, and CD319 expression in M1 macrophages (Fig. 5B). Second, we stimulated human macrophages with the TLR2 agonist Pam2CSK4. Pam2CSK4 induced IL1β, TNFα, and IL6 expression to levels similar to MM1 macrophages (Fig. 5C) but did not elicit the type I interferon response (Toshchakov et al., 2002) (Fig. 5D) or CD38, CD274, and CD319 expression (Fig. 5E). In addition, Pam2CSK4 treatment did not elevate ABCA1, CD36, and PLIN2, suggesting that this agonist does not stimulate metabolic activation (Fig. 5F).
Fig. 5. Metabolic activation fails to induce the type I interferon response.
Panel A: Heatmap depicts regulation of interferon-regulated proteins across human MM0, MM1, and MMe cells (blue = down-regulated, yellow = up-regulated). Panel B: Comparison of M1 cell surface marker levels in MM1 macrophages made from wt and Ifnar−/− mice. Panels C-F: Human MM0 macrophages were classically activated or treated with Pam2CSK4, a selective agonist for TLR2. Panel C: Pro-inflammatory cytokine expression. Panel D: IRF7 expression. Panel E: M1 cell surface markers were quantified by flow cytometry. Panel F: qRT-PCR analysis of MMe markers. Panels G-H: Human MM0 macrophages were classically or metabolically activated. Panel G: Immunoblotting for total and phosphorylated (P-Ser172) IKKε. Panel H: IRF7 expression. Panels I-J: Male C57BL/6 mice were place on a low-fat diet (LFD) or high-fat diet (HFD) for 16 weeks and ATMs from epididymal fat were isolated. Panel I: Immunoblotting for total and phosphorylated (P-Ser172) IKKε. Panel J: IRF7 expression in purified ATMs. Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to MM0; N=4–6. Flow cytometry results are expressed as signal-to-noise ratios in the mean fluorescence intensity (MFI). See also Fig. S5.
Given that the type I interferon pathway controls CD38, CD274, and CD319 expression in M1 macrophages, and that MMe cells express different cell surface proteins, we postulated that that the type I interferon response would not be activated in these cells. As expected, neither human nor murine MMe macrophages exhibited IRF7 gene expression and IKKε phosphorylation (Fig. 5G–H, Fig. S5), two key drivers of the type I interferon response (Honda et al., 2005). Pro-inflammatory ATMs from obese mice also did not exhibit elevated IKKε phosphorylation or IRF7 gene expression relative to lean mice (Fig. 5I–J).
Since MMe macrophages fail to engage the type I interferon response, and activation of this pathway is a key feature of TLR4 activation, we investigated whether surface markers of MMe macrophages were dependent upon the presence of TLRs. Ablation of Tlr2 or Tlr4, which mediate the pro-inflammatory effects of palmitate on macrophages (Himes and Smith, 2010; Shi et al., 2006), attenuated Il1β expression but did not alter Plin2 expression in MMe macrophages (Fig. 6A). Collectively, these studies demonstrate that unlike MM1 macrophages, cell surface markers of MMe macrophages are independent of the type I interferon pathway and TLR activation.
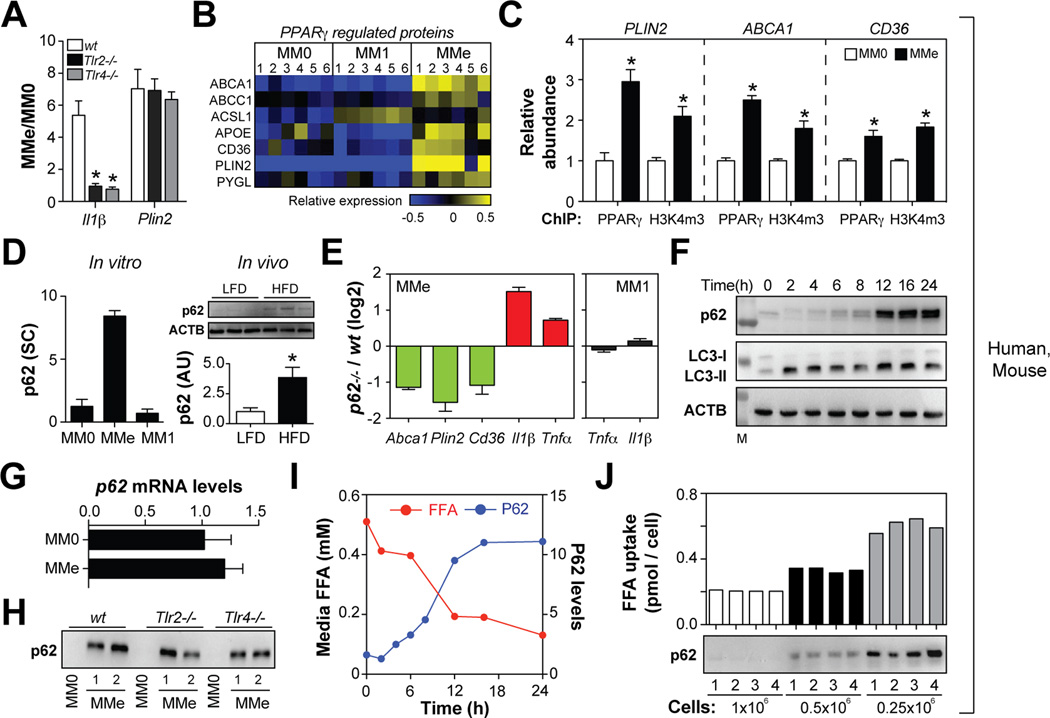
Fig. 6. p62 and PPARγ promote MMe marker expression and limit inflammation.
Panel A: Comparison of gene expression in MMe macrophages made from wild-type (wt), Tlr2−/−, and Tlr4−/− mice. Panel B: Heatmap depicts regulation of PPARγ-regulated proteins across human MM0, MM1, and MMe cells (blue = down-regulated, yellow = upregulated). Panel C: PPARγ binding to its target gene promoters is increased in MMe relative to MM0 macrophages, and correlated with abundance of the open chromatin mark H3K4m3. Relative abundance of MM0 ChIP levels were arbitrarily set to 1. Panel D: Protein levels of p62 in human macrophages in vitro (spectral counts, SC) and in vivo (arbitrary units, AU) in murine ATMs isolated from mice fed a low-fat (LFD) or high-fat diet (HFD). Panel E: Comparison of gene expression in MMe and MM1 macrophages from wild-type and p62−/− mice. Panel F: Kinetics of p62 accumulation and LC3-I/-II levels in human MMe macrophages. Panel G: p62 mRNA levels. Panel H: p62 levels in MM0 and MMe macrophages made from wild-type (wt), Tlr2−/−, and Tlr4−/− mice. Panel I: Kinetics of p62 accumulation and FFA uptake by human MMe macrophages. Panel J: Varying numbers of human macrophages were metabolically activated and palmitate uptake and p62 levels were determined. Where applicable, results are means and SEMs; *, denotes p<0.05, t-test relative to MM0 or LFD; N=3–6. See also Figs. S6, S7.
p62 and PPARγ promote MMe surface marker expression
What molecular mechanisms drive cell surface marker expression in MMe macrophages? We used two approaches to exploit our proteomics data to identify signaling pathways controlling cell surface marker expression in MMe macrophages.
First, we used bioinformatics analysis and found that surface proteins selectively induced in MMe macrophages were involved in lipid metabolism and regulated by PPARγ (Fig. 6B, p=10−7). Consistent with the informatics data, chromatin immunoprecipitation (ChIP) studies demonstrated increased binding of PPARγ to the promoters of PLIN2, ABCA1, and CD36 in human and murine MMe macrophages (Fig. 6C, Fig. S6) and PPARγ promoter binding correlated with histone H3K4 trimethylation (H3K4m3), an epigenetic mark for open chromatin (Fig. 6C). Furthermore, a PPARγ antagonist (T0070907; 1µM) reduced ABCA1, PLIN2 and CD36 expression in MMe macrophages (Fig. S6).
Second, we searched the membrane proteome for proteins with well-established roles in cell signaling that were selectively induced in MMe macrophages. This analysis identified a strong and selective induction of sequestome-1 (p62) (Fig. 6D). Moreover, p62 was significantly elevated in ATMs from obese relative to lean mice (Fig. 6D). p62 is a scaffolding protein that regulates a variety of cell signaling cascades (Komatsu et al., 2007). Previous studies suggest that p62 regulates adipocyte lipid metabolism and adiposity in mice (Muller et al., 2013; Rodriguez et al., 2006). To determine if p62 regulates the MMe phenotype, we compared macrophages from wild-type and p62-deficient (p62−/−) mice and found Plin2, Abca1, and Cd36 expression was reduced in p62−/− cells (Fig. 6E). Thus, p62 and PPARγ regulate expression of CD36, ABCA1, and PLIN2 in MMe macrophages.
p62 and PPARγ limit inflammation during ‘metabolic activation’
The induction of PPARγ in pro-inflammatory MMe macrophages seemed surprising given that saturated FFAs are poor PPARγ ligands (Wahli and Michalik, 2012) and that PPARγ exerts anti-inflammatory effects (Odegaard et al., 2007). We examined the effect of PPARγ on the inflammatory phenotype of MMe macrophages by treating cells with the PPARγ antagonist T0070907. Consistent with an anti-inflammatory role of PPARγ (Odegaard et al., 2007), blocking PPARγ with T0070907 increased expression of IL1β in MMe cells (Fig. S6). The p62 induction identified in MMe macrophages (Fig. 4D) also had an anti-inflammatory effect, as p62−/− macrophages markedly over-produced Tnfα and Il1β in response to metabolic, but not classical activation (Fig. 6E). These data indicate that p62 and PPARγ attenuate pro-inflammatory cytokine production during metabolic activation, which may in part explain why the sterile inflammation associated with metabolic disease is often of lower intensity than that induced by bacterial infection.
Palmitate uptake induces P62 accumulation by inhibiting autophagy
What mechanism induces p62 during metabolic activation? Whereas p62 is normally degraded by the autophagolysosome, it accumulates under conditions of failed autophagy (Komatsu et al., 2007). Consistent with a blockade in autophagy in MMe macrophages, p62 protein accumulated in a time dependent fashion (Fig. 6F) and its accumulation could not be explained by increased transcription (Fig. 6G). Similarly, LC3-II protein, another marker of failed autophagy (Shpilka et al., 2011), also accumulated in MMe macrophages (Fig. 6F). Moreover, treatment with chloroquine (an inhibitor of autophagy) induced LC3-II accumulation in MM0 macrophages, but not in MMe macrophages (Fig. S7).
How could metabolic activation inhibit autophagy? One possibility is that this inhibition of autophagy is linked to the pro-inflammatory state. Arguing against this possibility, MM1 macrophages did not accumulate p62 (Fig. 6D), and inactivation of Tlr2 or Tlr4 in MMe macrophages abrogated pro-inflammatory cytokine expression (Fig. 6A), but had minimal effect on p62 (Fig. 6H).
In addition to binding to TLRs, palmitate is also internalized by macrophages (Fig. 6I), and the kinetics of palmitate internalization and p62 accumulation in human macrophages was well correlated (Fig. 6I). Furthermore, experiments in which we increased the amount of palmitate ingested per cell by varying the palmitate: MMe macrophage ratio in culture markedly increased p62 accumulation (Fig. 6J). Interestingly, ablation of Cd36 had no effect on palmitate uptake by BMDMs in vitro (Fig. S4), and previous studies demonstrated normal palmitate uptake in Cd36−/− ATMs (Nicholls et al., 2011). Collectively, these findings suggest that CD36-independent palmitate uptake has a greater effect in suppressing autophagy and inducing p62 accumulation than palmitate recognition by cell surface TLRs.
Manuscript re-write end.
DISCUSSION
ATM inflammation plays a key role in obesity-associated insulin resistance and type 2 diabetes (Chawla et al., 2011; Lumeng and Saltiel, 2011; Olefsky and Glass, 2010; Wellen and Hotamisligil, 2005). A common observation reported in studying macrophages in obese mice and humans, is that ATMs overexpress pro-inflammatory cytokines via signaling pathways that are operative in M1 macrophages, yet they cannot be identified when using cell surface markers of classical activation.
One possible explanation for these discordant findings is that currently available surface markers for M1 macrophages, particularly in humans, may not be as specific as previously anticipated (Geissmann et al., 2010). To circumvent this problem, we used a proteomics approach to define a set of cell surface proteins that i) report classical activation in human monocyte-derived macrophages differentiated in the presence of M-CSF or GM-CSF (two well-established paradigms for generating macrophages in vivo) ii) are maintained in macrophages exposed to mixed M1/M2 stimuli, iii) are conserved between human and mouse macrophages, and iv) are validated in vivo in a setting of chronic bacterial infection (CF patients).
Our findings show that markers identified by such stringent approaches still fail to characterize pro-inflammatory ATMs from obese subjects, both in omental and subcutaneous adipose depots. Similar findings were obtained in our analyses of ATMs isolated from obese mice in a DIO model. Collectively, these data provide strong evidence that pro-inflammatory ATMs do not adopt the classically activated M1 phenotype.
Our demonstration that cell surface markers of classical activation are primarily regulated by the type I interferon response may have important implications for how pro-inflammatory ATM phenotypes (and macrophage phenotypes in other diseases) are conceptualized in vivo. Thus, dissociation of mechanisms that control cell surface marker expression (how we detect macrophages) from pathways that control cytokine production (how we detect inflammation) enables these processes to be regulated independently. In a setting of classical activation, both pathways are co-regulated by LPS (Beutler, 2004).
However, our data show that metabolic stimuli promote inflammation without activating the type I interferon response, a finding that is consistent with the inability of M1 markers to identify MMe macrophages in vitro and pro-inflammatory ATMs in vivo. Moreover, this concept may not be restricted to settings of sterile inflammation as bacterial products that are primarily recognized by TLR2 also produce inflammation without activating the type I interferon response (Toshchakov et al., 2002), an observation that we confirmed experimentally. Based on these findings, it is likely that the repertoire of cell signaling pathways induced by a particular pro-inflammatory stimulus, and not necessarily the extent of inflammation, determines the cell surface markers that will ultimately be useful for interrogating that cell type in vivo.
Thus, we propose that cell surface markers of macrophages be used not to identify ‘cellular phenotypes’, but rather as robust indicators of specific cell signaling pathways that are activated in a given microenvironment. Our approach for using disease-specific conditions to define novel surface markers and delineate mechanisms driving their expression can therefore provide insights into the signaling pathways engaged and the functional properties of the macrophages. In this respect, proteins regulated by the type I interferon response, which identify M1 macrophages, modulate host anti-viral and anti-bacterial responses (Honda et al., 2005). On the other hand, metabolic dysfunction and nutrient excess impose unique challenges for ATMs. Insulin resistance in the adipocyte leads to a continuous and excessive exposure of ATMs to FFA, particularly saturated FFA such as palmitate, which our data suggest plays a key role in triggering metabolic activation of macrophages. Thus, the induction of proteins involved in lipid metabolism, which delineate MMe macrophages, may enable these macrophages to appropriately buffer their environment from excessive lipids while maintaining cell health. Importantly, saturated FFAs up-regulate pro-inflammatory cytokine expression in ATMs and fail to induce the M2 markers CD163, CD206, TFRC, and TGFBI (Becker et al., 2012; Martinez et al., 2006), properties that clearly distinguish these cells from M2 macrophages, which also induce lipid metabolism through PPARγ activation (Odegaard et al., 2007).
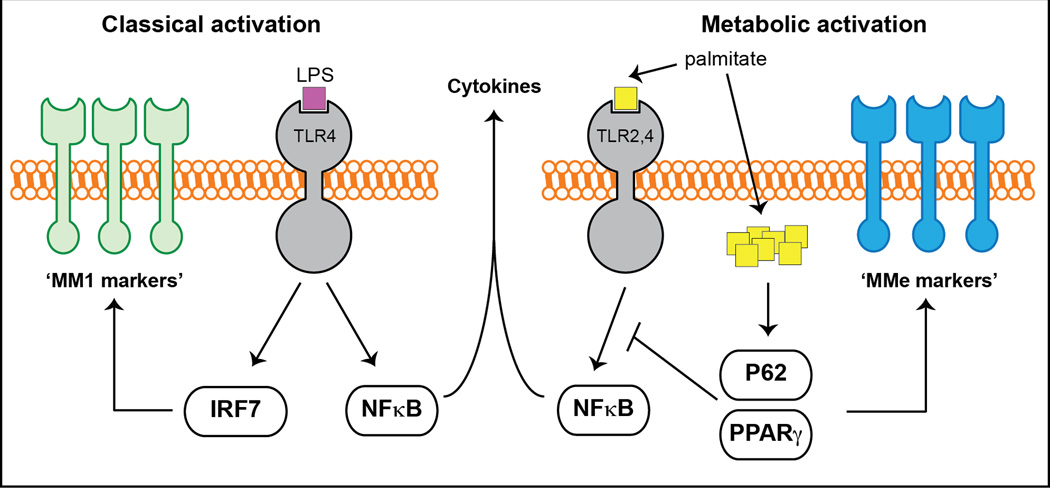
On a molecular level, the metabolically activated macrophage phenotype is mediated by at least two independent mechanisms: a pathway involving palmitate binding to cell surface TLRs that drives pro-inflammatory cytokine production, and a pathway mediated by palmitate internalization which activates p62 and PPARγ, thereby promoting lipid metabolism and limiting inflammation (Fig. 7). The balance between these two processes determines the overall response of macrophages to metabolic dysfunction (ie. pro- or anti-inflammatory), and can produce complex macrophage phenotypes spanning the spectrum between the ‘M1-like’ and ‘M2-like’ states. Indeed, our analyses demonstrate strong correlations between cell surface CD36 and ABCA1 levels on ATMs (markers often associated with M2 macrophages, (Martinez et al., 2006)) and metabolic dysfunction in obese humans and mice. Concordant with our findings, previous studies suggested the presence of mixed macrophage phenotypes in obese human subjects and mice (Sica and Mantovani, 2012; Zeyda et al., 2007).
Fig. 7. A molecular model for metabolic and classical activation of macrophages.
See text for details.
Our model further predicts that excessive accumulation of palmitate within the macrophage would further attenuate or silence inflammation via p62 and PPARγ activation, and promote a more ‘M2-like’ ATM phenotype characterized by increased lipid metabolism. Factors influencing the amount of FFAs ingested by macrophages include adipose tissue mass, adipocyte size, and extent of insulin resistance in the adipocyte, all of which would increase with the degree of metabolic dysfunction in patients with type 2 diabetes. In mice, we would expect that duration of high-fat feeding regimens and the mouse model of obesity used (Lepob/ob develop more severe metabolic dysfunction than DIO) to greatly influence the ATM phenotype observed. Consistent with these assumptions, previous studies demonstrated that obesity-induced shifts in ATMs from a ‘M2-like’ to a ‘M1-like’ phenotype (Lumeng et al., 2007) are reversed when mice are fed a high-fat diet for extended periods of time (Shaul et al., 2010). Similarly, recent work by Ferrante and colleagues showed that ATMs of obese Lepob/ob mice (obese BW = 50–70g) are not inflamed, but rather are characterized by increased lipid metabolism (Xu et al., 2013). In our analyses of more mildly obese and insulin resistant mice (DIO, obese BW = 30–35g) we observed up-regulated cytokine expression in ATMs. However, we also noted increases in genes involved in lipid metabolism suggesting that the antiinflammatory pathway is operative, but not yet dominant.
In summary, our findings demonstrate that metabolic dysfunction leads to a macrophage phenotype that is mechanistically distinct from classical activation, suggesting that disease-specific environments drive macrophage inflammation via different mechanisms. Importantly, our data do not imply that treating macrophages with adipose tissue or mixtures of glucose, insulin, and palmitate can holistically model the ATM phenotype since conditioning environments in vivo are more complex and dynamic. However, our data suggest that metabolic activation is a suitable model for conceptualizing ATM phenotype and function, as it reconciles the seemingly contradictory findings of elevated pro-inflammatory cytokine expression in a cell expressing markers of an ‘M2-like’ phenotype. Future gene knockout studies that specifically attenuate signaling pathways driving metabolic activation will be required to more carefully delineate its contribution to ATM inflammation and metabolic disease phenotypes in vivo.
EXPERIMENTAL PROCEDURES
Ethics
All human studies were approved by the Institutional Review Boards at the University of Washington (IRB#13293), Seattle Children’s Hospital (IRB#31279), and Fred Hutchinson Cancer Research Center (IRB#6874). Animal studies were approved by the Institutional Animal Care and Use Committee (ACUP#72209) at the University of Chicago.
Differentiation and activation of human monocyte-derived macrophages
Peripheral blood monocytes were isolated from healthy donors and differentiated to macrophages in the presence of M-CSF (125ng/mL) or GM-CSF (10ng/mL) as previously described (Martinez et al., 2006). Treatments and activations were performed for 24h as follows: MM1: LPS (100ng/mL) and IFNγ (20ng/mL), MM2: IL4 (20ng/mL), MMe: glucose (30mM) and insulin (10nM) and palmitate (0.4mM); human adipose tissue conditioned media (1/10 dilution); Pam2CSK4 (100ng/mL); Chloroquin (100mM). Resultant macrophage populations were subjected to analysis by flow cytometry, qRT-PCR, immunoblotting, and plasma membrane proteomics.
Plasma membrane proteomics
Macrophage plasma membrane proteins were isolated using a membrane-impermeable, biotinylation reagent (N-hydroxysulfosuccinimide-SS-biotin; Pierce) as previously described (Becker et al., 2012). Mass spectrometric analyses and statistical analyses of proteomics data were performed essentially as previously described (Becker et al., 2010). Full details are provided in the online supplement.
Bioinformatics analysis
Functional analyses of the proteomics data were generated through the use of IPA (Ingenuity® Systems, www.ingenuity.com). Informatics data were analyzed by the hypergeometric test with Benjamini-Hochberg correction.
Subject recruitment (elective abdominal surgeries)
Human adipose tissue was obtained from anonymous donors undergoing abdominoplasty (N=7) at Seattle Plastic Surgery, or from study participants undergoing laparoscopic intraabdominal surgeries (N=7) at the Puget Sound Surgery Center in Edmonds, WA. Exclusion criteria included smoking; abuse of alcohol or other drugs; pregnancy; history of cardiovascular, autoimmune, or other chronic inflammatory disease; or current or recent use of anti-diabetic, anti-inflammatory, or steroid hormone drug.
Adipose tissue collection and processing
Human adipose tissue was collected by surgeons conducting abdominoplasty (subcutaneous only) or intraabdominal surgery (subcutaneous and omental) and processed within 10 minutes by study staff. Stromavascular cells were isolated as previously described (Hagman et al., 2012). In some instances, a 100mg piece of freshly harvested adipose tissue was cultured in 1mL media for 24h to obtain human adipose tissue conditioned media.
Subject recruitment (Cystic Fibrosis)
Spontaneously expectorated sputum was collected in clinic, or sent overnight on ice from the patient’s home. Study participants were older than 18 years of age and had a confirmed diagnosis of CF. Exclusion criteria included significant hemoptysis within 2 weeks of collection or a prior lung transplant.
Sputum processing
To solubilize sputum, samples were mixed with an equal volume of 0.1% DTT in PBS and placed on ice for 30min with intermittent vortexing. Samples were passaged through sterile cotton gauze and filtered through 70 µm nylon gauze cell strainers. Cells were centrifuged at 300×g for 10 min at 4°C, and washed 4 times with 50ml RPMI to reduce debris. Cells were counted and assessed for viability using trypan blue exclusion. If contaminated with saliva, salivary components were aspirated from the sample prior to sputum processing.
Mouse studies
Male C57BL/6 mice were obtained from Jackson laboratories. BMDMs were isolated as previously described (Becker et al., 2012) and activated along the MM1 and MMe pathways using identical conditions as for the human macrophages. For DIO studies, mice were fed a low-fat (Harlan Tekland; 2918) or high-fat (Research Diets Inc.; D12451) diets for 16 weeks. Epididymal adipose tissue was collected, washed, subjected to collagenase digestion to isolate stromavascular cells, and ATMs were purified using anti-CD11b coupled magnetic beads (Miltyeni).
Flow cytometry
Fluorochrome labeled cells were analyzed according to the workflows presented in Fig. S2, S3. Analyses were conducted using a Canto-II or LSRII flow cytometer (BD Biosciences) and data were analyzed using FlowJo software v.9.4.11. Protein levels were quantified by mean fluorescence intensity and normalized to isotype to facilitate comparison between patients.
Antibodies
Antibodies for flow cytometric measurements of ABCA1, CD1c, CD11c, CD14, CD15, CD16, CD36, CD38, CD45, CD274, and CD319 were purchased from BD Biosciences (San Jose, CA), Beckman Coulter (Danvers, MA), Novus Biologicals (Littleton, CO) BioLegend (San Diego, CA), eBioscience (San Diego, CA), or Miltyeni (Auburn, CA).
Chromatin Immunoprecipitation
ChIP experiments were performed as previously described (Mutskov et al., 2002) using specific primers (Table S3) designed based on previously published PPARγ ChIP-Seq data (Mikkelsen et al., 2010) and the RXR-binding sites from the ENCODE project. Full details are provided in the online supplement.
Supplementary Material
Research highlights.
-
❖
Stimuli associated with metabolic disease promote macrophage metabolic activation.
-
❖
Metabolic activation involves distinct mechanisms and surface markers.
-
❖
ATMs of obese humans/mice overexpress metabolic activation markers, not M1 markers.
-
❖
Markers of metabolically activated macrophages are positively correlated with BMI.
ACKNOWLEGEMENTS
We would like to acknowledge Drs. Lisa L. Sowder, Phil Haeck, Mary Lee Peters, and Sharam Salemy at Seattle Plastic Surgery for providing adipose tissue from patients undergoing abdominoplasty. This research was supported by grants from the National Institutes of Health (P30DK089507, RO1HL110879, R21CA143248, P30DK017047, P30CA015704), American Heart Association (10SDG3600027), and American Diabetes Association (7-09-CT-36). Mass spectrometry experiments were performed through the Proteomics Core at Northwestern University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS:
Conceived and designed the experiments: LB, MK, BRC, KH, SH. Performed the experiments: BRC, KH, DH, EP, KQS, VM, JNK. Recruited and coordinated patients undergoing abdominoplasty and bariatric surgery: IL, PSB, RWL, MC. Performed abdominal surgeries and collected adipose tissue samples: PSB, RWL, MC. Critically reviewed the manuscript: All authors. Wrote the manuscript: LB.
REFERENCES
- Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M. IKK-beta links inflammation to obesity-induced insulin resistance. Nature medicine. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- Becker L, Gharib SA, Irwin AD, Wijsman E, Vaisar T, Oram JF, Heinecke JW. A macrophage sterol-responsive network linked to atherogenesis. Cell Metab. 2010;11:125–135. doi: 10.1016/j.cmet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Liu NC, Averill MM, Yuan W, Pamir N, Peng Y, Irwin AD, Fu X, Bornfeldt KE, Heinecke JW. Unique proteomic signatures distinguish macrophages and dendritic cells. PLoS One. 2012;7:e33297. doi: 10.1371/journal.pone.0033297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- Calay ES, Hotamisligil GS. Turning off the inflammatory, but not the metabolic, flames. Nature medicine. 2013;19:265–267. doi: 10.1038/nm.3114. [DOI] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Hagman DK, Kuzma JN, Larson I, Foster-Schubert KE, Kuan LY, Cignarella A, Geamanu E, Makar KW, Gottlieb JR, Kratz M. Characterizing and quantifying leukocyte populations in human adipose tissue: impact of enzymatic tissue processing. J Immunol Methods. 2012;386:50–59. doi: 10.1016/j.jim.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MS, Jung DY, Morel C, Lakhani SA, Kim JK, Flavell RA, Davis RJ. JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science. 2013;339:218–222. doi: 10.1126/science.1227568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes RW, Smith CW. Tlr2 is critical for diet-induced metabolic syndrome in a murine model. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:731–739. doi: 10.1096/fj.09-141929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller TD, Lee SJ, Jastroch M, Kabra D, Stemmer K, Aichler M, Abplanalp B, Ananthakrishnan G, Bhardwaj N, Collins S, et al. p62 links beta-adrenergic input to mitochondrial function and thermogenesis. J Clin Invest. 2013;123:469–478. doi: 10.1172/JCI64209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes & development. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls HT, Kowalski G, Kennedy DJ, Risis S, Zaffino LA, Watson N, et al. Hematopoietic cell-restricted deletion of CD36 reduces high-fat diet-induced macrophage infiltration and improves insulin signaling in adipose tissue. Diabetes. 2011;60:1100–1110. doi: 10.2337/db10-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annual review of physiology. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Patsouris D, Li PP, Thapar D, Chapman J, Olefsky JM, Neels JG. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 2008;8:301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven GM, Hollenbeck C, Jeng CY, Wu MS, Chen YD. Measurement of plasma glucose, free fatty acid, lactate, and insulin for 24 h in patients with NIDDM. Diabetes. 1988;37:1020–1024. doi: 10.2337/diab.37.8.1020. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. The New England journal of medicine. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Saberi M, Woods NB, de Luca C, Schenk S, Lu JC, Bandyopadhyay G, Verma IM, Olefsky JM. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009;10:419–429. doi: 10.1016/j.cmet.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul ME, Bennett G, Strissel KJ, Greenberg AS, Obin MS. Dynamic, M2-like remodeling phenotypes of CD11c+ adipose tissue macrophages during high-fat diet--induced obesity in mice. Diabetes. 2010;59:1171–1181. doi: 10.2337/db09-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagyrelated ubiquitin-like protein family. Genome biology. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nature immunology. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends in endocrinology and metabolism: TEM. 2012;23:351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW., Jr Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyda M, Farmer D, Todoric J, Aszmann O, Speiser M, Gyori G, Zlabinger GJ, Stulnig TM. Human adipose tissue macrophages are of an antiinflammatory phenotype but capable of excessive pro-inflammatory mediator production. Int J Obes (Lond) 2007;31:1420–1428. doi: 10.1038/sj.ijo.0803632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.