Abstract
Elongation factor G (EF-G) promotes the movement of two tRNAs and the mRNA through the ribosome in each cycle of peptide elongation. During translocation, the tRNAs transiently occupy intermediate positions on both small (30S) and large (50S) ribosomal subunits. How EF-G and GTP hydrolysis control these movements is still unclear. We used fluorescence labels that specifically monitor movements on either 30S or 50S subunits in combination with EF-G mutants and translocation-specific antibiotics to investigate timing and energetics of translocation. We show that EF-G–GTP facilitates synchronous movements of peptidyl-tRNA on the two subunits into an early post-translocation state, which resembles a chimeric state identified by structural studies. EF-G binding without GTP hydrolysis promotes only partial tRNA movement on the 50S subunit. However, rapid 30S translocation and the concomitant completion of 50S translocation require GTP hydrolysis and a functional domain 4 of EF-G. Our results reveal two distinct modes for utilizing the energy of EF-G binding and GTP hydrolysis and suggest that coupling of GTP hydrolysis to translocation is mediated through rearrangements of the 30S subunit.
Keywords: molecular machines, movement, protein synthesis, ribosome, translation
Introduction
The elongation phase of protein synthesis entails repetitive cycles of mRNA decoding by aminoacyl-tRNA, peptide bond formation, and translocation of tRNAs and mRNA through the ribosome, which exposes the next codon in the decoding site. Each time after an amino acid has been transferred to the growing peptide chain, the ribosome carries a peptidyl-tRNA in the A site and a deacylated tRNA in the P site (pre-translocation state, PRE). Translocation is a function inherent to the ribosome (Gavrilova & Spirin, 1972; Fredrick & Noller, 2003). Spontaneous translocation is very slow (in the time range of minutes to hours) and is driven in either direction by the differences in the free energy of tRNA–mRNA–ribosome interactions (Shoji et al, 2006; Konevega et al, 2007; Bock et al, 2013). In the cell, translocation is promoted by elongation factor G (EF-G) at the cost of GTP hydrolysis and proceeds in the milliseconds time range. EF-G, a five-domain translational GTPase, binds to the complex and facilitates the movement of tRNAs to the P and E site, respectively, which frees the A site for accepting of the next aminoacyl-tRNA, thereby completing the translation elongation cycle. Translocation is inhibited by a number of antibiotics, such as tuberactinomycins (e.g., viomycin), aminoglycosides (hygromycin B or paromomycin), as well as spectinomycin, thiostrepton, and fusidic acid (Wilson, 2013). The antibiotics inhibit translocation in diverse ways, by blocking peptidyl-tRNA in the A site, stabilizing particular intermediate conformations of the ribosome, or affecting EF-G binding to the ribosome. Therefore, antibiotics are useful tools to dissect the process of translocation.
The PRE complex is dynamic (Blanchard et al, 2004; Cornish et al, 2008) and fluctuates between the classical state, where the tRNAs are located in the A and P sites on both 30S and 50S subunits (A/A and P/P states), and hybrid states, where the acceptor domains of the tRNAs are moved toward the P and E sites, while the anticodon domains remain bound in the A and P sites (A/P and P/E states) (Moazed & Noller, 1989). At the same time, the subunits rotate with respect to one another (Frank & Agrawal, 2000; Agirrezabala et al, 2008; Julian et al, 2008). Structurally, the hybrid/rotated state is not a single intermediate, because a variety of distinct hybrid states have been identified. The classical and different hybrid/rotated substates [denoted as classical C and hybrid H1 and H2 states (Munro et al, 2007), GS1 and GS2 (Fei et al, 2008), MSI and MSII (Frank & Gonzalez, 2010), classes 2, 4A, 4B, 5 and 6 (Agirrezabala et al, 2012), R1 and R2 (Zhang et al, 2009), or PRE1-5 states (Fischer et al, 2010)] differ in the orientation of the tRNAs, the degree of subunit rotation, the conformation of the 30S subunit, and the position of the L1 stalk. Moreover, crystal and cryo-electron microscopy (cryo-EM) structures indicate that binding of EF-G with a non-hydrolyzable GTP analog to a ribosome with a tRNA in the P site induces the formation of further intermediate states of translocation on the 30S subunit (Ratje et al, 2010; Pulk & Cate, 2013; Tourigny et al, 2013; Zhou et al, 2013). Due to a swiveling motion of the 30S head domain, the tRNA anticodon domain is shifted to a position between the P and E sites on the 30S subunit, resulting in a pe/E state of the tRNA; presumably, the A-site tRNA may also attain a transient ap/P state. Further intermediates, containing both deacylated and peptidyl-tRNA and EF-G, were isolated using the antibiotics viomycin (Brilot et al, 2013) and fusidic acid (Ramrath et al, 2013). All these different structures show tRNAs in somewhat different chimeric positions, which we collectively name chimeric (CHI) states. However, the position of such chimeric intermediates on the time axis of translocation is uncertain, and the timing of 50S translocation to the final post-translocation (POST) state is unknown.
One controversial issue concerns the mechanism by which hybrid/rotated state formation contributes to EF-G-facilitated translocation. It was suggested that EF-G binding is restricted to the hybrid/rotated state and that the rate of conversion from the classic to the hybrid state determines the rate of EF-G-dependent translocation (Zavialov & Ehrenberg, 2003; Munro et al, 2010). On the other hand, EF-G binding to the ribosome and subsequent GTP hydrolysis are largely independent of the conformational state of the ribosome (Rodnina et al, 1997; Walker et al, 2008; Chen et al, 2011a; Wintermeyer et al, 2011). Recent single-molecule data suggest that EF-G can sample both states (Chen et al, 2013). In some cases (e.g., with initiator tRNAfMet in the P site), the hybrid intermediate is only short-lived and induced by EF-G binding (Chen et al, 2011a; Fei et al, 2011).
The spontaneous movements of the tRNA acceptor ends on the 50S subunit between the classical and hybrid states are rapid, taking place in the milliseconds to seconds range. Binding of EF-G in complex with a non-hydrolyzable GTP analog favors the formation of hybrid/rotated states by slowing down the transition from the hybrid to the classical state, whereas the rate of transition from the classical to hybrid/rotated state remains unaltered (Kim et al, 2007; Spiegel et al, 2007; Cornish et al, 2008; Fei et al, 2009, 2011; Munro et al, 2010; Chen et al, 2011a, 2013); Mg2+ ions have the opposite effect (Kim et al, 2007; Chen et al, 2013). However, in the hybrid A/P state, the peptidyl transferase reactivity of peptidyl-tRNA (as measured with the indicator puromycin (Pmn) reaction) remains low (Semenkov et al, 1992; Sharma et al, 2004), indicating that it has not reached the final post-translocation (POST) state on the 50S subunit. In comparison with the high tRNA mobility on the 50S subunit, the translocation of the tRNA anticodon domains together with the mRNA on the 30S subunit is intrinsically very slow (Shoji et al, 2006; Konevega et al, 2007; Fischer et al, 2010); EF-G accelerates 30S translocation by several orders of magnitude (Rodnina et al, 1997; Savelsbergh et al, 2003; Seo et al, 2006). The ribosome is intrinsically dynamic and appears to function as a Brownian machine that undergoes spontaneous conformational fluctuations driven by thermal energy, which can be coupled to directed motion (Wintermeyer et al, 2004; Spirin, 2009; Fischer et al, 2010; Frank & Gonzalez, 2010; Munro et al, 2010). EF-G is a GTP-binding protein that combines the energy regime characteristic for switch GTPases with that of motor proteins (Cunha et al, 2013). The interactions of EF-G with the ribosome and GTP hydrolysis lead to conformational changes that may (i) facilitate tRNA movement, for example by remodeling structural elements of the ribosome that limit the movement (Savelsbergh et al, 2003; Schuwirth et al, 2005; Khade & Joseph, 2011); (ii) allow EF-G to act as a Brownian pawl providing directionality of tRNA movement (Savelsbergh et al, 2000); or (iii) induce a power stroke-like movement of EF-G, thereby actively transporting tRNAs through the ribosome (Taylor et al, 2007; Chen et al, 2013). The contribution of GTP hydrolysis to the formation of translocation intermediates or the movement to the POST state on the 30S and 50S subunits is not clear. Here, we address these questions by rapid kinetic analysis using reporter groups attached to the amino terminus of peptidyl-tRNA and to the 3′ end of the mRNA. To identify reaction intermediates, we utilized translocation-specific antibiotics and EF-G mutants, including a GTPase-deficient EF-G mutant. The results reveal the timing of tRNA translocation on 30S and 50S subunits and suggest how the energy of EF-G binding and GTP hydrolysis is utilized to promote tRNA movement through the ribosome.
Results
Direct observation of 30S and 50S translocation
Previously, movement on the 30S subunit has been observed using various environmentally sensitive fluorescence reporters, such as pyrene or fluorescein attached to the 3′end of the mRNA (Studer et al, 2003; Peske et al, 2004; Dorner et al, 2006; Ermolenko & Noller, 2011). In contrast, due to the lack of suitable reporters, the movement of the tRNA acceptor end on the 50S subunit during translocation has not been examined kinetically. To study tRNA translocation on the 50S subunit, we utilized BODIPY-FL (Bpy) attached to the N-terminal methionine of the nascent peptide on peptidyl-tRNA (Bpy-MetPhe-tRNAPhe). Bpy does not interfere with translation and appears to be an ideal fluorophore due to its small size and advantageous, well-characterized spectroscopic properties (Ellis et al, 2008). The translocation of tRNA and mRNA on the 30S and 50S subunits of Escherichia coli ribosomes was monitored using reporter groups both in Bpy-MetPhe-tRNAPhe and the 3′ end of the mRNA (Alexa405, Alx) in the same complex (Fig1A, inset). The latter fluorophore was chosen because its fluorescence can be monitored selectively also in the presence of Bpy (Supplementary Fig S1). When translocation on double-labeled PRE complexes with mRNA-Alx, deacylated tRNAfMet in the P site and Bpy-MetPhe-tRNAPhe in the A site was initiated by rapid mixing with EF-G and GTP in a stopped-flow apparatus, the fluorescence of both reporters decreased with similar rates (Fig1A). The translocation efficiency was close to 80%, as verified biochemically using the Pmn test (Supplementary Fig S2). Two-exponential fitting of the traces obtained with the two labels yielded a rapid phase, which contributed >90% of the fluorescence change (referred to as amplitude) for Bpy and >80% for Alx and a slow phase (about 1 s−1), which may be due to a small portion of inactive ribosomes and was not considered further. Comparison of the rates of the predominant rapid phases of 50S and 30S translocation showed that the movements on the two subunits took place at the same rate, about 28 s−1 at saturation with EF-G (Table1).
Figure 1. Kinetics of 50S and 30S translocation.
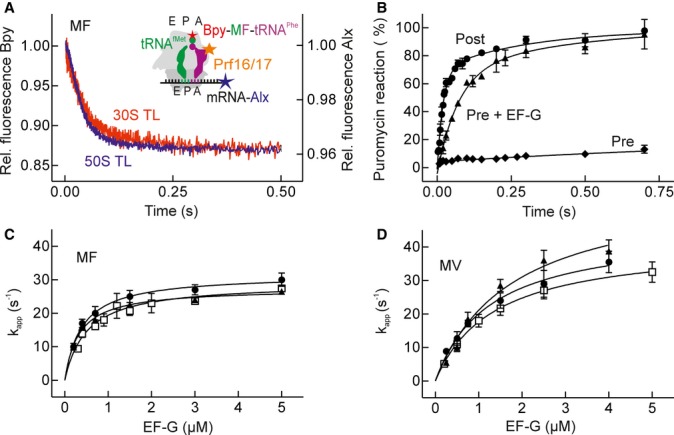
- Time courses of Bpy-MetPhe-tRNAPhe translocation on the 50S subunit (50S TL; blue trace) and mRNA-Alx translocation on the 30S subunit (30S TL; red trace) after rapid mixing of EF-G (4 μM, final concentration after mixing) with the PRE complex double-labeled with Bpy and Alx. The inset shows positions of the fluorescence reporters in the PRE complex, including the position of the proflavin (Prf) label in the D loop, which was used for comparison with previous data.
- Time-resolved Pmn reaction with PRE and POST complexes double-labeled with Bpy and Alx. PRE complexes were mixed with Pmn in the absence of EF-G (Pre; ♦) or with Pmn and EF-G–GTP (Pre + EF-G; ▴) (kPRE = 14 ± 4 s−1). POST complexes reacted with Pmn at a rate of kPOST = 48 ± 7 s−1 (Post; •). The intrinsic rate of translocation into the Pmn-reactive POST intermediate (kTL Pmn; Table1) was calculated from kPRE and kPOST (Materials and Methods). Error bars represent standard deviations (s.d.) obtained from three independent experiments.
- Concentration dependence of the apparent rate constants of Bpy-MetPhe-tRNAPhe (MF) translocation (kapp) monitored by Bpy (▴) and Alx (•). Translocation rates measured with the Prf label in fMetPhe-tRNAPhe(Prf16/17) are shown for comparison (□).
- As (C), but with Bpy-MetVal-tRNAVal (MV). Time courses of translocation (kapp) were monitored by Bpy (▴), Alx (•), or Prf in fMetVal-tRNAVal(Prf) (□).
Data information: Error bars (s.d.) in (B) and (C) were obtained from at least two independent experiments with 7 technical replicates each.
Table 1.
Rate constants of 50S and 30S translocation
| EF-G/antibiotic | 50S translocationa | 30S translocationa | ||
|---|---|---|---|---|
| k50S fast, s−1 | k50S slow, s−1 | kTl Pmn, s−1 | k30S, s−1 | |
| wtb/no antibiotic | 28 ± 3 | –c | 20 ± 6d | 27 ± 2d |
| wt/Vio | 25 ± 1 | – | ˜0 | ˜0 |
| wt/Spc | 61 ± 5 | – | n.d.e | 0.17 ± 0.01 |
| wt/Str | 11 ± 1 | – | n.d.e | 11 ± 1 |
| wt/HygB | 28 ± 2 | 0.06 ± 0.02 | 0.16 ± 0.02 | 0.08 ± 0.01 |
| EF-G(H91A) | 9 ± 1 | 0.7 ± 0.1 | 1.5 ± 0.2 | 0.9 ± 0.2 |
| EF-G(H583K) | 25 ± 1 | 0.7 ± 0.1 | 1.7 ± 0.2 | 1.0 ± 0.2 |
| EF-G(Δ4/5) | – | 0.06 ± 0.01 | ˜0.01f | 0.04 ± 0.02 |
Rate constants determined at 4 μM EF-G, 37°C.
wt, wild-type EF-G.
–, not observed, that is, the amplitude of the signal change is < 10% of the total amplitude observed with wt EF-G in the absence of antibiotics.
The reported rates for kTl Pmn and k30S pertain to the steps that constitute 80% or more of the total reaction amplitudes.
n.d., not determined. The rates could not be determined, because binding of Spc and Str significantly reduced the rate of Pmn reaction in the POST complex, precluding the deconvolution of the two rates.
From (Savelsbergh et al, 2000).
To further validate the use of Bpy as a reporter of 50S translocation, we developed a time-resolved Pmn assay to investigate the movement of the 3′ end of Bpy-MetPhe-tRNAPhe into the POST state. The Pmn reaction with peptidyl-tRNA in the P site is completed within milliseconds, which is comparable to the rate of translocation itself; hence, to derive the rate of translocation from Pmn kinetics, the two reactions have to be deconvoluted. This difficulty probably explains why the Pmn reaction was so far mostly used to study slow translocation kinetics, as observed in the presence of antibiotics (Peske et al, 2004; Pan et al, 2007). To measure the rate of authentic, unperturbed translocation with the help of Pmn, we performed experiments in a time-resolved fashion using the quench-flow technique (Fig1B). The rate of the Pmn reaction was measured upon addition of EF-G to the PRE complex and compared with that of the POST complex. The time required for the PRE complex to react includes the times for both translocation and Pmn reaction of the resulting POST state, which allowed us to elucidate the intrinsic rate of 50S translocation (designated as kTL Pmn) (Materials and Methods), about 20 s−1 (Table1). This rate is within the standard deviation of the translocation rate measured with Bpy, suggesting that the latter reporter in fact monitors the movement of the peptidyl-tRNA to the POST state; for comparison, the rate of the Pmn reaction of peptidyl-tRNA in the hybrid PRE state (A/P) is in the range 0.002–0.02 s−1 (Peske et al, 2004; Sharma et al, 2004).
For peptidyl-tRNAPhe, a translocation rate of about 25 s−1 was measured not only by the Bpy and Alx labels, as described above, but was also observed when translocation was monitored by a proflavin label attached to the elbow region of the tRNA (Fig1C), a well-characterized label previously used to study the kinetics of translocation (Rodnina et al, 1997; Savelsbergh et al, 2003; Pan et al, 2007). 50S and 30S translocations were also synchronous at 23°C, about 7 s−1 (Supplementary Fig S3A), in agreement with values obtained with the proflavin label (Pan et al, 2007). Similar translocation rates were obtained with a PRE complex containing Bpy-MetVal-tRNAVal (Fig1D, Supplementary Fig S3B), which in the absence of EF-G is prone to undergo spontaneous reverse translocation (Konevega et al, 2007). Thus, EF-G–GTP binding and GTP hydrolysis promote the synchronous forward movement of both acceptor and anticodon domains of the tRNAs together with the mRNA, and the elbow region of peptidyl-tRNA on the two ribosomal subunits, regardless of the intrinsic thermodynamic gradient of tRNA affinities, which would favor the POST state for fMetPhe-tRNAPhe and the PRE state for fMetVal-tRNAVal (Konevega et al, 2007).
Previous work using the proflavin reporter in tRNAfMet(Prf20) suggested that the displacement of the P site-bound deacylated tRNA may precede the movement of the A-site peptidyl-tRNA (Pan et al., 2007). The time courses of the reaction showed the formation of a short-lived high-fluorescence intermediate, called INT (Pan et al, 2007), followed by a slower reaction (Supplementary Fig S3C and D). At 25°C, INT formation was faster (15–25 s−1) than 50S and 30S translocations (6 s−1), as reported previously (Pan et al, 2007). However, at 37°C, INT formation took place at a rate (30 s−1) which is not significantly different from that of peptidyl-tRNA movement on the 50S and 30S subunits (28 s−1) and was followed by a slower step (13 s−1), which probably reflects tRNA movement through the E site (Cunha et al, 2013). Thus, the formation of the INT state, that is, the EF-G-facilitated movement of the elbow of deacylated tRNA on the 50S subunit, may precede or accompany full translocation of tRNAs, depending on experimental conditions.
50S translocation on ribosomes in classical or hybrid states
Previous cryo-EM studies have shown that the ribosomes in classical and rotated/hybrid conformations are in equilibrium with each other under experimental conditions similar to those used here (Blanchard et al, 2004; Munro et al, 2007; Agirrezabala et al, 2008; Cornish et al, 2008; Fei et al, 2008; Julian et al, 2008; Fischer et al, 2010; Chen et al, 2013). Single-molecule FRET studies suggested that increasing the concentration of Mg2+ ions increased the population of the classical state by increasing its lifetime, whereas the lifetime of the hybrid state was not affected (Kim et al, 2007; Lee et al, 2007). The effect of Mg2+ on the conversion from the hybrid (H) to the classical (C) state can be described by an equilibrium dissociation constant KHC (Fig2A). EF-G binding shifts the equilibrium toward the hybrid/rotated state by reducing the rate of the conversion from the hybrid to classical state (Cornish et al, 2008; Fei et al, 2008; Munro et al, 2010). If translocation preferentially proceeded through the hybrid/rotated state, then translocation should become slower when most of the complexes are trapped in the C state (Fig2A). To test the relationship between hybrid-state formation, EF-G binding, and EF-G-induced translocation, we examined whether shifting the dynamic equilibrium between hybrid and classical states by varying the Mg2+ concentration affected 50S translocation. The amplitude of 50S translocation did not change in the whole range of Mg2+ concentrations from 3.5 to 20 mM (37°C), which suggests that the step monitored by Bpy does not represent hybrid/rotated state formation; otherwise, the amplitude of the fluorescence change would be expected to be smaller at lower Mg2+ concentrations, which favor the hybrid state. The rate of translocation decreased with increasing Mg2+ concentrations in a logarithmic manner from 30 to 15 s−1 (Fig2B). Thus, EF-G-driven translocation on ribosomes that are predominantly in the classical state (at 20 mM Mg2+) proceeds at a similar rate (within a factor of two) as on those predominantly in the hybrid state (at 3.5 mM Mg2+).
Figure 2. Effect of hybrid-to-classical state transition on 50S translocation.
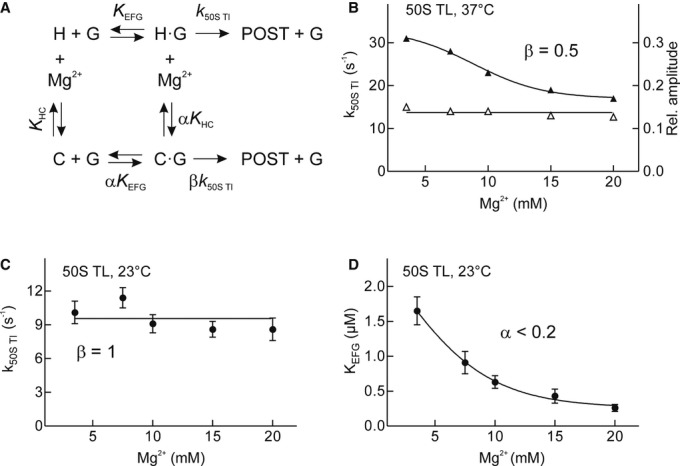
- Dynamic equilibria between hybrid (H) and classical (C) states of the ribosome and the effect of EF-G (G). Mg2+ shifts the equilibrium from H toward C [equilibrium dissociation constant KHC; (Kim et al, 2007)]. This equilibrium is affected by EF-G, which favors the H–G complex (Cornish et al, 2008), resulting in an equilibrium dissociation constant αKHC. H binds EF-G with equilibrium dissociation constant KEFG; according to the law of mass balance, C must bind EF-G with the constant αKEFG. The rates of translocation in the H–G and C–G complexes (k50S Tl) may be the same or differ by a factor β.
- Mg2+ dependence of 50S translocation rates, k50S Tl (▴), and reaction amplitudes (△) at 37°C. β is the ratio of the lowest and highest k50Tl values.
- Mg2+ dependence of 50S translocation rate at 23°C at buffer conditions used in Kim et al (2007).
- Mg2+ dependence of KEFG at conditions as in (C). The values are from the experiment shown in Supplementary Fig S4A. α is the ratio of the lowest and highest KM values.
Data information: Error bars (s.d.) in (B–D) were obtained from at least two independent experiments with 7 technical replicates each.
If EF-G shifts the equilibrium between classical and hybrid states (Cornish et al, 2008; Fei et al, 2008; Munro et al, 2010), then—in the simplest model (Fig2A)—the law of mass balance would predict that the affinities of EF-G for states C and H differ by the same factor, that is, the affinity of EF-G to the classical state should be lower than to the hybrid state. To test this prediction, we measured the kcat and KM values of EF-G-dependent 50S translocation at different Mg2+ concentrations (Supplementary Fig S4A); these experiments were carried out at 23°C for better comparison with single-molecule data obtained at buffer conditions very similar to those used in this work (Kim et al, 2007). At these conditions, the rate of 50S translocation was independent of Mg2+ (Fig2C). Surprisingly, the KM values decreased with increasing Mg2+ concentration ((Fig2D, α < 0.2), rather than increased as would be predicted by a simple model (Fig2A). Notably, because the KM value includes all microstates that contribute to the apparent affinity of EF-G, the effect on the KM value should be independent of the detailed kinetic mechanism of EF-G binding and the exact nucleotide state of EF-G at the onset of 50S translocation. These data indicate that EF-G can bind to the classical state very efficiently and promote rapid translocation, in stark contrast to previous claims (Zavialov & Ehrenberg, 2003). The linear replot of the Mg2+ dependence (Kim et al, 2007) indicates that the stabilization of the classical state C depends on one or two net Mg2+ ions (Supplementary Fig S4B). This fully accounts for the observed Mg2+ dependence of the KM values, which involves one net Mg2+, supporting the notion that the observed change in the KM values is due to the conversion of the ribosomes from the classical to the hybrid state, although an additional effect of Mg2+ on the EF-G dwell time (Chen et al, 2013) may also contribute.
Uncoupling of 50S and 30S translocation
To relate EF-G-promoted 50S translocation to conformational states of the ribosome, we used a number of well-characterized antibiotics, including viomycin, spectinomycin, streptomycin, and hygromycin B, that affect translocation in different ways. Viomycin (Vio) binds at the interface between the two subunits, promotes the formation of the rotated-state conformation of the ribosome, suppresses tRNA dynamics, perturbs EF-G dynamics when sampling the PRE state of the ribosome, and completely blocks translocation (Modolell & Vazquez, 1977; Peske et al, 2004; Ermolenko et al, 2007; Kim et al, 2007; Stanley et al, 2010; Chen et al, 2013). The addition of Vio to the PRE complex resulted in a small decrease in Bpy fluorescence, followed by an additional signal change brought about by EF-G (Fig3A). This additional amplitude indicates that the expected increase in the fraction of the rotated states, as induced by Vio (Ermolenko et al, 2007; Spiegel et al, 2007), alone does not lead to full 50S translocation. The rate of 50S translocation was the same as in the absence of antibiotic (Table1). No translocation was observed on the 30S subunit when Vio was present (Fig3B), and the reactivity of peptidyl-tRNA with Pmn remained low, as expected (Modolell & Vazquez, 1977; Peske et al, 2004). Thus, in the presence of Vio, partial 50S translocation takes place independent of 30S translocation, and the resulting intermediate is frozen in a state which is not populated during translocation without the inhibitor.
Figure 3. Uncoupling of 50S and 30S translocation by antibiotics.
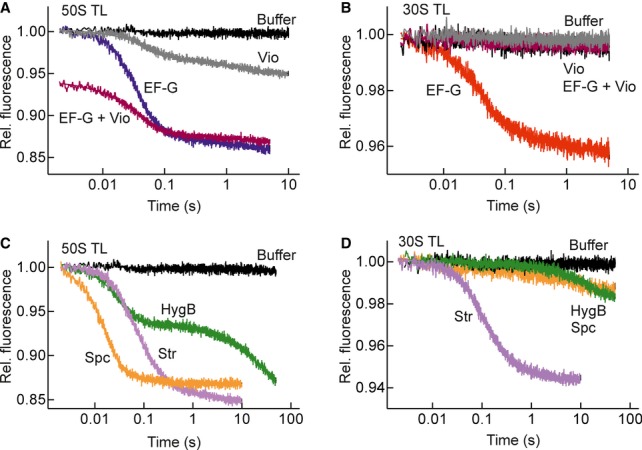
- Fluorescence change of Bpy-MetPhe-tRNAPhe upon addition of Vio alone (gray), EF-G alone (blue), EF-G to the PRE complex pre-incubated with Vio (EF-G+Vio) (magenta), or buffer (black).
- 30S translocation at conditions as in (A) or in the absence of antibiotics (EF-G, red).
- Time courses of 50S translocation in the presence of Spc (orange), HygB (green), Str (lilac), and control in the absence of EF-G (Buffer, black).
- 30S translocation with antibiotics as indicated in (C).
Spectinomycin (Spc), which binds to helix 34 in the head domain of the 30S subunit and traps a distinct swiveling state of the head domain (Borovinskaya et al, 2007), increased the rate of 50S translocation and uncoupled it from 30S translocation, which was very slow (Fig3C and D; Table1). Binding of streptomycin (Str) has a dual effect on translocation: on one hand, it traps the 30S subunit in a conformation that is both error-prone (Carter et al, 2000) and inherently more prone to rapid translocation as manifested by lowering of the transition state barrier (Peske et al, 2004). On the other hand, Str stabilizes the tRNA in the A site, which in effect disfavors translocation by decreasing the energy of the ground state (Peske et al, 2004). Str reduced the rate of both 30S and 50S translocations to a similar extent (Fig3C and D; Table1). Hygromycin B (HygB) strongly inhibited 30S translocation, whereas 50S translocation proceeded to completion. HygB binds between A and P sites at the decoding center of the 30S subunit and stabilizes nucleotide A1493 of 16S rRNA in a position which causes a steric block to the movement of tRNAs (Borovinskaya et al, 2008). With HygB present, the fluorescence change due to rapid tRNA movement on the 50S subunit was about 50% of the control, whereas the rate of the following slow step coincided with the rate of 30S translocation (Fig3C and D; Table1). The rate of tRNA movement into the Pmn-reactive state was similar to that of 30S translocation and of the second phase of 50S translocation monitored by fluorescence (Supplementary Fig S2; Table1). This result suggests the existence of an intermediate state, which is not Pmn-reactive and is stabilized by HygB, during the stepwise movement of peptidyl-tRNA on the 50S subunit from the A/A or A/P to the P/P state.
Role of GTP hydrolysis
GTP hydrolysis increases the overall rate of translocation by about 30-fold (Rodnina et al, 1997; Katunin et al, 2002; Pan et al, 2007; Cunha et al, 2013). To assess whether or not the effect is subunit-specific, we used a GTPase-deficient EF-G mutant with His91 replaced with Ala, by analogy to the H84A substitution in E. coli EF-Tu, which decreased the rate of GTP hydrolysis by six orders of magnitude (Daviter et al, 2003). EF-G(H91A) binds GTP with unchanged affinity, but does not hydrolyze GTP on the ribosome; the mutant factor supports only a single round of translocation, because its release from the ribosome is blocked (Cunha et al, 2013). When double-labeled pre-translocation complexes were mixed with EF-G(H91A), the fluorescence of Bpy and Alx decreased to similar levels as with wild-type (wt) EF-G (Fig4A and B); however, the rates were lowered substantially (Table1). The rate of 30S translocation was reduced to about 1 s−1, 30-fold lower than with wt EF-G, in agreement with previous results with non-hydrolyzable GTP analogs, such as GTPγS or caged GTP (Rodnina et al, 1997; Katunin et al, 2002). The kinetics of 50S translocation became biphasic (Fig4A), with a fast step of approximately 10 s−1 (Fig4C), which was only three times slower than with wt EF-G, and an additional slow step which contributed about 50% of the amplitude and had a rate of 1 s−1 at EF-G saturation. The latter rate coincided with the rate of 30S translocation (Fig4D) and with the movement of the peptidyl-tRNA into the Pmn-reactive POST state (Supplementary Fig S2; Table S1). Thus, when GTP hydrolysis was blocked, the two steps of 50S translocation were uncoupled. The formation of the intermediate 50S translocation state required EF-G binding, but not GTP hydrolysis, whereas the transition from the intermediate to the 50S POST state was linked to 30S translocation, which, in turn, was coupled to GTP hydrolysis.
Figure 4. Inhibition of translocation by mutations in EF-G.
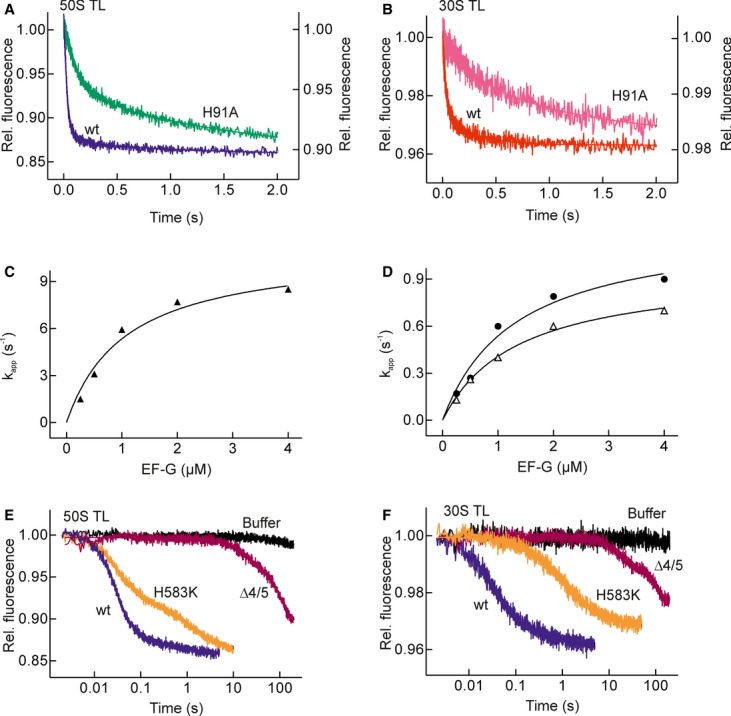
- Time courses of 50S translocation in the presence of GTPase-deficient EF-G(H91A) (green, right Y-axis) compared with wt EF-G (blue, left Y-axis).
- Time courses of 30S translocation in the presence of EF-G(H91A) (pink, right Y-axis) compared to wt EF-G (red, left Y-axis).
- Concentration dependence of apparent rate constants of the fast step of 50S translocation as induced by EF-G(H91A) (▴).
- Concentration dependence of apparent rate constants of the slow step of 50S translocation (△) and 30S translocation (•).
- Time courses of 50S translocation with wt EF-G (blue), EF-G(H583K) (orange), EF-G(Δ4/5) (magenta) or without factor (Buffer, black).
- Time courses of 30S translocation with wt EF-G and EF-G mutants (colors as in (E)).
Role of domain 4 of EF-G
Domain 4 of EF-G is known to play an important role in translocation by coupling the conformational rearrangements of EF-G to forward movement of the tRNAs (Rodnina et al, 1997; Peske et al, 2000, 2004; Savelsbergh et al, 2000; Taylor et al, 2007). With an EF-G mutant lacking domains 4 and 5 (EF-G(Δ4/5)) both 50S and 30S translocations were severely inhibited (Fig4E and F; Table1). Because EF-G(Δ4/5) binds to the ribosome and hydrolyzes GTP as rapidly as wt EF-G (Savelsbergh et al, 2000), translocation with EF-G(Δ4/5) must be inhibited directly after the GTP hydrolysis step, presumably by an inhibition of a rate-limiting rearrangement of the ribosome that controls tRNA translocation and Pi release (Savelsbergh et al, 2003).
The replacement of the conserved His583 at the tip of domain 4 with Lys (EF-G(H583K)) inhibits translocation, but does not interfere with GTP hydrolysis, Pi release, or the dissociation of EF-G from the ribosome (Savelsbergh et al, 2000). Similar to the effect of HygB (Fig3C and D) or the H91A mutation (Fig4A and B), 50S translocation in the presence of EF-G(H583K) was biphasic (Fig4E). The rapid step was independent of 30S translocation and led to an intermediate that was not reactive with Pmn (Supplementary Fig S2). The slower step proceeded at the rate of 30S translocation and led to a Pmn-reactive POST state (Table1). The rate of 30S translocation was reduced substantially, suggesting a specific effect of domain 4 on mRNA–tRNA movement on the 30S subunit.
Discussion
Role of rotational PRE substates for translocation
The present kinetic analysis of correlated tRNA movements during translocation on both 50S and 30S subunits provides new insights into translocation catalysis by EF-G (Fig5). Peptide bond formation, which results in deacylated tRNA in the P site and peptidyl-tRNA in the A site, promotes the formation of the hybrid/rotated states and accelerates EF-G-dependent translocation (Semenkov et al, 2000; Horan & Noller, 2007; Marshall et al, 2008; Walker et al, 2008), and there are several ways of how this acceleration may be achieved. One possibility is that the initial recruitment of EF-G is restricted to the hybrid/rotated state of the ribosome (Valle et al, 2003; Zavialov & Ehrenberg, 2003; Munro et al, 2010). This model is not supported by the present data (Fig2; Supplementary Fig S4), which rather suggest that EF-G can bind to the pre-translocation complex in either state, hybrid or classical, and—by inference—to any substate between the two. This is in line with reports that indicated similar association rate constants (or arrival rates) for EF-G–GTP and binding affinities of EF-G to ribosomes in different conformational states (Rodnina et al, 1997; Walker et al, 2008; Chen et al, 2011a, 2013; Wintermeyer et al, 2011). Interestingly, a recent single-molecule study reported that the average dwell times for EF-G–GTP sampling rotated and non-rotated states were similar, whereas EF-G–GDPNP dissociated 7.5 times more rapidly from the non-rotated state (Chen et al, 2013). This finding may suggest that the GTP or GDPNP-bound states of EF-G have different preferences to the ribosome sub-states, which should be considered when analyzing EF-G recruitment to the ribosome. It also may explain why in cryo-EM only a complex of EF-G–GDPNP with rotated ribosomes was observed (Valle et al, 2003), as the low concentration of complexes used in cryo-EM would probably not allow the isolation of readily dissociating complexes. Furthermore, initial sampling (Chen et al, 2013) and the following EF-G engagement (Munro et al, 2010) appear to be distinct steps, which may favor different substates of the ribosome. Finally, the identity of the tRNA bound to the ribosome appears to influence the distribution of conformational states of the ribosome and EF-G dynamics (Fei et al, 2011).
Figure 5. Schematic of translocation.
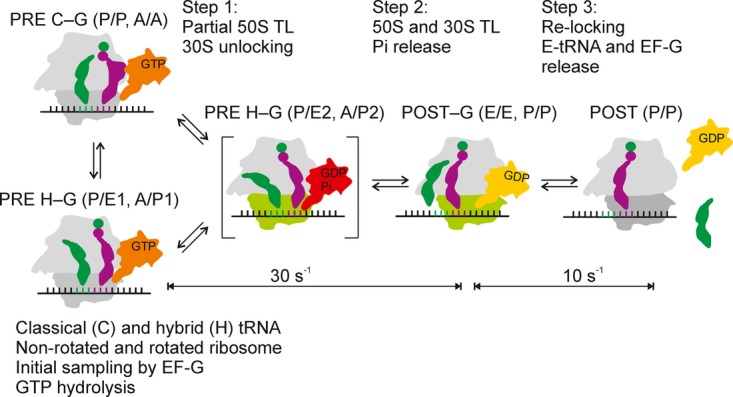
EF-G–GTP binds to the PRE complex in the classical (C) state (with tRNAs in the P/P and A/A orientations) or in the hybrid/rotated state (H) (with tRNA positions designated as P/E1 and A/P1) (Walker et al, 2008; Chen et al, 2011a), resulting in PRE C–G or H–G intermediates, followed by rapid GTP hydrolysis.
Step 1, 30S unlocking (indicated by green subunit) (Savelsbergh et al, 2003) concomitant with the movement of the P-site tRNA toward the E site (INT position (Pan et al, 2007); designated here as P/E2) and the A-site tRNA into the intermediate position (A/P2). A/P2 state is observed with HygB, EF-G(H91A), and EF-G(583K) prior to 30S translocation. The intermediate shown in square brackets normally is short-lived and accumulates when there is no GTP hydrolysis.
Step 2, translocation on the 30S subunit and the completion of translocation on the 50S subunit, resulting in the POST complex with tRNAs in the classical E/E and P/P states and EF-G occupying the A site (POST–G).
Step 3, relocking of the 30S subunit and dissociation of EF-G and deacylated tRNA, leading to the POST complex.
Movements of individual elements of the ribosome (e.g., proteins L1, L11, and L12, or the 30S subunit head) and various chimeric states of 30S are not shown. The rate of unperturbed 50S and 30S translocation is 30 s−1 (this paper), the rate of the following rearrangements, which probably involves back-rotation of the 30S subunit head, is 10 s−1 (Cunha et al, 2013).
Another model that links hybrid state formation to translocation suggests that the transition from the classical to the hybrid/rotated state comprises the rate-limiting state of translocation (Valle et al, 2003; Zavialov & Ehrenberg, 2003; Munro et al, 2010). We show here that the rate of 50S translocation is largely independent of whether the ribosome assumes the classical or hybrid/rotated state. Similarly, mutations in 23S rRNA or tRNA (e.g., initiator tRNAfMet) that disfavor hybrid state formation only moderately (within a factor of five) affect the rate of 30S translocation (Dorner et al, 2006; Walker et al, 2008; Fei et al, 2011), consistent with the reported differences in EF-G arrival times (Chen et al, 2013). Furthermore, recent molecular dynamics simulations suggest that spontaneous subunit rotation and movements within the 30S subunit during the interconversion of classic and hybrid states are very rapid, taking place in the sub-microseconds time range (Bock et al, 2013). Taken together, these results challenge the notion that the formation or stabilization of the hybrid/rotated state provides the rate-limiting step of EF-G-dependent translocation. Rather, our results are consistent with a model in which the initial reversible EF-G binding to the ribosome (“sampling”) (Chen et al, 2013) is followed by GTP hydrolysis and the rapid formation of a short-lived hybrid/rotated intermediate (Chen et al, 2011a; Fei et al, 2011), which is then stabilized by EF-G docking on the PRE state (Cornish et al, 2008; Munro et al, 2010; Fei et al, 2011; Chen et al, 2013), followed by further conformational adjustments upon formation of the POST state (Gao et al, 2009; Tourigny et al, 2013). Thus, whereas the hybrid/rotated states are authentic intermediates of translocation that reflect the intrinsic dynamics of the ribosome and are crucial for translocation, the kinetics of these fluctuations are unlikely to be rate-limiting for EF-G recruitment or tRNA translocation.
Substates of EF-G-dependent translocation
Binding of EF-G–GTP and GTP hydrolysis induces rapid (at about 30 s−1) concerted movements of the tRNAs on both subunits to a state which is likely to be an early POST state (Fig5). We assign this state as POST, rather than PRE, because peptidyl-tRNA in that state rapidly reacts with Pmn. This early POST complex (POST–G, because EF-G is still bound in this complex) undergoes further rearrangements into a late POST complex, which entail further movement of deacylated tRNA through the E site, backward rotation of the subunit head, and dissociation of EF-G (Guo & Noller, 2012; Chen et al, 2013; Cunha et al, 2013) and takes place at a rate of about 10 s−1 (Cunha et al, 2013). The rates of tRNA and mRNA movement into the POST–G state coincide with the rate of Pi release (Savelsbergh et al, 2003). Although the intrinsic rates of translocation on the 50S and 30S subunits and of Pi release may be different, they are synchronized by the preceding rate-limiting step of 30S unlocking (Savelsbergh et al, 2003). In comparison, the movement of tRNAs through the unlocked state of the ribosome appears intrinsically rapid (Savelsbergh et al, 2003; Wen et al, 2008; Munro et al, 2010). EF-G might accelerate translocation by opening of the groove where the codon–anticodon complexes reside and displacing ribosome elements that act as hurdles for 30S translocation (Schuwirth et al, 2005; Zhang et al, 2009). Alternatively, EF-G may facilitate 30S head domain movements, consistent with recent findings suggesting that the movement of the 30S head coincides with mRNA movement (Ratje et al, 2010; Guo & Noller, 2012). In keeping with this notion, the translocation rate can be increased by manipulating the decoding center of the 30S subunit toward higher conformational flexibility, for example by cleavage of 16S rRNA between nucleotides A1493 and G1494 (Lancaster et al, 2008), by disrupting the interactions between the A-site codon and ribosomal residues in the decoding site (Khade & Joseph, 2011), or by altering intersubunit bridges B1a, B4, B7a, and B8 (Liu & Fredrick, 2013).
The concerted movement of the tRNA acceptor and anticodon domains on 50S and 30S subunits, respectively, can be uncoupled by antibiotics or mutations in EF-G. This allowed us to isolate discrete substates of translocation that cannot be observed otherwise. The results obtained with HygB, EF-G(H91A), and EF-G(H583K) suggest that 50S translocation proceeds in at least two steps (PRE A/P1→ PRE A/P2→ POST–G; Fig5). During the first step (Step 1), which is independent of 30S translocation, the 3′ end of peptidyl-tRNA moves from the classic (C) or hybrid (A/P1) state into the A/P2 state; in that state, the reaction with Pmn remains slow (Table1), suggesting that the A/P2 state differs from the final POST state. It is likely that the P-site tRNA concomitantly moves to the state designated as INT (Pan et al, 2007). EF-G–GTP binding—independent of GTP hydrolysis—is sufficient to promote this movement, probably by re-shaping the landscape of spontaneous thermal motions within the ribosome. On the other hand, the energy of EF-G–GTP binding alone is not sufficient to promote rapid movement on the 30S subunit. Rather, the second step (Step 2, Fig5), which entails both 30S translocation and tRNA movement on the 50S subunit from the PRE A/P2 state into the early POST–G state, is driven by GTP hydrolysis, which couples conformational rearrangements of EF-G to the engagement of domain 4 with the 30S codon-anticodon complex. In the final step, deacylated tRNA spontaneously dissociates from the ribosome (Semenkov et al, 1996; Uemura et al, 2010; Chen et al, 2011b, 2013), probably coupled to the backward swiveling of the 30S head, counter-rotation of the 30S body, and dissociation of EF-G (Ermolenko & Noller, 2011; Guo & Noller, 2012; Chen et al, 2013; Cunha et al, 2013), resulting in the final POST (P/P) state.
Some antibiotics stabilize intermediates that are distinct from the A/P2 state. With Spc, 30S translocation is strongly inhibited, while the 3′ end of peptidyl-tRNA moves into a state which—according to the extent of the fluorescence change we observe—is very close to the POST state. This notion is consistent with the reported high Pmn reactivity of the Spc-blocked translocation intermediate, which is two orders of magnitude faster than that of the PRE state and only tenfold slower than that of the POST state (Pan et al, 2007). An EF-G mutant lacking domain 1, which promotes 50S translocation to the Pmn-reactive state, but not 30S translocation (Borowski et al, 1996), may stabilize a similar intermediate. Also in the presence of Vio, the 3′ end of peptidyl-tRNA appears to occupy a position close to the P site; however, the Pmn reactivity of that state remains low and 30S translocation is blocked; the structure of that state was recently determined by cryo-EM (Brilot et al, 2013). Yet another intermediate of translocation was recently characterized, using the antibiotic fusidic acid and GDP for stabilization (Ramrath et al, 2013). In that state, the tRNAs were in chimeric ap/P and pe/E states, and the 30S subunit assumed a conformation with swiveled head. Because fusidic acid blocks EF-G in a state where the mRNA has already moved by one codon (Spiegel et al, 2007) and the 3′ end of peptidyl-tRNA is reactive with Pmn (Savelsbergh et al, 2009), that state may be classified as an early POST state, although it is not clear whether it is identical to the POST–G state described here. Together, kinetic and structural data suggest that, in addition to several distinct PRE and POST states, there are also several chimeric states where the 3′ ends of tRNA have moved partially toward the authentic POST state, or the translocation on the 30S subunit is incomplete due to the swiveled conformation of the 30S head domain.
The role of EF-G and GTP hydrolysis
EF-G is a GTP-binding protein, which combines the characteristics of a switch GTPase, which upon Pi release changes to a low-affinity conformation allowing for the dissociation of the factor from the ribosome, with those of a motor that accelerates movement by a conformational change induced by GTP hydrolysis (Rodnina et al, 1997; Cunha et al, 2013). GTP hydrolysis is utilized to accelerate mRNA and tRNA movements on the 30S subunit, thereby synchronizing tRNA and mRNA movements on the two ribosomal subunits. The rate of 30S translocation closely correlates with the ability of domain 4 of EF-G to engage with the A site (Munro et al, 2010), presumably by interactions of the tip of domain 4 with the A-site codon-anticodon complex (Savelsbergh et al, 2000; Taylor et al, 2007; Gao et al, 2009; Ratje et al, 2010; Ramrath et al, 2013). The rate of EF-G engagement is controlled by conformational changes of EF-G (Peske et al, 2000; Wriggers et al, 2000; Taylor et al, 2007) which are driven by GTP hydrolysis (Rodnina et al, 1997; Katunin et al, 2002; Pan et al, 2007; Munro et al, 2010) and coupled to ribosome rearrangements through the delayed release of Pi (Savelsbergh et al, 2003, 2005; Pan et al, 2007). Domain 4 may promote 30S translocation by altering the conformation of the ribosome, for example by opening the mRNA-binding cleft or by stabilizing the open conformation of the E-site gate (Borovinskaya et al, 2008; Ratje et al, 2010; Pulk & Cate, 2013; Tourigny et al, 2013). EF-G domain 4 may disrupt the interactions of the mRNA–tRNA duplex with two universally conserved bases (A1492, A1493) in the ribosomal decoding center (Taylor et al, 2007; Ramrath et al, 2013) or restrict backward movement (Frank & Agrawal, 2000; Gao et al, 2009; Pulk & Cate, 2013; Tourigny et al, 2013; Zhou et al, 2013), thus coupling GTP hydrolysis to 30S translocation in a way that resembles the power stroke of motor proteins. This view is supported by the recent crystal structures of EF-G trapped on the ribosome in the pre-hydrolysis state (Pulk & Cate, 2013; Tourigny et al, 2013; Zhou et al, 2013). These structures predict that GTP hydrolysis is associated with a rearrangement of EF-G that is initiated at the nucleotide binding pocket, pivots around domain 3, and results in a movement of domain 4 that affects the interactions with the 30S subunit. One attractive possibility suggested by those structures is that GTP hydrolysis is required to allow for the backward rotation of the 30S subunit head, which would complete the transition to the POST state, and there is kinetic evidence in favor of this suggestion (Cunha et al, 2013). Conformational dynamics of EF-G that links GTP hydrolysis, Pi release, and factor dissociation to conformational rearrangements of the ribosome may account for the multiple kinetically significant steps prior to the counter-rotation of the subunits, which completes the translocation process (Chen et al, 2013). Recent force measurements suggested that EF-G generates a power stroke of 89 pN during ribosome translocation (Yao et al, 2013), also in line with a motor function of the factor. Thus, the translocating ribosome–EF-G complex combines features of a Brownian machine and a power-stroke motor. EF-G orchestrates the rapid synchronous progression of tRNAs and mRNA through the ribosome by combining the two energy regimes.
Materials and Methods
Buffers and reagents
Ribosomes, translation factors, and tRNAs were from E. coli, if not stated otherwise. The experiments were carried out in buffer A (50 mM Tris–HCl, pH 7.5, 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2) at 37°C.
Ribosomes, mRNAs, tRNAs, and translation factors
Chemicals were from Roche Molecular Biochemicals, Sigma Aldrich, or Merck. Radioactive compounds were from Hartmann Analytic. Ribosomes from E. coli MRE 600, f[3H]Met-tRNAfMet, [3H]Met-tRNAfMet, [14C]Phe-tRNAPhe, EF-Tu, and initiation factors were prepared as described (Rodnina et al, 1997, 1999; Savelsbergh et al, 2003). Proflavin-labeled tRNAPhe (yeast) and tRNAVal (E. coli) were prepared according to the published protocols (Wintermeyer et al, 1979; Milon et al, 2007). The following mRNA constructs (IBA, Göttingen) were used (start codon and first elongation codon are underlined):
mMFAlx405: GUUAACAGGUAUACAUACUAUGUUUGUUAUUAC-Alx405
mMVAlx405: GUUAACAGGUAUACAUACUAUGGUUUUCAUUAC-Alx405
Bpy-Met-tRNAfMet
[3H]Met-tRNAfMet was prepared and purified by HPLC as described (Milon et al, 2007), except that N10-formyltetrahydrofolate was omitted. Modification of [3H]Met-tRNAfMet at the α-amino group with BODIPY-FL sulfosuccinimidyl ester (Bpy-SSE; Invitrogen, D6140) was carried out by incubating [3H]Met-tRNAfMet (30 μM) with a 130-fold excess of Bpy-SSE (4 mM) in 20 mM HEPES buffer (pH 8.5) for 4 min at 0°C. The reaction was stopped by adding potassium acetate (pH 5) to 0.2 M, and the tRNA was precipitated by adding 2.5 volumes of ice-cold ethanol. Excess dye was removed by four additional precipitation steps. The resulting tRNA pellet was dried, dissolved in H2O and stored at −80°C. The concentration of tRNA was determined by radioactivity counting, and the extent of Bpy modification was assessed photometrically, using extinction coefficients of 75,000 M−1 cm−1 for Bpy (505 nm), and 575,000 M−1 cm−1 for the tRNA (260 nm), yielding labeling efficiencies of >80%. Bpy-[3H]Met-tRNAfMet was fully active in initiation complex formation.
Cloning, expression, and purification of EF-G and EF-G mutants
The gene coding for EF-G containing a C-terminal His tag was cloned into pET24a(+). The H583K mutation was introduced using the QuikChange protocol. The gene coding for EF-G was cloned into pTXB1, the H91A mutation was introduced by the QuikChange protocol, and the EF-G(Δ4/5) construct was produced by deletion of amino acids R475 to K704 using the Phusion polymerase deletion protocol (NEB). EF-G and EF-G mutants were overexpressed in BL21(DE3) and purified as described (Cunha et al, 2013). EF-G(H91A) and EF-G(Δ4/5) were expressed using plasmid pTXB1 with an N-terminal intein cleavage site, a chitin-binding domain, and a His tag (Cunha et al, 2013). Concentrations were determined either by spectrophotometry at 280 nm, using an extinction coefficient of 64,300 M−1 cm−1 for EF-G (Nguyen et al, 2010) or by SDS–PAGE and densitometry using a reference protein of known concentration.
Ribosome complexes
To prepare initiation complexes, ribosomes (1 μM) were incubated with a threefold excess of mRNA and a 1.5-fold excess each of IF1, IF2, IF3, and f[3H]Met-tRNAfMet, or Bpy-[3H]Met-tRNAfMet, in buffer A containing 1 mM GTP for 30 min at 37°C. Ternary complex EF-Tu–[14C]Phe-tRNAPhe–GTP was prepared by incubating EF-Tu (2 μM) with GTP (1 mM), phosphoenolpyruvate (3 mM), and pyruvate kinase (0.5 mg ml−1) for 15 min at 37°C and then with [14C]Phe-tRNAPhe (1 μM) for 1 min. To form the PRE complex with fMetPhe-tRNAPhe in the A site, initiation complex and ternary complex were mixed and incubated for 1 min at 37°C.
To form ternary complex containing [14C]Phe-tRNAPhe(Prf) or [14C]Val-tRNAVal(Prf), deacylated proflavinated tRNA (25 μM) was incubated in buffer A with the respective purified aa-tRNA-synthetase, GTP (1 mM), ATP (3 mM), DTT (1 mM), [14C]Val or [14C]Phe (45 μM), pyruvate kinase (0.1 mg ml−1) and EF-Tu (48 μM) for 30 min at 37°C. Ternary complex was purified by size-exclusion chromatography on a Biosuite 250 HR column using an Alliance HPLC system (Waters). Fractions containing ternary complex were mixed with initiation complex and incubated for 1 min. Resulting PRE complexes were purified by centrifugation through a 1.1 M sucrose cushion in buffer A with 21 mM MgCl2. Pellets were dissolved in buffer A with 21 mM MgCl2, and tRNA binding was verified by nitrocellulose filtration. The functional activity of pre-translocation complexes was tested by the Pmn assay (Rodnina et al, 1997). For the kinetic experiments, the concentration of MgCl2 was lowered to 7 mM.
Rapid kinetic methods
Fluorescence experiments were carried out in buffer A containing 1 mM GTP at 37°C, if not stated otherwise, using a stopped-flow apparatus (SX-20MV; Applied Photophysics). Bpy fluorescence was excited at 470 nm and detected after passing a KV500 cut-off filter (Schott). Alx405 fluorescence was excited at 400 nm and measured after passing a KV418 cut-off filter (Schott). Prf fluorescence was excited at 470 nm and measured after passing a KV500 cut-off filter (Schott). Equal volumes of pre-translocation complexes (80 nM) were rapidly mixed with EF-G or EF-G(H91A) at different concentrations as indicated. Experiments with the EF-G mutants H583K and Δ4/5 and translocation experiments in the presence of antibiotics (Vio 200 μM, Spc 900 μM, Str 20 μM, and HygB 20 μM) were performed at saturating EF-G concentration (4 μM). Time courses were evaluated using TableCurve software by exponential fitting using two exponential terms with two apparent rate constants, kapp1 and kapp2 and the fraction of the fast step in the overall reaction amplitude (% fast). EF-G titrations were fitted to hyperbolic functions. Average kapp values and standard deviations (SD) were obtained from at least seven time courses.
The time-resolved Pmn reaction was performed in a quench-flow apparatus (KinTek) by mixing Bpy/Alx-labeled PRE or POST complexes (0.2 μM, final concentration after mixing) with Pmn (10 mM) and EF-G (4 μM) in buffer A at 37°C. The reactions were quenched with 50% formic acid and mixed with 1.5 M sodium acetate saturated with MgSO4. Bpy-[3H]Met[14C]Phe-Pmn was extracted into ethyl acetate and quantified by radioactivity counting (Rodnina et al, 1997). The time required for the PRE complex to react (apparent rate constant kPRE) includes the time needed for translocation (kTL Pmn) and for the Pmn reaction of the resulting POST state (kPOST). Deconvolution of the translocation rate from the two values (1/kTl Pmn = 1/kPRE − 1/kPOST) gives the rate of tRNA translocation. Mean rates and standard deviations were derived from three independent experiments.
Acknowledgments
We thank Anna Bursy, Olaf Geintzer, Sandra Kappler, Theresia Uhlendorf, Tanja Wiles, and Michael Zimmermann for expert technical assistance. The work was supported by a grant of the Deutsche Forschungsgemeinschaft in the framework of Sonderforschungsbereich 860 (M.V.R.) and by RFBR, research project No. 13-04-40212-H comfi (to A.L.K.).
Author contributions
All authors conceived the research, designed experiments, analyzed the data, and wrote the paper. WH, CEC, ALK, and FP prepared materials and conducted experiments.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information for this article is available online: http://emboj.embopress.org
References
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci USA. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LV, Blau C, Schroder GF, Davydov II, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmuller H. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol. 2013;20:1390–1396. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Fredrick K, Cate JH. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008;14:1590–1599. doi: 10.1261/rna.1076908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski C, Rodnina MV, Wintermeyer W. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc Natl Acad Sci USA. 1996;93:4202–4206. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, Cooperman BS. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011a;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Stevens B, Kaur J, Smilansky Z, Cooperman BS, Goldman YE. Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc Natl Acad Sci USA. 2011b;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013;1:e24315. doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JP, Bakke CK, Kirchdoerfer RN, Jungbauer LM, Cavagnero S. Chain dynamics of nascent polypeptides emerging from the ribosome. ACS Chem Biol. 2008;3:555–566. doi: 10.1021/cb800059u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL., Jr Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Fei J, Richard AC, Bronson JE, Gonzalez RL., Jr Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol. 2011;18:1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova LP, Spirin AS. Mechanism of translocation in ribosomes. II. Activation of spontaneous (nonenzymic) translocation in ribosomes of Escherichia coli by p-chloromercuribenzoate. Mol Biol Rep. 1972;6:248–254. [PubMed] [Google Scholar]
- Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- Lancaster LE, Savelsbergh A, Kleanthous C, Wintermeyer W, Rodnina MV. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol Microbiol. 2008;69:390–401. doi: 10.1111/j.1365-2958.2008.06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fredrick K. Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucl Acids Res. 2013;41:565–574. doi: 10.1093/nar/gks1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Konevega AL, Peske F, Fabbretti A, Gualerzi CO, Rodnina MV. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Modolell J, Vazquez D. The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B, Ticu C, Wilson KS. Intramolecular movements in EF-G, trapped at different stages in its GTP hydrolytic cycle, probed by FRET. J Mol Biol. 2010;397:1245–1260. doi: 10.1016/j.jmb.2010.02.039. [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W. Thiostrepton inhibits turnover but not GTP hydrolysis by elongation factor G on the ribosome. Proc Natl Acad Sci USA. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Semenkov Y, Shapkina T, Makhno V, Kirillov S. Puromycin reaction for the A site-bound peptidyl-tRNA. FEBS Lett. 1992;296:207–210. doi: 10.1016/0014-5793(92)80380-y. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS. The ribosome as a conveying thermal ratchet machine. J Biol Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2·80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2013;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Savelsbergh A, Konevega AL, Peske F, Katunin VI, Semenkov YP, Fischer N, Stark H, Rodnina MV. Function of elongation factor G in translocation and ribosome recycling. In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes. Structure, Function, and Dynamics. Wien, New York: Springer; 2011. pp. 329–338. (eds) [Google Scholar]
- Wintermeyer W, Schleich HG, Zachau HG. Incorporation of amines or hydrazines into tRNA replacing wybutine or dihydrouracil. Methods Enzymol. 1979;59:110–121. doi: 10.1016/0076-6879(79)59073-9. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Agrawal RK, Drew DL, McCammon A, Frank J. Domain motions of EF-G bound to the 70S ribosome: insights from a hand-shaking between multi-resolution structures. Biophys J. 2000;79:1670–1678. doi: 10.1016/S0006-3495(00)76416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Li Y, Tsai TW, Xu S, Wang Y. Noninvasive Measurement of the Mechanical Force Generated by Motor Protein EF-G during Ribosome Translocation. Angew Chem Int Ed Engl. 2013;52:14041–14044. doi: 10.1002/anie.201307419. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


