SUMMARY
The maintenance of nuclear compartmentalization by the nuclear envelope and nuclear pore complexes (NPCs) is essential for cell function; loss of compartmentalization is associated with cancers, laminopathies and aging. We uncovered a pathway that surveils NPC assembly intermediates to promote the formation of functional NPCs. Surveillance is mediated by Heh2, a member of the LEM (Lap2-emerin-MAN1) family of integral inner nuclear membrane proteins, which binds to an early NPC assembly intermediate, but not to mature NPCs. Heh2 recruits the Endosomal Sorting Complex Required for Transport (ESCRT) – III subunit Snf7 and the AAA-ATPase Vps4 to destabilize and clear defective NPC assembly intermediates. When surveillance or clearance is compromised, malformed NPCs accumulate in a Storage of Improperly assembled Nuclear Pore Complexes compartment, or SINC. The SINC is retained in old mothers to prevent loss of daughter lifespan, highlighting a continuum of mechanisms to ensure nuclear compartmentalization.
INTRODUCTION
The genome is compartmentalized by the nuclear envelope (NE), which establishes a double-membrane barrier to most molecules. The integrity of the NE is disrupted in several cancer cells (Vargas et al., 2012), and in cells expressing “laminopathy” (Capell and Collins, 2006) mutations (De Vos et al., 2011) suggesting that loss of nuclear compartmentalization is an input to human disease. Consistent with this idea, oxidative damage of nuclear pore complexes (NPCs), which control the traffic of molecules across the NE, can lead to nuclear protein aggregates in old neurons (D'Angelo et al., 2009) – a hallmark of neurodegeneration (Woulfe, 2007). Moreover, there is a disruption of nuclear transport in cells expressing a dominant-negative form of lamin A, which causes Hutchinson-Gilford progeria (Kelley et al., 2011). Cumulatively, these observations underscore the importance of nuclear compartmentalization to cellular homeostasis and predict mechanisms that maintain NE integrity and NPC function.
Loss of NE integrity could be triggered by defects in pathways that remodel the NE like the assembly of the NPC. NPC assembly during interphase requires the local fusion of the inner and outer nuclear membranes (INM and ONM) to form a pore in the NE (Fernandez-Martinez and Rout, 2009); fusion is coupled to the recruitment of ~30 distinct NPC constituents (nucleoporins or nups). Since nups are found in multiple copies in a mature NPC, NPC assembly requires at least 450 nup protomers in yeast (Alber et al., 2007a; 2007b) and many more in human cells (Bui et al., 2013). The spatiotemporal regulation of these events and the biochemical intermediates in this process remain ill defined, although integral INM proteins of the LEM (Lap2-emerin-MAN1)(Yewdell et al., 2011) and SUN (Sad1, UNC84) families (Talamas and Hetzer, 2011) might help form early NPC assembly intermediates.
The complexity of coupling the recruitment of hundreds of proteins with membrane remodeling events that, if compromised, could lead to a loss of NE integrity predicts the existence of mechanisms to ensure NPC assembly fidelity. Moreover, defects in NPC assembly would likely result in the accumulation of oligomeric protein assemblies, potentially embedded in the NE. How cells might deal with these putative protein aggregates remains unknown but it has been hypothesized that nuclear protein aggregates might be cleared through a vesicular intermediate in the NE lumen (Rose and Schlieker, 2012). Such a mechanism would be analogous to the nuclear egress of several viruses (Mettenleiter et al., 2013) and large ribonucleoprotein (megaRNP) granules (Speese et al., 2012). The machinery that contributes to these events includes the AAA-ATPase torsins, which reside in the ER lumen (Jokhi et al., 2013), but might also include the Endosomal Sorting Required for Transport (ESCRT) complexes (Lee et al., 2012).
The ESCRT machinery is composed of five modules, ESCRT 0, I, II, III and the AAA-ATPase Vps4 and is best known for sorting ubiquitylated membrane protein receptors into multivesicular bodies (MVBs)(McCullough et al., 2013). The invagination and fission of endosomal membranes to form MVBs is thought to be the result of the polymerization of ESCRT-III components into a spiral and the action of Vps4, which likely also recycles ESCRT-III (Henne et al., 2013; Hurley and Hanson, 2010). ESCRT-III functions in several other cellular contexts, including in HIV budding (Garrus et al., 2001; Votteler and Sundquist, 2013), plasma membrane wound repair (Jimenez et al., 2014), cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007), centrosome duplication (Morita et al., 2010) and exosome secretion (Baietti et al., 2012). At least one additional study implicates ESCRTs in a nuclear function, although few nuclear binding partners have been identified (Stauffer et al., 2001).
Interestingly, both fission and budding yeast, which are divergent by about 500 million years (Rhind et al., 2011), share a genetic interaction between VPS4 and a membrane ring nup gene, POM152 (Fig. 1A), thus implicating the ESCRT machinery in a conserved function at NPCs (Costanzo et al., 2010; Frost et al., 2012). Consistent with this idea, compromised ESCRT-III/Vps4 function leads to the accumulation of misassembled NPCs into a NE compartment that we term the SINC for Storage of Improperly assembled Nuclear Pore Complexes. Our data support the interpretation that ESCRT-III is recruited to the NE by integral INM proteins to surveil and clear defective NPC assembly intermediates to ensure the fidelity of NPC assembly.
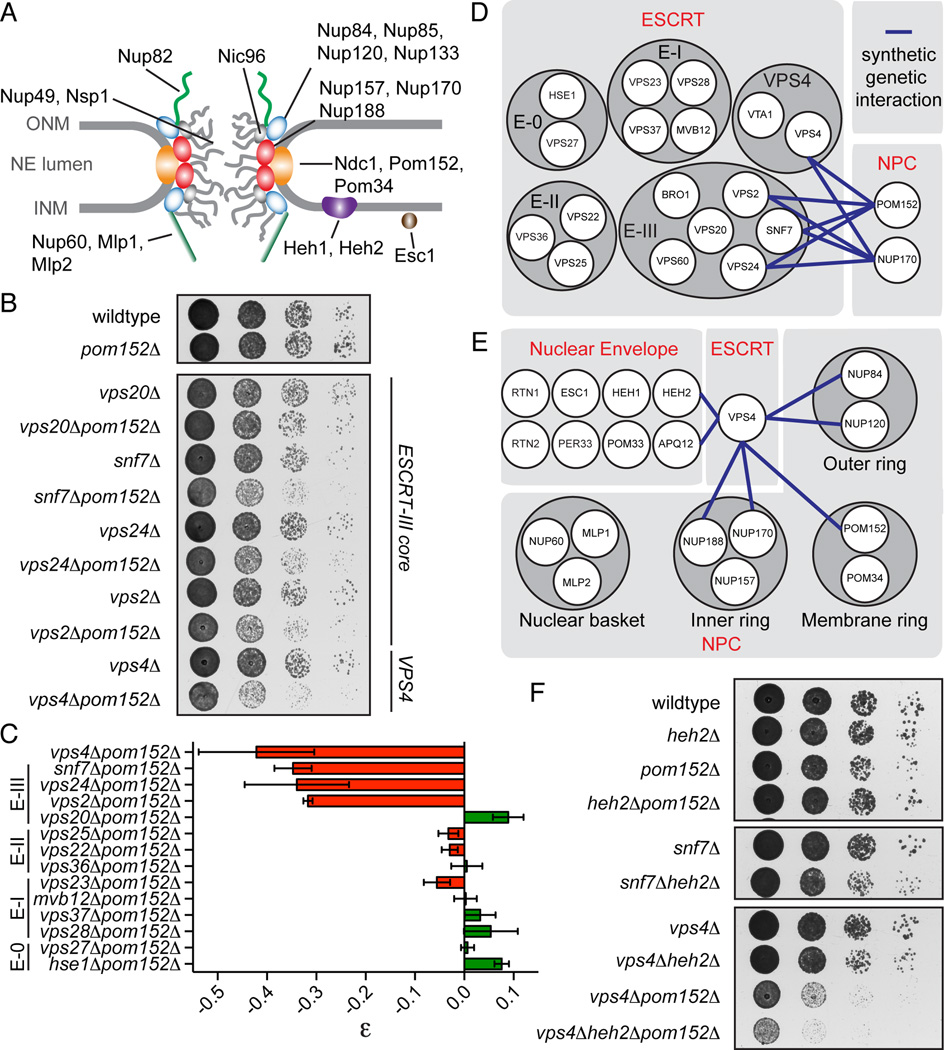
Figure 1. ESCRT-III/VPS4 specifically interact with nup genes.
(A) Schematic of the NPC. Nups shown in the inner (red oval), outer (blue oval), and membrane (orange oval) ring complexes; nuclear basket and cytoplasmic filaments in green. FG-nups are grey lines and linker nup Nic96 is grey oval. Integral (Heh1/2) and associated (Esc1) INM proteins shown.
(B) POM152 interacts with ESCRT-III genes and VPS4.
(C) Quantification of epistasis (ε) between POM152 and ESCRT genes. ε can range from −1 to 1 where negative numbers are synthetic sick interactions and positive numbers are suppressive interactions. Mean +/- s.d. from 3 experiments. “E” abbreviates ESCRT.
(D and E) Schematic of all tested genetic interactions.
(F) VPS4 and SNF7 genetically interact with HEH2.
See Figure S1.
RESULTS
ESCRT-III and VPS4 genetically interact with the NPC
The evolutionary conservation of the genetic interaction between VPS4 and POM152 in fission and budding yeast (Costanzo et al., 2010; Frost et al., 2012) predicts a broader interaction network between the NPC and ESCRT machinery. To test this, we crossed pom152Δ strains with strains containing gene deletions of ESCRT-0, I, II and III. This analysis revealed a striking network in which POM152 specifically interacted with VPS4 and the ESCRT-III genes SNF7, VPS24 and VPS2, but not VPS20 or genes encoding ESCRT-0, I or II (Fig. 1B–D, S1A). By measuring colony sizes, we calculated an epistasis value (ε) for the various gene pairings (Phillips, 2008) and determined that pom152Δsnf7Δ, pom152Δvps24Δ, and pom152Δvps2Δ cells had an average ε of −0.33 and vps4Δpom152Δ of −0.42 (Fig. 1C). Nup170Δ strains showed an identical epistasis profile as pom152Δ cells (Fig. 1D, S1B, C). VPS4 alleles encoding point mutants in the oligomerization (Gonciarz et al., 2008) or ATPase (Babst et al., 1998) domains were incapable of rescuing the loss of fitness of vps4Δpom152Δ cells, suggesting that these genetic interactions reflect a loss of Vps4’s catalytic function (Fig. S1D).
As summarized in Fig. 1E, we also observed genetic interactions between VPS4 and genes encoding the inner and outer ring nups, but not nuclear basket nups or a subset of NE proteins, with the exception of APQ12 (Fig. S1C). This set of interactions were similar to those we reported between two integral INM proteins (Heh1 and Heh2) and the NPC (Yewdell et al., 2011). Consistent with this, we observed a mild fitness loss of heh2Δsnf7Δ cells (Fig. 1F) and more strikingly, an enhancement of the vps4Δpom152Δ interaction after deletion of HEH2 (Fig. 1F).
Snf7 binds to Heh1 and Heh2 in vivo
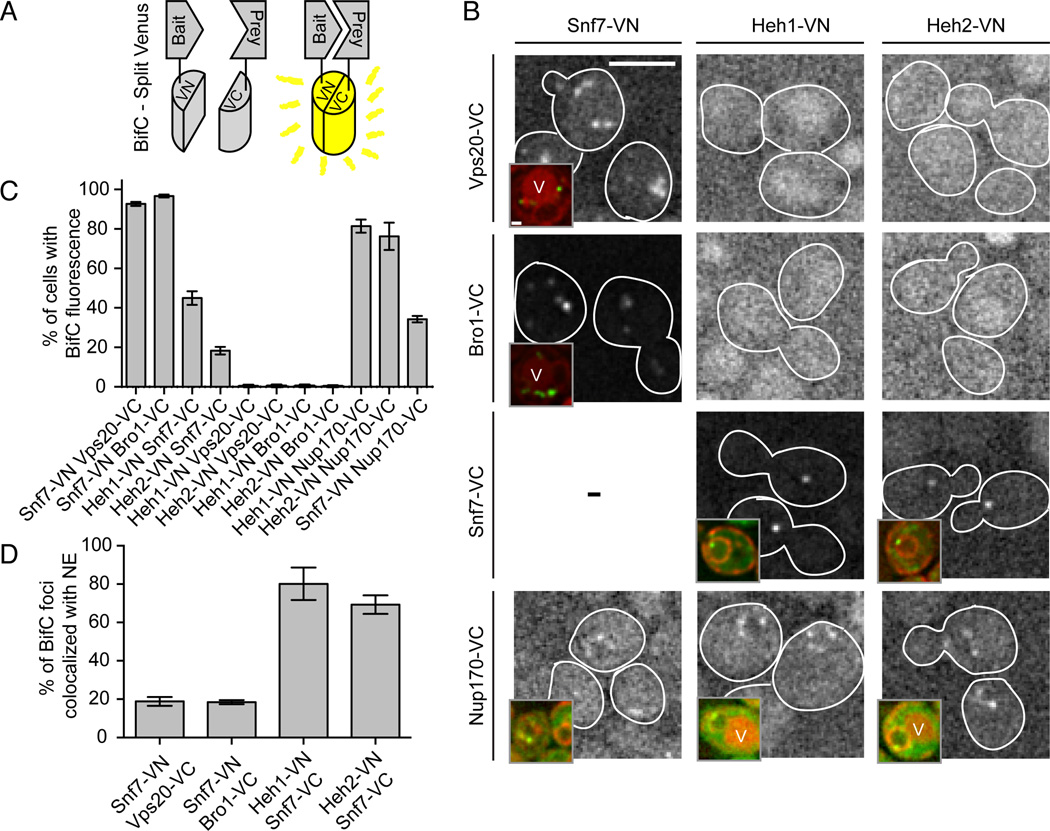
To evaluate if the genetic interactions between ESCRT-III/VPS4 and NE genes reflected a biochemical interaction at the NE, we employed bifunctional complementation (BifC)(Chun et al., 2007) by co-expressing bait and prey proteins as fusions to the N- and C-terminal domains of Venus (VN and VC; Fig. 2A). First, we confirmed interactions between the ESCRT-III components Snf7 and Vps20 with the adapter ALIX/Bro1 (Wemmer et al., 2011) by co-expressing Snf7-VN with Vps20-VC or Bro1-VC. In ~90% of these cells, several fluorescent puncta appeared in the cytoplasm consistent with their localization on endosomes (Fig. 2B, C). Strikingly, the combinations of Heh1-VN or Heh2-VN with Snf7-VC resulted in the appearance of one or two fluorescent spots (Fig. 2B) in ~45 and 18% of cells, respectively (Fig. 2C). These interactions were specific as we were unable to detect fluorescence when Heh1/2-VN were combined with Vps20-VC or Bro1-VC, even when background fluorescence was enhanced (Fig. 2B). Interestingly, the specificity of the ESCRT-III component Snf7 over its sister ESCRT-III (Vps20) for Heh1/2 was also reflected by examining the steady state distribution of ESCRT subunits in nup133Δ cells where NPCs are clustered to one side of the NE (Fig. S2)(Doye et al., 1994). In ~95% of these cells, a portion of Snf7-GFP (and Vps4-GFP) was found overlapping at the edge of the NPC cluster in an association that lasted for at least 10 min (Fig. S2B). We also observed the ESCRT-III subunit Vps24-GFP at the periphery of the NPC cluster but rarely other components of ESCRT-III, II, I or 0 (Fig. S2A, B).
Figure 2. Snf7 physically interacts with Heh1 and Heh2 at the NE.
(A) Diagram of BifC where bait and prey are co-expressed as VN and VC fragments of Venus. Interactions of bait and prey result in Venus fluorescence.
(B) Snf7 interacts with Heh1, Heh2 and Nup170 at the NE. Fluorescent micrographs of cells expressing VC and VN fusions. Background fluorescence has been enhanced to facilitate interpretation of the low signal to noise in some cells. Cell boundaries traced from transmitted light images (not shown). Bar is 5µm. Insets are a merge of fluorescent micrographs of Venus and the NE/ER marker HDEL-dsRed. In some cells vacuoles (V) are visible due to autofluorescence. Bar in inset is 1 µm.
(C) Plot of the percentage of cells expressing VN and VC fusions where Venus was observed. Plots are mean +/− s.d. from 3 experiments (n>200).
(D) Plot of the percentage of fluorescent spots seen colocalized with HDEL-dsRED at the NE. Mean +/− s.d. from 3 experiments (n>40).
See Figure S2.
Further support for ESCRT-III at the NE could be derived from our BifC experiments as the Heh1/2-Snf7 interaction spots colocalized with the NE fraction of a NE/ER marker, HDEL-dsRed, in at least 80% of cells (Fig. 2B, inset; 2D). In contrast, the majority of the Snf7-VN/Vps20-VC and Bro1-VC interaction foci surrounded the vacuole. We further confirmed that Heh1/2 interact with Nup170 at the NE (Yewdell et al., 2011) and although difficult to detect, we observed an interaction between Snf7-VN and Nup170-VC at the NE (Fig. 2B, C). Taken together, our BifC data suggest that the ESCRT-III-NPC genetic network might reflect a physical link between Snf7 and the INM proteins Heh1/2 and their NPC binding partner, Nup170.
Heh2 specifically binds Snf7
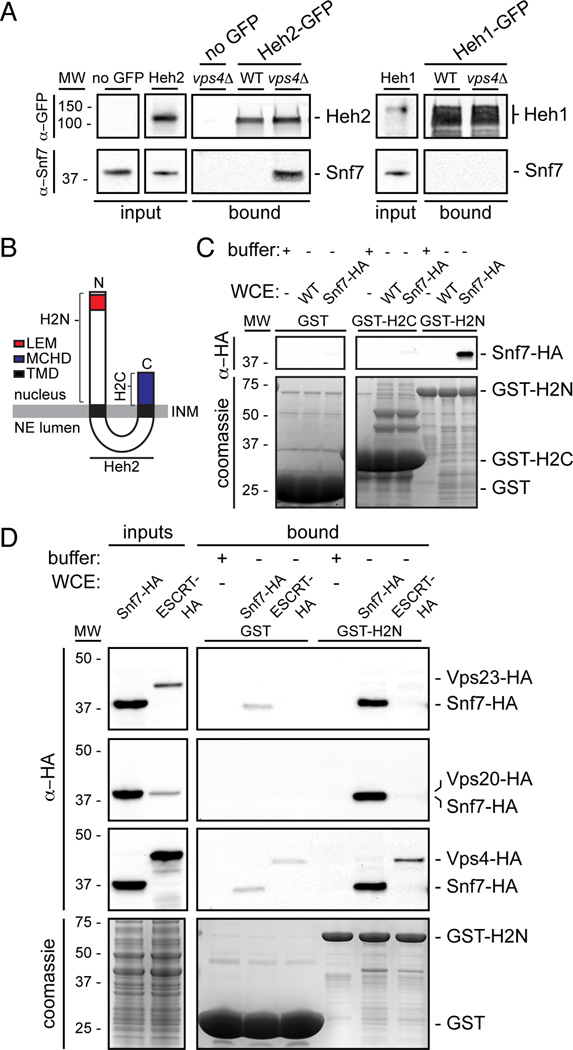
To evaluate the interactions between Heh1/2 and Snf7 biochemically, we affinity purified Heh1/2-GFP from whole cell extracts of WT cells using conditions that maintain interactions between the Heh proteins and Pom152 and Nup170 (Yewdell et al., 2011); we could not detect Snf7 bound to either Heh1- or Heh2-GFP (Fig. 3A). To reconcile these results with the BifC, we reasoned Heh1/2-Snf7 interactions might be highly dynamic. Snf7 interactions are stabilized in the absence of Vps4 (Babst et al., 2002), therefore we assessed whether Snf7 was capable of binding to Heh1 or Heh2 in vps4Δ cells. Strikingly, in vps4Δ cells we detected Snf7 bound to Heh2-GFP by Western blot (Fig. 3A). This interaction was specific to Heh2 as it was undetectable in Heh1-GFP pull outs and in no-GFP controls derived from vps4Δ cells. Consistent with this result, in vps4Δpom152Δ cells, we detected a fraction of Snf7-GFP (but not other ESCRTs) at the NE in association with a cluster of nups (Fig. S3A, B).
Figure 3. Heh2 specifically binds Snf7.
(A) Snf7 co-purifies with Heh2-GFP in vps4Δ cells. Western blots of proteins bound to Heh1- and Heh2-GFP from WT and vps4Δ cell extracts.
(B) Schematic of the topology and domain architecture of Heh2. LEM is Lap2-emerin-MAN1, MCHD is MAN1 C-terminal Homology Domain and TMD is transmembrane domain.
(C and D) Snf7 specifically interacts with the N-terminus of Heh2. GST-fusions of Heh2 domains were immobilized on GST-beads (bottom, coomassie panels) and incubated with buffer or whole cell extracts (WCE) as indicated in legend. HA fusions detected by Western blot.
See Figure S3.
We probed the specificity of the interaction between Heh2 and Snf7 by purifying Heh2 N- and C-terminal domains (H2N and H2C; Fig. 3B) as GST fusion proteins from E. coli extracts (Fig. 3C, coomassie panel). Purified GST-fusions were incubated with yeast whole cell extracts of a WT strain or one expressing Snf7 as a fusion to the HA epitope (Snf7-HA). Snf7-HA specifically bound to GST-H2N but not to GST-H2C or GST alone. We further tested interactions with an ESCRT-I (Vps23), another ESCRT-III (Vps20), and Vps4. These experiments were performed in tandem with extracts containing Snf7-HA as a control. While a small fraction of Vps4 was bound, we did not detect Vps23 or Vps20 (Fig. 3D), even after normalizing for the abundance of Snf7.
Deletion of ESCRT-III and VPS4 leads to clustering of defective NPCs
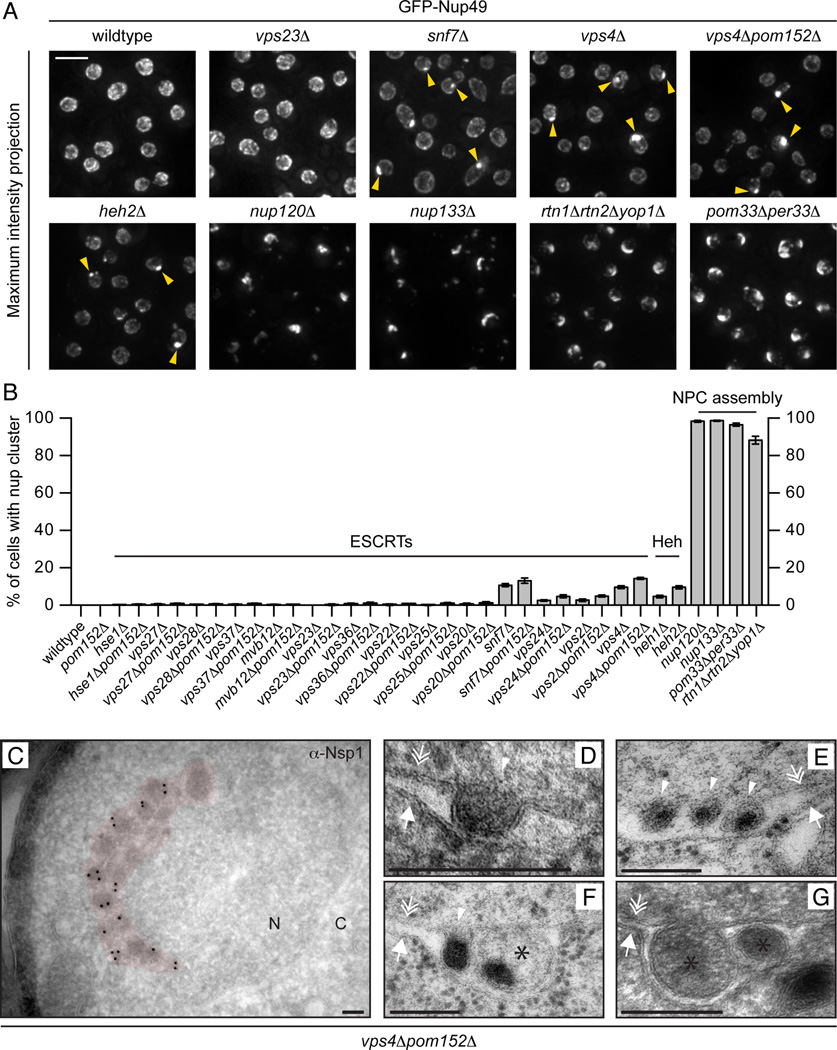
Our genetic data and the interaction between Snf7 and Heh2 (and associated nups (Yewdell et al., 2011)) predict a role for ESCRT-III/Vps4 in NPC function. To test this idea, we examined the distribution of GFP-Nup49 in ESCRTΔ strains. While in the majority of cases GFP-Nup49 was distributed at the NE in a punctate pattern representative of NPCs (Fig. 4A and B), it accumulated in a single cluster in ~10% of snf7Δ and vps4Δ cells (Fig. 4A, arrowheads, B) and ~14% of snf7Δpom152Δ and vps4Δpom152Δ cells. This nup cluster is morphologically distinct from the typical NPC clustering seen in genetic backgrounds where NPC assembly is perturbed like in nup133Δ (Doye et al., 1994), nup120Δ (Aitchison et al., 1995; Heath et al., 1995), rtn1Δyop1Δ (Dawson et al., 2009) and pom33Δper33Δ (Chadrin et al., 2010) cells (Fig. 4A). In these strains, the majority of the GFP-Nup49 is found in the NPC cluster consistent with a block in the assembly of all NPCs. In contrast, a variable fraction of the total GFP-Nup49 is found clustered in vps4Δ and snf7Δ cells, with a pool that remains uniformly distributed at the NE. This suggests that these nup clusters do not arise through the disruption of the assembly of every NPC. Interestingly, ~5% of heh1Δ and ~10% of heh2Δ cells had a morphologically similar nup cluster to vps4Δ/snf7Δ strains (Fig. 4A, B) further supporting that Heh2 functions in a pathway with ESCRT-III/Vps4.
Figure 4. Nups accumulate in a cluster at the NE in the absence of ESCRT-III/Vps4.
(A) GFP-Nup49 accumulates at one side of the NE in snf7Δ and vps4Δ strains. Maximum intensity projections shown. Arrowheads point to the nup cluster. Bar is 5µm.
(B) Plot of the percentage of cells in the indicated strains where GFP-Nup49 accumulates in a cluster. Mean +/− s.d. from 3 experiments (n>400).
(C-G) TEM micrographs of vps4Δpom152Δ cells showing an accumulation of NPC-like structures on one side of the NE (pseudo-colored red) in addition to intralumenal vesicles. All bars are 200 nm. In (C), an anti-Nsp1 antibody and 10nm-gold conjugated secondary antibodies label the NPC-like structures. “N” and “C” represent nucleoplasm and cytoplasm, respectively. Arrows, double arrows and arrowhead point to the ONM, INM and INM invaginations, respectively. Asterisks label putative vesicles in the NE lumen.
We examined the ultrastructure of the nup cluster in vps4Δpom152Δ by transmission EM. As shown in Fig. 4C, electron-dense structures morphologically similar to NPCs were crowded together on one side of the NE. Immunostaining with an antibody to the FG-nup, Nsp1, and gold-conjugated secondary antibodies is consistent with the conclusion that they contain nups (Fig. 4C). However, many of these densities were not uniform in size with diameters that often exceeded 100 nm, suggesting that they might not represent ‘normal’ NPCs. Interestingly, rarely (9/~1000 cell sections), we observed invaginations of the INM and vesicular structures in the NE lumen filled with protein density (Fig. 4D–G). These structures are reminiscent of perinuclear vesicles observed during viral (Mettenleiter et al., 2013) and megaRNP (Jokhi et al., 2013; Speese et al., 2012) nuclear egress.
The Storage of Improperly assembled Nuclear Pore Complexes
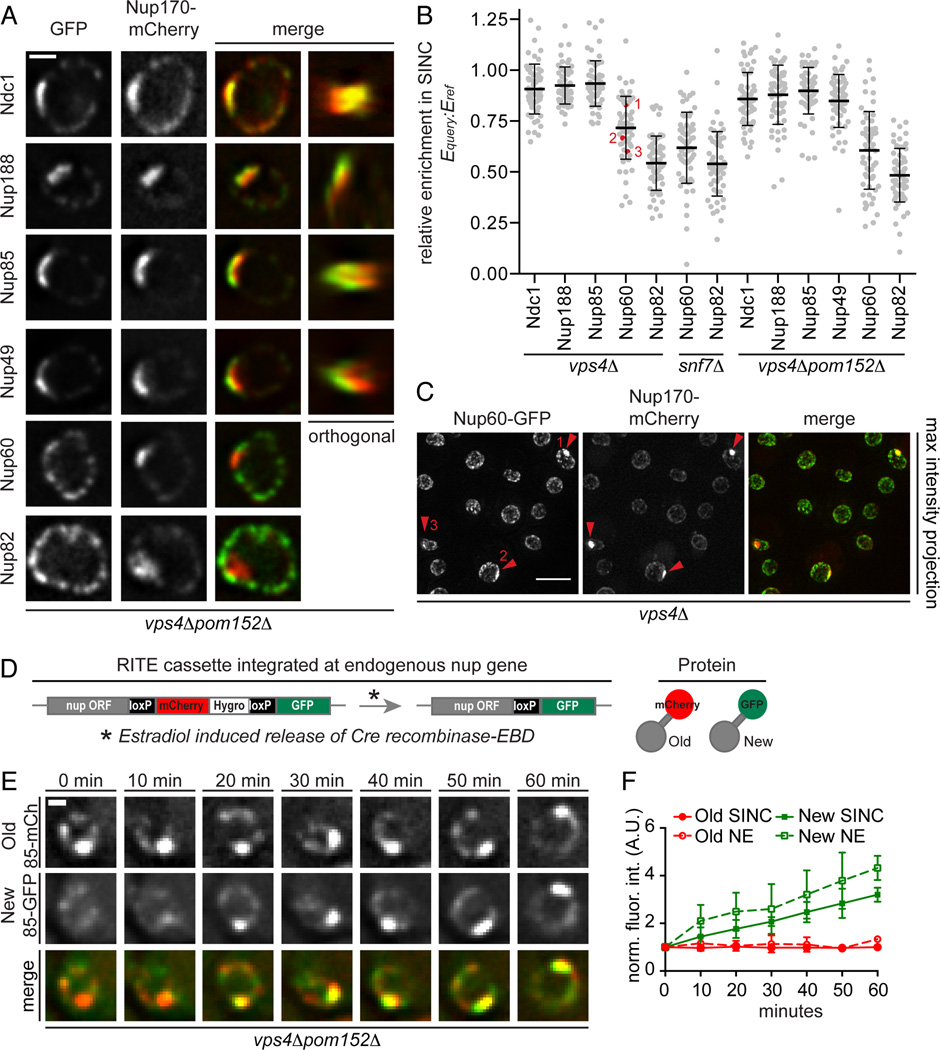
The lack of uniformity of the NPC-like densities in vps4Δpom152Δ cells prompted us to investigate whether the cluster contained fully formed NPCs by localizing GFP-tagged nups from each of the major nup subcomplexes (Fig. 1A; 5A, left “GFP” panels). We used Nup170-mCherry as a cluster reference to evaluate co-localization. Orthogonal views show a plaque-like accumulation on the surface of the NE (Fig. 5A, far right panel). Since the cluster varies in size, we also related the level of enrichment of total nup-GFP levels (Equery) that were proportioned into the cluster compared to the enrichment of Nup170-mCherry (Eref) by calculating a Equery:Eref ratio. These analyses showed that components of the membrane (Ndc1), inner (Nup188), outer (Nup85) ring complexes, and FG nups (Nup49) were localized to a cluster at the NE. Interestingly, while the mean value of relative enrichment for these nups was ~0.9, there was a broad range (0.65–1.25) at the single cell level, suggesting variability in the accumulation of individual nups, which would not be predicted if the cluster contained mature NPCs (Fig. 5B). Most strikingly, the relative enrichment of the cytoplasmic and nuclear basket nups varied much more widely (0.1–1.05) with mean values of ~0.6 and ~0.5, respectively. A similar trend for all tested nups was observed in the nup clusters of snf7Δ and vps4Δ cells (Fig. 5B) and examples of vps4Δ cells in the same field showing different Equery:Eref ratios are displayed in Fig. 5C. Taken together, our data suggest that the cluster is predominantly filled with variable relative amounts of scaffold nups that are severely depleted of the peripheral elements of the NPC. We therefore interpret the cluster of nups observed in vps4Δ/snf7Δ cells to be composed of malformed NPCs of irregular nup composition and term this structure the SINC for Storage of Improperly assembled Nuclear Pore Complexes.
Figure 5. The SINC is a product of NPC misassembly.
(A) The SINC contains an irregular nup composition depleted of cytoplasmic and nucleoplasmic nups. vps4Δpom152Δ cells expressing GFP-tagged nups and Nup170-mCherry. The 4th column is an orthogonal (x,z or y,z) view of the SINC. Bar is 1 µm.
(B) Plot of the relative nup-GFP enrichment (Equery) in the SINC compared to Nup170-mCherry (Eref) expressed as a Equery:Eref ratio. Line is mean, error bars are s.d., grey circles are individual cells, and red circles with numbers correspond to the indicated cells in (C). n=20 in each of 3 experiments.
(C) Variability in the relative accumulation of Nup60-GFP and Nup170-mCherry in the SINC. Arrowheads point to SINCs, numbers refer to individual cells in (B). Bar is 5µm.
(D) Schematic of RITE where estradiol induces a genetic switch from the production of Nup-mCherry to Nup-GFP. EBD is estradiol binding domain.
(E) Newly synthesized Nup85-GFP accumulates in the SINC. Fluorescent micrographs of the nucleus of a vps4Δpom152Δ cell showing Nup85-GFP (New/85-GFP), Nup85-mCherry (Old/85-mCh) and merged images at 10 min intervals in the presence of estradiol. Bar is 1 µm.
(F) Quantification of Nup85-GFP and mCherry fluorescence (normalized fluorescence intensity in arbitrary units (A.U.)) at the NE and SINC after estradiol addition, values normalized to t=0. Mean +/− s.d. from 3 experiments (n=3, 5, 5).
See Figure S4.
The SINC accumulates due to NPC misassembly and not disassembly
To evaluate whether the accumulation of malformed NPCs in the SINC is a result of NPC breakdown or defects in NPC assembly, we tested whether NPC assembly factors accumulate in the SINC. We localized several GFP-tagged (GFP panels, Fig. S4) integral membrane proteins implicated in early NPC assembly events including Heh2 (Yewdell et al., 2011), Pom33, Per33 (Chadrin et al., 2010), Rtn1 (Dawson et al., 2009) and the SUN-protein Mps3 (Talamas and Hetzer, 2011) with Nup170-mCherry as a SINC reference (Fig. S4). All of these proteins were found enriched in the SINC. In contrast, we did not observe colocalization with spindle pole body proteins (Spc42-GFP) or with other NE membrane proteins like Hmg1 (Fig. S4). These data support a model in which the SINC forms during NPC assembly.
We next simultaneously assessed the distributions of an assembled (mCherry) and newly synthesized (GFP) version of the scaffold nup, Nup85, using recombination induced tag exchange (RITE) in vps4Δpom152Δ cells (Fig. 5D)(Verzijlbergen et al., 2010). In cells in which the genetic switch from the mCherry to GFP version was induced, the intensity of the “old” Nup85-mCherry at the SINC and at the NE remained constant over at least 60 minutes (Fig. 5E, F). In striking contrast, we observed the appearance and accumulation of “new” Nup85-GFP in the SINC. Importantly, not all Nup85-GFP is targeted to the SINC, as we also observed uniform NE accumulation. Taken together, these data are consistent with the interpretation that some NPC assembly events result in the formation of the SINC, which is representative of an accumulation of aberrant NPC assembly products rather than NPC breakdown.
The SINC is enriched in old mother cells
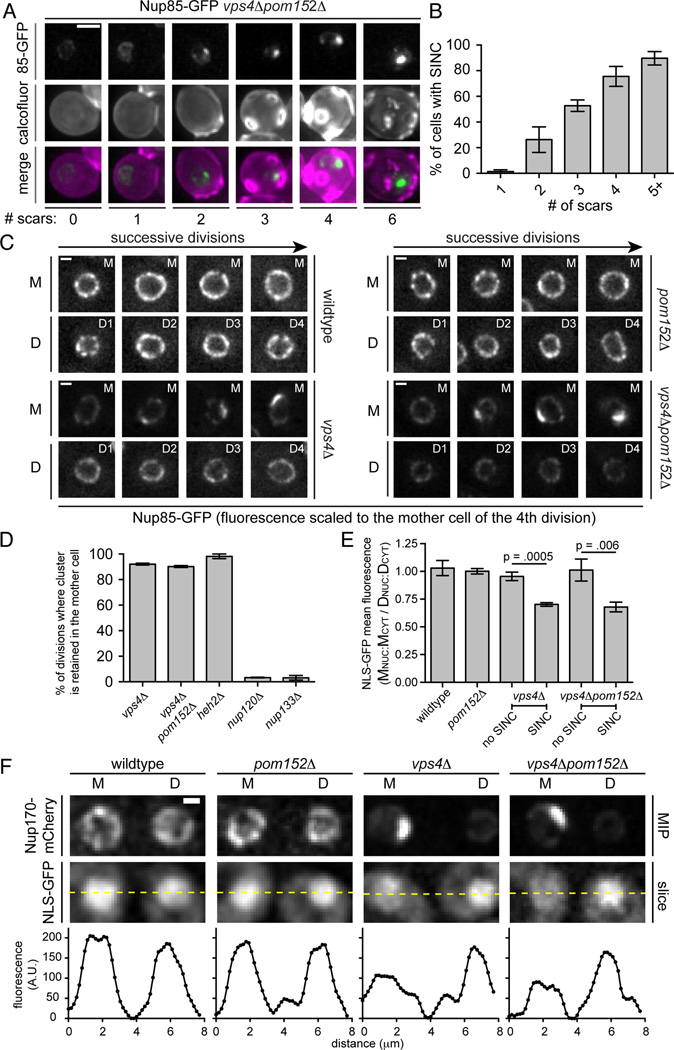
The accumulation of malformed NPCs in the SINC evoked a quality-control mechanism analogous to the incorporation of misfolded proteins into compartments like the juxta-nuclear quality control compartment (JUNQ) and insoluble protein deposit (IPOD), which are subsequently retained in mother cells (Kaganovich et al., 2008; Spokoini et al., 2012). We therefore wondered whether the low penetrance of the SINC in 10–14% of vps4Δ cells (Fig. 4B) reflected its enrichment in aged mother cells. To test this possibility, we assessed the replicative age of vps4Δpom152Δ cells by counting bud scars using calcofluor staining. Remarkably, the proportion of cells containing the SINC increased with bud scar number such that it was visible in virtually all cells containing >5 bud scars (Fig. 6A, B). To directly visualize SINC fate, we monitored its appearance and segregation through multiple divisions in vps4Δ (Fig. 6C) and vps4Δpom152Δ (Fig. 6C, Movie S1) cells. Strikingly, in ~90% of mitoses the SINC was retained in the mother cell (Fig. 6D). Interestingly, we observed similar mother retention of the nup cluster of heh2Δ cells (Fig. 6D; Movie S2), suggesting that this cluster might also be a SINC, whereas daughters inherited virtually all nup133Δ/nup120Δ NPC clusters (Fig. 6D, Movies S3 and S4).
Figure 6. The SINC is not transmitted to daughter cells.
(A) The SINC accumulates in old mothers. Fluorescence micrographs of Nup85-GFP in vps4Δpom152Δ cells with the indicated number of bud scars stained with calcofluor. Bar is 1 µm.
(B) Plot of percentage of cells with Nup85-GFP in SINC in reference to bud scar number. Mean +/− s.d. from 3 experiments, n>60.
(C) The SINC is retained in mother cells. Images of Nup85-GFP at a mother (M) nucleus and of its four daughter nuclei (D1-D4) in the indicated strains. Fluorescence in each image series is normalized to the fluorescence of the mother cells after the final division to avoid saturating the SINC signal. Bar is 1µm.
See Movie S1.
(D) NPC clusters of nup120A and nup133A strains are not retained in mothers. Plot of the percentage of divisions that result in the retention of nup clusters in mother. Mean +/− s.d. from 3 experiments.
(E) Nuclear compartmentalization is lost in vps4A mother cells. Plot of the quantification of the relative nuclear accumulation of NLS-GFP between individual mother (M) and daughter (D) cells expressed as a ratio of MNUC:MCYT to DNUC:DCYT. Eleven divisions (mother and daughter) analyzed from 3 experiments. Mean +/− s.d. P values from an unpaired, two-tailed t-test.
(F) Daughter cells retain nuclear transport. Fluorescence micrographs of NLS-GFP of a mother (M) and its daughter (D) after division. Fluorescence intensities along indicated lines are plotted. MIP is maximum intensity projection. Bar is 1µm.
Nuclear transport defects segregate with SINC
In a model in which the SINC is a repository for defective NPCs that is restricted from being transmitted to daughters, we wondered whether nuclear transport would also be asymmetrically inherited. We monitored the localization of a ‘classical’ NLS reporter (NLS-GFP) in WT, pom152Δ, vps4Δ and vps4Δpom152Δ cells through multiple cell divisions and related a nuclear:cytoplasmic ratio for mother and daughter cells just after cytokinesis. In WT and pom152Δ cells, there were no differences between the relative nuclear accumulation of the NLS-GFP reporter (Fig. 6E, F). In contrast, in SINC-containing vps4Δ or vps4Δpom152Δ cells, there was a clear reduction of NLS-GFP nuclear accumulation specific to mother cells, which was reversed in daughters (Fig. 6E, F).
Increased levels of nups in vps4Δ mother cells
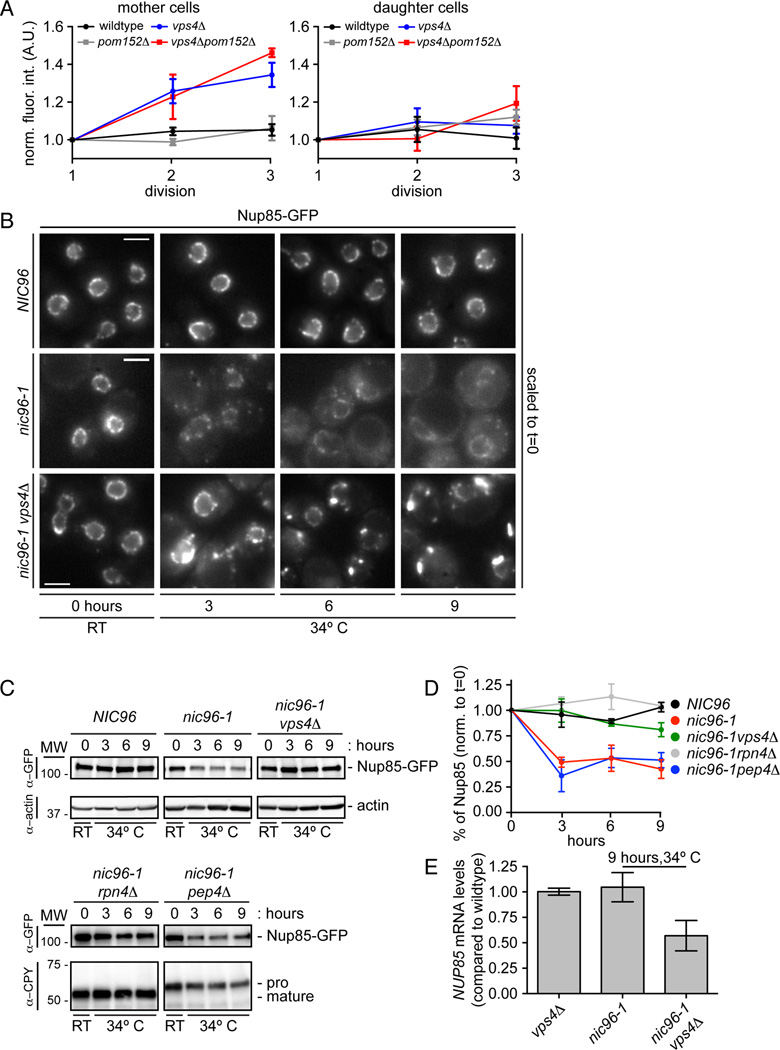
SINC formation occurs in cells lacking Heh2, ESCRT-III or Vps4, suggesting that the ESCRT machinery counteracts the accumulation of misassembled NPCs. Our data are therefore consistent with two potential models of ESCRT function. In one, ESCRT-III/Vps4 directly contributes to NPC biogenesis. In another, it prevents aberrant NPC assembly. To differentiate between these possibilities, we measured nup levels by quantifying the total fluorescence of Nup85-GFP at the NE in mother and daughter cells through several divisions. In WT and pom152Δ cells, Nup85-GFP levels did not change, suggesting that there are similar NPC numbers in mothers and daughters after each division (Fig. 7A). A model in which Vps4 functions in NPC assembly predicts that the total levels of nups would be unchanged. Remarkably, however, in vps4Δ and vps4Δpom152Δ cells there was a dramatic rise in Nup85-GFP fluorescence at the NE through each division that was restricted to mother cells; the levels of Nup85-GFP in vps4Δ and vps4Δpom152Δ daughter cells did not appreciably change (Fig. 7A). These results suggest a model in which the SINC contains defective NPCs that would not otherwise form in the presence of Vps4, and implicate Vps4 in clearing or preventing the accumulation of defective nups at the NE.
Figure 7. Vps4 destabilizes misassembled nups.
(A) An extra pool of Nup85 accumulates in the SINC of vps4Δ mothers. Plot of total Nup85-GFP fluorescence in mother (left) and daughter (right) nuclei measured after each of three divisions (normalized to first). Mean +/− s.d. from 3 experiments in which 5 cells (mother and daughter) were measured.
(B) Nup85-GFP is stabilized in vps4Δ cells when NPC assembly is inhibited. Micrographs of Nup85-GFP at either RT or at 34°C. Bars are 5µm.
(C) Nup85-GFP is degraded by the proteasome when NPC assembly is inhibited. Western blot of Nup85-GFP levels (with actin or CPY load controls) from experiments identical to that shown in B.
(D) Plot of the quantitation of Nup85-GFP levels in the indicated strains. Mean +/− s.d. from 3 experiments.
(E) NUP85 transcript levels decline in nic96-1vps4Δ cells. RT-qPCR analysis of NUP85 transcript compared to WT cells.
See Figure S5.
Vps4 destabilizes defective NPC assembly intermediates
To gain more insight into the mechanism of ESCRT-III/Vps4 function, we modeled a defective NPC assembly step by introducing a temperature sensitive allele of the essential nup Nic96 (nic96-1); at the non-permissive temperature (34°C, Fig. S5A) NPCs are not formed in these cells (Grandi et al., 1995). Consistent with this, we observed a decline of Nup85-GFP fluorescence at the NE and a concomitant increase in cytoplasmic Nup85-GFP in nic96-1 cells grown at 34°C for several hours (Fig. 7B). Interestingly, we observed a ~40% reduction in the total levels of Nup85-GFP after 9 hours at 34°C by Western blot (Fig. 7C, D). These results suggest either a reduced synthesis or degradation of Nup85 when NPC assembly is inhibited. Strikingly, the deletion of VPS4 resulted in the accumulation of Nup85-GFP in plaques reminiscent of the SINC (Fig. 7B). The fluorescence intensity of these plaques suggested a build up of Nup85-GFP levels. Indeed, when we examined protein levels by Western blot, the nic96-1vps4Δ cells showed a stabilization of Nup85-GFP to WT levels (Fig. 7C, D), which could not be explained by differences in growth rate (Fig. S5B) or transcript levels as measured by reverse transcriptase-quantitative PCR (Fig. 7E).
We next assessed whether the degradation of Nup85 was driven by the proteasome or vacuolar peptidases. As shown in Figure 7C and D, Nup85 levels were stabilized when proteasome function was attenuated through deletion of Rpn4 (Johnson et al., 1995) and unaffected in pep4Δ strains where vacuolar proteases are not activated (as monitored by the accumulation of the pro-form of carboxypeptidase Y (CPY)(Ammerer et al., 1986)(Fig. 7C)). Similar results were obtained by treating cells with either the proteasome inhibitor MG132 or the vacuolar peptidase inhibitor AEBSF (Lee and Goldberg, 1996)(Fig. S5C, D). Together, our results suggest a role for Vps4 in the clearance and proteasome-mediated degradation of Nup85 that is not assembled correctly.
DISCUSSION
Budding yeast ensure the formation of functional NPCs through: 1) an ESCRT-III/Vps4 dependent mechanism that plays a role in potentially recognizing and clearing defective NPC assembly intermediates and, 2) a compartment that we term the SINC that sequesters defective NPCs to prevent their passage to daughter cells. Together these mechanisms create a robust quality control system that ensures nuclear compartmentalization is maintained in progeny and supports the notion that defective NPCs might reduce replicative lifespan (Colombi et al., 2013; Lusk and Colombi, 2014; Makio et al., 2013; Shcheprova et al., 2008).
Our data implicate the ESCRT machinery in preventing the formation of nonfunctional NPCs. We propose a model in which ESCRT-III/Vps4 helps recognize and clear defective NPC assembly intermediates before a step in which they are committed to maturing into a malformed NPC destined for the SINC. The nature of this commitment step remains unclear but it is likely related to the fusion of the INM and ONM; it is difficult to imagine removing a double-membrane spanning complex without threatening nuclear integrity. Consistent with this assertion, in some post-mitotic cells the scaffold of the NPC is extremely long-lived, suggesting that there are no turnover mechanisms capable of removing NPCs (Savas et al., 2012; Toyama et al., 2013). Indeed, even pathways like piece-meal autophagy of the nucleus do not degrade NPCs (Pan et al., 2000).
While intact NPCs might not be degraded, we observe the proteasome-mediated degradation of at least Nup85 when NPC assembly is inhibited, supporting a role for ubiquitin modification of nups in the surveillance process; several studies have identified that nups are ubiquitylated (Hayakawa et al., 2012; Hitchcock et al., 2003; Peng et al., 2003). Interestingly, unlike the canonical ESCRT-mediated degradation of ubiquitylated membrane receptors that are recognized by ESCRT-0, I and II, the pathway here invokes a model in which ubiquitylation would occur after ESCRT-III/Vps4 function. This might explain why we do not observe vacuolar degradation of nups, which would be more typical of an ESCRT-mediated pathway.
The absence of the ubiquitin-binding arm of the ESCRT-machinery and the known endosomal ESCRT-III recruitment mechanism, which includes Vps25 and the ALIX homologue Bro1(Saksena et al., 2009; Teis et al., 2010), raises the question of how ESCRT-III is recruited to the NE. Our data support that NE-specific adaptors like Heh2 and Nup170 (either alone or in complex) are the most likely candidates to recruit Snf7. Moreover, a physical interaction between Snf7 and Heh2 provides an attractive hypothesis for how defective NPC assembly intermediates are recognized. Our prior data support that Heh2 is capable of binding to Pom152 and Nup170, a likely early intermediate in the NPC assembly pathway (Yewdell et al., 2011). Since Heh2 is not a stable component of NPCs, we suggest that it has the inherent capacity to distinguish between an assembly intermediate and fully formed NPCs. The molecular mechanism of this potential switch might underlie the commitment step between surveillance and the formation of an NPC, or the SINC.
The interaction of Snf7 with a putative Heh2-nup complex suggests that a step in surveillance is the removal of a membrane-associated complex. The general role of AAA-ATPases including Vps4 in remodeling protein complexes might indicate a direct role for Vps4 in removing nups prior to their degradation (Hanson and Whiteheart, 2005; Scott et al., 2005). Since ESCRT-III is Vps4’s primary substrate (Obita et al., 2007; Shestakova et al., 2010), this scenario further emphasizes the importance of Snf7’s interactions with Heh2 and nups in contributing to their ultimate clearance. It should be considered that, a priori, such surveillance need not occur at the NE, yet we favor that model because we localize Snf7 interactions at the NE using BifC.
A unifying theme of ESCRT-III function is the formation of helical filaments that contribute to membrane deformation and scission (Henne et al., 2013). Since there is evidence that NPC assembly intermediates accumulate at the INM during the initial steps of NPC assembly (Doucet et al., 2010; Funakoshi et al., 2011; Makio et al., 2009; Marelli et al., 2001; Talamas and Hetzer, 2011; Yewdell et al., 2011), we wonder whether clearance of defective NPC assembly intermediates might occur through the formation of an intralumenal vesicle derived from the INM that would be delivered to the proteasome in the cytoplasm. While we see intralumenal vesicles in vps4Δ cells, their rarity made it unfeasible to identify their protein constituents by immuno-EM. Nonetheless, this model remains attractive as ESCRT-III function at the INM might support nuclear egress of herpesvirus (Mettenleiter et al., 2013) and megaRNPs (Speese et al., 2012). Last, we wonder whether the INM invaginations observed in nup116Δ (Wente and Blobel, 1993), gle2-1 (Murphy et al., 1996), and apq12Δ (Scarcelli et al., 2007) strains might reflect ESCRT-III function, as the machinery that drives these phenotypes remains poorly understood.
In eukaryotes that undergo mitotic NE breakdown, the surveillance mechanism described here is likely most relevant during NPC biogenesis in interphase or in post-mitotic cells. Interestingly, there is precedence for mitotic surveillance of the assembly of the nuclear basket that delays abscission (Mackay et al., 2010). Remarkably, co-depletion of the Snf7 orthologue, Chmp4C, alongside Nup153, overrides this checkpoint (Carlton et al., 2012), exemplifying another link between the NPC and ESCRT-III. Together, these results emphasize the importance of NPC assembly quality control at all steps of the cell cycle and highlight the critical importance of ensuring nuclear compartmentalization.
EXPERIMENTAL PROCEDURES
Yeast strains and epistasis analysis
All strains are listed in Table S1; see Extended Experimental Procedures for their generation. Epistasis was determined by comparing colony sizes on plates containing 10-fold serially diluted single and double deletion strains after 48 h at RT for at least three independent experiments. We quantified epistasis by measuring colony size using the formula Wxy = αxαy + ε, where Wxy represents the fitness of the double knockout strain, αx and αy are the fitness of the single knockouts, and ε is the deviation due to epistasis (Phillips, 2008).
Fluorescence Microscopy
All images were acquired on a Deltavision widefield deconvolution microscope (Applied Precision/GE Healthcare) with an Evolve™ EMCCD camera (Photometrics) or a CoolSnap HQ2 CCD camera (Photometrics)(Fig. 2B, 5A, 5C, 6A, S2B, and S4). Cells were either placed on 1.4% agarose pads containing CSM and 2% dextrose and sealed with VALAP (1:1:1, vaseline:lanolin:paraffin) or in microfluidic plates (Y04C/ CellASIC™)(Fig. 6C, F, and S5B).
Affinity purification/binding experiments
Heh1/2-GFP were purified from cell extracts using GFP-Trap_M magnetic beads (Chromotek) and bound proteins eluted with SDS-PAGE sample buffer. Similarly, for binding assays using recombinant GST-fusions as bait, GST-fusions were purified on GT-sepharose from E. coli extracts before incubation with yeast cell extracts. Bound proteins were eluted with SDS-PAGE sample buffer and processed for Western blotting.
Electron microscopy
For immuno-EM, cells were fixed with 4% PFA and 0.1% glutaraldehyde and processed for immunolabeling with anti-Nsp1 antibody (32D6, Abcam), followed by 10 nm gold-labeled secondary antibodies (Fig. 4C). For high pressure freezing (Fig. 4D–G) cells were frozen using a Leica HMP101 and freeze substituted using a Leica Freeze AFS unit. Grids were viewed in a FEI Tecnai Biotwin TEM at 80 kV.
Quantification of Western blots
HRP-conjugated secondary antibodies and ECL were used to detect proteins on Western blots. For quantification of relative protein levels, the integrated density of an area surrounding the protein band was measured using Fiji/ImageJ (Schindelin et al., 2012) and normalized to the mean integrated density of the entire lane visualized using Ponceau S.
Supplementary Material
HIGHLIGHTS.
There is a surveillance mechanism that ensures proper nuclear pore complex assembly
A quality control compartment prevents the inheritance of malformed NPCs
ESCRT-III is recruited to the nuclear envelope by inner nuclear membrane proteins
Vps4 clears defective NPC intermediates via proteasomal degradation
ACKNOWLEDGEMENTS
We are grateful to the Mather’s foundation for their support and to A. Frost for sharing unpublished data. Thank you to C. Burd and D. Katzmann for advice and reagents, and to M. King, T. Carrol, C. Schlieker and M. Hochstrasser for comments on the manuscript. We appreciate reagents from E. Hurt, M. Rout and T. Walther and the EM expertise of M. Graham and X. Liu. We are indebted to R. Christiano, Z. Hakhverdyan and M. Rout for help during revisions. All authors are supported by NIH R01 GM105672 with additional funds to CPL and PC from NIH R21HG006742. BMW was also supported by NIH 5T32GM007223.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
BMW and CPL conceived of and designed experiments; they also analyzed and interpreted all data and wrote the manuscript; BMW carried out all of the experiments with the exception of those that lead to Fig. 3A (JJ) and 3B–C (PC).
REFERENCES
- Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. The Journal of Cell Biology. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. The EMBO Journal. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Developmental Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bui KH, Appen von A, DiGuilio AL, Ori A, Sparks L, Mackmull M-T, Bock T, Hagen W, Andrés-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. The Journal of Cell Biology. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun W, Waldo GS, Johnson GVW. Split GFP complementation assay: a novel approach to quantitatively measure aggregation of tau in situ: effects of GSK3β activation and caspase 3 cleavage. J Neurochem. 2007;103:2529–2539. doi: 10.1111/j.1471-4159.2007.04941.x. [DOI] [PubMed] [Google Scholar]
- Colombi P, Webster BM, Fröhlich F, Lusk CP. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. The Journal of Cell Biology. 2013;203:215–232. doi: 10.1083/jcb.201305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. The Genetic Landscape of a Cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. The Journal of Cell Biology. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EMM, Verstraeten VLRM, van Steensel MAM, Marcelis CLM, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 2011;20:4175–4186. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell Cycle-Dependent Differences in Nuclear Pore Complex Assembly in Metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. The EMBO Journal. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Current Opinion in Cell Biology. 2009;21:603–612. doi: 10.1016/j.ceb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, Weibezahn J, Hein MY, Poser I, Mann M, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol. Biol. Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and structural studies of yeast Vps4 oligomerization. Journal of Molecular Biology. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. The EMBO Journal. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hayakawa A, Babour A, Sengmanivong L, Dargemont C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. The Journal of Cell Biology. 2012;196:19–27. doi: 10.1083/jcb.201108124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. The Journal of Cell Biology. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. Molecular Mechanisms of the Membrane Sculpting ESCRT Pathway. Cold Spring Harbor Perspectives in Biology. 2013;5:a016766–a016766. doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AL, Auld K, Gygi SP, Silver PA. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT Machinery Is Required for Plasma Membrane Repair. Science. 2014:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JB, Datta S, Snow CJ, Chatterjee M, Ni L, Spencer A, Yang C-S, Cubeñas-Potts C, Matunis MJ, Paschal BM. The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol. Cell. Biol. 2011;31:3378–3395. doi: 10.1128/MCB.05087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-P, Liu P-T, Kung H-N, Su M-T, Chua H-H, Chang Y-H, Chang C-W, Tsai C-H, Liu F-T, Chen M-R. The ESCRT Machinery Is Recruited by the Viral BFRF1 Protein to the Nucleus-Associated Membrane for the Maturation of Epstein-Barr Virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- Lusk CP, Colombi P. Toward a consensus on the mechanism of nuclear pore complex inheritance. Nucleus. 2014;5:97–102. doi: 10.4161/nucl.28314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. The Journal of Cell Biology. 2010;191:923–931. doi: 10.1083/jcb.201007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K, Wozniak RW. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. The Journal of Cell Biology. 2009;185:459–473. doi: 10.1083/jcb.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T, Lapetina DL, Wozniak RW. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. The Journal of Cell Biology. 2013;203:187–196. doi: 10.1083/jcb.201304047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. The Journal of Cell Biology. 2001;153:709–724. doi: 10.1083/jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Müller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 2013;15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung H-Y, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. The EMBO Journal. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Schlieker C. Alternative nuclear transport for cellular protein quality control. Trends in Cell Biology. 2012;22:509–514. doi: 10.1016/j.tcb.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional Reconstitution of ESCRT-III Assembly and Disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely Long-Lived Nuclear Pore Proteins in the Rat Brain. Science. 2012;335:942–942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. The Journal of Cell Biology. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Chung H-Y, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, et al. Structural and mechanistic studies of VPS4 proteins. The EMBO Journal. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–735. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang Y-T, Li Q, et al. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. Journal of Cell Science. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. The Journal of Cell Biology. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. The EMBO Journal. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijlbergen KF, Menendez-Benito V, van Welsem T, van Deventer SJ, Lindstrom DL, Ovaa H, Neefjes J, Gottschling DE, van Leeuwen F. Recombination-induced tag exchange to track old and new proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:64–68. doi: 10.1073/pnas.0911164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus Budding and the ESCRT Pathway. Cell Host and Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer M, Azmi I, West M, Davies B, Katzmann D, Odorizzi G. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. The Journal of Cell Biology. 2011;192:295–306. doi: 10.1083/jcb.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. The Journal of Cell Biology. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe JM. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 2007;33:2–42. doi: 10.1111/j.1365-2990.2006.00819.x. [DOI] [PubMed] [Google Scholar]
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol. Biol. Cell. 2011;22:1375–1388. doi: 10.1091/mbc.E10-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.