Abstract
While substrates with nanopillars (NPs) have emerged as a promising platform for isolation of circulating tumor cells (CTCs), the influence of diameter and spacing of NPs on CTC capture is still unclear. In this paper, CTC-capture yield and cell behaviors have been investigated by using antibody functionalized NPs of various diameters (120–1100 nm) and spacings (35–800 nm). The results show a linear relationship between cell capture yield and effective contact area of NP substrate where NP array of small diameter and reasonable spacing is preferred; however, spacing that is too small or too large adversely impairs capture efficiency and specificity, respectively. In addition, the formation of pseudopodia between captured cells and substrate is found to depend not only on cell adhesion status but also on eluting strength and shear direction. These findings provide essential guidance on designing NP substrates for more efficient capture of CTCs and manipulation of cytomorphology in the future.
1. Introduction
Circulating tumor cells (CTCs) are cells that shed from a tumor mass and circulate in peripheral blood. Detection and analysis of CTCs can guide cancer diagnosis, prognosis and treatment.1–3 Isolation of CTCs from patients' blood attracts significant attention as a first step toward CTCs analysis. A variety of techniques have been developed in the past decades, which rely mainly on physical properties (e.g., size, density and deformability)4–6 or expressions of biomarkers7–11. However, most of these methods failed to show clinical validity or utility, thus remained in the laboratory settings due to their limited isolation efficiency and the rarity of CTCs, i.e., as few as 1 CTC per 1 mL peripheral blood. Therefore, improvement of CTC isolation efficiency and specificity is a pressing issue.
Recently, nanostructured substrates have emerged as a promising CTCs detection and isolation platform since they can provide high isolation efficiency (80–95%, normally ~90%).12–16 The cell-nanostructure interaction has also been reviewed comprehensively and several basic design principles has been delivered.17 For the selection of nanomaterials, silicon nanomaterials are a good candidate for CTC study due to their biocompatibility and unique electronic properties.18 Compared to flat surfaces, nanostructured substrates provide larger surface area for the immobilization of anti-epithelial cell adhesion molecule (anti-EpCAM), increasing the binding odds between antibody and membrane receptor and also total binding forces, thus enhancing CTC-isolation efficiency. Moreover, the natural extracellular matrix or basement membrane mimicking nanostructured substrates offer a suitable environment for CTCs attachment. In previous studies, nanostructures with size ranging from 30 to 1150 nm have been employed for CTCs isolation, and high efficiency was reported in all cases. For instance, 88% of cancer cells were captured onto antibody immobilized silicon nanowires (SiNWs) with diameter of 50–160 nm;19 high isolation efficiency up to 95% was reported on antibody grafted silicon nanopillars (NPs) with diameter in the range of 100–200 nm;20 cancer cell isolation efficiency on antibody grafted spherical particles with diameter of 232 nm was 4–5 times higher than on a flat surface.21 Nanostructured surfaces with halloysite nanotubes was also demonstrated to enhance the CTC recruitment with a three-fold increase for MCF7 cells, while nonspecific leukocytes adhesion was prevented.22 Meanwhile, efficient CTC capture with high selectivity has also been reported by using functionalized graphene oxide nanosheets23 or soft polystyrene nanotube substrates24. Recently, developing multifunctional platforms modified with nanostructures comes to be of significant research interests for biomedicine applications. A nanodendritic silica coating has been developed to not only efficiently capture CTCs, but also own the ability to directly monitor the results due to its transparency under water.25 Controlled CTC release after cell capture has also aroused more attention since it is very important for subsequent cell analysis.26 A thermoresponsive substrate with NPs was developed to capture targeted CTCs and then release CTCs in a prescribed manner.27 Efficient CTC capture and release process has also been achieved by using programmable fractal gold nanostructures through an electrochemical method.28 Nevertheless, preferred nanostructure geometry is still unclear for CTCs isolation. A quantitative study of the relationship between capture efficiency and nanostructure geometry is necessary for the optimization of substrate topographical design for CTC isolation. At the same time, morphology of captured cancer cells on nanostructured substrates is not conclusive either. In previous reports, the captured cells were found to be flatter on aptamer grafted nanostructured substrates than that on planar surfaces: cells changed from a globular shape to a semielliptical one.9, 29 The flatter shape of captured cells indicated that more receptors on the cell membrane make contact with immobilized antibodies; meanwhile, the decreased cell height further prevented them from being eluted under high shear stress.29 Contrarily, it was reported that the high aspect ratio of bare SiNWs and polystyrene (PS) NPs support cell adhesion but restrict cell spreading due to insufficient area for development of micron-scale focal adhesion.30, 31 Furthermore, nanostructured substrates may also influence the pseudopodia formation. Captured cells showed either significant lamellipodia or filopodia on nanostructured substrates with certain aspect ratios.32
In this paper, we investigated the effects of NP geometry on cell capture yield, cytomorphology and pseudopodia formation. Capture yield is defined as the ratio of cells captured on the NP substrate to total number of cells initially loaded. Our results showed that capture yield is linear to effective contact area of the NP substrate. Dense NPs with smaller diameter yield larger effective contact area, thus were preferred. However, once the spacing between NPs was smaller than the size of a microvillus (<100 nm) or was too large (>800 nm), cell capture yield and specificity were significantly impaired. We also confirmed that the formation of either lamellipodia or filopodia correlates with adhesion status and eluting strength. Specifically, under the same shear stress, lamellipodia were generally formed on NPs with high aspect ratio (>10) where captured cells were firmly attached; furthermore, significant filopodia formation was observed on NPs with low aspect ratio where captured cells were more prone to detach, indicating filopodia might obtain newborn adhesive force to resist shear stress mediated detachment.33
2. Experimental details
2.1 Fabrication of silicon wafers covered with NPs
Silicon wafers covered with NPs with different diameters were fabricated by metal assisted chemical etching.34 In detail, the silicon surface (Si (100), B-doped, 0.004–0.007 Ωcm) was cleaned by a RCA-I approach. Deionized water, ammonia solution (25%) and hydrogen peroxide (30%) were mixed in ratio of 5:1:1. The solution was heated up to 80 degrees and silicon chips were cleaned for 15 mins. Afterwards nanospheres (polystyrene (PS) with diameters: 1.39 μm, 722 nm and 390 nm) were deposited by a modified Langmuir-Blodgett technique in monolayer for lithographic structuring. In the next step the PS-spheres were etched in O2-plasma to reduce their diameters. Following silver (30–60 nm) was evaporated onto the surface. PS-spheres were lifted off by dichlormethane in an ultra-sonic bath and the chips were rinsed with acetone, ethanol and deionized water. Thus a metal film on silicon with specific openings was created. The size of the openings correlates with the size of the plasma-etched PS-spheres. After that etching in a hydrofluoric acid and hydrogen peroxide based solution was carried out. The solution consists of 37 mL deionized water, 12.5 mL hydrofluoric acid (40%) and 0.5 mL hydrogen peroxide. The chips were etched for 5 mins. The metal film sinks into the silicon, which is solved by the etching solution. In the end, around 1 μm long NPs were obtained and silver was removed by concentrated nitric acid for 2 mins. The diameter of the NPs is equal to the size of the opening in the metal film and is also equal to the diameter of the PS-spheres after plasma-etching. Finally, 5 x 5 mm wafer substrates with NPs of diameter ranging from 120 nm to 1100 nm were attained.
2.2 Surface functionalization of anti-EpCAM
Prior to the surface functionalization, all the wafer substrates were washed using a three-step cleaning process (acetone, isopropyl alcohol and DI water), and dried under nitrogen.7 Then, wafer substrates were fixed in 35x10 mm petri dishes and treated with oxygen plasma to confer the hydroxyl groups on the substrate surface. In order to chemically modify the substrate, a three-step surface functionalization process was immediately applied. They were firstly pretreated with 5% (v/v) 3-mercaptopropyl trimethoxysilane in ethanol for 30 minutes at room temperature (RT), followed by incubation with 0.01mM N-y-maleimidobutyryloxysuccinimide ester (GMBS) in 200 proof ethanol for 30 minutes at RT. NeutrAvidin was then immobilized to GMBS by incubating the substrates with 10 μg/ml NeutrAvidin in phosphate buffered saline (PBS) overnight at 4°C. Within 24 hours of the experiment, 10 μg/ml biotinylated goat antihuman EpCAM solution in PBS containing 1% (w/v) bovine serum albumin (BSA) were added to the substrate for 2 hours at RT. After each reaction, PBS or ethanol, depending on the solvent used in the previous step, was used to remove unbounded molecules. One hour prior to running the cell test, the substrates were purged with 3% BSA with 0.05% Tween20.
2.3 Cell culture and cell-test sample preparation
PC3 prostate cancer cells was selected as an appropriate platform for optimization study of capture of CTCs, as lower concentration of EpCAM, namely 51667 molecules per cell, was expressed for PC3 cells.7 PC3 cells were cultured at 37 °C in 5% CO2 in F-12K growth medium containing 1.5 mM L-glutamine supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin with media change every 2–3 days. Cells were then released through incubation in 0.05% Trysin-0.53 mM EDTA at 37°C for around 5 minutes. Hemocytometer was used to count cells and according dilution was subsequently pursued to reach a cell concentration around 105/mL.19 The cell concentration was selected also for ensuring the yield of a large result data pool to reach a reliable conclusion.
2.4 Cell-capture yield test and cell detachment test
For the cell-capture yield test, 20 μL of cell solutions was added to the wafer substrates and immediately followed by incubation at 37 °C in 5% CO2 for different incubation time mentioned in the manuscript. Next, 2 mL of PBS buffer solution was added to the petri dish, which was then fixed on the shaker. Different shaking speeds and different lasting time were applied to detach PC3 cells, followed by removing all suspensions. Both regular microscope and scanning electron microscopy (SEM) were applied to analyze the results including cell amount, geometry and morphology.
For SEM specimen preparation, a standard procedure reported in our previous work29 was followed. Briefly, Cells were firstly fixed with 4% paraformaldehyde at 4 °C for 1 hour. Next, paraformaldehyde was removed and cells were rinsed by PBS buffer solution twice. Cells were then dehydrated in ethanol with a concentration gradient of 20%, 30%, 50%, 70%, 85%, 95%, and 100% (15 minutes in each solution). After treatment of 100% ethanol, all cell samples were lyophilized overnight. Finally, dehydrated cells were sputter coated with gold and imaged by Zeiss 1550 SEM. Images of cells from both upright view and tilted view were scanned for post-analysis.
2.5 Quantitative analysis of cell-capture yield, cell morphology and substrate wettability
To calculate cell capture yield, SEM images were taken section by section throughout the whole substrate and amount of captured cells was then counted. As for the quantitative analysis of cell morphology, SEM images of top view were used for calculating the apparent contact area by tracing the boundary of captured cells using ImageJ software (NIH). Meanwhile, SEM images of tilted view were used for measuring the height of the captured cells, which was modified by the functionalized substrates was evaluated by measuring their contact angles. 1.5 μL, 3 μL and 4.5 μL of water droplet was added to the substrates and images were then analyzed using "contact angle" plug-in in ImageJ.
3. Results and discussion
3.1 Fabrication and characterization of NP arrays
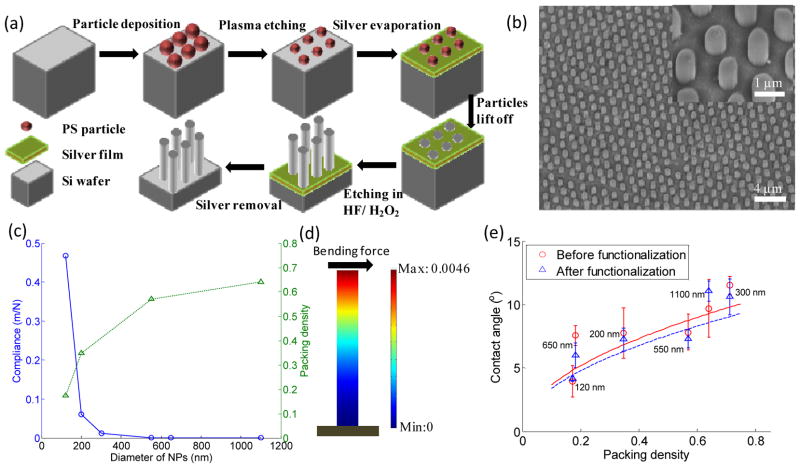
Anti-EpCAM coated NPs with diameter ranging from 100 to 1100 nm, spacing ranging from 30 to 800 nm, and fixed height of 1 μm were prepared (see Methods section). A schematic of the fabrication process and a tilted SEM image of fabricated NPs are shown in Fig. 1(a) and 1(b), respectively. The average diameter, spacing and standard deviation (SD) of each group (n=30) are summarized in Tab. 1. The variations of diameter and spacing from each group are within 2–8%, arising from the size distribution of PS-spheres and different O2 plasma treatment time. To avoid truncated cone structures or collapse of NPs all groups were etched for 5 minutes only, and thus NPs with same length of 1 μm were obtained.
Fig. 1.
(a) Fabrication process of NP arrays by metal assisted chemical etching method. (b) A typical SEM image of NP arrays with average diameter of 650 nm from tilted view. The inset in (b) shows the magnification of NPs from tilted view. (c) Mechanical and geometrical properties of NP substrates: compliance and packing density. (d) A finite element method (FEM) analysis of a single bending NP showing the displacement of an NP with diameter of 120 nm. The unit of the color bar is nanometer. (e) Wettability of different NP substrates before and after functionalization. The solid line and dashed line stand for fitted curves using Cassie's law for non-functionalized and functionalized NP substrates, respectively. Error bars in (e) stand for standard derivation from three independent experiments.
Tab. 1.
Geometrical Properties of NP arrays. Different groups are named as "NP" followed by their average diameters.
| Group | Diameter of NPs [nm] | Spacing [nm] |
|---|---|---|
| NP120 | 118.2±9.4 | 154.3±17.5 |
| NP200 | 204.8±7.3 | 154.9±14.9 |
| NP300 | 292.7±11.3 | 38.7±2.5 |
| NP550 | 571.5±36.5 | 144.0±10.5 |
| NP650 | 649.6±38.1 | 797±126.1 |
| NP1100 | 1147.8±23.1 | 189.2±25.0 |
We highlighted the observation of a linear relationship between cell capture yield and effective contact area on nanopillar substrates. Meanwhile, cell cytomorphology was demonstrated to depend on the substrate nanotopography and cell adhesion status.
We first characterized various NPs' compliance, packing density and wettability. Bending compliance affects the adhesion, spreading and migration of attached cells,32, 35 and was calculated as:
| (1) |
where h and D are the height and diameter of NPs, respectively; E is Young's modulus of NPs with a value of 70 GPa.36 Fig. 1(c) shows NPs with smaller diameter yield a larger compliance, indicating a potential of a larger deformation. As such, NP120 with smallest diameter were considered for deformation analysis. When an individual NP120 is subjected to both focal complex force (0.8–0.9 nN/μm2)37 and ligand-receptor bond force (6.7×10–6 dyn for each bond38 and approximately 51667 EpCAM per PC3 cell39), its maximal tip displacement is 0.0046 nm (Fig. 1(d) and see Supporting Information S1 for more details). With such negligible deformation, all NPs were considered as rigid. SEM images further confirm that most of NPs stand straightly (Fig. 3(b-c) and Fig. 5(f-g)). With a hexagonal NP array, packing density η was calculated as:
| (2) |
where Dg is the spacing between two neighboring NPs. As shown in Fig. 1(c), NPs with larger diameter were more densely packed. For comparison purpose, a denser (η = 0.71) and a sparser (η =0.18) packing pattern were obtained from NP300 and NP650, respectively, due to their extraordinary spacing (Table. 1). We then investigated the wettability of NPs by studying the contact angles of water droplets on substrates. On a heterogeneous surface, nanostructured patterns have been reported to enhance surface hydrophobicity or hydrophilicity.40 As a result, SiNP substrates were enhanced to be superhydrophilic due to its hydrophilic nature of oxidized surface in air with contact angle less than 10°. The measurement results are well fitted with the values calculated by applying Cassie's law (Fig. 1(e) and see Supporting Information S2 for more details). It should be noted that surface functionalized with antibodies insignificantly increased wettability due to the inherent superhydrophilic property of NPs.
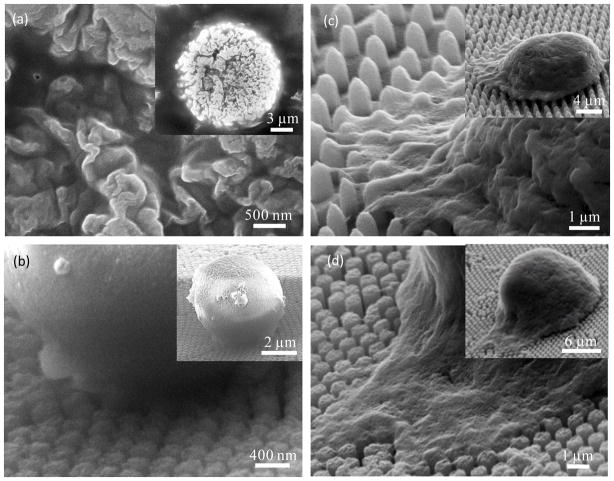
Fig. 3.
(a) Zoomed-in and zoomed-out SEM images reveal details of microvilli on the cell membrane. (b)-(d) Cell behaviors in NP substrates with a spacing of 38 nm for NP300 in (b), 790nm for NP650 in (c), 140 nm for NP550 in (d). All cells were incubated for 1 hour followed by 10 minutes of shaking in 60 rpm.
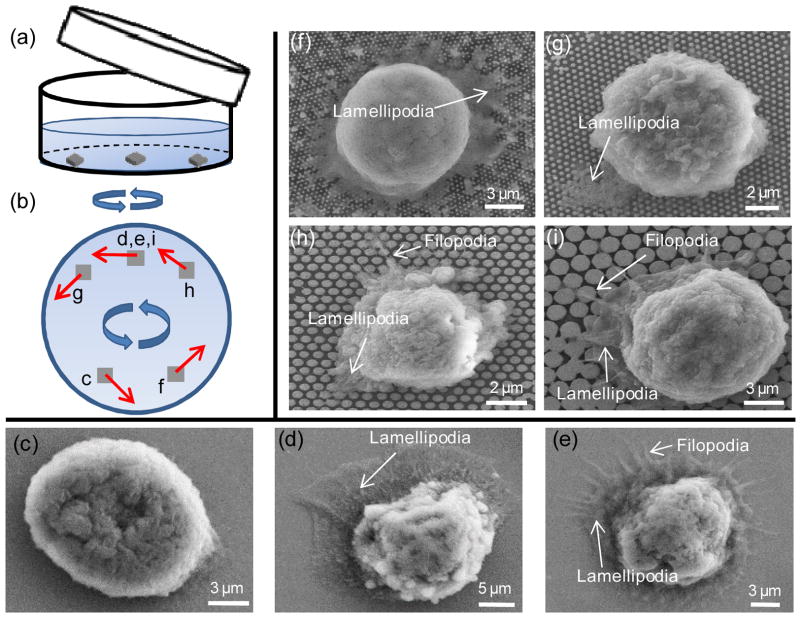
Fig. 5.
Schematics of experimental setup and SEM images showing the cell-NP interactions (lamellipodia and filopodia) for different NP substrates. (a) A schematic of the experimental setup during shaking. (b) shows the corresponding locations on wafer substrates for (c)–(i), with corresponding flow directions marked in red arrows. (c)–(e) show cell behaviors on planar wafers with shaking speeds of 0, 60 rpm and 400 rpm, respectively. (f)–(i) show cell behaviors in NP120, NP200, NP 550 and NP1100 with an average spacing of 140 nm, respectively. Typical lamellipodia and filopodia are annotated in each figure. Orientations of (c)-(i) have been adjusted to be the same as that in (b).
3.2 Spacing and diameter effects of NPs on cell capture yield and cell behaviors
The cell-NPs incubation time and orbit shear stress for eluting were optimized on planar wafers first (see Supporting Information S3 for more details), and the selected conditions were applied throughout the study in all NPs groups. The capture yield reached plateau after one hour (Fig. 2(a)). Shaking at 60 rpm for 10 minutes effectively detached nonspecifically attached PC3 cells while maintaining the maximal capture yield (Fig. 2(b)).
Fig. 2.
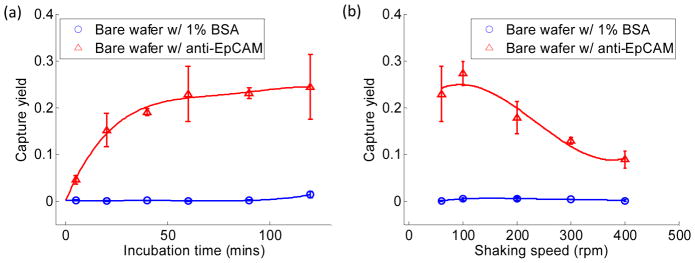
Quantitative evaluation of capture yield for cases with 1% BSA coated and with anti-EpCAM coated (a) at different incubation times followed by 10 minutes of shaking in 60 rpm and (b) after 1 hour of incubation followed by 10 minutes of shaking under various shaking speeds. Each error bar represents a standard derivation and is obtained from three independent experiments.
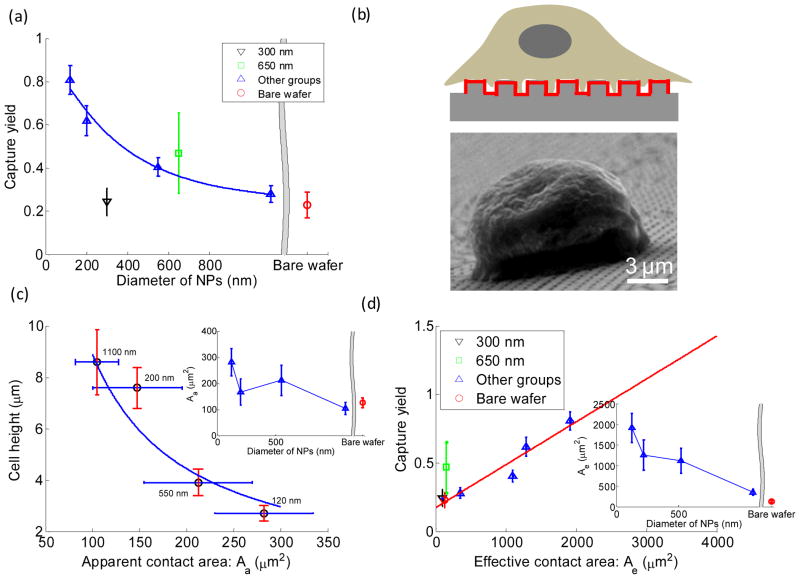
With the proper experimental conditions determined, we next examined spacing effects on capture yield and cell behaviors. Fig. 3(a) shows the average diameter of a microvillus was 105.8 ±14.8 nm measured from zoomed-in SEM images of a single cell sitting on a bare wafer, which is in agreement with previous reports.41 6 NPs groups were thus divided into 3 different categories: NP300 with extremely small spacing (~40 nm), NP650 with extremely large spacing (~800 nm), and the remaining 4 groups with approximately similar spacing (140–190 nm). Low capture yield of 24.3%±6.3% was found in NP300 group, which was close to 22.9% obtained on a planar wafer (Fig. 4(a)). We speculate that such low capture yield is likely due to the small spacing that prevents microvilli or pseudopodia from penetrating through the gap and contacting with the lateral surface of NP300. Thus cells were unable to gain sufficient adhesion force to resist the following eluting shear stress. As cells only contacted with the top surface of NP300 (Fig. 3(b)), cells on NP300 showed a globular shape which was unlike the flat cell morphology in other groups. On the contrary, cells showed a very flat shape on NP650, where microvillus or pseudopodia easily filled into the 797 nm spacing (Fig. 3(c)). Compared to cells on NP300 these cells had larger contact area and gained bigger adhesion force. Hence, relatively higher capture yield of 46.95%±18.7% was obtained on NP650; however, due to its sparse packing pattern (Fig. 1(c)) and relatively smaller effective contact area (discussed later), capture yield on NP650 was still low (Fig. 4(a)) in comparison to NP120 and NP200. The significantly higher SD on NP650 was also likely to be caused by larger spacing in which cells can physically situate in various ways. Thus, the nonspecific settlement may contribute to the large capture yield fluctuation. In the remaining 4 groups with 140–190 nm spacing, cells spread moderately and effectively interacted with NPs’ lateral walls (Fig. 3(d)); the capture yield ranged from 28.0% to 80.8% along with the increased effective contact area of each group (discussed later). The above results indicated that spacing is an important factor that is directly related to capture yield. Dense NPs with very small spacing (~ 50 nm) adversely impairs cell capture yield. In one previous study, dense silica bead (diameter ranges from 100 to1150 nm) were deposited closely onto a glass slide without any spacing, cell capture yield was only 1.2–1.6 times higher than that on a planar surface.42 This result also indicates the spacing plays an important role in capture yield. On the contrary, increased nonspecific settlement may appear on sparse NP surface when the spacing is larger than 500 nm, i.e., diameter of filopodia. Considering the spacing of NP120 and its over 80% capture yield (Fig. 4(a)), 140–200 nm spacing probably well suits the needs of efficient and specific capture.
Fig. 4.
(a) Cell capture yield measured on substrates with different diameters of NPs after 1 hour of incubation and 10 minutes of shaking in 60 rpm. (b) Schematics (not in scale) of a captured cell on an NP substrate with red bold line marked as the effective contact area, and a typical SEM image of a captured cell on NP1100 from tilted view. (c) Cell morphology consisting of cell height and apparent contact area for different NP substrates. The inset of (c) shows apparent contact areas for NP substrates with various diameters and planar wafer. (d) Cell capture yield measured on NP substrates with various diameters and planar wafer. The inset in (d) shows effective contact areas for NP substrates with various diameters and planar wafer. The solid line in (d) stands for a linear fit. Error bars for capture yield in (a) and (d) stand for standard derivation from three independent experiments. Error bars in (c) stand for standard derivation from 20 arbitrary cells' measurements.
We then examined the effects of diameter of NPs on capture yield and cell behaviors. In the remaining 4 groups, diameter of NPs was considered as the major varying factor. The spacing ranged from 144.0 nm to 189.2 nm with the maximal difference 45.2 nm was smaller than the diameter of a single microvillus, thus was treated as a minor factor here. The average spacing of 160.6 nm (SD: 19.7 nm) of the 4 groups was generally adopted for the following studies and discussion. Fig. 4(a) shows that capture yield decreased with increasing diameter of NPs, reaching 28.0%±3.9% in NP1100 group which was almost the same as that in planar wafer case (22.9%). We explained this trend by adhesion strength induced by NP-cell interaction and thus introduced two parameters: apparent contact area Aa and effective contact area Ae . Aa was defined as the projection area of a captured cell from top view; Ae was defined as the area of the NP substrate with chances to touch the captured cell. It was calculated as the whole surface area of the NP substrate consisting of top, lateral and bottom surfaces beneath the captured cell as shown in Fig. 4(b). This is because microvilli (diameter: 100–120 nm; length: 2–4 μm) and filopodia (diameter of 100–500 nm; length from few microns to >50 μm) can fit into the spacing of NPs, and can even reach the bottom of the substrate. Through SEM images of the captured cells, we measured apparent contact area Aa (Fig. 3(a)) and cell height (Fig. 4(b)) from top and tilted view, respectively, and summarized in Fig. 4(c); all data points were well fitted by a rectangular hyperbola, indicating that estimating the average volume of cells as a simplified cylinder-like shape was consistent in each group. According to the definition of effective contact area, Ae was further calculated as:
| (3) |
Fig. 4(e) showed Ae decreased with increasing diameter as expected, indicating NPs with smaller diameter have larger surface area. Fig. 4(d) revealed that capture yield decreased linearly with decreasing Ae . Furthermore, the linear function also applied to planar wafer and NP300 cases, where Ae is exactly or approximately equal to Aa . However, the linear function did not apply for NP650, mainly due to cells’ nonspecific settlement as discussed above. Contrarily, we did not observe an apparent relationship between capture yield and Aa (Fig. S1). It was concluded that nanostructured surfaces can provide larger surface area for enhanced antibody immobilization; larger antibody functionalized area can significantly increase the odds of antibody-receptor binding and thus the cell capture yield.12 Another previous report also validated that cell capture yield can be improved by increasing the total area of gold clusters on SiNWs for antibody immobilization.19 Nevertheless, we clearly showed for the first time that capture yield was linearly associated with Ae instead of Aa on substrates with nano-patterns. Therefore, with suitable spacing of 140–200 nm, higher capture yield can be obtained with smaller NPs of larger surface area for more antibody immobilization.
In addition to the effects of diameter of NPs on capture yield, we also observed different cell morphologies and spreading effects on antibody-coated NPs with different diameters. First of all, cells had relatively flat shape on the remaining 4 NPs groups (Fig. 5). As shown in Fig. 4(c), we observed an overall decreasing of Aa and increasing of cell height with increased NPs diameter. As a result, captured cells were able to endure higher shear force on smaller NPs, thus leading to higher capture yield. Our previous work has also verified that decreased cell height further prevented captured cells from being washed away under high flow velocity.43 Additionally, the inset of Fig. 4(c) shows that antibody-coated NPs did not necessarily inhibit the cell spreading compared to planar wafers, which is different from a previous study.31 In our previous report, epidermal growth factor receptor (EGFR) overexpressed cancer cells became flatter and reshaped to cover as large of an anti-EGFR aptamer functionalized area as possible.9 On the contrary, cells on mutant aptamer substrates (control group) maintained a globular shape. Such spreading and flatness of cancer cells on aptamer surfaces have been proven to be a potential modality for discrimination of cancer cells from healthy cells.44 Similarly, on anti-EpCAM functionalized surfaces in our case, PC3 cells also spread well due to the intense interactions between antibodies and receptors; cells gradually spread onto the substrate which was facilitated by the newly generated affinity bonds at the contact edge of cell-substrate. In the other scenario of planar wafers, cells have to develop micron-scale focal adhesions independently; NPs with high aspect ratio might fail to provide sufficient areas for development of focal adhesion and thus cell spreading was inhibited on bare NPs. It was also noted that cells on NP550 showed larger Aa with smaller height compared to that on NP200. We speculated that it might be due to the complementary size match between the diameter of NPs (550 nm) and focal complex (generally with size around 500 nm), although the exact mechanism behind this phenomenon is still unknown.
Finally, we studied the formation of subcellular adhesion structures under shear flow. It was observed that the formation of pseudopodia under shear is closely correlated with cellular adhesion status, shear force and shear direction. Fig. 5(a) and (b) show a schematic of the experimental setup and locations of each wafer substrate corresponding to (c)–(i). We first examined cell behaviors on anti-EpCAM functionalized planar wafers at shaking speeds of 0, 60 and 400 rpm for 10 minutes after one hour of incubation. When shear force increases, a continuous transition of cell behaviors is observed, from being intact, dominant expression of lamellipodia to significant formation of filopodia (Fig. 5(c)–(e)). Next, we examined cell behaviors on NP substrates by exertion of the same shear force (60 rpm for 10 min) on captured cells. Lamellipodia were more frequently observed in NP120 and NP200 cases (Fig. 5 (f) and (g)) where the total adhesion force between the substrate and cell is relatively large, while filopodia were dominant in NP500 and NP1100 cases (Fig. 5(h) and (i)) where adhesion force is relatively small. Based on these results, we speculated that the final formation of either lamellipodia or filopodia depends on the relative strength of cell adhesion force and applied shear force. Under shear force, cells were inclined to roll on the substrate; the larger the shear force, the greater the effect until final detachment. In our case, during the eluting process these unwashed cells are inclined to form new adhesive bonds at the leading edge and dissociate old ones at the trailing edge.45 If the adhesive force generated between the cell and substrate is much larger than the applied shear force (cells on NP120 and NP200 substrates; cells on planar surface washed by 60rpm), the flowing fluid just gently aligns cells in the flow direction, and cells have sufficient time to form lamellipodia. In comparison, attached cells with less adhesion force in NP500 and NP1100 cases or suffering large shear force on planar surface are more susceptible and easier to roll on the surface. These cells develop filopodia during such dynamics, which is in agreement with a previous study showing that filopodia primarily acted as the traction force during spreading on a microstructured pattern.33 It was also observed that the direction of the formation of lamellipodia and filopodia matched with the flow direction induced by shaking in Fig. 5(b), indicating that lamellipodia and filopodia formed initially in the leading edge, which is in agreement with a previous study.46 These findings indicate the possibility to control the formation of pseudopodia type and direction by applying shear force on attached cells.
4. Conclusion
In summary, we found a linear relationship between capture yield and effective contact area of NP substrate with diameter ranging from 120 to 1100 nm. NPs with smaller diameter are preferred to achieve higher capture yield due to their large effective contact area. On the other hand, very small spacing below 100 nm (typically size of microvilli) significantly impairs the capture yield. The effect of shear force on cell behaviors on an NP substrate was also studied for the first time. We found that the formation of filopodia or lamellipodia depends on the adhesion status of the captured cell, eluting strength and its direction. These findings provide guidance on the design of NP geometry for efficient CTC isolation, and possibilities to tune cytomorphology by manipulating shear forces.
Supplementary Material
Acknowledgments
This work was supported in part by National Science Foundation (NSF) grant CBET-1113040, CBET-1264808, and National Institute of Health (NIH) grant EB015105 (to Y. Liu) and by ECNA PD-130011, UniSA (to Y. Wan). The authors thank Max Planck Institute of Microstructure Physics for the technical support of preparing NPs. We also thank Dr. Dadhichi Paretkar for helpful discussions.
Footnotes
These authors declare no competing financial interests.
Additional details about calculation of maximal displacement of an individual NP; additional study of substrate wettability through Cassie's Law; additional details about selection of incubation time and shaking speeds. This material is available via the Internet or from the author.
Contributor Information
Yuan Wan, Email: yuan.wan@unisa.edu.au.
Yaling Liu, Email: yal310@lehigh.edu.
References
- 1.Wicha MS, Hayes DF. J Clin Oncol. 2011;29:1508. doi: 10.1200/JCO.2010.34.0026. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Brakenhoff RH, Brandt B. Nat Rev Cancer. 2008;8:329. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 3.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Chem Eng Sci. 2011;66:1508. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, Siewert J. In Molecular Staging of Cancer. 2003;162:149. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 5.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Am J Pathol. 2000;156:57. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan SJ, Lakshmi RL, Chen P, Lim WT, Yobas L, Lim CT. Biosens Bioelectron. 2010;26:1701. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 7.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Natl Acad Sci. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Kim Y-t, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Cancer Res. 2010;70:9371. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Wang S, Song Y, Yang J. Micro Nanosyst. 2012;4:254. [Google Scholar]
- 11.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA. Proc Natl Acad Sci. 2012;109:19626. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Asghar W, Demirci U, Wan Y. Nano Today. 2013;8:374. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RH, Macoska JA, Merajver SD, Fu J. ACS nano. 2012;7:566. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei Ki, Sun J, Sherman DJ, Behrenbruch CP, Wu H. Angew Chem. 2009;121:9132. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Angew Chem. 2011;123:11973. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin S, Wu YL, Hu B, Wang Y, Cai P, Tan CK, Qi D, Zheng L, Leow WR, Tan NS. Adv Mater Interfaces. 2014;1:1300043. [Google Scholar]
- 17.Liu X, Wang S. Chem Soc Rev. 2014;43:2385. doi: 10.1039/c3cs60419e. [DOI] [PubMed] [Google Scholar]
- 18.Peng F, Su Y, Zhong Y, Fan C, Lee ST, He Y. Acc Chem Res. 2014;47:612. doi: 10.1021/ar400221g. [DOI] [PubMed] [Google Scholar]
- 19.Park GS, Kwon H, Kwak DW, Park SY, Kim M, Lee JH, Han H, Heo S, Li XS, Lee JH. Nano lett. 2012;12:1638. doi: 10.1021/nl2045759. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK. Angew Chem, Int Ed. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Adv Mater. 2011;23:4788. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell MJ, Castellanos CA, King MR. J Nanomater. 2012;2012:831263. doi: 10.1155/2012/831263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF. Nat Nanotechnol. 2013;8:735. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Chen L, Liu H, Yang G, Zhang P, Han D, Wang S, Jiang L. NPG Asia Mater. 2013;5:e63. [Google Scholar]
- 25.Yang G, Liu H, Liu X, Zhang P, Huang C, Xu T, Jiang L, Wang S. Adv Healthcare Mater. 2014;3:460. doi: 10.1002/adhm.201300233. [DOI] [PubMed] [Google Scholar]
- 26.den Toonder J. Lab Chip. 2011;11:375. doi: 10.1039/c0lc90100h. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S. Adv Mater. 2013;25:922. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Chen L, Xu T, Liu H, Liu X, Meng J, Yang G, Jiang L, Wang S. Adv Mater. 2013;25:3566. doi: 10.1002/adma.201300888. [DOI] [PubMed] [Google Scholar]
- 29.Wan Y, Mahmood M, Li N, Allen PB, Kim Yt, Bachoo R, Ellington AD, Iqbal SM. Cancer. 2012;118:1145. doi: 10.1002/cncr.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi S, Yi C, Ji S, Fong CC, Yang M. ACS Appl Mater Interfaces. 2008;1:30. doi: 10.1021/am800027d. [DOI] [PubMed] [Google Scholar]
- 31.Hu W, Crouch AS, Miller D, Aryal M, Luebke KJ. Nanotechnology. 2010;21:385301. doi: 10.1088/0957-4484/21/38/385301. [DOI] [PubMed] [Google Scholar]
- 32.Kim DJ, Seol JK, Lee G, Kim GS, Lee SK. Nanotechnology. 2012;23:395102. doi: 10.1088/0957-4484/23/39/395102. [DOI] [PubMed] [Google Scholar]
- 33.Guillou H, Depraz-Depland A, Planus E, Vianay B, Chaussy J, Grichine A, Albigès-Rizo C, Block MR. Exp Cell Res. 2008;314:478. doi: 10.1016/j.yexcr.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Geyer N, Fuhrmann B, Huang ZP, de Boor J, Leipner HS, Werner P. J Phys Chem C. 2012;116:13446. [Google Scholar]
- 35.Kim DJ, Lee G, Kim GS, Lee SK. Nanoscale Res Lett. 2012;7:1. doi: 10.1186/1556-276X-7-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaccodine R, Schlegel W. J Appl Phys. 2004;37:2429. [Google Scholar]
- 37.Galbraith CG, Yamada KM, Sheetz MP. J Cell Biol. 2002;159:695. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng J, Soper SA, McCarley RL, Murphy MC. Proc SPIE. 2004:278. [Google Scholar]
- 39.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK. Proc Natl Acad Sci. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cha TG, Yi JW, Moon MW, Lee KR, Kim HY. Langmuir. 2010;26:8319. doi: 10.1021/la9047402. [DOI] [PubMed] [Google Scholar]
- 41.Mooseker MS, Tilney LG. J Cell Biol. 1975;67:725. doi: 10.1083/jcb.67.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Weldon AL, Kumnorkaew P, Xu B, Gilchrist JF, Cheng X. Langmuir. 2011;27:11229. doi: 10.1021/la2015868. [DOI] [PubMed] [Google Scholar]
- 43.Wan Y, Tan J, Asghar W, Kim Y-t, Liu Y, Iqbal SM. J Phys Chem B. 2011;115:13891. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 44.Mahmood MA, Arafat C, Kim Y-t, Iqbal SM. 2013 35th Annual International Conference of the IEEE; 2013; p. 4164. [DOI] [PubMed] [Google Scholar]
- 45.Krasik EF, Hammer DA. Biophys J. 2004;87:2919. doi: 10.1529/biophysj.104.039693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abercrombie M. Proc R Soc London, Ser B. 1980:129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.