Abstract
Approximately 25% of childhood B-cell precursor acute lymphoblastic leukemia have an ETV6/RUNX1 (E/R) gene fusion that results from a t(12;21). This genetic subgroup of leukemia is associated with near-triploidy, near-tetraploidy, and trisomy 21 as rather specific types of secondary changes. Here, we show that, unlike various controls, E/R-expressing Ba/F3 clones acquire a tetraploid karyotype on prolonged culture, corroborating the assumption that E/R may attenuate the mitotic checkpoint (MC). Consistent with this notion, E/R-expressing diploid murine and human cell lines have decreased proportions of cells with 4N DNA content and a lower mitotic index when treated with spindle toxins. Moreover, both RUNX1 and E/R regulate mitotic arrest-deficient 2 L1 (MAD2L1), an essential MC component, by binding to promoter-inherent RUNX1 sites, which results in down-regulation of MAD2L1 mRNA and protein in E/R-expressing cells. Forced expression of E/R also abolishes RUNX1-induced reporter activation, whereas E/R with a mutant DNA-binding site leads to only minor effects. Our data link for the first time E/R, MC, and MAD2L1 and provide new insights into the function of the E/R fusion gene product. Although tetraploidy is an almost exclusive feature of E/R-positive leukemias, its rarity within this particular subgroup implies that further yet unknown factors are required for its manifestation.
Keywords: ETV6/RUNX1, t(12;21), acute lymphoblastic leukemia, mitotic checkpoint MAD2L1, tetraploidy
Introduction
The t(12;21)(p13;q22) with its molecular counterpart, the ETV6/RUNX1 (E/R) (also known as TEL/AML1) gene fusion, characterizes approximately 25% of childhood B-cell precursor ALL cases. E/R is unique among the fusion genes involving RUNX1, as it is associated with acute lymphoblastic leukemias rather than myeloid leukemias (Speck and Gilliland, 2002). Accumulating evidence suggests that this fusion gene can initiate leukemia development, and although it is not sufficient to cause overt leukemia per se, it is necessary for its maintenance (Zelent et al., 2004; Diakos et al., 2007; Hong et al., 2008). E/R encodes a chimeric protein that is composed of the N-terminal non-DNA-binding region of ETV6 and almost the entire RUNX1 protein. It retains the runt domain of RUNX1 that is required for DNA binding and heterodimerization and, therefore, also provides an essential function of RUNX1 fusion genes (Hiebert et al., 1996; Morrow et al., 2007; Roudaia et al., 2009; Wolyniec et al., 2009). The E/R protein acts as an aberrant transcription factor and represses or disrupts the regulation of RUNX1 target genes in a cellular context-dependent manner (Pui et al., 2004; Zelent et al., 2004; Wotton et al., 2008).
Approximately 15% of E/R-positive leukemias acquire an extra chromosome 21 as a secondary change (Attarbaschi et al., 2004). Furthermore, near-tetraploid and near-triploid karyotype patterns in childhood B-cell precursor ALL were recently recognized as rather specific secondary abnormalities, although they occur in only approximately 5% of E/R-positive cases (Attarbaschi et al., 2006; Raimondi et al., 2006).
On the basis of these observations, we reasoned that E/R can affect chromosome segregation, most likely by compromising the surveillance mechanisms of the mitotic checkpoint (MC) (Weaver and Cleveland, 2005; Peters, 2007). The MC guarantees that all sister chromatids are properly attached to the spindles before cell-cycle progresses through mitosis. A central component of this checkpoint is the ‘mitotic arrest-deficient 2’ (MAD2) protein. Partial loss of function as well as over-expression of MAD2L1 leads to the abrogation of MC, chromosome mis-segregation, aneuploidy, and failure to arrest mitosis in the presence of microtubule poisons (Sotillo et al., 2007). The unexpected finding of a tetraploid karyotype of Ba/F3 cells on stable expression of E/R prompted us to pursue this issue further. We have, therefore, investigated the influence of E/R on MC and especially on its major component MAD2L1.
Results and discussion
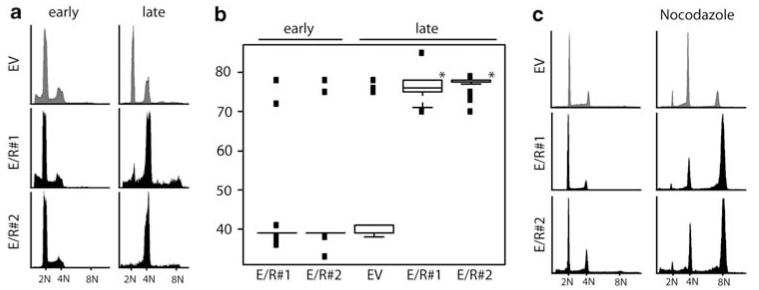
To assess the function of E/R, we used Ba/F3, a murine IL-3-dependent putative pro B-cell line, that is frequently used as a model cell line to study the effects of genes involved in the development of B-cell precursor ALL. For this purpose, we have created stable E/R-expressing Ba/F3 clones as reported earlier (Diakos et al., 2007) and selected those with low expression of the chimeric protein. After continuous culture for approximately 2 months, all E/R-expressing clones (n = 5) acquired a tetraploid karyotype, whereas cells containing plasmids encoding either full-length ETV6, a truncated form of ETV6 (ETV6-F, with dominant-negative effect over ETV6 wt function) (Sasaki et al., 2004), RUNX1, GFP, or an empty vector remained diploid. This change in ploidy was confirmed by DNA content analysis and cytogenetics of several individual clones (Figures 1a and b). To corroborate the specificity of this effect, E/R-positive Ba/F3 clones and controls were synchronized by a short exposure to low doses of nocodazole and accumulated—unlike control cells—at 8N on a second exposure to the spindle toxin (Figure 1c). These findings indicate that tetraploidization is a specific effect of E/R-expressing Ba/F3 cells in long-term cultures.
Figure 1.
E/R expression induces a tetraploid karyotype in Ba/F3 cells. Representative histograms visualizing DNA content analysis (PI) of two E/R + clones (#1 and #2) and empty vector (EV) control clones at early and late passages. (b) Box blot analysis depicting chromosome numbers of E/R-expressing or EV Ba/F3 cells from cultures as in (a). Welch’s t-test, *P≤0,05. (c) DNA content analysis of untreated (left part) and synchronized E/R and EV Ba/F3 clones on nocodazole treatment (right part). Cells were released from the first arrest (nocodazole 75 ng/ml for 14 h) for 3 days before they again were exposed to nocodazole. One of at least three independent experiments is shown.
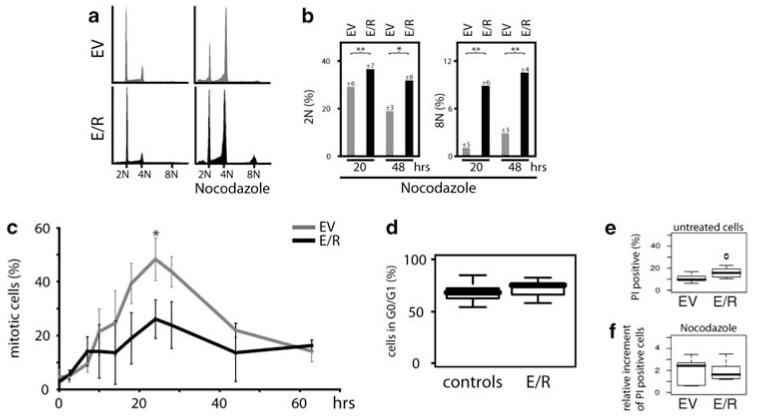
Hence, we used diploid E/R-expressing Ba/F3, and several E/R-positive and E/R-negative leukemia cell lines to investigate the influence of the chimeric E/R protein on MC. Consistent with the notion that cells with an attenuated MC are unable to fully arrest their cell cycle in mitosis when treated with spindle poisons (Kops et al., 2005), the proportion of E/R-positive cells with a 2N and 8N DNA content was significantly increased under the influence of nocodazole as compared with controls (Figures 2a and b; Supplementary Figures 1a and b). The higher percentage of E/R-positive cells with 8N implies that these cells did not accurately segregate their chromosomes and/or terminate cytokinesis. Determination of mitotic cells with the phospho-histone H3 staining, which is tightly correlated with chromosome condensation during mitosis, revealed consistently lower mitotic indexes in nocodazole-treated E/R-expressing Ba/F3 cell lines and leukemias, but not in E/R-negative model and leukemia cell lines (Figure 2c; Supplementary Figure 1c).
Figure 2.
E/R attenuates the MC. Cell-cycle distribution of a mixture of three Ba/F3 clones that stably expressed E/R or contained an empty vector (EV) by DNA content analysis. (a) Shown are representative histograms in the absence (left) and presence (right) of nocodazole (333 ng/ml for 48 h). (b) Bar graphs indicate percentages of E/R-expressing and EV Ba/F3 cells with 2N and 8N after treatment with nocodazole for 20 and 48 h. The median and standard deviation (indicated at the top of each bar) are taken from five different experiments. Welch’s t-test, *P≤0.05; **P≤0.01. (c) Mitotic index of stably E/R-expressing and EV Ba/F3 cells was determined by phospho-histone H3 staining on MC activation by nocodazole for the indicated times. Experiments depict the average and standard errors of the mean of three individual experiments. Welch’s t-test, *P≤0.05. (d) Cell-cycle distribution of Ba/F3 clones stably expressing E/R or various control vectors (ETV6, truncated ETV6, RUNX1, GFP, or EV) by DNA content analysis with PI. Box blots depict the percentage of cells in G0/G1 of E/R + clones and controls from 5 and 7 independent experiments, respectively. (e/f) Apoptosis rates were assessed by PI staining in stably E/R-expressing and EV Ba/F3 cells. Percentages of PI-positive cells are depicted under optimal culture conditions (e) and on nocodazole treatment for 24 h relative to untreated cells (f). Welch’s t-test, P = 0.8.
Given that the cell-cycle distribution assessed by DNA content analysis of E/R-positive Ba/F3 cell lines showed only a minor—statistically not significant—G0/1 increase compared with several controls (empty vector, expression vectors for RUNX1, ETV6, ETV6-F, or GFP) (Figure 2d), its deregulation is unlikely to be the cause of the low mitotic index, especially as cells were also measured after a long exposure to nocodazole. These findings accord with the observed reduction of G1 phase cells by RUNX1 and its increase by CBF oncoproteins (Lou et al., 2000; Strom et al., 2000; Ford et al., 2009).
Of note and in line with a latent MC destabilization (Kops et al., 2005), the spontaneous apoptosis rate of both E/R-positive Ba/F3 as well as E/R-positive leukemic cell lines were already slightly, but not significantly, higher than those of the respective parental Ba/F3 and E/R-negative control cell lines under optimal culture conditions (Figure 2e; Supplementary Figure 1d). We also excluded the possibility that nocodazole per se might selectively increase the apoptotic rate of mitotic E/R-positive cells by showing that the proportion of PI-positive cells did not increase considerably over basic levels (Figure 2f; Supplementary Figure 1e). The E/R-dependent MC impairment was further confirmed by the reduced levels of the checkpoint-associated protein securin, whose degradation at the start of anaphase initiates sister chromatid separation (Pines, 2006), and thus concords with the lower mitotic index (Supplementary Figure 1f). These findings strikingly resemble those recently obtained in similar experiments using acute myeloid leukemia samples and cell lines with a truncated RUNX1/ETO, a fusion protein that is closely related to E/R (Boyapati et al., 2007; Wolyniec et al., 2009).
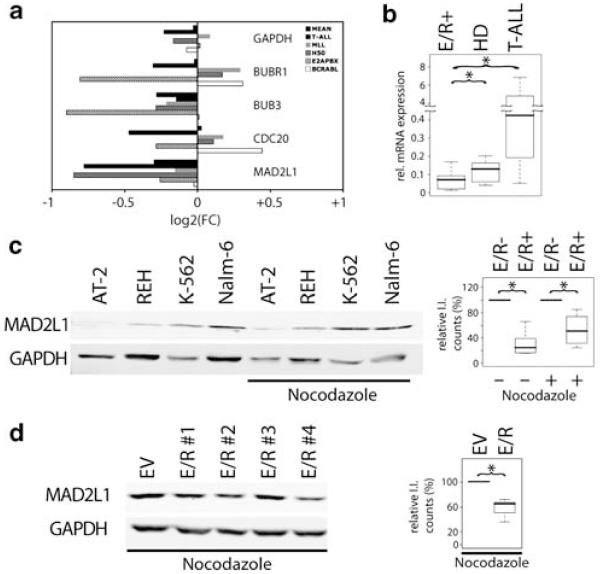
Having shown the influence of E/R on MC function, we set out to unravel its potential molecular mechanism. For this purpose, we interrogated publicly available Affymetrix data sets from childhood ALL (Ross et al., 2003) for the expression of genes implicated in MC function. MAD2L1 and BUB3 (budding uninhibited by benzimidazole 3), two key players of MC, were consistently repressed in E/R-positive leukemias compared with all other ALL subgroups (Figure 3a). The finding that the MAD2L1 promoter region contained four perfect matches for RUNX binding (TGT/CGGT) and 13 sites with 85% homology within 3 kb upstream of the transcription start site makes it a likely candidate for a direct target of RUNX1 and also E/R. In contrast, no perfect RUNX-binding site was present within 3 kb of the promoter region of BUB3.
Figure 3.
E/R represses MAD2L1 expression. (a) Micro-array analysis of MAD2L1 expression in various subgroups of childhood ALL. Published data were analyzed for M-values (log2(FC)) of probe sets from genes instrumental in MC function. Shown is the MAD2L1 expression from E/R-positive leukemias relative to their E/R-negative counterparts in each leukemic subgroup. Mean, the mean of all leukemic groups; T-ALL, T-ALL; MLL, ALL with MLL fusion genes; H50, high hyperdiploid ALL; E2APBX, TCF3/PBX1 (E2A/PBX1) gene rearrangement positive ALL; BCRABL, ALL with BCR/ABL fusion gene. (b) MAD2L1 mRNA was quantified by TaqMan qRQ-PCR and the ABI Prism 7900 detection system (Applied Biosystems, Foster City, CA, USA) in 10 cases each of the after distinct childhood ALL subgroups: E/R +, high hyperdiploid (HD) and T-cell precursor (T-ALL). The ABL gene was used as a standard reference for normalization. Box plots display the relative expression of MAD2L1 using STS 2.2 software. Shown are the median and interquartile range, 95% of the values are within the range of a whisker. Median values were used for statistical analysis; Welch’s t-test, *P≤0.05. (c) Western blot analysis was performed from whole cell lysates of E/R-positive (REH, AT-2) and E/R-negative leukemic cell lines (K562 and Nalm-6). The MAD2L1 protein was detected with a specific anti-MAD2L1 antibody (ab 3632, Abcam, Cambridge, UK). Anti-GAPDH (6C5, Santa Cruz Biotechnology, CA, USA) was used as loading control. Left: protein levels under standard culture conditions and on nocodazole treatment (50 ng/ml nocodazole, 18 h). Right: box plots show the quantification of MAD2L1 protein in the respective groups from four independent experiments. MAD2L1 protein was normalized to GAPDH expression using the LICOR software (I.I., Integrated Intensity counts). Welch’s t-test, *P≤0.05. (d) Western blot analysis (left) and quantification (right) of MAD2L1 in E/R + Ba/F3 clones and EV controls. One of three independent experiments is shown. Welch’s t-test, *P≤0.05.
We, therefore, focused our further work on MAD2L1 and validated the expression of MAD2L1 by quantitative RT–PCR in primary childhood ALL samples and confirmed a significant difference between E/R-positive and E/R-negative leukemias (Figure 3b). In accordance with mRNA expression, protein expression of MAD2L1 was less abundant in E/R-positive leukemic and model cell lines (Figures 3c and d). Conversely, MAD2L1 mRNA expression was up-regulated after E/R silencing in AT-2 and REH leukemic cell lines, further emphasizing its regulation by E/R (our unpublished observation).
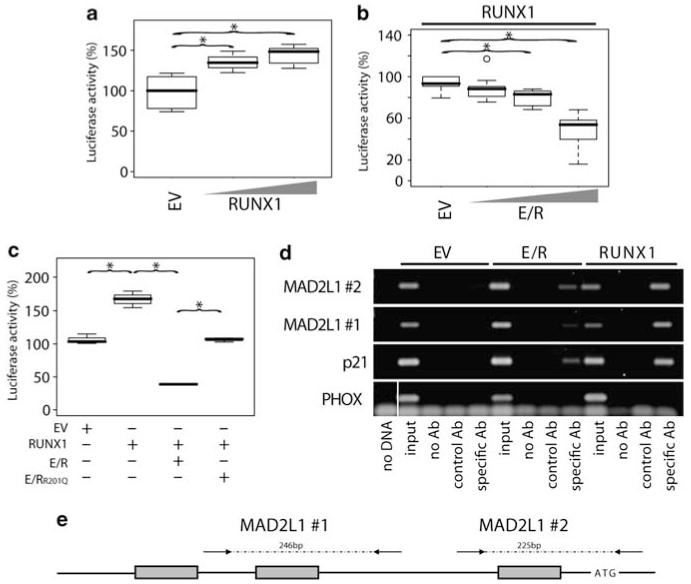
To test the possibility that MAD2L1 expression is directly regulated, we determined its transcriptional activity in the context of RUNX1 and E/R. Two constructs that contained endogenous promoter sequences with one or three perfect RUNX1 consensus sites were used for luciferase-based reporter assays (Guardavaccaro et al., 2008). Both promoter constructs revealed a dose-dependent activation on RUNX1 transfection and E/R-associated repression of RUNX1-induced MAD2L1 activity in NIH3T3 cells (Figures 4a and b). Similarly, forced expression of RUNX1 led to induction of MAD2L1 promoter activation and coexpression of E/R to its abrogation in HEK293 cells (data not shown). In contrast, transfection of NIH3T3 cells with an expression vector with a point mutation in the runt domain (E/RR201Q) that had been earlier shown to diminish DNA binding, but not heterodimerization with CBFβ (Song et al., 1999; Li et al., 2003), resulted in only partial reduction of RUNX1-induced activation (Figure 4c). These data accord with earlier studies and indicate that DNA binding is essential for E/R activity and that E/R modulates RUNX1-induced gene regulation (Hiebert et al., 1996; Friedman, 1999; Morrow et al., 2007; Wotton et al., 2008; Roudaia et al., 2009).
Figure 4.
Direct regulation of MAD2L1 by RUNX1 and abrogation of this effect by E/R. Luciferase assays were performed using a 1.2 kb MAD2L1 genomic reporter fragment containing three perfect RUNX1-binding sites (a–c). NIH3T3 cells were transfected (JetPEI Polyplus transfections, Illkirch, France) with RUNX1, E/R, E/RR201Q, or a combination thereof. Cells were cotransfected with CBFβ (12.5 ng) to enhance RUNX1 binding and TK Renilla for transfection control. The total amount of DNA for each experiment was kept constant. Values obtained from EV control samples were set to 100%. Luciferase activity is depicted using (a) increasing amounts of RUNX1 pcDNA3.1-expression vector (100 and 500 ng) or (b) E/R (200, 400, and 800 ng) in addition to RUNX1 (400 ng) to show dose-dependent reporter activation by RUNX1 and its repression by E/R, respectively. Box plots of three representative experiments are depicted. Welch’s t-test, *P≤0.05. (c) NIH3T3 cells were transfected using various combinations of 267 ng empty vector (EV), E/R, or a DNA-binding mutant E/RR201Q, and/or 100 ng RUNX1-expression vector. One of three representative experiments is depicted. Welch’s t-test, *P≤0.05. (d) RUNX1 and E/R occupation of MAD2L1 promoter region in vivo was assessed by ChIP. Chromatin from Myc-tagged E/R, Myc-RUNX1, or Myc-tagged EV stably expressing HEK293 cells was immunoprecipitated with anti-c-Myc (9E11, Abcam) or control antibody (anti-N cadherin, 610920, BD). Aliquots of the isolated DNA fragments (without antibody (noAb), with control or specific Ab and input DNA) were subjected to PCR with specific primers that amplify regions of the MAD2L1 promoter (Supplementary Methods) and analyzed by DNA gel electrophoresis. Specific primers were used to amplify specific regions of p21WAF1 and PHOX promoters as positive and negative controls, respectively. The gel from one of two independent experiments is shown. A vertical line has been inserted to indicate where the gel lane was cut. These gels came from the same experiment. (e) Schematic diagram of the 1.2 kb of the promoter region of the MAD2L1 gene cloned into the pGL3. The three perfect RUNX1-binding motifs (TGT/CGGT) are indicated by gray boxes. Arrows flanking the two RUNX1 sites (#1 and #2) indicate primer positions used for PCR amplification of isolated DNA fragments after ChIP. Numbers refer to the size of the expected PCR product. Graph is not to scale.
We then went on to show a direct in vivo interaction between E/R and the MAD2L1 promoter by ChIP analysis. Using Myc-RUNX1 or Myc-E/R stably expressing HEK293 cells, we were able to prove that both RUNX1 and E/R bind to the chromatin at two consensus RUNX1 sites, which were also present in the constructs used for the reporter assays (Figures 4d and e).
Collectively, these data suggest that direct suppression of MAD2L1 by E/R binding specifically adds to the destabilization of the MC (Perez de Castro et al., 2007). However, as only a small subgroup of E/R-positive leukemias acquires non-random-specific numerical chromosomal abnormalities (Attarbaschi et al., 2004, 2006; Raimondi et al., 2006), it seems unlikely that MAD2L1-associated MC attenuation alone is responsible for this phenomenon. Furthermore, it remains to be investigated whether the E/R-induced deregulation of MAD2L1 or any other MC components might also contribute in other ways to leukemogenesis, such as perhaps p53 activation or deregulation of the DNA damage response pathways (Michel et al., 2004; Kops et al., 2005; Fang et al., 2006; Ha et al., 2007; Perez de Castro et al., 2007).
Supplementary Material
Acknowledgements
We thank O Williams for expression vectors containing E/R or the mutant E/R, D-E Zhang for the RUNX1-expression vector, A Friedman for CBFβ-expression vector, D Guardavaccaro for the luciferase reporter plasmid for the MAD2L1 promoter in pGL3, JD Rowley for AT-2 cell line, L Orel for the Myc-ETV6/RUNX1 vector, Idriss M Bennani-Baiti for stimulating discussions, and Marion Zavadil for proofreading the paper. This study was supported in part by a grant from the FWF P17551-B14 and the Austrian Ministry for Education, Science and Culture (GENAU-Ch.I.L.D) and the St Anna Kinderkrebsforschung to ER P-G. A Kilbey is funded by the Leukaemia Research Fund. GK participated in the design of the study, performed experiments, interpreted data, and wrote the paper. UK, GF, AI, RJ performed research and interpreted data. AK performed and interpreted ChIP experiments. JCN participated in study design and interpretation of ChIP data. GM contributed patient samples. OAH participated in the interpretation of data and writing of the paper. ER P-G designed and supervised research and wrote the paper.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Attarbaschi A, Mann G, Konig M, Dworzak MN, Trebo MM, Muhlegger N, et al. Incidence and relevance of secondary chromosome abnormalities in childhood TEL/AML1+ acute lymphoblastic leukemia: an interphase FISH analysis. Leukemia. 2004;18:1611–1616. doi: 10.1038/sj.leu.2403471. [DOI] [PubMed] [Google Scholar]
- Attarbaschi A, Mann G, Konig M, Steiner M, Dworzak MN, Gadner H, et al. Near-tetraploidy in childhood B-cell precursor acute lymphoblastic leukemia is a highly specific feature of ETV6/RUNX1-positive leukemic cases. Genes Chromosomes Cancer. 2006;45:608–611. doi: 10.1002/gcc.20324. [DOI] [PubMed] [Google Scholar]
- Boyapati A, Yan M, Peterson LF, Biggs JR, Le Beau MM, Zhang DE. A leukemia fusion protein attenuates the spindle checkpoint and promotes aneuploidy. Blood. 2007;109:3963–3971. doi: 10.1182/blood-2006-09-045583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakos C, Krapf G, Gerner C, Inthal A, Lemberger C, Ban J, et al. RNAi-mediated silencing of TEL/AML1 reveals a heat-shock protein- and survivin-dependent mechanism for survival. Blood. 2007;109:2607–2610. doi: 10.1182/blood-2006-04-019612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liu T, Wang X, Yang YM, Deng H, Kunicki J, et al. BubR1 is involved in regulation of DNA damage responses. Oncogene. 2006;25:3598–3605. doi: 10.1038/sj.onc.1209392. [DOI] [PubMed] [Google Scholar]
- Ford AM, Palmi C, Bueno C, Hong D, Cardus P, Knight D, et al. The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells. J Clin Invest. 2009;119:826–836. doi: 10.1172/JCI36428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. Leukemogenesis by CBF oncoproteins. Leukemia. 1999;13:1932–1942. doi: 10.1038/sj.leu.2401590. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha GH, Baek KH, Kim HS, Jeong SJ, Kim CM, McKeon F, et al. p53 activation in response to mitotic spindle damage requires signaling via BubR1-mediated phosphorylation. Cancer Res. 2007;67:7155–7164. doi: 10.1158/0008-5472.CAN-06-3392. [DOI] [PubMed] [Google Scholar]
- Hiebert SW, Sun W, Davis JN, Golub T, Shurtleff S, Buijs A, et al. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Li Z, Yan J, Matheny CJ, Corpora T, Bravo J, Warren AJ, et al. Energetic contribution of residues in the Runx1 Runt domain to DNA binding. J Biol Chem. 2003;278:33088–33096. doi: 10.1074/jbc.M303973200. [DOI] [PubMed] [Google Scholar]
- Lou J, Cao W, Bernardin F, Ayyanathan K, Rauscher IF, Friedman AD. Exogenous cdk4 overcomes reduced cdk4 RNA and inhibition of G1 progression in hematopoietic cells expressing a dominant-negative CBF—a model for overcoming inhibition of proliferation by CBF oncoproteins. Oncogene. 2000;19:2695–2703. doi: 10.1038/sj.onc.1203588. [DOI] [PubMed] [Google Scholar]
- Michel L, Benezra R, Diaz-Rodriguez E. MAD2 dependent mitotic checkpoint defects in tumorigenesis and tumor cell death: a double edged sword. Cell Cycle. 2004;3:990–992. [PubMed] [Google Scholar]
- Morrow M, Samanta A, Kioussis D, Brady HJ, Williams O. TEL-AML1 preleukemic activity requires the DNA binding domain of AML1 and the dimerization and corepressor binding domains of TEL. Oncogene. 2007;26:4404–4414. doi: 10.1038/sj.onc.1210227. [DOI] [PubMed] [Google Scholar]
- Perez de Castro I, de Carcer G, Malumbres M. A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- Peters JM. Cell biology: the checkpoint brake relieved. Nature. 2007;446:868–869. doi: 10.1038/446868a. [DOI] [PubMed] [Google Scholar]
- Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Raimondi SC, Zhou Y, Shurtleff SA, Rubnitz JE, Pui CH, Behm FG. Near-triploidy and near-tetraploidy in childhood acute lymphoblastic leukemia: association with B-lineage blast cells carrying the ETV6-RUNX1 fusion, T-lineage immunophenotype, and favorable outcome. Cancer Genet Cytogenet. 2006;169:50–57. doi: 10.1016/j.cancergencyto.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, et al. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Nakamura Y, Maki K, Waga K, Nakamura F, Arai H, et al. Functional analysis of a dominant-negative DeltaETS TEL/ETV6 isoform. Biochem Biophys Res Commun. 2004;317:1128–1137. doi: 10.1016/j.bbrc.2004.03.172. [DOI] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Strom DK, Nip J, Westendorf JJ, Linggi B, Lutterbach B, Downing JR, et al. Expression of the AML-1 oncogene shortens the G(1) phase of the cell cycle. J Biol Chem. 2000;275:3438–3445. doi: 10.1074/jbc.275.5.3438. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wolyniec K, Wotton S, Kilbey A, Jenkins A, Terry A, Peters G, et al. RUNX1 and its fusion oncoprotein derivative, RUNX1-ETO, induce senescence-like growth arrest independently of replicative stress. Oncogene. 2009;28:2502–2512. doi: 10.1038/onc.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton S, Terry A, Kilbey A, Jenkins A, Herzyk P, Cameron E, et al. Gene array analysis reveals a common Runx transcriptional programme controlling cell adhesion and survival. Oncogene. 2008;27:5856–5866. doi: 10.1038/onc.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.