Abstract
FK506 inhibits the Ca2+/calmodulin-dependent protein phosphatase calcineurin, which plays a critical role in yeast subjected to salt stress. A chemical genetic screen for small molecules that suppress growth inhibition by high NaCl plus FK506 identified a structurally related class of suppressors of FK506 (SFKs) named SFKs 2–4. To identify possible protein targets for these small molecules, a genome-wide screen of ≈4,700 haploid yeast deletion strains was undertaken for strains showing resistance to high NaCl plus FK506. This screen yielded a number of genes not previously implicated in salt stress, including ALD6, which encodes an NADP+-dependent aldehyde dehydrogenase, and UTR1, which encodes an NAD+ kinase. Transcriptional profiling of yeast treated with SFK2 indicated that the SFKs target the Ald6p pathway. In addition, screening of the deletion strains for hypersensitivity to SFK2 yielded ZWF1, encoding glucose-6-phosphate dehydrogenase, which has been shown to play an overlapping role with Ald6p in NADPH production. Furthermore, the SFKs inhibited the activity of Ald6p in vitro. Having established that the SFKs target Ald6p, they were used as tools to implicate systematically other gene products in the Ald6p pathway, including Utr1p, which may function by supplying Ald6p with its NADP+ cofactor. Furthermore, growth improvement by the SFKs on high NaCl plus FK506 was shown to require GPD1, which encodes an NADH-dependent glycerol-3-phosphate dehydrogenase that is important for the production of glycerol in response to osmotic stress.
Small molecules are useful tools for the exploration of biology, allowing instantaneous modulation of protein function across diverse biological systems. The small-molecule immunosuppressant FK506, which inhibits the Ca2+/calmodulin-dependent protein phosphatase calcineurin, has greatly facilitated the study of calcineurin biology in many different organisms, from yeast to humans (1). The discovery of new small-molecule modulators is hindered by a lack of means for rapid target identification. The model organism Saccharomyces cerevisiae offers a number of tools for target identification, including well annotated microarrays for transcriptional profiling, as well as a collection of almost every viable deletion strain. Recent work has illustrated the use of these tools in linking compounds with target pathways (2–8). Herein, we demonstrate the power of small-molecule screening in the yeast system by first identifying the target of a class of small-molecule suppressors of FK506 (SFKs) and then using the SFKs to identify other genes involved in the signaling network of the target.
In yeast, calcineurin plays a critical role during a number of stresses including Na+/Li+ stress (9). During Na+/Li+ stress, a rise in cytosolic Ca2+ activates calmodulin, which in turn stimulates calcineurin. Calcineurin induces, via the transcription factor Tcn1p/Crz1p, the transcription of ENA1, which encodes a P-type Na+-ATPase that pumps Na+ from the cell (10, 11). In addition, calcineurin converts the Trk1/2p K+ channel from a K+/Na+ transporter to a high-affinity K+ transporter, thereby reducing the amount of Na+ that enters the cell (10). Because calcineurin is required for the Na+/Li+ stress response, FK506 greatly inhibits yeast growth under salt-stress conditions.
High concentrations of Na+/Li+ subject yeast to both ion toxicity and osmotic stress. Whereas the calcineurin pathway is important for Na+/Li+ stress specifically, the high-osmolarity glycerol mitogen-activated protein kinase pathway is important for osmotic stresses in general (12). One of the primary responses of yeast to external osmotic stress is to increase internal levels of the osmoprotectant glycerol. Glycerol is produced from the glycolytic intermediate dihydroxyacetone phosphate by the enzymes glycerol-3-phosphate dehydrogenase and glycerol-3-phosphatase. Two isozymes of glycerol-3-phosphate dehydrogenase, encoded by GPD1 and GPD2, have been identified. Gpd1p is critical for osmoadaptation, whereas Gpd2p is critical for redox balance during anaerobic growth (13, 14). Under conditions of high osmolarity, the high-osmolarity glycerol mitogen-activated protein kinase pathway stimulates the expression of GPD1, thereby increasing glycerol production and protecting the cell from external osmotic stress (13).
In this study, we explore the biology of calcineurin in yeast by using a chemical genetic screen for small-molecule SFKs. To identify possible targets of the SFKs, we perform a parallel screen of the yeast deletion set. We then are able to implicate the aldehyde dehydrogenase Ald6p as the target of SFKs 2–4 by (i) transcriptionally profiling yeast treated with SFK2 and (ii) identifying haploid deletion strains that show hypersensitivity to SFK2. Having identified Ald6p as the target of the SFKs, we then use the SFKs to identify other genes in the Ald6p signaling network. We use SFK2 to knock down Ald6p function systematically in a number of NaCl/FK506-resistant deletion strains, effectively creating a series of double deletions, to determine which of the deleted genes are required for the activity of SFK2. From this we implicate Utr1p in the Ald6p pathway. In addition, we show that growth improvement by the SFKs on high NaCl/FK506 requires GPD1, which encodes an enzyme important for glycerol production.
Materials and Methods
Yeast Strains and Media. The yeast deletion strain sets were obtained from the Saccharomyces Genome Deletion Project (Research Genetics, Huntsville, AL). The identities of strains BY4742 Δald6, BY4741 Δzwf1, BY4742 Δutr1, and BY4742 Δgpd1 were confirmed by PCR. Rich medium [yeast extract/peptone/dextrose (YPD)] contained 1% yeast extract (Difco), 2% Bacto peptone (Difco), and 2% glucose. NaCl and LiCl were added as indicated. Agar (2%) was added for plates.
Screening of the Yeast Deletion Set. Approximately 4,700 haploid deletion strains in the BY4742 background were screened for resistance to NaCl plus FK506. Strains were pinned from 96-well stock plates by using a 96-pin tool into 96-well plates containing YPD plus 0.8 M NaCl and 125 nM FK506. The plates were incubated at room temperature for 3 days. The OD600 of each well was read by using a Spectramax Plus 384 plate reader (Molecular Devices). Those strains with an OD600 that was >3 SDs above the mean OD600 of all strains were collected in a 96-well plate for additional retesting. Retesting is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Approximately 4,700 haploid deletion strains in the BY4741 background were screened for hypersensitivity to SFK2, as described in Supporting Materials and Methods.
Transcriptional Profiling. Yeast were grown in YPD to midlog phase, diluted to OD600 0.125, treated with either DMSO or 10 μM SFK2, and grown to an OD600 of 1.2 at 30°C. For NaCl/FK506 treatment profiles, yeast were grown in YPD to an OD600 of 1 and treated with H2O or 0.7 M NaCl by using a 5 M NaCl stock, plus 125 nM FK506, plus either DMSO or 10 μM SFK2, for 30 min at 30°C. mRNA isolation, reverse transcription, labeling, hybridization, and scanning have been described (5).
ALD6 Overexpression, Recombinant Ald6p Purification, and Activity Assay. Plasmids for ALD6 overexpression and expression of GST-Ald6p, as well as the purification strategy for the GST-Ald6p and the Ald6p activity assay, are described in Supporting Materials and Methods.
Results
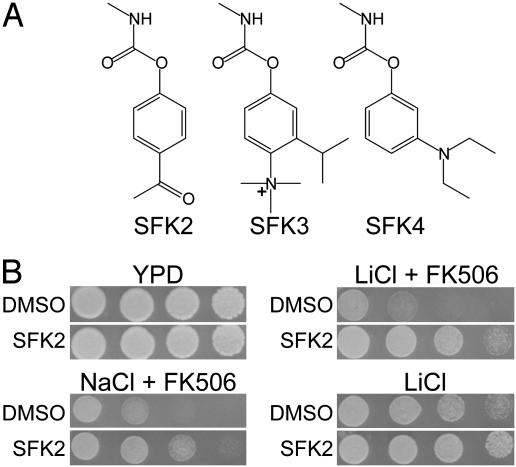
Identification of a Structurally Related Class of SFKs. A chemical genetic screen was performed previously to identify small molecules that improve the growth of FK506-treated yeast in high-salt media (5). In the current study we focus on a structurally related class of SFKs (SFKs 2–4). SFKs 2–4 improve growth maximally on high NaCl/FK506 when used at a concentration of 5–10 μM. As shown in Fig. 1A, SFKs 2–4 all contain a carbamate moiety. The carbamate is necessary for activity, because a version of SFK2 without the carbamate, 4-hydroxyacetophenone, is unable to improve growth on high salt (data not shown).
Fig. 1.
Chemical structures and characterization of the SFKs. (A) The chemical structures of SFK2, SFK3, and SFK4. (B) Wild-type yeast were serially diluted in 5-fold increments and spotted on YPD (rich medium) or YPD with 0.8 M NaCl plus 125 nM FK506, 0.15 M LiCl plus 125 nM FK506, or 0.2 M LiCl.
SFK2 not only improves growth on high NaCl plus FK506 but also on high LiCl plus FK506 (Fig. 1B). SFK2 does not improve growth on high KCl, a nontoxic salt, or high sorbitol, an osmotic stress (data not shown). SFK2 improves growth on high NaCl (data not shown) and high LiCl (Fig. 1B) even in the absence of FK506.
A Genome-Wide Screen for Deletion Strains with Improved Growth on High NaCl Plus FK506. To identify possible targets of the SFKs, we screened the set of haploid deletion strains from the Saccharomyces Genome Deletion Project for resistance to 0.8 M NaCl plus 125 nM FK506 (similar conditions were used in our small-molecule screen). Presumably, if the SFKs were improving growth by causing a loss of function in a protein encoded by a nonessential gene, then such a screen should identify the majority of possible protein targets for the SFKs. Large-scale growth experiments have been performed previously in which pooled yeast strains were subjected to a variety of stresses including NaCl and their growth was monitored by using DNA barcodes and Affymetrix chips (15). However, although this pooled method accurately identified strains that were hypersensitive to growth conditions, it did not accurately identify strains that were resistant to growth conditions. Strains that show resistance to NaCl/FK506 are listed in Table 1, with functional annotations (16). Of the 29 strains identified, only 7 had been shown previously to play some role in NaCl/FK506 resistance and/or osmotic stress resistance (see Table 1 legend).
Table 1. Haploid deletion strains showing resistance to NaCl/FK506 that represent potential targets for the SFKs.
| Gene | Name | Score | Function |
|---|---|---|---|
| YDR300C | PRO1 | 6+ | Glutamate 5-kinase, catalyzes first step in proline biosynthesis |
| YLR362W* | STE11 | 5+ | Mitogen-activated protein kinase kinase kinase, component of high-osmolarity response pathway |
| YMR216C* | SKY1 | 5+ | Ser/Thr protein kinase that regulates polyamine transport and ion homeostasis |
| YGL025C | PGD1 | 5+ | Component of RNA polymerase II holoenzyme and mediator subcomplex |
| YDR392W | SPT3 | 5+ | Component of the SAGA histone acetyltransferase complex |
| YGL066W | SGF73 | 5+ | Component of the SAGA histone acetyltransferase complex |
| YGL024W | 4+ | Unknown function, coding region overlaps with PGD1 | |
| YNL229C* | URE2 | 4+ | Regulator of nitrogen utilization and salinity response, Gln3p cytoplasmic anchor |
| YDL005C | MED2 | 4+ | Component of RNA polymerase II holoenzyme and mediator subcomplex |
| YPL062W | LPE8 | 4+ | Unknown function, coding region overlaps with promoter of ALD6 |
| YPL061W | ALD6 | 4+ | Cytosolic acetaldehyde dehydrogenase, functions in NADPH regeneration |
| YOL050C | 4+ | Unknown function | |
| YLR055C | SPT8 | 4+ | Component of the SAGA histone acetyltransferase complex |
| YNL135C* | FPR1 | 3+ | FK506-binding protein, homolog of human FKBP12 |
| YPL161C | BEM4 | 3+ | Interacts with Rho-type GTPases |
| YML016C* | PPZ1 | 3+ | Ser/Thr phosphatase involved in ion homeostasis |
| YGL007W | 3+ | Unknown function, coding region lies upstream of PMA1 | |
| YPL137C | 3+ | Unknown function, coding region overlaps with YPL136W | |
| YJL159W | HSP150 | 3+ | Secreted O-glycosylated protein required for tolerance to heat shock |
| YDL025C | 3+ | Ser/Thr protein kinase related to the NPR1 subfamily | |
| YJR059W* | PTK2 | 2+ | Ser/Thr kinase that activates of Pma 1p, the plasma membrane proton pump |
| YJR049C | UTR1 | 2+ | NAD+ kinase |
| YIL038C | NOT3 | 2+ | Component of the CCR4-Not complex, regulator of transcription |
| YKL032C | IXR1 | 2+ | Transcription factor regulating the transcription of COX5B |
| YPL136W | 2+ | Unknown function, interacts with Pma2p in a high-throughput two-hybrid assay | |
| YDR146C | SWI5 | 2+ | Transcription factor that regulates the transcription of HO |
| YEL009C* | GCN4 | 2+ | Transcription factor that regulates the response to amino acid starvation |
| YML048W | GSF2 | 2+ | Unknown function, involved in glucose repression |
| YER007C-A | 1+ | Unknown function, homologous to human oncogene MCTS1 |
NaCl/FK506-resistant strains were initially identified in 96-well plates in liquid culture. For retesting on agar plates, NaCl/FK506-resistant strains were serially diluted in 10-fold increments and spotted on YPD or YPD with 0.8 M NaCl plus 125 nM FK506 (see Supporting Materials and Methods). Strains with an asterisk (*) have been shown previously to play a role in NaCl/FK506 resistance and/or osmotic stress resistance.
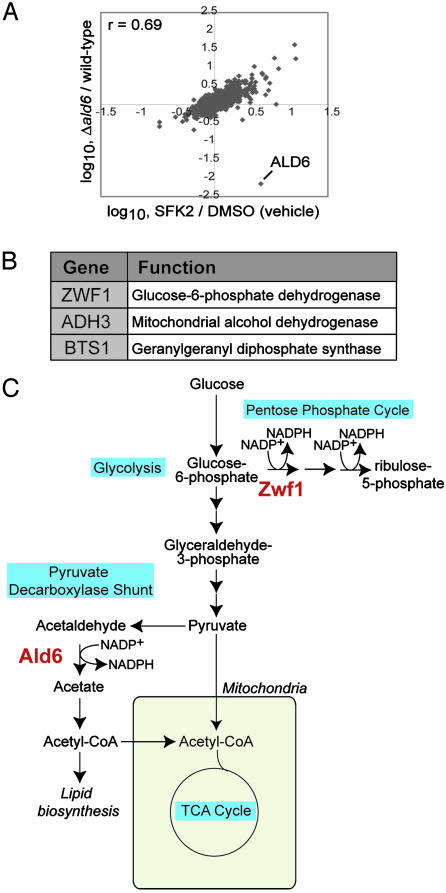
Transcriptional Profiling of Yeast Treated with SFK2. To characterize the SFKs further, we transcriptionally profiled wild-type yeast after SFK2 treatment. SFK2 induces the up-regulation of a number of aldehyde dehydrogenase genes including ALD4 (+4.7), ALD6 (+4.1), and ALD5 (+2.7). Given that the Δald6 strain appeared in the list of NaCl/FK506-resistant strains (Table 1), we hypothesized that the SFKs might target the Ald6p pathway. Ald6p is a component of the pyruvate decarboxylase shunt, which plays a critical role in providing acetyl-CoA to the cytosolic compartment, as well as in helping to maintain redox balance (see Fig. 2C) (17). Comparison of the profile of wild-type yeast treated with SFK2 to that of the Δald6 deletion mutant revealed that the two profiles are highly similar, further suggesting that the SFKs might target the Ald6p pathway (Fig. 2 A). The overall correlation coefficient (r) between the two profiles is 0.69.
Fig. 2.
Transcriptional profiling and hypersensitivity screening indicate that the SFKs target the Ald6p pathway. (A) Correlation plot comparing the transcriptional profile of wild-type yeast treated with 10 μM SFK2 to that of the Δald6 strain. (B) List of gene deletion strains that showed hypersensitivity to 10 μM SFK2, starting with the most hypersensitive (ZWF1). (C) Scheme showing the interrelationship between the pentose phosphate cycle and the pyruvate decarboxylase shunt. Both Zwf1p of the pentose phosphate cycle and Ald6p of the pyruvate decarboxylase shunt play important roles in the production of NADPH.
To identify transcriptional changes that are associated with SFK2 treatment under conditions of high NaCl/FK506, we compared the transcriptional profile of yeast treated with NaCl/FK506 to that of yeast treated with NaCl/FK506 plus SFK2 (see Fig. 6, which is published as supporting information on the PNAS web site). Under the conditions of NaCl/FK506, SFK2 treatment results in the up-regulation of ACS1, encoding an acetyl-CoA synthetase, which converts the acetate generated by aldehyde dehydrogenase to acetyl-CoA (18). In addition, SFK2 treatment results in the up-regulation of a number of genes involved in the β-oxidation of fatty acids to acetyl-CoA, including POT1, POX1, FAA2, FOX2, ECI1, and DCI1 (19). The transcriptional changes induced under NaCl/FK506 stress by SFK2 treatment thus are consistent with an up-regulation of functions downstream of aldehyde dehydrogenase, such as acetyl-CoA production. Interestingly, ALD6 itself is not up-regulated by SFK2 under conditions of NaCl/FK506.
A Genome-Wide Screen for Deletion Strains That Are Hypersensitive to SFK2. To further characterize the SFKs, the set of haploid deletion strains from the Saccharomyces Genome Deletion Project were screened for hypersensitivity to SFK2 treatment. If a deletion strain is hypersensitive to compound treatment, it suggests that the deletion is in a pathway that shares overlapping roles with that of the target of the compound. The deletion strains showing the most hypersensitivity to SFK2 are listed in Fig. 2B.
The strain deleted for ZWF1, which encodes the first enzyme in the pentose phosphate cycle, was the most sensitive to treatment with SFK2 (Fig. 2B), suggesting that SFK2 targets a pathway with functions that overlap with those of the pentose phosphate cycle. One of the central functions of the pentose phosphate cycle is the production of NADPH (20) (see Fig. 2C). Ald6p uses almost exclusively NADP+ (rather than NAD+) as a cofactor (21). Furthermore, the Δald6 and Δzwf1 deletions have been shown to be synthetically lethal (22). Presumably, this synthetic lethality is caused by the overlapping functions of the two genes in NADPH generation. The hypersensitivity of the Δzwf1 strain to SFK2 thus further supports the idea that the SFKs inhibit the Ald6p pathway.
Two other deletion strains, Δadh3 and Δbts1, were shown to be sensitive to SFK2 treatment (Fig. 2B). Similar to Ald6p, the mitochondrial alcohol dehydrogenase Adh3p is involved in carbon metabolism and redox balance. Under anaerobic conditions, Adh3p is thought to participate in an ethanol-acetaldehyde shuttle, which shuttles NADH out of the mitochondria for oxidation by glycerol-3-phosphate dehydrogenase, generating glycerol (23). Intriguingly, Bts1p, a geranylgeranyl diphosphate synthase (24), is linked genetically to the calcineurin pathway. The Δbts1 strain was shown recently to be synthetically lethal with the calcineurin deletion strain and hypersensitive to either FK506 or cyclosporin A (3).
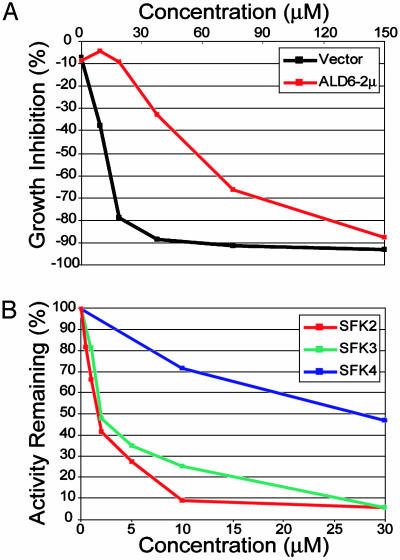
Verification of Ald6p as a Target of the SFKs. To verify that Ald6p is a relevant protein target of the SFKs, we determined whether ALD6 overexpression could suppress the effects of the SFKs on yeast growth. At high concentrations, SFKs 2–4 inhibit growth in YPD, and this growth inhibition can be suppressed by overexpression of ALD6 (Fig. 3A and data not shown). In addition, SFKs 2–4 improve growth on high NaCl/FK506, and ALD6 overexpression can suppress this growth improvement. ALD6 overexpression shifts the EC50 of each of the SFKs for growth improvement on NaCl/FK506 by ≈2-fold (data not shown). The SFKs also are able to inhibit the activity of recombinant Ald6p in an in vitro activity assay, although to different degrees (Fig. 3B).
Fig. 3.
The SFKs target Ald6p. (A) Overexpression of ALD6 suppresses growth inhibition by SFK2. Percent growth inhibition represents growth of either control or ALD6-overexpressing yeast in the presence of different concentrations of SFK2 as a percentage of growth in the presence of vehicle (DMSO). (B) SFKs 2–4 inhibit Ald6p enzymatic activity in vitro.
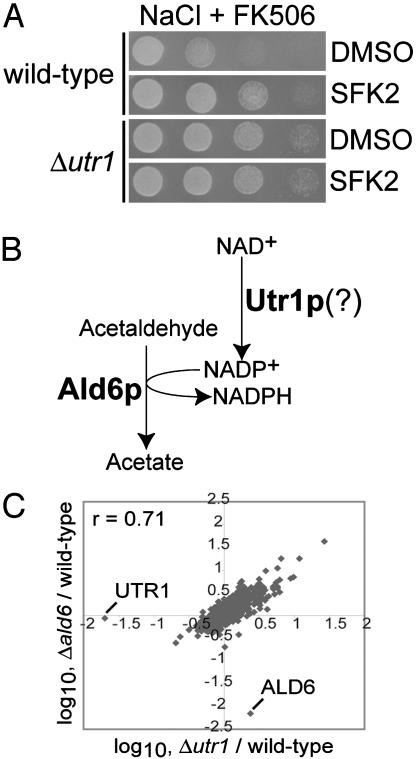
Ald6p and Utr1p May Act in the Same Biochemical Pathway. Having established that the SFKs target Ald6p, we next used the SFKs as tools to implicate other gene products in the Ald6p pathway. In yeast genetics, the analysis of the phenotype of a double deletion of two genes is often illuminating. By using the SFKs to inhibit Ald6p activity, in a sense creating the Δald6 mutation, in any deletion strain of our choosing, we were able to make “double deletions” in a highly efficient manner. We reasoned that if the SFKs have no activity (e.g., cannot improve growth on high NaCl/FK506) in a particular deletion strain, then the gene deleted in that strain might be linked to the Ald6p pathway. In several of the NaCl/FK506-resistant strains (from Table 1), SFK2 was not able to improve growth on NaCl/FK506 (data not shown). These strains included Δutr1 (see Fig. 4A). Utr1p has been identified as an NAD+ kinase based on its sequence similarity to other NAD+ kinases and its biochemical activity in vitro (25). Because Ald6p utilizes NADP+, we hypothesized that Utr1p might participate in the Ald6p pathway, possibly by supplying Ald6p with its cofactor (Fig. 4B).
Fig. 4.
SFK2 implicates the NAD+ kinase Utr1p in the Ald6p pathway. (A) SFK2 is not able to improve growth on NaCl/FK506 in the Δutr1 strain. Either wild-type or Δutr1 yeast cells were serially diluted 5-fold and spotted on 0.8 M NaCl plus 125 nM FK506 containing either vehicle (DMSO) or 10 μM SFK2. (B) Model for how the NAD+ kinase Utr1p might supply the NADP+-dependent Ald6p with cofactor. (C) Correlation plot comparing the transcriptional profile of the Δutr1 strain with that of the Δald6 strain.
The transcriptional profile of the Δald6 deletion strain and that of the Δutr1 deletion strain are highly similar, further suggesting that the two genes might participate in the same pathway. The overall correlation coefficient between the two profiles is 0.71 (Fig. 4C). In addition, the transcriptional profile of wild-type yeast treated with SFK2 and that of the Δutr1 deletion strain are highly similar with a correlation coefficient of 0.68.
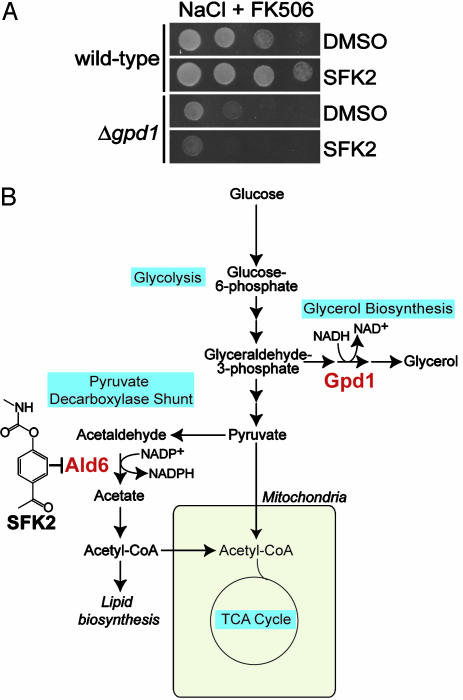
Growth Improvement by the SFKs on High NaCl Plus FK506 Requires GPD1. From metabolite analysis of fermenting yeast, the Δald6 deletion mutation has been shown to result in an increased intracellular glycerol content (17, 26). Because increased glycerol production is an important response to osmotic stress, we wanted to test whether the increased glycerol content of the Δald6 deletion mutant could explain the improved growth of the mutant on high NaCl/FK506. Because Gpd1p is critical for glycerol production during osmotic stress, we tested whether the SFKs could still improve growth on high NaCl/FK506 in the Δgpd1 background. As shown in Fig. 5A, SFK2 can no longer improve growth on high NaCl/FK506 in the Δgpd1 strain, suggesting that the SFKs improve growth via Gpd1p.
Fig. 5.
Growth improvement by SFK2 on NaCl/FK506 requires GPD1.(A) SFK2 is not able to improve growth on NaCl/FK506 in the Δgpd1 strain. Either wild-type or Δgpd1 yeast cells were serially diluted 5-fold and spotted on 0.7 M NaCl plus 125 nM FK506 containing either vehicle (DMSO) or 10 μM SFK2. (B) Scheme showing the interrelationship between the glycerol biosynthetic pathway and the pyruvate decarboxylase shunt.
Discussion
In this study, we identified the protein target of a class of small molecules (SFKs 2–4) discovered through a forward chemical genetic screen for compounds that improve growth on high NaCl/FK506. To identify possible protein targets of the compounds from our chemical genetic screen, we performed a parallel genetic screen of the yeast deletion strains from the Saccharomyces Genome Deletion Project for strains showing improved growth on high NaCl/FK506. Assuming that our small molecules cause a loss of function in a nonessential gene product, this parallel genetic screen should have identified many of the possible protein targets for our small molecules. To link SFKs 2–4 to one of the possible targets, we further characterized the compounds by transcriptionally profiling yeast cells treated with SFK2 and identifying deletions strains that showed hypersensitivity to SFK2. The transcriptional profile of yeast cells treated with SFK2 was highly similar to the transcriptional profile of the Δald6 strain. Furthermore, the screen for SFK2-hypersensitive deletion strains identified Δzwf1, which had been shown previously to be synthetically lethal with Δald6. Taken together, these data suggested that the SFKs targeted the Ald6p pathway.
To verify that Ald6p was the target of the SFKs, we showed that overexpression of ALD6 suppresses growth inhibition by the SFKs in YPD and that overexpression of ALD6 supresses growth improvement by the SFKs on high NaCl/FK506. In addition, SFKs 2–4 were able to inhibit Ald6p in an in vitro activity assay. Although we have provided significant evidence that the SFKs target Ald6p, it is possible that the SFKs target other aldehyde dehydrogenases in addition to Ald6p. In fact, we have been able to show that SFK2 also inhibits the in vitro activity of Ald4p, the aldehyde dehydrogenase most closely related to Ald6p that uses both NAD+ and NADP+ (data not shown). The fact that the SFKs also may target other aldehyde dehydrogenases should be considered, for example, when interpreting the results of the genome-wide screen for SFK2-hypersensitive strains.
Having verified that the SFKs target Ald6p, we next used the SFKs as tools to implicate other gene products in the Ald6p signaling network. With the SFKs, we were able to inhibit Ald6p activity, in a sense creating the Δald6 mutation, in any deletion strain of our choosing. In this way, we could make the double deletion of Δald6 and any other deletion in a highly efficient manner. If the SFKs have no activity (e.g., cannot improve growth on high NaCl/FK506) in a particular deletion strain, then it suggests that the gene deleted in that strain might be linked to the Ald6p pathway. Through systematic screening of the NaCl/FK506-resistant strains (Table 1) for strains in which SFK2 had no activity, we were able to implicate the NAD+ kinase Utr1p in the Ald6p pathway. Although Utr1p had been shown previously to be an NAD+ kinase through sequence homology and biochemical analysis, our results implicate Utr1p in the Ald6p pathway. We link Utr1p to the Ald6p pathway by showing that (i) both ALD6 and UTR1 are involved in salt stress (see Table 1), (ii) SFK2 does not improve growth on high NaCl/FK506 in the Δutr1 strain, and (iii) deletion of either ALD6 or UTR1 results in a similar transcriptional response.
Utr1p may function by supplying Ald6p with its NADP+ cofactor. NAD+ kinases have been shown to facilitate NADP+-dependent processes. In yeast, Utr1p is thought to contribute to the NADPH-dependent ferrireductase system by supplying it with NADP+ (25, 27). The ferrireductase system reduces extracellular ferric chelates to their ferrous counterparts for uptake by the cell. During fertilization in sea urchin eggs, an NAD+ kinase stimulates an NADP+-dependent enzyme, glucose-6-phosphate dehydrogenase. Fertilization causes an influx in Ca2+, activating calmodulin, which in turn binds to and activates the NAD+ kinase (28). Once activated, the NAD+ kinase provides NADP+ to glucose-6-phosphate dehydrogenase, thereby increasing the production of NADPH via the pentose phosphate cycle (29). As in the fertilization of sea urchin eggs, salt stress in yeast is accompanied by a Ca2+ influx that activates calmodulin. Activated calmodulin could potentially stimulate Utr1p, which in turn may generate NADP+, for use by Ald6p.
By inhibiting Ald6p, the SFKs likely improve growth on high NaCl/FK506 via Gpd1p, an enzyme that is critical for survival during osmotic stress. This hypothesis is supported by our finding that SFK2 is no longer able to improve growth on high NaCl/FK506 in the Δgpd1 strain (Fig. 5A). As a defense against osmotic stress, yeast cells increase production of glycerol, thereby elevating the osmolarity of the cytosol. The Δald6 strain grown under fermenting conditions is known to have a somewhat elevated intracellular glycerol concentration over wild-type yeast (17, 26). In addition, we have been able to show that the Δald6 strain grown to log phase under aerobic conditions also has an elevated glycerol concentration (data not shown). Glycerol production via Gpd1p requires NADH (see Fig. 5B). It has been postulated that the deletion of the gene encoding the NADP+-dependent Ald6p results in an increased glycerol concentration as a result of the activity of other NAD+-dependent aldehyde dehydrogenases (Ald2p and Ald3p) or NAD+/NADP+-dependent aldehyde dehydrogenases (Ald4p) (17). It is unclear, however, whether the elevated glycerol concentration of the Δald6 strain is sufficient to explain its improved growth on high NaCl/FK506. We were unable to show that the SFKs elevate glycerol concentration (data not shown). However, given that the increase in glycerol concentration seen in the Δald6 strain is slight and given that the growth improvement on high NaCl/FK506 by the SFKs is weaker than that by deletion of ALD6, we may not be able to detect an increase beyond the statistical error of our methods. Although the SFKs likely improve growth on high NaCl/FK506 through Gpd1p, they may improve growth on LiCl/FK506 through additional mechanisms. Indeed, SFK2 treatment is able to improve the growth of a Δgpd1 strain on high LiCl/FK506 (data not shown).
In this study, as well as in earlier work (5), we identified small molecules that suppress the effects of FK506 in yeast. To identify potential targets for SFKs 2–4, we screened the yeast deletion set. We then were able to link the SFKs to a specific target pathway (the Ald6p pathway) by transcriptionally profiling yeast treated with SFK2 and identifying the chemical–genetic interactions of SFK2 with the yeast deletion set. In this study and in earlier work (5), we thus have established a framework for target identification that should facilitate target identification for other small-molecule SFKs. Furthermore, by using the SFKs to implicate other gene products in the Ald6p pathway, we have demonstrated the utility of small molecules in network dissection. In theory, the SFKs could be used to screen the entire genome for other gene products in the Ald6p target pathway. In addition, if the SFKs are active in other, higher organisms, they could be used to probe the calcineurin pathway in other systems.
Supplementary Material
Acknowledgments
We kindly thank Harvard University's ICCB for use of small-molecule libraries and facilities for screening; Paul Clemons for providing the FK506; Daniel Tessier for providing the pVT100-U plasmid; Paul Doetsch for providing the pYEX4T-1 plasmid; and Alykhan Shamji for a careful reading of this manuscript. R.A.B. was supported by a fellowship from the National Science Foundation. S.L.S. is an Investigator with the Howard Hughes Medical Institute and a Donald W. Reynolds Foundation Cardiology Scholar. This research was supported by National Institute of General Medical Sciences Grant GM38627 and the Donald W. Reynolds Foundation, Cardiovascular Clinical Research Center.
Abbreviations: SFK, suppressor of FK506; YPD, yeast extract/peptone/dextrose.
References
- 1.Liu, J., Farmer, J. D., Jr., Lane, W. S., Friedman, J., Weissman, I. & Schreiber, S. L. (1991) Cell 66, 807–815. [DOI] [PubMed] [Google Scholar]
- 2.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 3.Parsons, A. B., Brost, R. L., Ding, H., Li, Z., Zhang, C., Sheikh, B., Brown, G. W., Kane, P. M., Hughes, T. R. & Boone, C. (2004) Nat. Biotechnol. 22, 62–69. [DOI] [PubMed] [Google Scholar]
- 4.Chan, T. F., Carvalho, J., Riles, L. & Zheng, X. F. (2000) Proc. Natl. Acad. Sci. USA 97, 13227–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher, R. A. & Schreiber, S. L. (2003) Chem. Biol. 10, 521–531. [DOI] [PubMed] [Google Scholar]
- 6.Gupta, S. S., Ton, V. K., Beaudry, V., Rulli, S., Cunningham, K. & Rao, R. (2003) J. Biol. Chem. 278, 28831–28839. [DOI] [PubMed] [Google Scholar]
- 7.Lum, P. Y., Armour, C. D., Stepaniants, S. B., Cavet, G., Wolf, M. K., Butler, J. S., Hinshaw, J. C., Garnier, P., Prestwich, G. D., Leonardson, A., et al. (2004) Cell 116, 121–137. [DOI] [PubMed] [Google Scholar]
- 8.Giaever, G., Flaherty, P., Kumm, J., Proctor, M., Nislow, C., Jaramillo, D. F., Chu, A. M., Jordan, M. I., Arkin, A. P. & Davis, R. W. (2004) Proc. Natl. Acad. Sci. USA 101, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura, T., Liu, Y., Hirata, D., Namba, H., Harada, S., Hirokawa, T. & Miyakawa, T. (1993) EMBO J. 12, 4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendoza, I., Rubio, F., Rodriguez-Navarro, A. & Pardo, J. M. (1994) J. Biol. Chem. 269, 8792–8796. [PubMed] [Google Scholar]
- 11.Matheos, D. P., Kingsbury, T. J., Ahsan, U. S. & Cunningham, K. W. (1997) Genes Dev. 11, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Rourke, S. M., Herskowitz, I. & O'Shea, E. K. (2002) Trends Genet. 18, 405–412. [DOI] [PubMed] [Google Scholar]
- 13.Albertyn, J., Hohmann, S., Thevelein, J. M. & Prior, B. A. (1994) Mol. Cell. Biol. 14, 4135–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansell, R., Granath, K., Hohmann, S., Thevelein, J. M. & Adler, L. (1997) EMBO J. 16, 2179–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 16.Csank, C., Costanzo, M. C., Hirschman, J., Hodges, P., Kranz, J. E., Mangan, M., O'Neill, K., Robertson, L. S., Skrzypek, M. S., Brooks, J., et al. (2002) Methods Enzymol. 350, 347–373. [DOI] [PubMed] [Google Scholar]
- 17.Remize, F., Andrieu, E. & Dequin, S. (2000) Appl. Environ. Microbiol. 66, 3151–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Berg, M. A., de Jong-Gubbels, P., Kortland, C. J., van Dijken, J. P., Pronk, J. T. & Steensma, H. Y. (1996) J. Biol. Chem. 271, 28953–28959. [DOI] [PubMed] [Google Scholar]
- 19.van Roermund, C. W., Waterham, H. R., Ijlst, L. & Wanders, R. J. (2003) Cell. Mol. Life Sci. 60, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voet, D. & Voet J. G. (1995) Biochemistry (Wiley, New York).
- 21.Wang, X., Mann, C. J., Bai, Y., Ni, L. & Weiner, H. (1998) J. Bacteriol. 180, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabowska, D. & Chelstowska, A. (2003) J. Biol. Chem. 278, 13984–13988. [DOI] [PubMed] [Google Scholar]
- 23.Bakker, B. M., Bro, C., Kotter, P., Luttik, M. A., van Dijken, J. P. & Pronk, J. T. (2000) J. Bacteriol. 182, 4730–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, Y., Proteau, P., Poulter, D. & Ferro-Novick, S. (1995) J. Biol. Chem. 270, 21793–21799. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, S., Suzuki, S., Mori, S. & Murata, K. (2001) FEMS Microbiol. Lett. 200, 181–184. [DOI] [PubMed] [Google Scholar]
- 26.Eglinton, J. M., Heinrich, A. J., Pollnitz, A. P., Langridge, P., Henschke, P. A. & de Barros Lopes, M. (2002) Yeast 19, 295–301. [DOI] [PubMed] [Google Scholar]
- 27.Lesuisse, E., Casteras-Simon, M. & Labbe, P. (1996) J. Biol. Chem. 271, 13578–13583. [DOI] [PubMed] [Google Scholar]
- 28.Epel, D., Patton, C., Wallace, R. W. & Cheung, W. Y. (1981) Cell 23, 543–549. [DOI] [PubMed] [Google Scholar]
- 29.Schomer, B. & Epel, D. (1998) Dev. Biol. 203, 1–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.