Abstract
In its prion form, Ure2p, a regulator of nitrogen catabolism in Saccharomyces cerevisiae, polymerizes into filaments whereby its C-terminal regulatory domain is inactivated but retains its native fold. The filament has an amyloid fibril backbone formed by the Asn-rich, N-terminal, “prion” domain. The prion domain is also capable of forming fibrils when alone or when fused to other proteins. We have developed a model for the fibril that we call a parallel superpleated β-structure. In this model, the prion domain is divided into nine seven-residue segments, each with a four-residue strand and a three-residue turn, that zig-zag in a planar serpentine arrangement. Serpentines are stacked axially, in register, generating an array of parallel β-sheets, with a small and potentially variable left-hand twist. The interior of the filament is mostly stabilized not by packing of apolar side chains but by H-bond networks generated by the stacking of Asn side chains: charged residues are excluded. The model is consistent with current biophysical, biochemical, and structural data (notably, mass-per-unit-length measurements by scanning transmission electron microscopy that gave one subunit rise per 0.47 nm) and is readily adaptable to other amyloids, for instance the core of Sup35p filaments and glutamine expansions in huntingtin.
Various normally innocuous and soluble proteins polymerize to form insoluble amyloid fibrils in several serious neuro-degenerative diseases, including Alzheimer's disease, Huntington's disease, and the transmissible prion diseases (1–4). Although amyloidogenic precursor proteins vary with respect to amino acid sequence and native fold, the resulting amyloid fibrils share similar generic properties: they are typically straight, rigid, and between 4 and 13 nm in diameter; and they are thermostable, protease-resistant, and rich in β-structure (5–8). Furthermore, the fibrils bind and align the dye Congo red, producing apple-green birefringence (9). The incidence of amyloid fibrils in important human diseases has attracted much effort toward investigating their structures and pathways of fibrillogenesis. Recently, the scope of these studies has broadened with the discovery that many proteins that are not normally amyloidogenic may be induced to form amyloid fibrils in vitro (2, 10).
Despite considerable progress, the exact nature of amyloid fibril structure and even the extent to which it is uniquely defined remain unclear. This dilemma may be attributed to the fact that methods of high resolution structure determination (x-ray crystallography and NMR spectroscopy) cannot be used because of the polymeric character and insolubility of the fibrils. Accordingly, x-ray fiber diffraction, electron microscopy (EM), optical spectroscopy, and other biophysical approaches have been the principal sources of data underlying current models. In particular, x-ray fiber diffraction data showed that some amyloid fibrils have “cross-β” structures with their β-strands arranged perpendicular to the fibril axis (7, 8). Several cross-β models have been proposed (7, 11–14). Initially, models with antiparallel β-sheets were favored. However, solid-state NMR measurements revealed that fibrils of β-amyloid peptide (residues 10–35) contain β-strands in a parallel, in-register arrangement (15), and this conformation was subsequently detected in other studies (16–19). Several models containing parallel β-sheets have been formulated (20–23). However, to date, no conclusive model has emerged.
Ure2p is a prion-forming protein of the yeast S. cerevisiae (24, 25). Ure2p is a cytoplasmic homodimeric protein (2 × 40 kDa) whose C-terminal domain [residues 91–354, a globular protein with a GST-like fold (26–28)] interacts with the GATA transcription factor Gln3p, preventing its entry into the nucleus (29, 30). In the prion state, Ure2p aggregates into filaments (25, 31, 32), and its effect on Gln3p is suppressed (24). Upon entering the filamentous state, Ure2p is reported to monomerize (33). Recently, EM, biophysical, and biochemical studies have begun to clarify the relationship between fibrillogenesis and inactivation of Ure2p (32, 34–36). The prion domain, residues 1–90, is responsible for filament formation (25, 34). In soluble Ure2p, this domain is unstable, protease-sensitive, and probably unfolded (37, 38). However, it readily forms fibrils that are stable, rigid, and protease-resistant (34). Very similar 4-nm fibrils are formed when the prion domain is alone, in Ure2p, or in fusion proteins. Ure2p1-65 fibrils contain >60% β-structure, according to Raman spectroscopy (34). In Ure2p filaments, a fibrillar core is surrounded by globular moieties, giving a diameter of ≈22 nm (34, 36). Proteolytic digestion experiments indicate that residues ≈71–90 are not in the core but link it to the C-terminal appendages (36). Scanning transmission electron microscopy mass measurements showed that filaments have one subunit per 0.45 ± 0.03 nm, regardless of which appendage is attached to the prion domain (36). These observations underlie the “amyloid core fibril” model of Ure2p filaments (32, 36). In prionogenesis, the Ure2p C-terminal domain retains its fold but is inactivated by steric blocking or monomerization.
Here we present a detailed model of the core fibril. The model incorporates structural principles manifested in the parallel β-helix class of proteins (39) but invokes an entirely different topology. Preliminary analysis also suggests that this fold and packing may be adapted to account for amyloid formation by many other proteins.
Materials and Methods
Molecular Modeling. The initial model for Ure2p10-39 was built as described in Results, using the biopolymer module of insight ii (Accelrys, San Diego) (40). The resulting structure was subjected to 300 steps of minimization based on the steepest descent algorithm with the H-bond distance constraints set at K = 50, to improve the geometry of H-bonds. The next 500 steps of refinement were performed by using the conjugate gradients algorithm. To allay concern that the H-bond constraints generated significant tensions in the minimized structure, the last calculation was performed without any restrictions. The discover module of insight ii, the consistent valence force field, and the distance-dependent dielectric constant were used. These calculations produced a compact structure without steric tension and with all donors and acceptors of H-bonds interacting with each other or having the possibility to bind water molecules (Fig. 6, which is published as supporting information on the PNAS web site). In accordance with procheck output (41), the overall average G-factor of the model is –0.50, a value typical for a good quality model.
Electron Microscopy. In shadowing experiments, the specimens were freeze-dried, and tantalum/tungsten was deposited from an elevation angle of 45° in a Baltec (Technotrade, Manchester, NH) BAF060 freeze–fracture machine. Negative staining and cryo-electron microscopy were performed as described (42). Micrographs were recorded on CM120 or CM200-FEG electron microscopes (FEI, Eindhoven, The Netherlands).
Results and Discussion
Formulation of the Model. The length of the Ure2p1-65 polypeptide suggests that, to fit into a 4-nm fibril in a cross-β conformation, it should be folded several times. Accordingly, we began to think in terms of serpentine arrangements. The next questions to address were as follows. (i) How can the highly hydrophilic prion domain fold into a stable fixed structure? (ii) Where are the turns of the β-strands located and how many of them are there? (iii) How do the “serpentine” folds pack in the fibril?
Usually, stable protein structures have a hydrophobic core formed by apolar side chains. About 25% apolar residue content may be suggested as a threshold below which such a structure would not form (43, 44). The Asn-rich prion domain with its ≈20% of apolar residues that is unfolded in native soluble Ure2p (37, 38) is below this threshold. However, the same peptide can form a stable fibrillar polymer: accordingly, the fibril core should be partially occupied by Asn and other polar residues. Among known structures, the only exceptions to the “minimal hydrophobic content” rule are some β-helical proteins (44–46). These proteins have repetitive three-dimensional structures (coils of the helix), which are occasionally reflected in detectable sequence repeats (“overt” repeats) but usually are not detectable as such (“covert” repeats) (47). The difficulty of detecting such repeats in sequences is compounded by the fact that they may vary in length through the insertion of loops at turn sites. However, adjacent coils tend to have conserved positions occupied by polar residues (Ser, Thr, Asn, and His) that engage in H-bonding inside the structure, thus playing a role equivalent to that of apolar residues inside globular proteins. A common motif of parallel β-helices is the “Asn-ladder” (39, 48), whereby Asn residues are found at the same position in successive coils and their side chains stack, forming hydrogen bonds with each other; additional stabilization is conferred by the NH group of each Asn interacting with the backbone. On this basis, we hypothesized that the Asn and Gln side chains of the Ure2p prion domain form similar hydrogen-bonded networks in the fibril interior. The most favorable H-bonding requires the same type of side chains in positions that are adjacent along the fibril axis. This arrangement is provided by stacking the serpentines in parallel and in register.
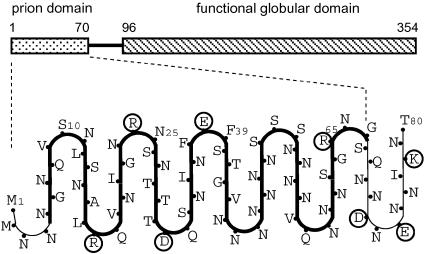
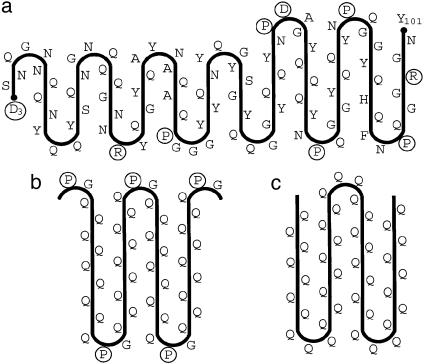
To locate the turns of the β-serpentine, we used the following considerations. The main issue was where to place the charged residues: in contrast to Asn, Gln, and apolar residues, charged residues should be unfavorable for a parallel in-register stacking of β-strands because of electrostatic repulsion. This repulsion would be especially strong if charged side chains of the same sign were placed one over another inside the fibril [it is about +26 kcal/mol at dielectric constant ε = 3 (49) and distance r = 0.47 nm, according to Coulomb's law]. However, if these side chains are on the fibril surface, they can avoid each other by lateral displacements; moreover, insertion of water molecules between charged groups would reduce the charge repulsion considerably (about +0.6 kcal/mol at ε = 80 and r = 0.8 nm). In this context, we did not find charged residues inside known β-helical proteins, with the sole exception of serralysin (50) where Asp coordinates with a Ca2+ ion. However, this ion coordination absolutely requires Gly residues in certain neighboring positions and this motif is absent in the Ure2p sequence. Thus, this consideration suggests that charged residues are located in the turns of the serpentine and on the outside of the fibril. The Ure2p prion domain excluding the linker region has four charged residues (Arg-17, Arg-24, Asp-31, and Glu-38) that are separated by equal increments of seven residues (Fig. 1). The regular distribution of charged residues led us to this arrangement whereby each seven-residue repeat consists of one strand and one turn.
Fig. 1.
Diagram of proposed β-serpentine fold for the Ure2p prion domain. The model has nine β-strands. Charged residues are circled. Because residues 1–10 are protease-sensitive for some Ure2p constructs (36), the first strand may detach relatively easily from the core.
The next step was to develop this scheme into an atomic model to test whether the proposed stacked serpentine topology can comply with constraints imposed by the covalent structure, van der Waals repulsions, and H-bonding requirements of the polypeptide chain. The turns have three residues between adjacent strands. A search among known β-helices found several possible templates for such turns. To build the prion domain model, we used one of the most typical turns with conformation ββpβpαLβ, which is found, e.g., in phage P22 tail-spike protein from position 184 to 189 (48) or P.69 pertactin from position 400 to 404 (51). The symbols αL, β, and βp denote, in order, residue backbone conformations close to the left-handed α-helix, the β conformation, and the polyproline conformation (underlined positions denote the termini of β-strands). Construction of the serpentine template was completed by connecting these turns by straight four-residue β-strands. Multiple copies of the template were then stacked in parallel and in-register apart from a slight twist of ≈1° per subunit (see below). We determined that the twist is left-handed by unidirectional shadowing EM (Fig. 2e). The initial orientation of the side chains was adjusted manually to alleviate apparent van der Waals repulsions and create an optimal network of H-bonds for interior polar residues.
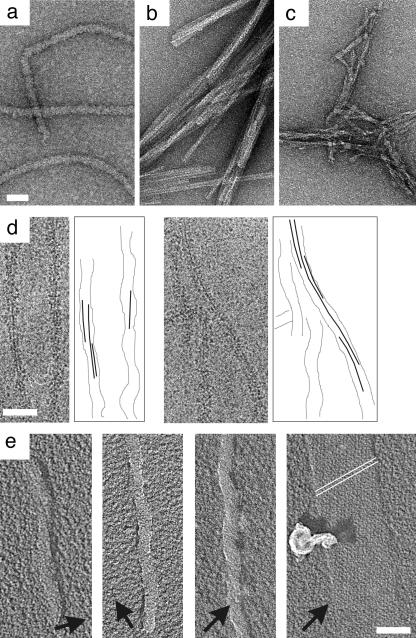
Fig. 2.
Electron micrographs of Ure2p-related filaments. (a) Ure2p filaments. (b) Bundled fibrils of Ure2p10-39. (c) Core fibrils produced by proteolytic digestion of Ure2p filaments (36). The specimens were negatively stained with uranyl acetate. (d) Cryo-micrographs of filaments of Ure2p1-80-GFP (courtesy of N. Cheng, National Institute of Arthritis and Musculoskeletal and Skin Diseases); alongside are shown graphical interpretations of one or two near-axial striations. (e) Unidirectional shadowing shows a long-pitch left-handed twist. Deposited metal is white, and shadows are black. Arrows indicate the shadowing direction. This experiment was performed with Ure2p1-80-GFP (35). We assume that this result extends to other prion domain-containing constructs, including native Ure2p. This assumption appears to be supported by AFM observations of Ure2p filaments (figure 6 of ref. 63). The rightmost panel of e shows the internal standard, bacteriophage T4 polyheads whose low-pitch helices (indexed as white lines) are right-handed (64). (Bar, 50 nm.)
To refine the model further, we focused on Ure2p10-39. This region is particularly conserved among different strains of yeast (52). In the model (Fig. 1), it corresponds to four strands and three pleats, residues 10 and 39 both being in turns. This peptide does indeed polymerize into long straight fibrils with a marked tendency to bundle. In Fig. 2 a–c, these fibrils are compared with Ure2p filaments and core fibrils exposed by proteolysis by negative staining EM. That Ure2p10-39 fibrils have a cross-β structure is consistent with detection of a 0.47-nm reflection by x-ray fiber diffraction (D. Sharma, H. Inouye, D. Kirschner, U.B., R.B.W., and A.C.S., unpublished results) and electron diffraction (N. Cheng, U.B., R.B.W., and A.C.S., unpublished results). A parallel β-strand orientation is supported by solid-state NMR data on Ure2p10-39 fibrils obtained by J. C.-C. Chan and R. Tycko (R. Tycko, personal communication).
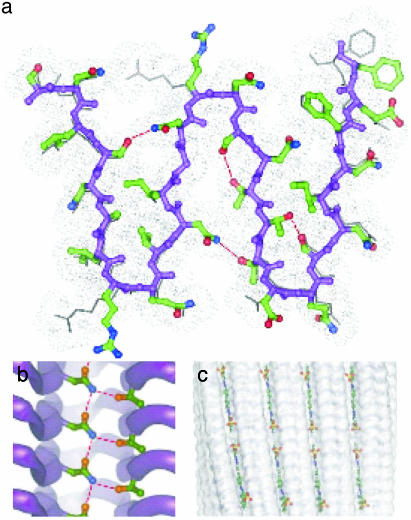
The model produced after energy minimization (see Materials and Methods) is shown in Fig. 3. We call it a “parallel superpleated β-structure” to distinguish it from the traditional “β-pleated sheet”: each superpleat involves 14 residues as compared to 2 residues in the traditional pleat (53), and the superpleats are oriented perpendicular to the traditional pleats. The peptide groups of the β-strands make H-bonds between adjacent chains. The pattern of H-bonding between the β-strands of adjacent serpentines is interrupted by the three-residue turns. The peptide group H-bonds in turns (between residues 10 and 11, 17 and 18, 24 and 25, 31 and 32, 38 and 39, 45 and 46, 52 and 53, 59 and 60, and 66 and 67), are oriented perpendicular to the fibril axis. In the fibril interior, several Asn side chains stack to form ladders as in pectate lyase (39): in addition, the NH group of each Asn bonds with a Ser, Thr, or Asn from an adjacent β-strand of the same chain. For example, the NH group of Asn-23 (Asn-20) forms an H-bond with the Oγ atom of Ser-13 (Thr-30) (Fig. 3a). In contrast to known β-helices, where polar side chains forming these H-bonds face the turns (39, 44), the β-serpentine stack has H-bonded side chains all along the strands. The resulting network of H-bonds between interior polar side chains (that we call “Asn/Gln locks”) should knit the structure together and increase the stability of the fibrils, making them protease-resistant (36) and thermostable (42). In terms of its spatial network of H-bonds, this structure is similar to polyglycine which forms stable insoluble aggregates (54, 55).
Fig. 3.
Parallel superpleated β-structure model of the Ure2p10-39 fibril. (a) Ball-and-stick representation of a transverse section. Some side chains (shown in skeletal representation) situated below the front serpentine layer may have orientations different from those of the neighboring layer because of electrostatic repulsion. The gray dotted van der Waals contour around the peptide shows close packing of the fibril structure. The backbone is in purple. Carbon, oxygen, and nitrogen atoms are in green, red, and blue, respectively. The H-bonds between the interior side chains are shown by red broken lines. (b) A lateral view along the β-strands. A ladder formed by H-bonding of Asn-20 and Thr-30 is shown. The other side chains are omitted for clarity. (c) Diagram showing proposed docking of Congo red molecules into surface grooves of the fibril.
We note that there is an alternative arrangement in which the sequence is shifted one residue toward the C terminus, along the serpentine template. In it, the charged residues remain on the outside, but it has several energetically unfavorable close contacts of interior side chains and, therefore, is less likely.
Features of the Model. The model of the Ure2p core fibril has nine β-sheets: in each sheet, the strands are parallel, and the sheets are packed parallel to each other with successive sheets in alternating orientations (Fig. 1). Its interior is topologically segmented into eight “bays,” each defined as the interface region between two adjacent sheets in a pleat. The bays form axially aligned grooves on the fibril surface (Fig. 3c) that might serve as binding sites for planar molecules of Congo red or other dyes of the same family or the Evans blue family that contain polyaromatic rings (9).
The segment beyond residue 70 is also quite rich in Asn but has a high incidence of charged residues (Fig. 1). This property is consistent with its serving as a linker connecting the C-terminal domain to the fibril backbone. If there were no such linker, it would not be possible to pack globular proteins of 3–4.5 nm in diameter together, because their contact points on the fibril backbone would then be only 0.47 nm apart. Instead, an extended linker allows them sufficient room to be accommodated around the filament periphery. In the event of elimination or shortening of the linker, additional strands might peel off the serpentine to serve as a surrogate linker.
It has been observed that substantial deletions are possible in the Ure2p prion domain while retaining the prion phenotype (ref. 52; M. J. Terry, H. Edskes, and R.B.W., unpublished results) and, by inference, the capability to form filaments. Thus, shortening the core fibril module (residues 1–70) from either end would progressively reduce the number of pleats while retaining essentially the same architecture. This trend raises the question of what is the minimum number of pleats to produce a stable fibril. As noted above, Ure2p10-39 (three pleats and four strands) fibrillizes readily and further truncation may be possible (unpublished work). Preliminary consideration of this model as applied to poly(Q)-type polypeptides (see below) suggests that three or four strands may be a minimum core in this system.
A key feature of the model is the in-register stacking of serpentines, one per layer. Here, the scanning transmission electron microscopy mass-per-unit-length data showing one subunit per ≈0.47 nm, regardless of which C-terminal appendage was attached (36), imposed a key constraint. This property explains how prion domain Ure2p1-65 is able to copolymerize readily with full-length Ure2p (34): the serpentines on both constructs match and stack together.
In the model, adjacent C-terminal moieties are attached on the same side of the filament. This means that, in a local segment of filament, one side is decorated with C-terminal moieties and the other side is bare. In consequence, some projections seen in cryo-electron micrographs should show the backbone as a dense striation that is not centered but on one side of the filament, and other projections will show a centered backbone: such is indeed the case (see figure 6 of ref. 36). In some segments, a second parallel striation is discernible (Fig. 2d) and can be explained as the stacking of C-terminal appendages.
This property also provides an explanation for the observed proclivity of prion domain constructs that have smaller C-terminal adjuncts to pair into double (type B) filaments (35). The side of the serpentine that is distal to the adjuncts has two pairs of oppositely charged residues (Arg-24 and Glu-38, and Arg-17 and Asp-31) facing outwards from turns (Fig. 1). If these sides are exposed, two oppositely oriented filaments could pair via the formation of two interfilament salt bridges at each level in the stack. As the size of the adjuncts increases, they are expected to be pushed, to an increasing extent, by mutual steric exclusion round to the “bare” side of the filament, where they should impede the interaction between filaments. Conversely, such salt bridge ladders would be strongly encouraged in adjunct-free prion domain constructs and indeed fibrils of such constructs exhibit a strong tendency to bundle (34, 35).
As noted above, it has been observed that the regularity of filaments depends on the adjunct and their helical repeat length may also vary for a given adjunct (35). The most regular construct to date is Ure2p1-80-GFP, which produced corkscrew filaments with axial repeats that were constant within a given filament but varied from 50 to 250 nm from filament to filament (35). A similar property has been observed for Sup35p filaments (56). The model suggests that the corkscrew morphology stems not from supercoiling of the filament axis, but from GFP appendages being locally clustered on the same side of the filament and slowly spiraling around a central fibril. We draw the following conclusions. (i) The axial twist per 0.47-nm step of the stack is small but variable [i.e., 360°/(250/0.47) to 360°/(50/0.47) = 0.7° to 3.4°]. (ii) This rotation angle can be affected by the packing of the adjuncts around the filament periphery or the stacking arrangement that happens to be initiated and is then propagated consistently along the growing fibril.
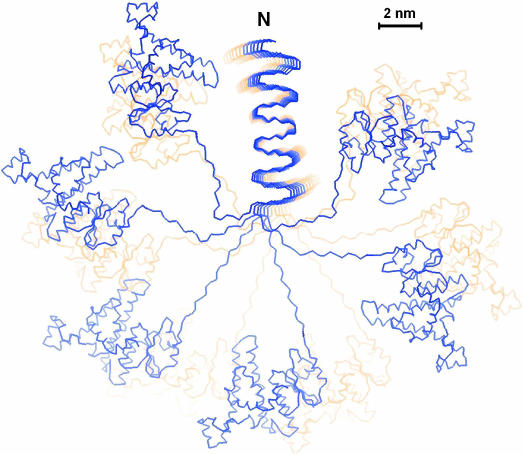
Constraints on Fibrillogenesis Imposed by Steric Exclusion of Globular Domains. To estimate the requirements imposed on linker length by having the exit points of the C-terminal adjuncts only ≈0.5 nm apart, we constructed a model of the Ure2p filament complete with globular domains (Fig. 4).
Fig. 4.
Model of the Ure2p prion filament. The model shows an axial view along the amyloid core fibril with 12 stacked β-serpentines, 6 blue and 6 yellow. The surrounding globular domains have the folds of C-terminal domain monomers and they are connected to the fibril backbone by the linker. No significance is attached to the conformation of the linkers other than that their length does not exceed that of a 25-residue polypeptide or to the positions and orientations of the C-terminal domains other than that they should not overlap with each other or with the fibril.
The minimum length for the linker corresponds to the closest possible packing of the globular domains and may be predicted if their size is known. With close packing, they should be arranged regularly in a pseudohelical manner with pitch equal to the axial projection of one globular domain (Gz). The number of globular domains (N) per turn is Gz/0.47 nm. The number of residues (n) in the minimal-length linker can be estimated by formula: n = ((N × Gθ + Fθ)/π – Gr)/2 × 0.34, where Gθ and Fθ are the azimuthal dimensions of the globular domain and the fibril, respectively; Gr is the radial dimension of the domain; and 0.34 nm is the length per residue of the extended linker polypeptide. For the Ure2p C-terminal domain, Gz ∼ 3 nm, Gθ ∼ 5.5 nm, Gr ∼ 3.5 nm, and Fθ∼ 3 nm, suggesting that n ∼ 13–14. In fact, Ure2p appears to have a linker of ≈25 residues (36), allowing a relatively loose packing of the globular domains that is consistent with EM data (36). One might imagine that shortening of the linker or increasing the size of the globular domain would be compensated by unfolding of the C-terminal portion of the serpentine until the remaining part is of subcritical size for fibril formation.
Variations on a Theme. Appraisal of the model indicates that the superpleated β-structure may be adjusted relatively easily to accommodate variations in the length of the β-strands and turns, the number of pleats, and the kinds of amino acids other than Asn that may be present in the interior (e.g., Gln, Thr, Ser, or some hydrophobic residues) and in the turns (e.g., Pro). Accordingly, we anticipate that a substantial range of variations in length and sequence substitution may be imposed on the Ure2p N-domain without abolishing its ability to fibrillize. From a present perspective, the insertion of charged residues into interior positions would appear to be the most subversive mutations. In the same vein, it appears likely that folds and packings of this kind may be applicable to a wide range of amino acid sequences.
Applicability of the β-Serpentine to Other Amyloids. From a preliminary appraisal of the sequence (57) of the yeast prion protein Sup35 (24, 56, 58), we infer that its fibril-forming sequence (N-domain) may also form a similar structure (Fig. 5a). In this case, there are five overt nine-residue repeats (59) as well as likely additional covert repeats, in comparison to the nine covert seven-residue repeats in Ure2p. Thus some β-strands and turns of the putative Sup35 serpentine may be longer than those of Ure2p, to place its charged and Pro residues in turns.
Fig. 5.
Possible β-serpentines for other proteins. (a) N-domain of Sup35. In the C-terminal portion, the Pro residues in the nine-residue pseudorepeats are assigned to turns. In the N-terminal portion, some hydrophobic residues are assigned to external positions, and they may be more efficiently sequestered by some other serpentine (different turn sites). (b) Poly(PGQ9). (c) Poly(Q) expansions. Because there are no charged nor Pro residues to punctuate turns, the length of the strands is not well defined: there may be several possibilities.
The amyloid fibrils formed by the poly(Q) tracts of huntingtin (3) may be explained in the same framework (Fig. 5c). In this case, the glutamine side chains should form a network of H-bonded “locks” in the filament interior. Interestingly, 46-residue peptides with repeats of PGQ9 and PGQ10 fibrillize as efficiently as Q45 (60). The corresponding PGQ7 and PGQ8 peptides assemble less readily, whereas introduction of one additional Pro residue (that is normally incompatible with the β-strand conformation) in the center of the Q9 element of PGQ9 completely blocks assembly (60). These observations suggest that Gln-rich structures consist of alternating β-strands and turns with an optimal repetitive unit of 11–12 residues (Fig. 5c). Thus, the minimal length of poly(Gln) to give fibrillization in vitro and disease in vivo, ≈40 residues (3), would correspond to a three- to four-pleat nucleus, and further expansion would add more pleats and, apparently, enhanced cooperativity.
Despite the anticipated general character of the described fold, we are aware of other possible arrangements. For example, it is known that short peptides of 5–10 residues can self-assemble into amyloid fibrils with antiparallel β-strands (61). Nevertheless, we expect that parallel superpleated β-structures may be quite widespread. Our preliminary analysis shows that the other more hydrophobic amyloidogenic sequences, for example, the NAC-region of α-synuclein (62), amylin, and even β-amyloid peptides, can also form structures of this kind.
The generation of amyloid filaments in vitro from a variety of proteins not known to have any connection with amyloid in vivo (2) has led to the suspicion that such a conformational transformation is possible for many, perhaps most, proteins. The in-register packing of serpentines, one per layer, suggests how this may come about. The stacking of like residues is a key to stabilization of these structures: in addition to ladders of Asn, Gln, Ser, and Thr, stacking of aliphatic residues such as Val, Ala, Ile, or Leu is observed in known β-helices (39): thus, regardless of sequence, like will stack over like. Natural protein sequences are likely to have local concentrations of charged residues where the stacking will be difficult to sustain, but they are also likely to have other local tracts that are amenable to this adaptable theme. The key requirements for generating homomeric ladders of like amino acids, regardless of sequence, are to have (i) one molecule per 0.47 nm layer and (ii) in-register stacking. The parallel β-serpentine model meets both constraints, and some other arrangements could also meet them, e.g., single-turn β-helices although the latter polypeptides would be confined to a narrow length range (20–30 residues).
Supplementary Material
Acknowledgments
We thank Dr. D. Winkler for help with metal-shadowing, Drs. N. Cheng and K. Grünewald for help with electron microscopy, Drs. W.-M. Yau and R. Tycko for synthesis of Ure2p10-39, and Mr. T. Cassese for helpful discussions. This work was supported in part by Fondation Recherche Médicale Grant INE20030307050 (to A.V.K.). The model coordinates are available on request.
Abbreviation: EM, electron microscopy.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobson, C. M. (1999) Trends. Biochem. Sci. 24, 329–332. [DOI] [PubMed] [Google Scholar]
- 3.Perutz, M. F. (1999) Trends. Biochem. Sci. 24, 58–63. [DOI] [PubMed] [Google Scholar]
- 4.Rochet, J. C. & Lansbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60–68. [DOI] [PubMed] [Google Scholar]
- 5.Shirahama, T. & Cohen, A. S. (1967) J. Cell. Biol. 33, 679–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eanes, E. D. & Glenner, G. G. (1968) J. Histochem. Cytochem. 16, 673–677. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner, D. A., Inouye, H., Duffy, L. K., Sinclair, A., Lind, M. & Selkoe, D. J. (1987) Proc. Natl. Acad. Sci. USA 84, 6953–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serpell, L. C., Fraser, P. E. & Sunde, M. (1999) Methods Enzymol. 309, 526–536. [DOI] [PubMed] [Google Scholar]
- 9.Puchtler, H. & Sweat, F. (1965) J. Histochem. Cytochem. 13, 693–694. [DOI] [PubMed] [Google Scholar]
- 10.Guijarro, J. I., Sunde, M., Jones, J. A., Campbell, I. D. & Dobson, C. M. (1998) Proc. Natl. Acad. Sci. USA 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake, C. & Serpell, L. (1996) Structure (London) 4, 989–998. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, J. W. (1997) Structure (London) 5, 595–600. [DOI] [PubMed] [Google Scholar]
- 13.Lazo, N. D. & Downing, D. T. (1998) Biochemistry 37, 1731–1735. [DOI] [PubMed] [Google Scholar]
- 14.Li, L., Darden, T. A., Bartolotti, L., Kominos, D. & Pedersen, L. G. (1999) Biophys. J. 76, 2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benzinger, T. L., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benzinger, T. L., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (2000) Biochemistry 39, 3491–3499. [DOI] [PubMed] [Google Scholar]
- 17.Antzutkin, O. N., Balbach, J. J., Leapman, R. D., Rizzo, N. W., Reed, J. & Tycko, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balbach, J. J., Petkova, A. T., Oyler, N. A., Antzutkin, O. N., Gordon, D. J., Meredith, S. C. & Tycko, R. (2002) Biophys. J. 83, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torok, M., Milton, S., Kayed, R., Wu, P., McIntire, T., Glabe, C. G. & Langen, R. (2002) J. Biol. Chem. 277, 40810–40815. [DOI] [PubMed] [Google Scholar]
- 20.Ma, B. & Nussinov, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14126–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetzel, R. (2002) Structure (London) 10, 1031–1036. [DOI] [PubMed] [Google Scholar]
- 23.Kishimoto, A., Hasegawa, K., Suzuki, H., Taguchi, H., Namba, K. & Yoshida, M. (2004) Biochem. Biophys. Res. Commun. 315, 739–745. [DOI] [PubMed] [Google Scholar]
- 24.Wickner, R. B. (1994) Science 264, 566–569. [DOI] [PubMed] [Google Scholar]
- 25.Masison, D. C. & Wickner, R. B. (1995) Science 270, 93–95. [DOI] [PubMed] [Google Scholar]
- 26.Coschigano, P. W. & Magasanik, B. (1991) Mol. Cell. Biol. 11, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousset, L., Belrhali, H., Janin, J., Melki, R. & Morera, S. (2001) Structure (London) 9, 39–46. [DOI] [PubMed] [Google Scholar]
- 28.Umland, T. C., Taylor, K. L., Rhee, S., Wickner, R. B. & Davies, D. R. (2001) Proc. Natl. Acad. Sci. USA 98, 1459–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drillien, R., Aigle, M. & Lacroute, F. (1973) Biochem. Biophys. Res. Commun. 53, 367–372. [DOI] [PubMed] [Google Scholar]
- 30.Lacroute, F. (1971) J. Bacteriol. 106, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edskes, H. K., Gray, V. T. & Wickner, R. B. (1999) Proc. Natl. Acad. Sci. USA 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speransky, V. V., Taylor, K. L., Edskes, H. K., Wickner, R. B. & Steven, A. C. (2001) J. Cell Biol. 153, 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bousset, L., Thomson, N. H., Radford, S. E. & Melki, R. (2002) EMBO J. 21, 2903–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, K. L., Cheng, N., Williams, R. W., Steven, A. C. & Wickner, R. B. (1999) Science 283, 1339–1343. [DOI] [PubMed] [Google Scholar]
- 35.Baxa, U., Speransky, V., Steven, A. C. & Wickner, R. B. (2002) Proc. Natl. Acad. Sci. USA 99, 5253–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxa, U., Taylor, K. L., Wall, J. S., Simon, M. N., Cheng, N., Wickner, R. B. & Steven, A. C. (2003) J. Biol. Chem. 278, 43717–43727. [DOI] [PubMed] [Google Scholar]
- 37.Perrett, S., Freeman, S. J., Butler, P. J. & Fersht, A. R. (1999) J. Mol. Biol. 290, 331–345. [DOI] [PubMed] [Google Scholar]
- 38.Thual, C., Bousset, L., Komar, A. A., Walter, S., Buchner, J., Cullin, C. & Melki, R. (2001) Biochemistry 40, 1764–1773. [DOI] [PubMed] [Google Scholar]
- 39.Yoder, M. D., Keen, N. T. & Jurnak, F. (1993) Science 260, 1503–1507. [DOI] [PubMed] [Google Scholar]
- 40.Dayring, H. E., Tramonato, A., Sprang, S. R. & Fletterick, R. J. (1986) J. Mol. Graphics 4, 82–87. [Google Scholar]
- 41.Laskowski, R. A., McArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 282–291. [Google Scholar]
- 42.Baxa, U., Ross, P. D., Wickner, R. B. & Steven, A. C. (2004) J. Mol. Biol., in press. [DOI] [PubMed]
- 43.Miller, S., Janin, J., Lesk, A. M. & Chothia, C. (1987) J. Mol. Biol. 196, 641–656. [DOI] [PubMed] [Google Scholar]
- 44.Kajava, A. V. (2001) J. Struct. Biol. 134, 132–144. [DOI] [PubMed] [Google Scholar]
- 45.Graether, S. P., Kuiper, M. J., Gagne, S. M., Walker, V. K., Jia, Z., Sykes, B. D. & Davies, P. L. (2000) Nature 406, 325–328. [DOI] [PubMed] [Google Scholar]
- 46.Liou, Y. C., Tocilj, A., Davies, P. L. & Jia, Z. (2000) Nature 406, 322–324. [DOI] [PubMed] [Google Scholar]
- 47.Kajava, A. V., Cheng, N., Cleaver, R., Kessel, M., Simon, M. N., Willery, E., Jacob-Dubuisson, F., Locht, C. & Steven, A. C. (2001) Mol. Microbiol. 42, 279–292. [DOI] [PubMed] [Google Scholar]
- 48.Steinbacher, S., Seckler, R., Miller, S., Steipe, B., Huber, R. & Reinemer, P. (1994) Science 265, 383–386. [DOI] [PubMed] [Google Scholar]
- 49.Roger, N. K. (1990) in Prediction of Protein Structure and the Principles of Protein Conformation, ed. Fasman, G. D. (Plenum, New York), pp. 357–390.
- 50.Baumann, U., Wu, S., Flaherty, K. M. & McKay, D. B. (1993) EMBO J. 12, 3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley, P., Charles, I. G., Fairweather, N. F. & Isaacs, N. W. (1996) Nature 381, 90–92. [DOI] [PubMed] [Google Scholar]
- 52.Edskes, H. K. & Wickner, R. B. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16384–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pauling, L. & Corey, R. B. (1953) Proc. R. Soc. London Ser. B 141, 21–33. [DOI] [PubMed] [Google Scholar]
- 54.Crick, F. H. & Rich, A. (1955) Nature 176, 780–781. [DOI] [PubMed] [Google Scholar]
- 55.Kajava, A. V. (1999) Acta Crystallogr. D 55, 436–442. [DOI] [PubMed] [Google Scholar]
- 56.Glover, J. R., Kowal, A. S., Schirmer, E. C., Patino, M. M., Liu, J. J. & Lindquist, S. (1997) Cell 89, 811–819. [DOI] [PubMed] [Google Scholar]
- 57.Kushnirov, V. V., Ter-Avanesyan, M. D., Telckov, M. V., Surguchov, A. P., Smirnov, V. N. & Inge-Vechtomov, S. G. (1988) Gene 66, 45–54. [DOI] [PubMed] [Google Scholar]
- 58.Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. (1997) Science 277, 381–383. [DOI] [PubMed] [Google Scholar]
- 59.Liu, J. J. & Lindquist, S. (1999) Nature 400, 573–576. [DOI] [PubMed] [Google Scholar]
- 60.Thakur, A. K. & Wetzel, R. (2002) Proc. Natl. Acad. Sci. USA 99, 17014–17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balbach, J. J., Ishii, Y., Antzutkin, O. N., Leapman, R. D., Rizzo, N. W., Dyda, F., Reed, J. & Tycko, R. (2000) Biochemistry 39, 13748–13759. [DOI] [PubMed] [Google Scholar]
- 62.Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. (2003) J. Biol. Chem. 278, 37530–37535. [DOI] [PubMed] [Google Scholar]
- 63.Jiang, Y., Li, H., Zhu, L., Zhou, J. M. & Perrett, S. (2004) J. Biol. Chem. 279, 3361–3369. [DOI] [PubMed] [Google Scholar]
- 64.Kistler, J., Aebi, U., Onorato, L., ten Heggeler, B. & Showe, M. K. (1978) J. Mol. Biol. 126, 571–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.