Abstract
Trigger factor (TF) and signal recognition particle (SRP) bind to the bacterial ribosome and are both crosslinked to protein L23 at the peptide exit, where they interact with emerging nascent peptide chains. It is unclear whether TF and SRP exclude one another from their ribosomal binding site(s). Here we show that SRP and TF can bind simultaneously to ribosomes or ribosome nascent-chain complexes exposing a SRP-specific signal sequence. Based on changes of the crosslinking pattern and on results obtained by fluorescence measurements using fluorescence-labeled SRP, TF binding induces structural changes in the ribosome–SRP complex. Furthermore, we show that binding of the SRP receptor, FtsY, to ribosome-bound SRP excludes TF from the ribosome. These results suggest that TF and SRP sample nascent chains on the ribosome in a nonexclusive fashion. The decision for ribosome nascent-chain complexes exposing a signal sequence to enter SRP-dependent membrane targeting seems to be determined by the binding of SRP, which is stabilized by signal sequence recognition, and promoted by the exclusion of TF due to the binding of the SRP receptor to ribosome-bound SRP.
Newly synthesized proteins emerging from the peptide exit tunnel of the ribosome (1, 2) interact with proteins that prevent premature folding, so-called chaperones (3, 4). In bacteria, the first chaperone that interacts with nascent peptide chains is trigger factor (TF), a 48-kDa protein that binds to ribosomes at 1:1 stoichiometry with moderate affinity (5–7). Subsequently, nascent polypeptides can interact with the chaperones DnaK, a member of the Hsp70 chaperone family, and DnaJ, and further downstream with the Hsp60 chaperonins GroEL/GroES (3, 4). Alternatively, proteins that are destined for insertion into the inner membrane of the bacterial cell are recognized by the signal recognition particle (SRP), which binds to nascent-chain ribosomes (RNC) and recognizes an SRP-specific signal sequence at the N terminus of the peptide emerging from the ribosome (8). In Escherichia coli, SRP consists of a 48-kDa protein, Ffh, that is responsible for binding to the ribosome and the nascent signal peptide, and a 114-nt RNA, 4.5S RNA. SRP binding initiates targeting of ribosomes synthesizing inner-membrane proteins to the translocation pore of the membrane. The committed step of membrane targeting is binding of the RNC–SRP complex to the SRP receptor, FtsY.
Both TF and SRP bind to the ribosome in the vicinity of the peptide exit, and crosslinking experiments have revealed ribosomal protein L23 as a common attachment site (9–11). Because protein L23 is rather small, 13 kDa, this finding raises the question whether there is binding competition due to overlap of the binding sites of TF and SRP. Whereas SRP binding to RNCs is greatly promoted by the signal sequence, TF probably binds to any nascent peptide, with some preference for sequences containing aromatic amino acids (12). TF is present in the cell in high concentration and in excess over ribosomes, such that every ribosome has TF bound, despite the moderate affinity (≈1 μM). On the other hand, SRP is present in the cell in substoichiometric amounts relative to ribosomes (1:4). Thus, if there were binding competition, the slow dissociation of TF from the ribosome (13) would preclude SRP binding to RNCs, raising the question as to how SRP could ever have a chance to bind to RNCs and, depending on the presence or absence of a signal sequence, initiate membrane targeting. In this work, we have addressed the issue by studying the binding of TF, SRP, and FtsY to ribosomes and RNCs by using UV-induced crosslinking, ultracentrifugation, and fluorescence.
Materials and Methods
Protein Purification, 4.5S RNA, and Fluorescence Labeling. Ffh and FtsY from E. coli, both carrying a C-terminal His-6 tag, were expressed and purified as described, as were cysteine mutants of Ffh (10). Cysteine mutants of Ffh and their azidophenacyl (AzP) derivatives used for UV-induced crosslinking (10) were fully active in forming SRP that was fully competent for ribosome binding. TF and TF mutants were prepared as described (6). Labeling of TF-D42C with benzophenone-4-iodoacetamide (BPIA) and crosslinking was performed as described (9). 4.5S RNA was prepared by transcription in vitro (14). Truncated 4.5S RNA(21–81) was transcribed from a plasmid (14) construct obtained by PCR mutagenesis. 4.5S RNA(21–81) was labeled at the periodate-oxidized 3′ end by coupling with the hydrazide derivative of Alexa 555 (Molecular Probes), following a previously published protocol (15). Cysteine mutants of Ffh were labeled by coupling with the maleimide derivative of Oregon green (OG) (Molecular Probes) using a standard protocol (unpublished data); labeling was quantitative, based on absorption measurements. Ffh–OG derivatives were fully active in binding to 4.5S RNA and to ribosomes.
Ribosome Nascent-Chain Complexes. Ribosomes from E. coli MRE600 and purified components of the translation system were prepared as described (16, 17). Truncated mRNA coding for 94 N-terminal amino acids of leader peptidase (lep) was translated and lep-RNCs were purified as described (10). A total of 60–65% of the ribosomes present in lep-RNC preparation carried a nascent peptide chain, as determined from the amount of f[3H]Met in trichloroacetic acid (TCA)-precipitable peptides after alkaline hydrolysis of peptidyl–tRNA. The peptide labeled by incorporation of [14C]Leu was homogeneous on SDS gel electrophoresis and migrated at a position consistent with the expected length of 94 aa (data not shown).
Results
Crosslinking of SRP and TF to Ribosomal Proteins. Crosslinking of SRP to ribosomal protein L23 and TF to proteins L23 and L29 has been shown by directed crosslinking from defined positions in TF (9) and SRP/Ffh (10) as well as by chemical crosslinking using a bifunctional crosslinker (10, 11). The observation that the addition of TF suppressed the crosslink between SRP/Ffh and ribosomal protein L23, and vice versa, was interpreted as to suggest competition between SRP and TF for a common binding site on the ribosome (10, 11). However, the alternative interpretation, that is, loss of crosslink due to a structural change induced by the simultaneous presence of SRP and TF on the ribosome, was not excluded (10, 11). To examine whether there is competition between TF and SRP for binding to the ribosome, we have studied the effects both of TF addition on SRP/Ffh crosslinking to ribosomal protein L23 and of SRP addition on TF crosslinking to proteins L23 and L29. Crosslinking from SRP was performed as described (10) with SRP carrying the UV-activatable AzP group at cysteine residues engineered into positions 17 and 25 of Ffh, which gave high-yield crosslinks with protein L23 (Fig. 1A). The addition of TF to the ribosome–SRP(AzP17) or ribosome–SRP(AzP25) complex at saturating concentration (10 μM, i.e., 30 times the Kd of 0.3 μM; see Fig. 3) decreased the crosslinking efficiency, but did not completely eliminate the crosslink (Fig. 1 A). The reverse experiment was performed by examining the UV-induced crosslinks between TF coupled with BPIA and ribosomal proteins L23 and L29 (9) in the absence and presence of SRP (Fig. 1B Left). Clearly, SRP had no influence on the TF crosslinks. However, a strong crosslink between Ffh and TF was observed when both SRP and TF–BPIA were present. This finding directly demonstrates simultaneous binding of SRP and TF to one ribosome and indicates that the crosslinker at position 42 in the ribosome-binding domain of TF is located in close vicinity to Ffh (within 10 Å, the length of the crosslinker BPIA) on the ribosome. No crosslinked TF*Ffh was detected in the supernatant (Fig. 1B Right), indicating that the crosslink between TF and SRP, which were present in large excess over ribosomes, is confined to the ribosome-bound species.
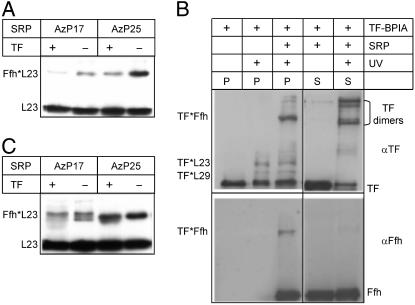
Fig. 1.
Ribosome binding of SRP and TF studied by crosslinking. (A) Crosslinking of SRP(AzP17/25) to vacant ribosomes. Ribosomes (1 μM), SRP(AzP17/25) (1 μM), and TF (10 μM) were irradiated with UV light, samples were centrifuged, and ribosomal pellets were analyzed by SDS gel electrophoresis and immunoblotting using antibodies against ribosomal protein L23 (10). Crosslinked proteins are indicated by asterisks, e.g., Ffh*L23. (B) Crosslinking of TF–BPIA to vacant ribosomes. Ribosomes (2 μM), SRP (10 μM), and TF–BPIA (10 μM) were mixed and irradiated with UV light; after ultracentrifugation, ribosomal pellets (P) and supernatants (S) were analyzed by SDS gel electrophoresis and immunoblotting (9). For staining, antibodies against TF (αTF) and Ffh (αFfh) were used. (C) Crosslinking from SRP to lep-RNC. UV-induced crosslinking was performed with SRP(AzP17/25) (10 nM) and lep-RNC (10 nM) (Materials and Methods) in the absence and presence of TF (1 μM), and samples were analyzed as in A.
Fig. 3.
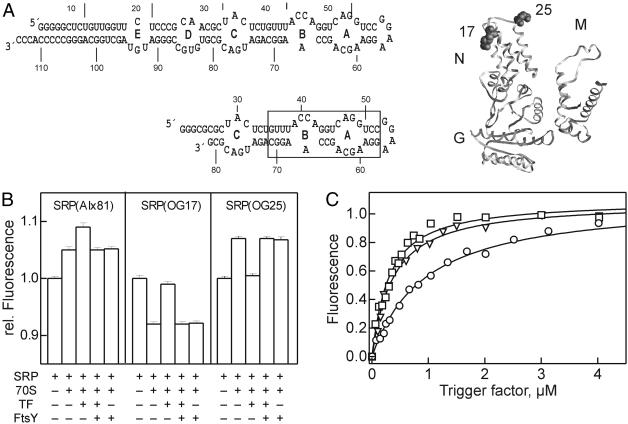
Ribosome binding of SRP, TF, and FtsY monitored by fluorescence. (A)(Left) Two-dimensional structure models of 4.5S RNA and 4.5S RNA(21–81). The approximate binding site of Ffh is boxed. (Right) Positions of OG labels in the N domain of Ffh. The arrangement of NG and M domains of Ffh is based on fluorescence data (I.B., unpublished data). (B) TF/FtsY binding to ribosome–SRP(Alx81) or ribosome–SRP(OG17/25) complexes. Fluorescence measurements were performed in buffer A containing 1 mM GDPNP at 20°C; excitation/emission was at 535/565 nm (Alx) or 470/517 nm (OG). To fluorescent SRP (0.1 μM) were added 70S ribosomes (0.1 μM), TF (1.2 μM), and FtsY (0.1 μM); the order of addition of TF and FtsY had no influence (data not shown). (C) TF titrations of ribosome–SRP(Alx81) complexes (0.1 μM) with TF (squares), TF (1–144) (triangles), or TF(FRK/AAA) (circles). Titrations were performed in buffer A at 20°C in the presence of 1 mM GDPNP. Data were evaluated by nonlinear fitting on the basis of 1:1 stoichiometric binding (20, 22); the following Kd values were obtained: TF, 0.33 ± 0.03 μM; TF(1–144), 0.30 ± 0.03 μM; TF(FRK/AAA), 1.0 ± 0.05 μM.
For signal peptide-specific binding of SRP to ribosomes, a ribosome nascent-chain complex was used that carried the first 94 aa of leader peptidase (lep-RNC) (Materials and Methods), an inner-membrane protein that harbors a strong SRP-specific signal anchor sequence at the N terminus. The affinity of SRP binding to lep-RNC is very high (Kd ≈ 0.1 nM; I.B., unpublished data), comparable to affinities reported for eukaryotic RNCs and SRP (18). Therefore, lep-RNC could be used at low concentration (10 nM), minimizing SRP binding to vacant ribosomes present in the RNC preparation. To examine TF binding to RNC–SRP complexes, SRP(AzP17) or SRP(AzP25) was bound to lep-RNC with or without added TF, and crosslinking was induced by UV irradiation. In the presence of TF, one of the two crosslinks from position 17 of Ffh (lower band) was diminished, whereas the other (upper band) was not; the crosslink from position 25 was not affected (Fig. 1C). The observation that only one of the SRP crosslinks was affected by TF, whereas the others were not, demonstrates that TF and SRP were bound to the RNC complex simultaneously and that TF binding influenced the SRP–RNC structure in a similar fashion as with vacant ribosomes.
The crosslinking experiments show that the crosslink from one position, AzP17 in Ffh, is diminished upon addition of TF, whereas the crosslinks from position 25 of Ffh, or from position 42 of TF, are less or not at all affected. The former result is analogous to the loss of crosslinking between SRP or TF and ribosomal protein L23 induced by adding excess TF or SRP to ribosome–SRP or ribosome–TF complexes, respectively, as reported (10, 11). The present results indicate that the latter observation is to be explained by an induced structural change, rather than by binding competition. Simultaneous binding of SRP and TF to one ribosome is also shown by the binding experiments described in the following.
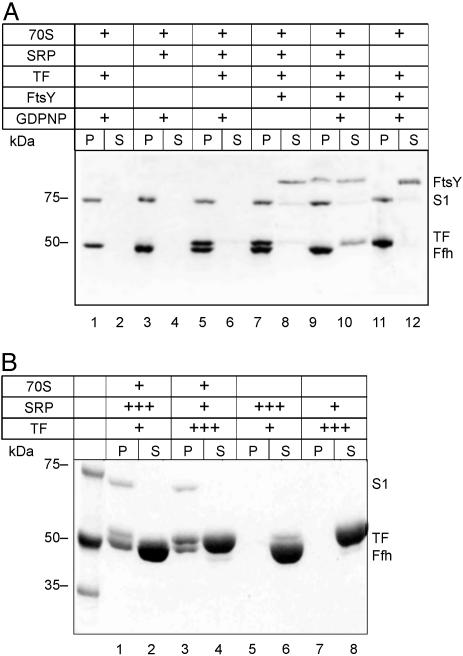
Ribosome Binding of SRP, TF, and FtsY Studied by Ultracentrifugation. Ribosome binding of SRP, TF, and FtsY was further examined by ultracentrifugation (Fig. 2). When SRP was added to vacant ribosomes in equimolar amount, all SRP (identified as Ffh) was found in the ribosome pellet (Fig. 2A, lanes 3 and 4), indicating 1:1 stoichiometric binding of SRP to the ribosome. Stoichiometric binding was also observed with TF and vacant ribosomes (Fig. 2A, lanes 1 and 2). The same extent of binding of both SRP and TF was observed when TF was added to ribosome–SRP complexes (Fig. 2A, lanes 5 and 6). Because ribosomes, SRP, and TF were present at the same concentration (1 μM) and all SRP and TF was bound, this result demonstrates simultaneous binding of SRP and TF to the ribosome. Binding of the SRP receptor, FtsY, to vacant ribosomes was not detected (data not shown), but there was efficient binding of FtsY to the ribosome–SRP complex, which completely abrogated TF binding (Fig. 2A, lanes 9 and 10). Some FtsY remained in the supernatant, indicating limited stability of the complex. No binding of FtsY and no displacement of TF was observed in the absence of GDPNP (Fig. 2A, lanes 7 and 8), in accordance with the known GTP (or GDPNP) requirement of FtsY binding to SRP (19). In the absence of SRP, FtsY did not bind to ribosomes and did not displace TF from ribosome–TF complexes (Fig. 2A, lanes 11 and 12), even when FtsY was added at 10-fold higher concentration (data not shown).
Fig. 2.
Ribosome binding of SRP, TF, and FtsY examined by ultracentrifugation. (A) Equimolar concentrations of ribosomes, SRP, TF, and FtsY. Components (1 μM each) were mixed in various combinations in buffer A (50 mM Tris·HCl, pH 7.5/70 mM NH4Cl/30 mM KCl/7 mM MgCl2), containing 1 mM GDPNP where indicated, and incubated for 10 min at 37°C. Samples (100 μl) were centrifuged at 436,000 × g (Sorvall M120GX) for 30 min at 4°C. Ribosomal pellets (P) and supernatants (S) were analyzed by SDS gel electrophoresis, and proteins were stained with Coomassie blue. S1, ribosomal protein S1 (69 kDa); the other ribosomal proteins migrated out of the gel. (B) Excess of SRP or TF over ribosomes. The experiment was performed as in A with ribosomes (1 μM, lanes 1–4), SRP (1 μM, lanes 3–6; 30 μM, lanes 1 and 2), and TF (1 μM, lanes 1, 2, 5, and 6; 30 μM, lanes 3, 4, 7, and 8); controls without ribosomes are shown in lanes 5–8. GDPNP (1 mM) was present in all samples.
The binding experiments were also performed at conditions where the competing ligand, SRP or TF, was added to the ribosome complex at very high concentrations, as used by Luirink and colleagues (10, 11). As shown in Fig. 2B, at these conditions, there was no competition between TF and SRP for binding to the ribosome, in keeping with simultaneous binding. The controls show that, even at very high concentrations of SRP or TF, no material appeared in the pellet fraction when ribosomes were not present.
Ribosome Binding of SRP, TF, and FtsY Studied by Fluorescence. Ribosome binding of SRP and TF was also studied by fluorescence, using fluorescence-labeled SRP. The fluorescent probe Alexa 555 (Alx) was introduced into SRP by labeling the 3′ end of 4.5S RNA(21–81) (Fig. 3A). The labeled RNA fragment comprised the Ffh-binding site and was fully active in binding Ffh to form SRP(Alx81) (data not shown). Binding of SRP(Alx81) to vacant ribosomes led to a fluorescence increase, and the addition of TF led to a further increase (Fig. 3B). No fluorescence change was observed when TF was added to SRP(Alx81) in the absence of ribosomes (data not shown). These results indicated that TF was bound to the ribosome–SRP complex and that there was no binding competition between TF and SRP. The addition of FtsY completely reversed the effect of TF, as expected if there was binding competition between FtsY and TF. Similar results were obtained by using fluorescent SRP reconstituted from full-length 4.5S RNA and Ffh labeled with OG at single cysteine residues that were engineered into positions 17 and 25 in the N domain of Ffh (Fig. 3A). Upon formation of the ribosome–SRP complex, the fluorescence of OG17 decreased and that of OG25 increased (Fig. 3B). Subsequent binding of TF increased the fluorescence of OG17 and decreased the fluorescence of OG25. Finally, the addition of the SRP receptor, FtsY, reversed the TF-induced signal changes of both OG17 and OG25 to the fluorescence levels observed in control experiments without TF. Based on crosslinking (10), positions 17 and 25 in the N domain of Ffh in the complex with the ribosome are close to protein L23, suggesting that a direct contact or an induced conformational change causes the fluorescence changes of OG17 and OG25 observed upon SRP binding to the ribosome. The fluorescence changes induced by TF binding may be attributed either to a direct interaction between TF and the labeled N domain of Ffh or to an indirect effect mediated by protein L23 to which TF is also bound (9).
The binding of TF to the ribosome–SRP complex was quantitated by titration, monitoring the fluorescence of ribosome-bound SRP(Alx81) (Fig. 3C). Titration with wild-type TF yielded a Kd of 0.3 μM. The same value (20) or slightly higher values (≈1 μM; refs. 12 and 13) were reported previously for the binding of TF to vacant ribosomes, indicating a small, if any, stabilizing effect of SRP on TF binding. At any rate, an interaction between TF and SRP, if any, must be weak because there was no indication from crosslinking (see above), fluorescence, or gel retardation experiments for complex formation between TF and SRP without ribosomes (data not shown). N-terminal fragments of TF, comprising only the N domain (TF1–144) or the NP domains (TF1–247), had a similar affinity, 0.3 μM, for binding to ribosome–SRP complexes, in agreement with previous results demonstrating that the N-terminal domain of TF comprises the ribosome-binding domain (21). Mutant TF in which three conserved amino acids (Phe-44, Arg-45, Lys-46) in the N-terminal domain were replaced with alanines (TF-FRK/AAA) exhibited a lower affinity, in line with previous crosslinking results (9).
Discussion
The present results show that TF and SRP bind simultaneously at the ribosomal peptide exit site, sharing protein L23 as a common attachment site. TF and SRP are bound in close proximity to each other, as indicated by BPIA crosslinking from position 42 of TF to Ffh (see Fig. 1C) and AzP crosslinking from Ffh to TF (S.-Q.G., unpublished data). Binding of TF induces structural changes in the ribosome–SRP complex, as evident from a change of crosslinks between SRP and protein L23 (Fig. 1) and signal changes of fluorescent groups attached to SRP (Fig. 3). Induced structural changes probably explain the reported loss of crosslinks between SRP and L23 induced by TF, which was interpreted to indicate competition between TF and SRP for binding to L23 (11). FtsY binding to ribosome-bound SRP displaces or excludes TF from the ribosome. The mechanism of displacement is unclear at present. It may be physical exclusion caused by overlapping binding sites. Alternatively, FtsY binding to ribosome-bound SRP may destabilize the interaction of TF with the ribosome, thereby accelerating the dissociation of TF, which is slow intrinsically (13).
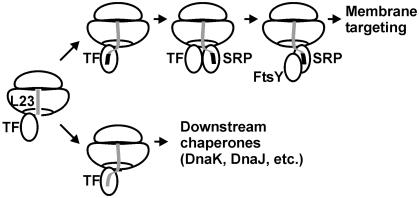
The intracellular concentrations of ribosomes and TF are high, such that practically all ribosomes have TF bound (20, 22), despite the moderate binding affinity. The slow dissociation of TF from the ribosome (13) raises the question as to how SRP could bind to ribosomes exposing an SRP-specific signal sequence if there were competition between TF and SRP. The present finding of simultaneous binding of TF and SRP resolves this issue, and the competition between FtsY and TF provides a mechanism for TF removal. Thus, the basic features of the interplay between TF and components of the SRP pathway can be described as follows (Fig. 4). TF is present on every ribosome and may bind any nascent chain, exerting its functions as a chaperone and, possibly, peptidyl–prolyl isomerase. The appearance of an SRP-specific signal anchor sequence in the nascent peptide promotes high-affinity binding of SRP. Unlike TF release from the ribosome, which is slow (13), peptide release from TF is rapid (23). Thus, it is possible that SRP binds to the signal sequence while TF remains bound to the ribosome and, possibly, other portions of the nascent peptide (24). Subsequently, FtsY binds to ribosome-bound SRP and displaces TF from the ribosome, thereby enabling membrane targeting of the RNC–SRP–FtsY complex. Ribosome-bound TF appears to sample nascent chains in a static manner, as suggested by low association and dissociation rates of the TF–ribosome complex (13), whereas sampling by SRP is dynamic, in accordance with rapid SRP binding to and dissociation from the ribosome observed in kinetic experiments (I.B., unpublished data). Rapid scanning of RNCs by SRP without interference by TF explains how SRP and FtsY, which in the cell are present in substoichiometric amounts relative to ribosomes, can efficiently target RNCs to the membrane for subsequent cotranslational membrane insertion. Only a small amount of SRP is required to bind firmly to the subset of SRP-specific RNCs during the targeting phase, leaving the remaining SRP for rapid scanning of RNCs for emerging SRP-specific signal sequences. It remains to be seen whether recruitment of TF and/or SRP to RNCs may be influenced by nascent signal peptides still residing in the exit tunnel (25, 26).
Fig. 4.
Interplay of TF, SRP, and FtsY on the ribosome. See text for the description of steps. The nascent peptide (gray line) is depicted as running through the exit tunnel of the large ribosomal subunit; the SRP-specific signal sequence is depicted as a black box. The peptide exit region of the ribosome is depicted in a highly simplified manner, showing only protein L23, which forms part of the attachment sites of both TF and SRP.
Acknowledgments
We thank Frank Peske for constructing cysteine mutants of Ffh and Georg Lentzen for the construct coding for truncated 4.5S RNA. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to E.D., B.B., and W.W.), the Human Frontier Science Program (to E.D.), the Alfried Krupp von Bohlen und Halbach-Stiftung (to M.V.R. and W.W.), and the Fonds der Chemischen Industrie (to M.V.R., B.B., and W.W.).
Abbreviations: AzP, azidophenacyl; BPIA, benzophenone-4-iodoacetamide; OG, Oregon green; RNC, nascent-chain ribosomes; SRP, signal recognition particle; TF, trigger factor.
References
- 1.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 2.Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 3.Frydman, J. (2001) Annu. Rev. Biochem. 70, 603–647. [DOI] [PubMed] [Google Scholar]
- 4.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- 5.Valent, Q. A., Kendall, D. A., High, S., Kusters, R., Oudega, B. & Luirink, J. (1995) EMBO J. 14, 5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesterkamp, T., Hauser, S., Lutcke, H. & Bukau, B. (1996) Proc. Natl. Acad. Sci. USA 93, 4437–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valent, Q. A., de Gier, J. W., von Heijne, G., Kendall, D. A., ten Hagen-Jongman, C. M., Oudega, B. & Luirink, J. (1997) Mol. Microbiol. 25, 53–64. [DOI] [PubMed] [Google Scholar]
- 8.Driessen, A. J., Manting, E. H. & van der Does, C. (2001) Nat. Struct. Biol. 8, 492–498. [DOI] [PubMed] [Google Scholar]
- 9.Kramer, G., Rauch, T., Rist, W., Vorderwulbecke, S., Patzelt, H., Schulze-Specking, A., Ban, N., Deuerling, E. & Bukau, B. (2002) Nature 419, 171–174. [DOI] [PubMed] [Google Scholar]
- 10.Gu, S. Q., Peske, F., Wieden, H. J., Rodnina, M. V. & Wintermeyer, W. (2003) RNA 9, 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ullers, R. S., Houben, E. N., Raine, A., ten Hagen-Jongman, C. M., Ehrenberg, M., Brunner, J., Oudega, B., Harms, N. & Luirink, J. (2003) J. Cell Biol. 161, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patzelt, H., Rudiger, S., Brehmer, D., Kramer, G., Vorderwulbecke, S., Schaffitzel, E., Waitz, A., Hesterkamp, T., Dong, L., Schneider-Mergener, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 14244–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier, R., Eckert, B., Scholz, C., Lilie, H. & Schmid, F. X. (2003) J. Mol. Biol. 326, 585–592. [DOI] [PubMed] [Google Scholar]
- 14.Jagath, J. R., Rodnina, M. V. & Wintermeyer, W. (2000) J. Mol. Biol. 295, 745–753. [DOI] [PubMed] [Google Scholar]
- 15.Lentzen, G., Dobberstein, B. & Wintermeyer, W. (1994) FEBS Lett. 348, 233–238. [DOI] [PubMed] [Google Scholar]
- 16.Rodnina, M. V. & Wintermeyer, W. (1995) Proc. Natl. Acad. Sci. USA 92, 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodnina, M. V., Savelsbergh, A., Matassova, N. B., Katunin, V. I., Semenkov, Y. P. & Wintermeyer, W. (1999) Proc. Natl. Acad. Sci. USA 96, 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan, J. J., Chen, J. C., Miao, Y., Shao, Y., Lin, J., Bock, P. E. & Johnson, A. E. (2003) J. Biol. Chem. 278, 18628–18637. [DOI] [PubMed] [Google Scholar]
- 19.Kusters, R., Lentzen, G., Eppens, E., van Geel, A., van der Weijden, C. C., Wintermeyer, W. & Luirink, J. (1995) FEBS Lett. 372, 253–258. [DOI] [PubMed] [Google Scholar]
- 20.Lill, R., Crooke, E., Guthrie, B. & Wickner, W. (1988) Cell 54, 1013–1018. [DOI] [PubMed] [Google Scholar]
- 21.Hesterkamp, T., Deuerling, E. & Bukau, B. (1997) J. Biol. Chem. 272, 21865–21871. [DOI] [PubMed] [Google Scholar]
- 22.Patzelt, H., Kramer, G., Rauch, T., Schonfeld, H. J., Bukau, B. & Deuerling, E. (2002) Biol. Chem. 383, 1611–1619. [DOI] [PubMed] [Google Scholar]
- 23.Maier, R., Scholz, C. & Schmid, F. X. (2001) J. Mol. Biol. 314, 1181–1190. [DOI] [PubMed] [Google Scholar]
- 24.Eisner, G., Koch, H. G., Beck, K., Brunner, J. & Muller, M. (2003) J. Cell Biol. 163, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatogawa, H. & Ito, K. (2002) Cell 108, 629–636. [DOI] [PubMed] [Google Scholar]
- 26.Liao, S., Lin, J., Do, H. & Johnson, A. E. (1997) Cell 90, 31–41. [DOI] [PubMed] [Google Scholar]