Abstract
The biosynthesis of pyrroloquinoline quinone (PQQ), a vitamin and redox cofactor of quinoprotein dehydrogenases, is facilitated by an unknown pathway that requires the expression of six genes, pqqA to -F. PqqC, the protein encoded by pqqC, catalyzes the final step in the pathway in a reaction that involves ring cyclization and eight-electron oxidation of 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic-acid to PQQ. Herein, we describe the crystal structures of PqqC and its complex with PQQ and determine the stoichiometry of H2O2 formation and O2 uptake during the reaction. The PqqC structure(s) reveals a compact seven-helix bundle that provides the scaffold for a positively charged active site cavity. Product binding induces a large conformational change, which results in the active site recruitment of amino acid side chains proposed to play key roles in the catalytic mechanism. PqqC is unusual in that it transfers redox equivalents to molecular oxygen without the assistance of a redox active metal or cofactor. The structure of the enzyme-product complex shows additional electron density next to R179 and C5 of PQQ, which can be modeled as O2 or H2O2, indicating a site for oxygen binding. We propose a reaction sequence that involves base-catalyzed cyclization and a series of quinone-quinol tautomerizations that are followed by cycles of O2/H2O2-mediated oxidations.
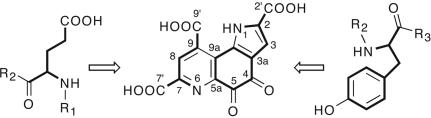
Pyrroloquinoline quinone [4,5-dihydro-4,5-dioxo-1H-pyrrolo-[2,3-f]quinoline-2,7,9-tricarboxylic acid; PQQ (Fig. 1)] is an aromatic, tricyclic ortho-quinone that serves as the redox cofactor for several bacterial dehydrogenases. Among the best known examples are methanol dehydrogenase and glucose dehydrogenase (1, 2). PQQ belongs to the family of quinone cofactors that has been recognized as the third class of redox cofactors following pyridine nucleotide- and flavin-dependent cofactors (3). Although plants and animals do not produce PQQ themselves, PQQ has invoked considerable interest because of its presence in human milk and its remarkable antioxidant properties (4–6). Recently, the first potential eukaryotic PQQ-dependent enzyme [aminoadipic 6-semialdehyde-dehydrogenase (AASDH; U26)] has been identified, indicating that PQQ may function as a vitamin in mammals as well (7).
Fig. 1.
Chemical structure of PQQ (4,5-dihydro-4,5-dioxo-1H-pyrrolo-[2,3-f]quinoline-2,7,9-tricarboxylic acid) with atom nomenclature. All carbon and nitrogen atoms of PQQ are derived from conserved tyrosine and glutamate residues of the PqqA peptide. R1 and R3 represent the N- and C-terminal portions of PqqA, respectively. R2 represent a three-amino-acid linker between Glu and Tyr.
Quinone cofactors are generally covalently linked to the polypeptide chain and derived posttranslationally from precursor amino acid residues encoded within their parental polypeptide chain. For example, in copper amine oxidases, topaquinone is formed by a “self-processing” oxidation of a specific Tyr residue in the presence of copper ion and molecular oxygen (8, 9). PQQ is distinct from the other quinone cofactors in that its biogenesis is independent of its site of action. PQQ is constructed from the amino acids glutamate and tyrosine, as shown in Fig. 1 (10, 11). The PQQ biosynthesis pathway in Klebsiella pneumoniae requires the expression of six genes, designated pqqABCDEF (12). PqqA encodes a 23-residue peptide with conserved glutamate and tyrosine residues that most likely serves as the precursor for PQQ biosynthesis (10, 13, 14). Transformed Escherichia coli cells carrying a plasmid that contains the PQQ operon from K. pneumoniae lacking pqqC, as well as a pqqC mutant strain of Methylobacterium extorquens AM1, accumulate an intermediate that can be converted to PQQ upon addition of PqqC (15, 16). These results demonstrate that PqqC catalyses the last step in PQQ biosynthesis.
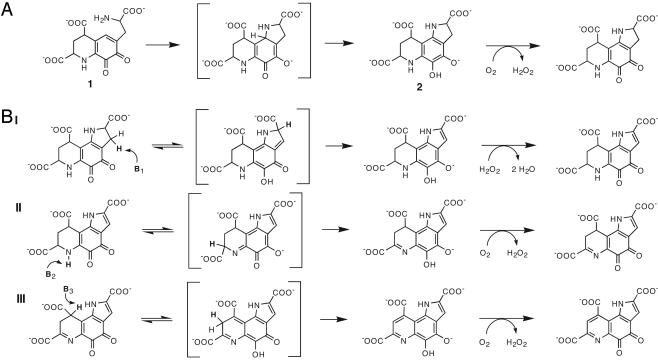
It has been shown that the PqqC reaction is accelerated in the presence of molecular oxygen (17) and that it requires NADPH and an uncharacterized activating factor (16) for sustained catalytic activity. Most recently, we elucidated the structure of the PqqC substrate, allowing the overall reaction catalyzed by PqqC to be inferred (18). The substrate is 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid (1 in Fig. 5A), a fully reduced derivative of PQQ, which has not yet undergone ring-cyclization at the pyrrole moiety. Herein, we present the structure of PqqC in complex with PQQ. Based on the enzyme active site environment, together with the stoichiometry of O2 uptake and H2O2 formation in a single-enzyme turnover, we propose a multistep catalytic mechanism for the reaction catalyzed by PqqC.
Fig. 5.
The proposed reaction mechanism of PqqC. (A) Ring cyclization of 3a-(2-amino-2-carboxy-ethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid (1) to an obligatory intermediate (2) and its subsequent oxidation. (B) Base-catalyzed tautomerization and the six electron H2O2/O2-mediated oxidation of 2. Tautomerization is initiated by enzyme-catalyzed proton abstraction at C3, N6, and C9, respectively. Oxidation of quinol intermediates presumably occurs by means of two single-electron transfer steps (not shown). The order of double-bond formation (I-III) is arbitrary and cannot be determined at this point. See Discussion for further details, including the identity of proposed catalytically important residues.
Methods
Expression and Purification of PqqC. PqqC from K. pneumoniae (NCBI accession no. X58778) was expressed in E. coli and purified as described elsewhere (19).
Single-Turnover Kinetics, Peroxide Formation, and Oxygen Uptake. The substrate for PqqC was purified from a pqqC knock-out mutant of M. extorquens AM1 as described elsewhere (17, 18). All experiments were performed in 0.1 M potassium phosphate buffer (pH 8.0) at 20°C, and all data fitting was done by using kaleidagraph (Synergy Software, Reading, PA).
Reactions were initiated by addition of substrate (12.4 μM) to a solution containing enzyme (5–100 μM). Reaction mixtures were quenched at designated time points by addition of HCl (0.5 M) and analyzed by reversed phase HPLC. A Beckman HPLC system equipped with a diode-array detection system was used, and substrate and product were separated by using a Vydac (Hesperia, CA) C18 column (5μ, 4.6 × 250 mm). A linear gradient of 0.1% trifluoroacetic acid from 0% to 80% CH3CN in 25 min was used. Substrate and product elute with retention times of 13.1 min and 14.6 min under these conditions, respectively. The amount of PQQ formed was determined by comparison with a standard curve of authentic material obtained from Fluka. The concentration of PQQ was determined spectrophotometrically in an aqueous solution at pH 7 (20). PQQ formation at low enzyme concentration (≤1 μM) was measured by using an enzymatic assay based on the activation of glucose dehydrogenase as described elsewhere (16).
The production of H2O2 was assessed with an enzymatic assay by using Amplex red (AR; 10-acetyl-3,7-dihydroxyphenoxazine) and horseradish peroxidase (HRP) obtained from Molecular Probes. The HRP-mediated oxidation of the colorless AR reagent by peroxide produces a chromophore, which can be measured either colorimetrically or by fluorimetry (21). Two different methods were used. In method A, H2O2 was measured in a continuous fashion by using 50 μM AR and 1 unit/ml HRP in samples containing 12.4 μM substrate and 90 μM PqqC. Reactions were initiated by the addition of substrate, and H2O2 formation was monitored spectrophotometrically. The amount of peroxide generated in these reactions was deduced by comparison with a standard curve generated from authentic H2O2 treated in the same manner as the samples above. In method B, aliquots were withdrawn from reaction mixtures containing substrate (12.4 μM) and PqqC (90 μM) and quenched in HCl (0.5 M) at designated time points. Samples from each quench were diluted 60-fold, and the amount of H2O2 produced was measured fluorimetrically upon treatment with AR/HRP and compared with an H2O2 standard (excitation at 530 nm, emission at 582 nm).
Oxygen consumption was measured by using a Clark oxygen electrode (YSI model 5300; YSI Inc., Yellow Springs, OH). Reactions were initiated by adding 5 μl of substrate (12.4 μM final concentration) to 995 μl of PqqC (90 μM) that had been equilibrated for 10–20 min at 20°C to obtain a stable baseline. The effective concentration of O2 in the protein solution was measured in a separate experiment by monitoring the consumption of oxygen during turnover of protocatechuic acid by protocatechuic dioxygenase as described elsewhere (22).
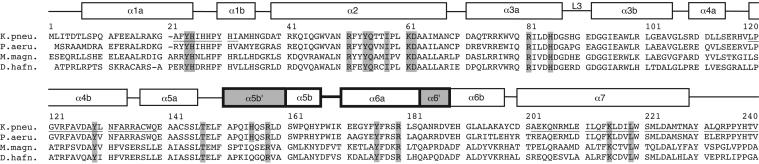
Sequence Alignment. The PqqC sequence alignment was obtained by using the program t-coffee (23) on a converged psi-blast (24) search of the microbial genome database at the National Center for Biotechnology Information (NCBI) with the PqqC sequence (gi 130800) from K. pneumoniae. Sequences were clustered at 80% identity and assembled into four different groups by using program cd-hit (25). A representative sequence of each cluster is shown to display conservation of active site residues.
Crystallization. Crystals were grown at 25°C by the sitting drop vapor diffusion method in droplets composed of one part protein solution (8 mg/ml in 20 mM Tris·HCl, at pH 8.0 and 1 mM DTT) and one part reservoir solution (1.2 M ammonium sulfate, Mes/NaOH at pH 6.0). The orthorhombic crystals are of space group P21212 and contain two molecules in the asymmetric unit. The PqqC/PQQ complex structure was obtained by soaking PqqC crystals in a crystallization solution containing 1 mM PQQ. For data collection, these crystals were transferred into cryobuffer [crystallization buffer with 25% (vol/vol) glycerol] and flash-cooled in liquid nitrogen.
Data Collection, Structure Solution, and Refinement. The PqqC structure was determined with a selenium-MAD experiment as described (19). Native and complex datasets were collected at beamline 9.1 of the Stanford Synchrotron Radiation Laboratory and processed with the hkl suite (26). The structure of the PqqC/PQQ complex was determined by molecular replacement by using molrep (27) with the native PqqC structure as a search model. Crystallographic refinement and model building was performed by using refmac5 (27) and o (28). The PqqC model comprises two protein monomers (residues 1–249) and 130 water molecules. In both chains, the region between residues 152 and 160 showed weak density and higher B-factors, indicative of partial disorder. The model for the PqqC/PQQ complex includes two PqqC monomers (residues 1–249), two PQQ molecules, two putative H2O2 molecules, and 119 water molecules. The native and complex structures were solved at 2.1 Å and 2.3 Å resolution, respectively. Further information concerning data collection and refinement statistics is available in Table 1, which is published as supporting information on the PNAS web site. Figures were drawn with pymol (DeLano Scientific, San Carlos, CA).
Coordinates. Coordinates and structure factors for PqqC and the PqqC/PQQ complex have been deposited with the Protein Data Bank (www.pdb.org/pdb) under accession codes 1OTV and 1OTW, respectively.
Results
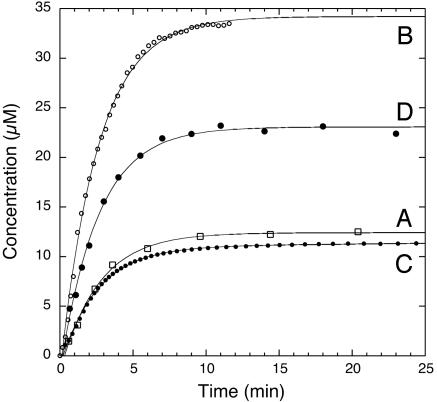
The PqqC Reaction. The PqqC reaction was analyzed for the production of PQQ by separation of substrate and product by HPLC. PqqC from K. pneumoniae produces 1 mol of PQQ per mol of enzyme in a single turnover. The reaction displays first-order kinetics for PQQ formation with regard to substrate under conditions of both excess and substoichiometric enzyme (data not shown). Even at low PqqC concentration (0.1 μM) in the presence of excess substrate, only one enzyme turnover could be detected by using a sensitive assay based on activation of the apo-form of glucose dehydrogenase. Under saturating conditions where all of the substrate is enzyme bound, the observed rate constant at 20°C is 0.38 (± 0.03) min–1 (Fig. 2, curve A).
Fig. 2.
Single-turnover kinetics of the PqqC reaction. Curve A illustrates formation of PQQ as determined by quantitative HPLC analysis after acid quenching at designated time points. Curve B shows dioxygen consumption measured polarographically using a Clark-type electrode. Curve C illustrates formation of H2O2 measured in a continuous coupled assay with AR/HRP. Curve D shows formation of H2O2 measured in a discontinuous assay using AR/HRP after acid quenching at designated time points. See Results for details.
To probe the putative role of O2 in the reaction, we looked for the consumption of molecular oxygen and production of hydrogen peroxide. As shown in Fig. 2, curve B, the rate of O2 uptake (kobs = 0.38 ± 0.02 min–1) is similar to the rate of PQQ formation. The measured stoichiometry of the reaction shows that the enzyme consumes ≈3 mol of O2 per mol of PQQ produced (2.77 ± 0.49). The production of H2O2 was measured by two different methods. With AR and HRP present in the PqqC assay mixture, approximately one equivalent (0.89 ± 0.12) of H2O2/PQQ was detected (Fig. 2, curve C). However, in a discontinuous assay, in which H2O2 was measured after acid denaturation of the protein, approximately two equivalents (1.86 ± 0.23) of H2O2/PQQ were formed (Fig. 2, curve D). Both methods yield identical first-order rate constants (within error) 0.40 ± 0.02 min–1 and 0.39 ± 0.03 min–1 for the continuous and discontinuous assays, respectively. The difference in the amount of H2O2 obtained by the two methods suggests that the enzyme tightly binds one equivalent of peroxide because denaturation of the protein is required to make the remaining peroxide accessible to detection. Note, however, that the data for the continuous assay (Fig. 2, curve C) were fit to two exponentials, in which the second slower and smaller amplitude phase may represent slow dissociation of the second equivalent of peroxide. In any case, the data show that PqqC consumes 3 mol of O2 per PQQ and generates 2 mol of H2O2 per PQQ in a single-turnover reaction.
Overall Structure of PqqC. PqqC is a 251-residue protein (molecular mass = 28.91 kDa) that forms a homodimer in solution, as evident from gel filtration experiments (data not shown). PqqC folds into a compact seven-helix bundle, with six circular aligned helices (α1, α2, α3, α4, α5, and α7), partly embracing a seventh hydrophobic helix (α6) (Figs. 3 and 4). Analysis of the PqqC structure shows that the seven α-helices provide the scaffold for an active site cavity (19). The cavity is lined with 42 mostly hydrophilic and aromatic residues that are highly conserved within PqqC proteins from different bacteria (Fig. 4). The cavity shows a distinct overall positive charge, measures 9 Å × 13 Å × 23 Å and embraces a molecular surface volume of 2,200 Å3 (29). Two openings connect it to the outside. A structural similarity search performed with the atomic coordinates of PqqC using the DALI server (30) yields human heme-oxygenase (31) with 9% sequence identity and a rms distance of 3.1 Å for the superposition of 194 Cα atoms.
Fig. 3.
Multiple sequence alignment of PqqC proteins from different bacterial sources. K.pneu., eubacterium K. pneumoniae (protein ID P27505); P.Aeru., Pseudomonas aeruginosa (protein ID NP_250677); M.magn., proteobacterium Magnetospirillum magnetotacticum (protein ID ZP_00052131), and D.hafn. the Gram-positive bacterium Desulfitobacterium hafniense (protein ID ZP_00101389). α-Helices and loops as observed in the structure of PqqC from K. pneumoniae are indicated with boxes above their sequences, where structural elements involved in the conformational change (α5b′, α5b, α6a, and α6′) are drawn in bold. Regions participating in the dimer interface are underlined. The helices formed upon ligand binding, as well as the active site residues coordinating the ligand, are highlighted in gray.
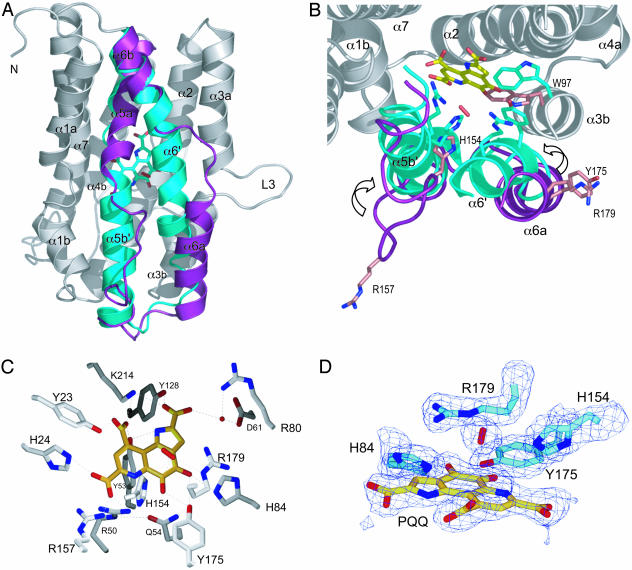
Fig. 4.
Active site analysis. (A) The conformational change upon formation of the PqqC/PQQ complex (see Results for further details). For comparison, structures of the unligated and ligated PqqC are superimposed and shown in ribbon representation (gray). The moving parts (residues 142–193) are depicted as colored ribbons: unligated (magenta) and ligated (cyan). The PQQ (cyan) and the putative H2O2 (red) molecule are shown in sticks. (B) Same as A but tilted 90° around the horizontal axis, showing the active site residues H154, R157, Y175, R179, and W97 as sticks in the open (magenta) and closed (cyan) structure. The PQQ molecule is depicted in yellow sticks, and the putative H2O2 is shown as a red stick. (C) The PqqC active site in the closed state with PQQ (yellow) and the putative H2O2 molecule (red). All PqqC active site residues, within a distance of 3.3 Å to PQQ, are shown in gray sticks. (D) The oxygen binding site. The PQQ molecule and active site residues H84, H154, Y175, and R179, as well as the putative H2O2 molecule, are depicted in sticks. The 2Fo-Fc electron density map is contoured at 1.5 σ.
Structure of the PqqC/PQQ Complex. The PqqC/PQQ complex structure was obtained by soaking PqqC crystals in a crystallization solution containing 1 mM PQQ. The structure shows that PQQ binds in the center of the active site cavity, accompanied by a large conformational change in the protein. The structural rearrangement is almost entirely executed in the region of helices α5a to α6b (residues 142–193) and includes the elongation of helix α5b to form α5b′ and the fusion of helices α6a and α6b into one long helix, α6′. Starting with α5a, a detailed description of the conformational change includes the following rearrangements depicted in Figs. 3 and 4. Helix α5a accommodates small shifts (≈0.7 Å) in the main-chain atoms, resulting in a slight compression of the helix. The coil region between residues 151 and 158 folds into α5b′ and closes the main entrance to the active site cavity. This rearrangement shifts H154 and R157 directly into the active site, ready to coordinate the carboxylic group C7′ of PQQ. The main chain in the region between residues 159 and 169, (from α5b to the beginning of α6a) shifts ≈1.8 Å upwards, keeping the overall main chain and side-chain conformation unchanged. The largest shift occurs in the region of helices α6a and α6b (residues 170–187, Figs. 3 and 4 A and B). Helix α6a rotates ≈90° around its long axis and shifts ≈3.5 Å upwards, 2 Å toward α5b′, and 4 Å toward the center of the molecule. This shift is facilitated by loop L6 winding up in a spring-like manner to connect helices α6a and α6b into one long helix, α6′. The Cα of Y175 moves 6.9 Å from a solvent exposed location to a position directly in the center of the molecule where it interacts with the oxygen atoms of C4 and C5 of the ligand. The new position forces the side-chain of W97 into another rotamer that is stabilized by H-bonds between the main-chain nitrogen of Y175 and a water molecule, which itself interacts with the carbonyl oxygen of A96. The same holds for R179, which moves ≈7.7 Å to interact with O4 of the substrate and the sidechains of E147 and D186. Thus, PQQ recognition in PqqC exhibits a classical induced fit mechanism, resulting in the ligand being completely buried from the solvent.
Active Site Interactions with the Ligand. PQQ binds in the center of the active site cavity coordinated by 18 highly conserved residues (Y23, H24, R50, Y53, Q54, I57, K60, D61, R80, H84, Y128, T146, H154, R157, Y175, R179, K214, and L218; see Fig. 4). Fig. 4C shows the coordination sphere of PQQ. The four residues H154, R157, Y175, and R179 move into the active site upon substrate binding (Fig. 4B). Critical sites of interaction between ligand and protein are the three carboxyl groups, the amino group in the pyrrole-ring N1, and the two quinone oxygens O4 and O5. The electron density maps at 2.3 Å resolution for the ligand show the tricyclic ring system and especially the C5—O5 bond in an approximately planar conformation. The three carboxylate groups of the ligand are twisted out of the plane of the ring system and form polar interactions with several active site residues. R157 interacts with the carboxyl group COO7′, which is also coordinated by two other positively charged residues, H24 and R50. Part of this network is a 12-Å-long and 3-Å-wide tunnel that bends from the COO7′ carboxyl group of PQQ, via R157 and five water molecules, out to the protein surface. Y175 coordinates the oxygen atoms of C5 and C4 of the ligand and creates another extended hydrogen bonding network with Q54 and R50. This site is connected to the outside via a 3 Å wide tunnel that goes 9 Å from PQQ-O5′ out to the protein surface near loop L3. The tube, blocked by water O31, is guided by Y175, R179, H84, Q182, and S178, all of which are in hydrogen bonding distance to each other. These multiple interactions lock the ligand into the active site and create a microenvironment that facilitates the complex enzymatic reaction.
An interesting feature of the PqqC/PQQ structure is the density located 3 Å above the C5 atom of PQQ (Fig. 4D). It is in close contact to R179 and H154 located at a distance of 2.8 Å and 3.3 Å, respectively. The elliptically shaped density is too strong to be modeled as a single water molecule, resulting in unrealistically low B-factors and strong positive density (3σ peak in 2FoFc map). Furthermore, it is too weak and does not have a sphere of coordinating residues as expected for a small ion, such as Na+ or Cl–, which are present in the crystallization buffer. Interestingly, the location of this density next to the reactive C5 of PQQ coincides with the substrate position in respect to PQQ in PQQ enzymes like glucose dehydrogenase and methanol dehydrogenase (32), strongly suggesting an important role in oxidation. Therefore and according to the PqqC reaction, which is oxygen dependent and produces hydrogen peroxide, the density most likely represents an O2 or H2O2 molecule, the latter being more probable. Modeling O2 or H2O2 into the density yields B-factors around 28 Å2 consistent with the range of B-factors displayed by surrounding atoms. Although these observations raise the possibility of a trapped O2 or H2O2 molecule, this interpretation clearly requires higher resolution data and additional experimental evidence. Currently there is no crystal structure of a protein with O2 or H2O2 bound at a non-metal site, but recent kinetic studies on several variants of the quino-protein copper amine oxidase have provided evidence for specific binding of O2 to a side-chain-generated pocket (33).
Discussion
PqqC facilitates cyclization and oxidation of 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid to PQQ in a reaction that occurs in the absence of any apparent cofactor. The product is bound and oriented by the fourteen conserved active site residues forming the positively charged active site cavity (Fig. 4C). By inference, substrate binding is concluded to induce a large conformational change, which closes the cavity and moves H154, R157, Y175, and R179 into the coordination sphere of the substrate to form an extended network of hydrogen bonds.
In our proposed mechanism, the first chemical step is the formation of the tricyclic ring system to yield 2 as shown in Fig. 5A. Cyclization occurs by a 1,4-addition of the primary amine to the conjugated ring structure, which may be facilitated by polar interactions of the substrate with several active site residues. Proton abstraction from N1 of the substrate and its subsequent activation for addition to C1a may be catalyzed by Y53, which is in close proximity to K214 (3.6 Å), making this a putative catalytic diad. Alternatively, the substrate may bind as the neutral amine, and the enzyme may simply provide a scaffold for the cyclization to occur, an event that should proceed rapidly once the amino group is deprotonated.
The next step involves the removal of eight electrons and eight protons from 2 to form PQQ, which means that PqqC needs to function as a dehydrogenase/desaturase. The PqqC/PQQ complex structure does not contain a redox active metal or cofactor, which raises the question of how oxidation is achieved. We propose a mechanism that leads to double bond formation by forming quinol intermediates, which are capable of the direct transfer of electrons to molecular oxygen and/or hydrogen peroxide (Fig. 5B) (see below). As shown, this mechanism involves sequential base-catalyzed tautomerizations to produce a reduced quinol from the oxidized quinone, with the introduction of three double bonds. The proposed mechanism predicts multiple proton abstractions occurring at different positions on the substrate. The structure of PqqC with PQQ identifies a number of amino acid residues important for catalysis. The product complex shows three histidine residues ideally placed for general base catalysis. These are H84, H24, and H154, which are all within 5 Å from C3, C9, and N6, respectively. Residues Y53/K214 could also facilitate proton abstraction because the oxygen of Y53 is within 4 Å of C9. Another important residue is Y175, which is within 3 Å of both O4 and O5 and is part of a hydrogen-bonding network that extends to the surface of the protein as discussed above. Y175 is therefore a likely candidate for the shuttling of protons to and from O4 and O5 during the postulated tautomerization and oxidation reactions. Unstable intermediates, which may collapse spontaneously due to the large driving force for aromatization of the quinol, are shown in brackets in the mechanism in Fig. 5B. Each time the quinol form is reached, it will transfer electrons to O2/H2O2, possibly in successive steps to produce a superoxide-semiquinone intermediate (not shown). We cannot determine the order of the proposed tautomerization steps at this point, and they are depicted in an arbitrary fashion in Fig. 5B.
The biochemical data indicate that molecular oxygen is not the exclusive oxidant in the reaction. The data show that three equivalents of O2 are consumed and two equivalents of H2O2 produced per mole of PQQ formed, respectively (Fig. 2). The stoichiometry can be explained if one equivalent of O2 is reduced by four electrons to form two equivalents of water whereas the remaining two equivalents of O2 undergo two-electron reduction to H2O2. Because the observed first-order rate constants for PQQ formation, oxygen consumption, and peroxide formation, respectively, are very similar, the first oxidation step seems to be the rate-limiting step in the overall reaction. Also, the release of peroxide after the first oxidation step is proposed to be slow compared with the rate of tautomerization, which would explain its complete reduction to water. The apparent slow release of peroxide at the end of the catalytic cycle, as implied by the different stoichiometries under continuous turnover vs. acid quenching (Fig. 2, curves C and D), may in part explain why the enzyme undergoes only a single turnover under in vitro conditions. Another contributing factor may be a slow release of bound PQQ, and experiments will be necessary to address these issues.
The crystal structure of the enzyme–product complex shows additional electron density inside the sealed cavity, next to the reactive C5 of PQQ. This location strongly points to a site of interaction between dioxygen and substrate. Because PQQ is present in the complex, the density may represent a H2O2 molecule. Although H2O2 was not included in the crystallization buffer, it could have been formed by means of a reaction with PQQH2. Although the crystals were soaked with PQQ, reduction of the cofactor to PQQH2 may have resulted from the presence of DTT in the buffer and/or from the electron beam of the synchrotron radiation. Thiols have been reported to be very efficient in reducing PQQ and derivatives thereof (34). Further support for H2O2 bound in the crystal comes from the biochemical studies discussed above, in which peroxide seems tightly bound to the enzyme in solution. We conclude that the structure most likely represents a ternary product complex, providing valuable information regarding the oxidative reaction. First, because the molecule is >3.0 Å away from both C4 and C5 of PQQ, the possibility of a covalent hydroperoxy intermediate is considered unlikely. Second, the close proximity of two basic residues, R179 (2.8 Å) and H154 (3.3 Å), would provide an ideal electrostatic environment to stabilize a superoxide anion produced upon electron transfer from the substrate. The formation of superoxide is often the rate-limiting step in these types of reactions, and electrostatic facilitation of superoxide formation seems to be emerging as a common theme in enzymes that reduce dioxygen (35). For example, the flavoprotein glucose oxidase uses a protonated histidine residue to reduce the activation barrier by ≈103-fold for electron transfer from reduced flavin to molecular oxygen (36). Finally, the positive electrostatic potential in the active site is expected to lower the pKa of the C4-C5 quinol, increasing the oxidation potential toward O2. The proposed structure-based mechanism introduces an “information-rich” framework for future investigations using site-directed mutagenesis and kinetic and spectroscopic analyses.
The reaction catalyzed by PqqC associates the enzyme with a rare family of cofactor-free oxidases and oxygenases (37). These enzymes include urate oxidase (38), 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (39), and ActVA-Orf6 monooxygenase (40). The common feature among these enzymes is their action on substrates that are well suited for direct reaction with dioxygen, i.e., in the absence of an organic cofactor or metal ion. It is known that reduced PQQH2 in aqueous solution is susceptible toward oxygen and produces H2O2 very rapidly (41).
The present x-ray crystal structures, in combination with biochemical data, provide a structural study of an enzyme involved in the PQQ-biosynthesis pathway. Our study identifies the final step in PQQ biosynthesis as a multistep reaction that includes cyclization and an overall eight-electron oxidation of 3a-(2-amino-2-carboxyethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid to PQQ. The structure of PqqC with PQQ identifies a number of important interactions between enzyme and product. These include K214, Y53, H24, H84, H154, and Y175 as possible participants in the proposed general base catalysis that leads to ring cyclization and tautomerization. Finally, a plausible dioxygen binding site is located next to H154, R179, and C5 of PQQ. Our data suggest a mechanism of oxidation whereby electrons are transferred directly to molecular oxygen and provide a structural foundation for further study of the mechanism of dioxygen activation without the assistance of a redox-active metal or cofactor.
Supplementary Material
Acknowledgments
We thank Constantina Bakolitsa and the staff at the Stanford Synchrotron Radiation Laboratory for help with data collection, and Jose Maria de Pereda and Charles Perrin for helpful suggestions. H.T. wants to thank Kimitoshi Sakamoto, Kazunobu Matsushita, and Osao Adachi for advice and support. R.S. was supported by Austrian Science Fund Fellowship J2209-B04, J.P.K. by National Institutes of Health Grant GM39296, and O.T.M. by a postdoctoral fellowship from the Miller Institute for Basic Research in Science, University of California, Berkeley.
Abbreviations: PQQ, pyrroloquinoline quinone; AR, Amplex red; HRP, horseradish peroxidase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1OTV and 1OTW).
References
- 1.Anthony, C. (2001) Antioxid. Redox Signal. 3, 757–774. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin, P. M. & Anthony, C. (1998) Adv. Microb. Physiol. 40, 1–80. [DOI] [PubMed] [Google Scholar]
- 3.Duine, J. A. (2001) Chem. Rec. 1, 74–83. [DOI] [PubMed] [Google Scholar]
- 4.He, K., Nukada, H., Urakami, T. & Murphy, M. P. (2003) Biochem. Pharmacol. 65, 67–74. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell, A. E., Jones, A. D., Mercer, R. S. & Rucker, R. B. (1999) Anal. Biochem. 269, 317–325. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg, F., Stites, T. E., Anderson, P., Storms, D., Chan, I., Eghbali, S. & Rucker, R. (2003) Exp. Biol. Med. (Maywood) 228, 160–166. [DOI] [PubMed] [Google Scholar]
- 7.Kasahara, T. & Kato, T. (2003) Nature 422, 832. [DOI] [PubMed] [Google Scholar]
- 8.Kim, M., Okajima, T., Kishishita, S., Yoshimura, M., Kawamori, A., Tanizawa, K. & Yamaguchi, H. (2002) Nat. Struct. Biol. 9, 591–596. [DOI] [PubMed] [Google Scholar]
- 9.Dove, J. E. & Klinman, J. P. (2001) Adv. Protein Chem. 58, 141–174. [DOI] [PubMed] [Google Scholar]
- 10.Houck, D. R., Hanners, J. L., Unkefer, C. J., van Kleef, M. A. & Duine, J. A. (1989) Antonie Van Leeuwenhoek 56, 93–101. [DOI] [PubMed] [Google Scholar]
- 11.Unkefer, C. J., Houck, D. R., Britt, B. M., Sosnick, T. R. & Hanners, J. L. (1995) Methods Enzymol. 258, 227–235. [DOI] [PubMed] [Google Scholar]
- 12.Meulenberg, J. J., Sellink, E., Riegman, N. H. & Postma, P. W. (1992) Mol. Gen. Genet. 232, 284–294. [DOI] [PubMed] [Google Scholar]
- 13.van Kleef, M. A. & Duine, J. A. (1988) Biofactors 1, 297–302. [PubMed] [Google Scholar]
- 14.Goosen, N., Huinen, R. G. & van de Putte, P. (1992) J. Bacteriol. 174, 1426–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velterop, J. S., Sellink, E., Meulenberg, J. J., David, S., Bulder, I. & Postma, P. W. (1995) J. Bacteriol. 177, 5088–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyama, H., Fukumoto, H., Saeki, M., Matsushita, K., Adachi, O. & Lidstrom, M. E. (2002) Biochem. Biophys. Res. Commun. 299, 268–272. [DOI] [PubMed] [Google Scholar]
- 17.Toyama, H., Chistoserdova, L. & Lidstrom, M. E. (1997) Microbiology 143, 595–602. [DOI] [PubMed] [Google Scholar]
- 18.Magnusson, O. Th., Toyama, H., Saeki, M., Schwarzenbacher, R. & Klinman, J. P. (2004) J. Am. Chem. Soc. 126, 5342–5343. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzenbacher, R., Stenner-Liewen, F., Stenner, H., Reed, J. C. & Liddington, R. C. (2004) Proteins, in press. [DOI] [PubMed]
- 20.Duine, J. A., Frank, J. & Jongejan, J. A. (1987) Adv. Enzymol. 59, 169–212. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, M., Diwu, Z., Panchuk-Voloshina, N. & Haugland, R. P. (1997) Anal. Biochem. 253, 162–168. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, B., Dove, J. E. & Klinman, J. P. (2000) Biochemistry 39, 3699–3707. [DOI] [PubMed] [Google Scholar]
- 23.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 24.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W., Jaroszewski, L. & Godzik, A. (2001) Bioinformatics 17, 282–283. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 27.CCP4. (1994) Acta Crystallogr. D 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 28.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 29.Liang, J., Edelsbrunner, H. & Woodward, C. (1998) Protein Sci. 7, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 31.Schuller, D. J., Wilks, A., Ortiz de Montellano, P. R. & Poulos, T. L. (1999) Nat. Struct. Biol. 6, 860–867. [DOI] [PubMed] [Google Scholar]
- 32.Oubrie, A., Rozeboom, H. J., Kalk, K. H., Olsthoorn, A. J., Duine, J. A. & Dijkstra, B. W. (1999) EMBO J. 18, 5187–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto, Y. & Klinman, J. P. (2002) Biochemistry 41, 13637–13643. [DOI] [PubMed] [Google Scholar]
- 34.Itoh, S., Kato, N., Ohshiro, Y. & Agawa, T. (1985) Chem. Lett. 135–136.
- 35.Klinman, J. P. (2001) J. Biol. Inorg. Chem. 6, 1–13. [DOI] [PubMed] [Google Scholar]
- 36.Roth, J. P. & Klinman, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fetzner, S. (2002) Appl. Microbiol. Biotechnol. 60, 243–257. [DOI] [PubMed] [Google Scholar]
- 38.Kahn, K. & Tipton, P. A. (1998) Biochemistry 37, 11651–11659. [DOI] [PubMed] [Google Scholar]
- 39.Fischer, F. & Fetzner, S. (2000) FEMS Microbiol. Lett. 190, 21–27. [DOI] [PubMed] [Google Scholar]
- 40.Sciara, G., Kendrew, S. G., Miele, A. E., Marsh, N. G., Federici, L., Malatesta, F., Schimperna, G., Savino, C. & Vallone, B. (2003) EMBO J. 22, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh, S., Ohshiro, Y. & Agawa, T. (1986) Bull. Chem. Soc. Jpn. 59, 1911–1914. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.