Abstract
The relationship between with-no-lysine [K] kinase 4 (WNK4) gene polymorphisms and hypertension has been widely investigated, However, the studies yielded contradictory results. To evaluate these inconclusive findings comprehensively, we therefore performed a meta-analysis. Ten articles encompassing 16 independent case-control studies with 6089 hypertensive cases and 4881 normotensive controls were selected for this meta-analysis. Four WNK4 gene polymorphisms were identified (G1155942T, G1156666A, T1155547C, and C6749T). The results showed statistically significant associations of G1155942T polymorphism (allelic genetic model: odds ration or OR = 1.62, 95% confidence interval or CI: 1.11–2.38, P = 0.01; dominant model: OR = 1.85, 95% CI: 1.07–3.19, P = 0.03) and C6749T polymorphism (allele contrast: OR = 2.04, 95% CI: 1.60–2.59, P<0.01; dominant model: OR = 2.04, 95%CI: 1.59–2.62, P<0.01; and homozygous model: OR = 5.01, 95% CI: 1.29–19.54, P = 0.02) with hypertension risk. However, neither C1155547T nor G1156666A was associated significantly with hypertension susceptibility. In conclusion, this meta-analysis suggested that WNK4 G1155942T and C6749T gene polymorphisms may contribute to the susceptibility and development of hypertension. Further well-designed studies with larger sample size are required to elucidate the association of WNK4 gene multiple polymorphisms with hypertension risk.
Hypertension is a worldwide public health problem; its overall prevalence is approximately 30–45% in the general population with a steep increase with aging1. The number of adults with hypertension are projected to be increased by 60% about 1.56 billion in 20252. Increased blood pressure is considered to be a major risk factor for cardiovascular, cerebrovascular and renal disease and it is closely related to both cardiovascular and cerebrovascular endpoints, including heart failure, myocardial infarction, and stroke3.According to the epidemiological data, nearly 16.5% of all deaths worldwide are attributed to hypertension, including 51% of deaths by strokes and 45% of deaths by coronary heart disease4. As the major controlled risk factor, prevention and treatment of hypertension might be of great significance to restrain the morbidity and mortality from cardio-cerebrovascular disease. However, the pathogenesis of hypertension is intricate and still has not been thoroughly understood. Epidemiological studies have suggested that 30–50% of blood pressure (BP) variation within a population is determined by the genetic origin5, however, the genes underlying the BP regulation have not been clearly elucidated.
BP is regulated by a complicated network, including vascular, renal, neuronal and endocrine mechanisms and is also influenced by environmental factors, genetic factors, and multifactorial interactions6. It is well established that sodium intake is positively associated, while potassium intake is inversely associated, with the hypertension prevalence7,8,9. Nonetheless, the BP response to dietary sodium and potassium intake seems to vary considerably among individuals because many individuals taking as much as 40 g of NaCl per day do not suffer from hypertension7.
Up to now, a serial of previously unrecognized kinases interacting each other in the distal nephron have been identified as playing important roles in sodium, potassium, and BP regulation. WNKs (with-no-lysine [K] kinases) are a novel family of serine-threonine protein kinases. They are different from other usual protein kinases in subdomain II of WNKs that lacks the common conserved catalytic lysine residue that is crucial for binding to ATP and protein phosphorylation10. There are four WNKs in mammalian11,12. Mutations of WNK1 and WNK4 are considered to be related with pseudohypoaldosteronism type II (PHA II), an autosomal dominant disease featuring hypertension, hyperkalemia, hyperchloremia and metablic acidosis13,14. Previous studies revealed that the pathphysiology of hypertension caused by mutations of WNK1 and WNK4 were involved in regulation of diverse ion transporters and channels of the distal nephron15,16,17,18. Mutations of WNK1 and WNK4 led to deregulated renal sodium absorption, potassium secretion, thereby giving rise to the PHA II phenotype19. Disease-causing mutations in WNK1 are large deletions of the first intron that lead to the increased expression of wildtype WNK1, mutations in the WNK4 gene are missense mutation and cluster within the highly conservative coding sequence outside the kinase domain11,20.
Subsequent numerous epidemiological studies were conducted to evaluate the relationship between WNKs gene variation and hypertension susceptibility, since Wilson FH et al. reported their potential relevance each other by studying a PHA II kindred11. Putku M et al. identified a significant association of WNK1 AluYb8 insertion with BP variation in the Estonian HYPEST cohort study and confirmed this result by a meta-analysis of three independent European samples21. Consistently, similar effects of WNK1 common and rare single nucleotide polymorphisms (SNPs) and haplotypes on BP have been found in both general population and hypertensives21,22,23,24. However, the association of WNK4 polymorphisms with hypertension susceptibility is ambiguous on account of the multiple potentially related gene loci and inconsistent results have been reported in different studies25,26,27,28. In addition, no meta-analysis has yet been conducted to assess the relationship between WNK4 polymorphisms and hypertension risk as we know. Therefore, in order to estimate the relationship between WNK4 gene polymorphisms and hypertension, especially G1155942T, G1156666A, T1155547C, and C6749T, we performed this meta-analysis.
Results
Characteristics of the studies
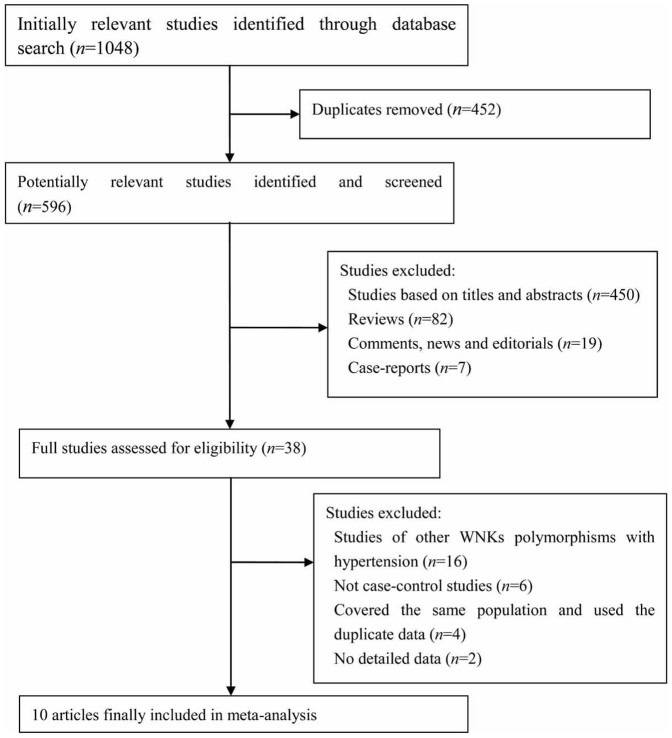
According to the inclusion criteria, 10 eligible studies25,26,28,29,30,31,32,33,34,35 were finally analyzed. The flow diagram of selection process of eligible studies is presented in Figure 1. Among all eligible studies, three articles by Erlich PM et al.25, Han Y et al.29, and Lu M et al.32 contained more than one study group, they were considered separately in the pooled analysis. Therefore, 16 independent case-control studies were finally collected in this meta-analysis to evaluate the relationship between the WNK4 gene polymorphisms and hypertension including 6089 cases and 4881 controls. Main characteristics of these included studies are shown in Table 1, and the genotype distribution, allele frequencies and Hardy-Weinberg equilibrium (HWE) of controls are presented in Table 2. Genotype distributions of examined polymorphisms in controls were consistent with HWE for each study. By comparing the data of genotype distributions and allele frequencies, we found that one study by Lu M et al.32 which concerned with the G1155942T and C1155547T polymorphisms in a Chinese minority ethnic population showed quite different findings from the others. To avoid the influence of the overall results by each single study and get more cautious and credible results, we performed sensitive analysis by omitting each study sequentially as well. The main results of this meta-analysis of the relationship between the WNK4 SNPs and hypertension risk are summarized in Table 3.
Figure 1. Flow diagram of the study selection procedure used for this meta-analysis of WNK4 Polymorphisms and hypertension.
Table 1. Characteristics of the eligible studies included in this meta-analysis.
| Diagnostic criteria(mmHg) | Mean blood pressure(mm Hg) | Mean age (years) | Gender (Female(%)) | BMI (kg/m2) | Cholesterol (mmol/L) | Triglycerides (mmol/L) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||||||||||||||
| Studied SNP and First author | Year | Country | Ethnicity | Genotyping method | Source of controls | Hypertensive | Normotensive | Systolic BP | Diastolic BP | Systolic BP | Diastolic BP | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls |
| G1155942T (Ala 589 Ser;exon8) | |||||||||||||||||||||
| Lv JY | 2003 | China | Caucasian | RT-PCR | NA | ≥140/90 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Erlich PM et al.(Population 2) | 2003 | America | Negroid | PCR/Homogeneous MassEXTEND | HB | ≥140/90a,b | <130/80c | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sun ZJ et al. | 2009 | China | Mongloid | PCR-RFLP | PB | ≥140/90a,b | <140/90 | 156.12 ± 25.56 | 96.88 ± 11.92 | 113.23 ± 12.73 | 73.78 ± 8.99 | 51.45 (12.09) | 49.50(13.97) | 141(54.4) | 136(57.9) | 25.37(6.54) | 22.02(3.57) | 5.09(1.02) | 4.39(1.08) | 1.68(1.36) | 1.06(0.86) |
| Lu M et al. | 2009 | China | Mongloid | TaqMan | PB | ≥140/90a,b | ≤ 120/80d | 159.7 ± 22.1 | 96.7 ± 13.2 | 112.2 ± 10.2 | 71.5 ± 6.8 | 49.0(7.5) | 48.7(10.8) | 334(73.7) | 291(64.0) | 28.3 (5.3) | 25.0(3.9) | 4.7(1.1) | 4.4(1.0) | 1.7(1.1) | 1.3(1.0) |
| Cao FF et al. | 2010 | China | Mongloid | TaqMan | PB | ≥140/90a,e | <140/90c | NA | NA | NA | NA | 48.86(10.47) | 46.17(10.18) | 309(54.9) | 207(59.8) | 26.36(4.20) | 24.49(3.74) | 5.17(1.52) | 4.72(1.59) | 1.39(0.94) | 1.17(0.86) |
| G1156666A (intron10) | |||||||||||||||||||||
| Erlich PM et al.(Population 1) | 2003 | America | Caucasian | PCR/Homogeneous MassEXTEND | HB | ≥140/90a,b | <130/80c | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Erlich PM et al.(Population 2) | 2003 | America | Negroid | PCR/Homogeneous MassEXTEND | HB | ≥140/90a,b | <130/80c | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Speirs HJ et al. | 2004 | Australia | Caucasian | Homogeneous MassEXTEND | PB | ≥140/90 | <130/90 | 173 ± 25 | 106 ± 15 | 119 ± 10 | 72 ± 8 | 52(12) | 44(12) | 112(61) | 92(42) | 27(5) | 25(4) | 4.9(0.1) | 4.8(0.07) | 1.9(0.1) | 1.3(0.05) |
| Zhang LP et al. | 2008 | China | Mongloid | PCR-RFLP | PB | ≥140/90b | <140/90 | 160.2 ± 18.6 | 96.1 ± 15.2 | 113.9 ± 13.2 | 75.0 ± 11.2 | 62.8(4.3) | 61.8(6.6) | 106(55.5) | 101(58.4) | 26.5(5.6) | 24.4(4.1) | 5.1(1.5) | 4.8(1.9) | 1.3(0.8) | 1.1(0.6) |
| Lu M et al. | 2009 | China | Mongloid | TaqMan | PB | ≥140/90a,b | ≤120/80d | 159.7 ± 22.1 | 96.7 ± 13.2 | 112.2 ± 10.2 | 71.5 ± 6.8 | 49.0(7.5) | 48.7(10.8) | 334(73.7) | 291(64.0) | 28.3(5.3) | 25.0(3.9) | 4.7(1.1) | 4.4(1.0) | 1.7(1.1) | 1.3(1.0) |
| T1155547C (Ala 535 Ala; exon7) | |||||||||||||||||||||
| Erlich PM et al.(Population 2) | 2003 | America | Negroid | PCR/Homogeneous MassEXTEND | HB | ≥140/90a,b | <130/80c | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Lu M et al. | 2009 | China | Mongloid | TaqMan | PB | ≥140/90a,b | ≤120/80d | 159.7 ± 22.1 | 96.7 ± 13.2 | 112.2 ± 10.2 | 71.5 ± 6.8 | 49.0 (7.5) | 48.7(10.8) | 334(73.7) | 291(64.0) | 28.3(5.3) | 25.0(3.9) | 4.7(1.1) | 4.4(1.0) | 1.7(1.1) | 1.3(1.0) |
| Cao FF et al. | 2010 | China | Mongloid | TaqMan | PB | ≥140/90a,e | <140/90c | NA | NA | NA | NA | 48.86(10.43) | 46.31(10.14) | 307(55.2) | 206(60.4) | 26.36(4.20) | 24.47(3.78) | 5.18(1.52) | 4.72(1.60) | 1.38(0.94) | 1.17(0.86) |
| rs9916754 (C6749T; extron7) | |||||||||||||||||||||
| Wang F | 2008 | China | Mongloid | TaqMan | PB | ≥140/90a,b,e | <140/90c | 165.71 ± 22.50 | 103.57 ± 12.00 | 118.43 ± 12.27 | 77.22 ± 6.99 | 48.86(10.43) | 46.31(10.14) | 307(55.2) | 206(60.4) | 26.36(4.20) | 24.47(3.78) | 5.18(1.52) | 4.72(1.60) | 1.38(0.94) | 1.17(0.86) |
| Han Y et al.(The first study) | 2011 | China | Mongloid | PCR-RFLP | PB | ≥140/90a,b,e | <130/85 | 154.2 ± 20.0 | 93.9 ± 11.2 | 119.0 ± 10.8 | 76.5 ± 6.9 | 58.3(8.4) | 56.3(7.9) | 533(65) | 500(64.8) | 25.7(3.5) | 24.0(3.3) | 5.46(1.11) | 4.98(1.08) | NA | NA |
| Han Y et al.(The second study) | 2011 | China | Mongloid | PCR-RFLP | PB | ≥140/90a,b,e | <130/85 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
NA: not available; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; RT-PCR: real-time PCR; HB: hospital based; PB: population based; BMI: Body Mass Index; a: and/or under antihypertensive treatment currently; b: and excluded secondary hypertension; c: and not being treated with antihypertensive medications; d: and not been diagnosed as hypertensive previously; e: and/or diagnosed as hypertensive in the past.*The continuous variables are expressed as means ± SD.
Table 2. Distribution of genotype and allele frequencies of each study included in this meta-analysis.
| Polymorphisms and First author | Year | Sample size | Genotypes | Allele frequencies (%) | HWE(P) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |||||||||
| G1155942T (Ala 589 Ser;exon8) | GG | GT | TT | GG | GT | TT | G | T | G | T | ||||
| Lv JY | 2003 | 896 | 451 | 864 | 30 | 2 | 447 | 4 | 0 | 98.1 | 1.9 | 99.6 | 0.4 | 0.92 |
| Erlich PM et al.(Population 2) | 2003 | 91 | 81 | 55 | 30 | 6 | 48 | 31 | 2 | 76.9 | 23.1 | 78.4 | 21.6 | 0.24 |
| Sun ZJ et al. | 2009 | 259 | 235 | 136 | 112 | 11 | 149 | 77 | 9 | 74.1 | 25.9 | 79.8 | 20.2 | 0.81 |
| Lu M et al. | 2009 | 451 | 454 | 0 | 24 | 427 | 0 | 31 | 423 | 2.7 | 97.3 | 3.4 | 96.6 | 0.45 |
| Cao FF et al. | 2010 | 563 | 346 | 526 | 36 | 1 | 338 | 8 | 0 | 96.6 | 3.4 | 98.8 | 1.2 | 0.83 |
| G1156666A (intron10) | GG | GA | AA | GG | GA | AA | G | A | G | A | ||||
| Erlich PM et al.(Population 1) | 2003 | 165 | 91 | 124 | 39 | 2 | 79 | 11 | 1 | 87 | 13 | 92.9 | 7.1 | 0.40 |
| Erlich PM et al.(Population 2) | 2003 | 113 | 94 | 109 | 4 | 0 | 90 | 4 | 0 | 98.2 | 1.8 | 97.9 | 2.1 | 0.83 |
| Speirs HJ et al. | 2004 | 184 | 219 | 152 | 29 | 3 | 176 | 43 | 0 | 90.5 | 9.5 | 90.2 | 9.8 | 0.11 |
| Zhang LP et al. | 2008 | 191 | 173 | 168 | 21 | 2 | 159 | 14 | 0 | 93.5 | 6.5 | 96 | 4 | 0.58 |
| Lu M et al. | 2009 | 452 | 452 | 415 | 36 | 1 | 399 | 53 | 0 | 95.8 | 4.2 | 94.1 | 5.9 | 0.19 |
| C1155547T (Ala 535 Ala; exon7) | CC | CT | TT | CC | CT | TT | C | T | C | T | ||||
| Erlich PM et al.(Population 2) | 2003 | 89 | 79 | 53 | 30 | 6 | 47 | 30 | 2 | 76.4 | 23.6 | 78.5 | 21.5 | 0.27 |
| Lu M et al. | 2009 | 451 | 454 | 0 | 24 | 427 | 0 | 31 | 423 | 2.7 | 97.3 | 3.4 | 96.6 | 0.45 |
| Cao FF et al. | 2010 | 556 | 341 | 518 | 37 | 1 | 333 | 8 | 0 | 96.5 | 3.5 | 98.8 | 1.2 | 0.83 |
| rs9916754 (C6749T; extron7) | CC | CT | TT | CC | CT | TT | C | T | C | T | ||||
| Wang F | 2008 | 556 | 341 | 518 | 37 | 1 | 333 | 8 | 0 | 96.5 | 3.5 | 98.8 | 1.2 | 0.83 |
| Han Y et al.(The first study) | 2011 | 801 | 767 | 672 | 120 | 9 | 701 | 65 | 1 | 91.4 | 8.6 | 95.6 | 4.4 | 0.69 |
| Han Y et al.(The second study) | 2011 | 271 | 303 | 228 | 41 | 2 | 272 | 30 | 1 | 91.7 | 8.3 | 94.7 | 5.3 | 0.86 |
HWE (P): the P-values of the Hardy-Weinberg equilibrium test of control group.
Table 3. The main results of the meta-analysis of the association between the WNK4 variant.
| Sample size | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Studied Polymorphisms | n | case | control | Genetic model | Statistical model | OR(95%CI) | Pz | I2 (%) | Pheterogeneity |
| C1155547T | 3 | 1096 | 874 | Allele contrast | Random model | 1.54(0.90,2.63) | 0.11 | 58.4 | 0.09 |
| Dominant | Random model | 1.70(0.57,5.13)* | 0.35 | 79.9 | 0.03 | ||||
| Homozygous | Fixed model | 2.49(0.57,10.78)* | 0.22 | 0 | 0.86 | ||||
| G1156666A | 5 | 1095 | 1029 | Allele contrast | Random model | 1.12(0.74,1.69) | 0.6 | 54.8 | 0.07 |
| Dominant | Random model | 1.08(0.68,1.71) | 0.74 | 59 | 0.05 | ||||
| Homozygous | Fixed model | 3.40(0.86,13.54)# | 0.08 | 0 | 0.8 | ||||
| G1155942T | 5 | 2260 | 1567 | Allele contrast | Random model | 0.01 | 57.1 | 0.05 | |
| Dominant | Random model | 0.03 | 64.7 | 0.04 | |||||
| Homozygous | Fixed model | 1.67(0.80,3.49) | 0.18 | 0 | 0.9 | ||||
| C6749T | 3 | 1628 | 1411 | Allele contrast | Fixed model | 0 | 0 | 0.37 | |
| Dominant | Fixed model | 0 | 0 | 0.42 | |||||
| Homozygous | Fixed model | 0.02 | 0 | 0.59 | |||||
*: for C1155547T, the study by Lu M et al. 2009 was excluded under the dominant and homozygous genetic model because of the absence of wild homozygote(CC) in both case and control group. #: for G1156666A, the study by Erlich PM et al. (population 2) 2003 was excluded under the homozygous genetic model because of the absence of mutational homozygote (AA) in both case and control group. : for G1155942T, the study by Lu M et al. 2009 was excluded under the dominant and homozygous genetic model because of the absence of wild homozygote(GG) in both case and control group.  : OR had statistical significance with corresponding 95% CI greater than 1.
: OR had statistical significance with corresponding 95% CI greater than 1.
Meta-analysis of WNK4 C1155547T polymorphism and hypertension
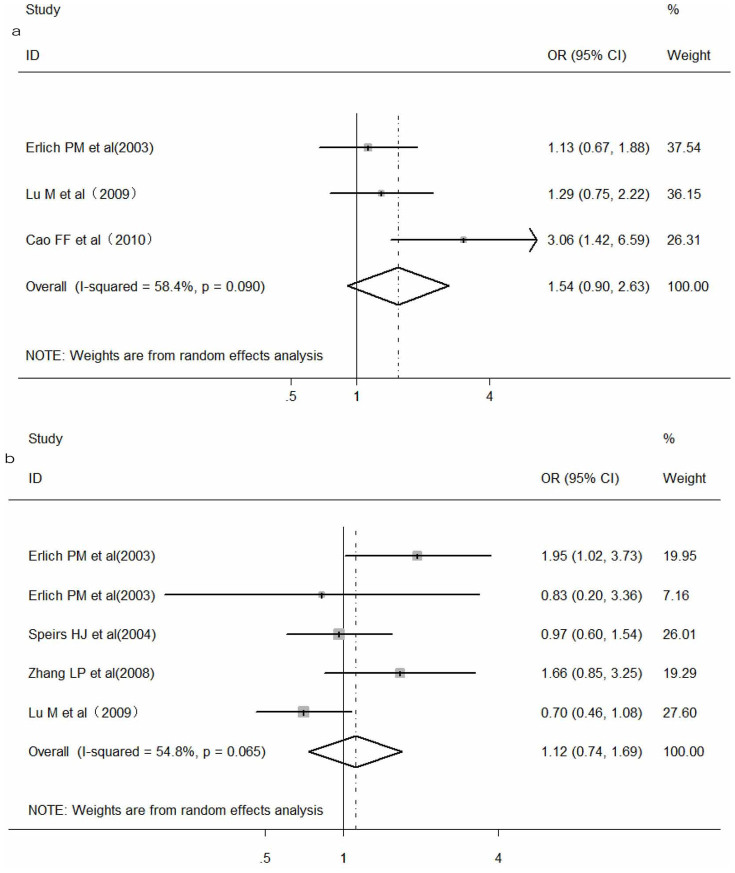
In total, three studies25,31,32 with 1096 cases and 874 controls were included in this meta-analysis to assess the relationship between C1155547T polymorphism and hypertension. No significant association was found between C1155547T polymorphism and the risk of hypertension under the allele contrast (odds ratio or OR = 1.54, 95% confidence interval or CI: 0.90–2.63, P = 0.11, Figure 2a), dominant model (OR = 1.70, 95% CI: 0.57–5.13, P = 0.35) and homozygous model (OR = 2.49, 95% CI: 0.57–10.78, P = 0.22). Because of the absence of wild homozygote (CC) in both case and control group, the study by Lu M et al.32 was excluded in the allele contrast and homozygous genetic model. Random-effects model was used in the allele contrast and dominant genetic model, for the heterogeneity between studies was significant (Pheterogeneity < 0.1, I2>50%), while fixed effects model was used in homozygous model.
Figure 2. Meta-analysis of the association of WNK4 C1155547T (a) and WNK4 G1156666A (b) polymorphisms with hypertension under allele contrast respectively.
Meta-analysis of WNK4 G1156666A polymorphism and hypertension
There were four articles25,26,32,35 including five studies focused on the correlation of WNK4 G1156666A polymorphism with hypertension. Heterogeneity between studies was significant in allele contrast and dominant model (Pheterogeneity < 0.1, I2>50%), and thus we used a random-effects model for pooled analysis. One study by Erlich et al.25 was excluded in the homozygous model because of the absence of mutational homozygote (AA) in both case and control group. There was no evidence to support any significant association between WNK4 G1156666A polymorphism and hypertension risk (allele genetic model: OR = 1.12, 95% CI: 0.74–1.69, P = 0.60, Figure 2b; dominant model: OR = 1.08, 95% CI: 0.68–1.71, P = 0.74; and homozygous model: OR = 3.40, 95% CI: 0.86–13.54, P = 0.08).
Meta-analysis of WNK4 G1155942T polymorphism and hypertension
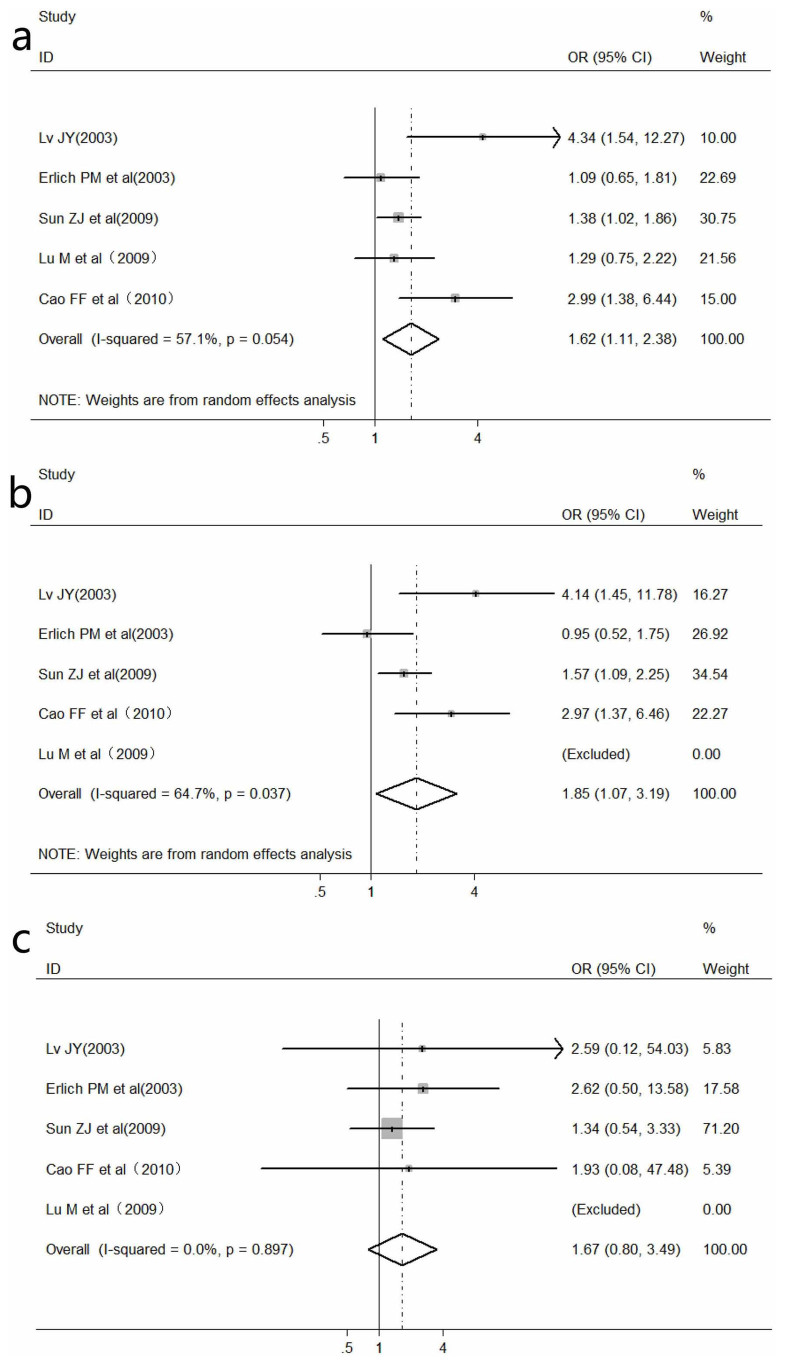
To evaluate the association of WNK4 G1155942T polymorphism with hypertension, five studies with 2260 cases and 1567 controls were pooled in this meta-analysis25,28,30,32,34. The study by Lu M et al.32 was excluded in pooled analysis in both dominant and homozygous genetic models, as a result of no existence of wild homozygote(GG) in both case and control group. Due to significant heterogeneity between the included studies, a random-effects model was used under allele contrast and dominant genetic model. Significant association between G1155942T polymorphism and hypertension was identified in both allele contrast (OR = 1.62, 95% CI: 1.11–2.38, P = 0.01) and dominant genetic model (OR = 1.85, 95% CI: 1.07–3.19, P = 0.03). However, no significant relation was found in homozygous genetic model (OR = 1.07, 95% CI: 0.80–3.49, P = 0.18) (Figure 3).
Figure 3. Meta-analysis of the association of WNK4 G1155942T polymorphism with hypertension.
a: under allele contrast; b: under dominant genetic model; c: under homozygote comparison.
Meta-analysis of WNK4 C6749T polymorphism and hypertension
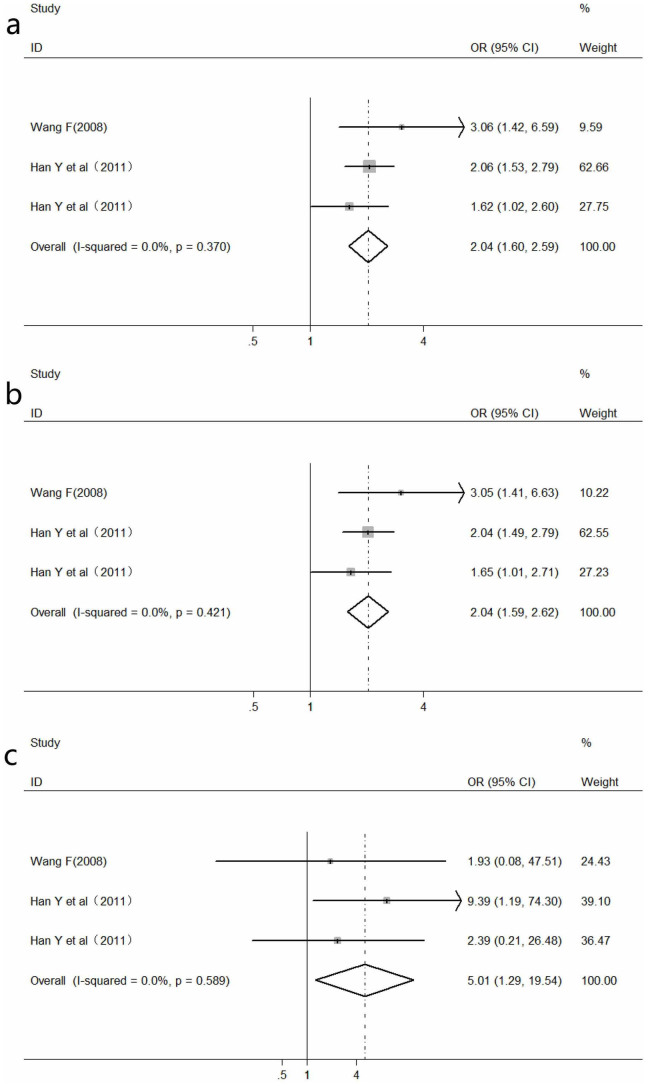
The association of WNK4 C6749T polymorphism with hypertension was found to be significant in allele contrast (OR = 2.04, 95% CI/: 1.60–2.59, P<0.01), dominant genetic model (OR = 2.04, 95% CI: 1.59–2.62, P<0.01) and homozygous genetic model (OR = 5.01, 95% CI: 1.29–19.54, P = 0.02) (Figure 4). As the heterogeneity between studies was not significant (Pheterogeneity > 0.1, I2<50%), a fixed-effects model was used.
Figure 4. Meta-analysis of the association of WNK4 C6749T polymorphism with hypertension.
a: under allele contrast; b: under dominant genetic model; c: under homozygote comparison.
Publication Bias
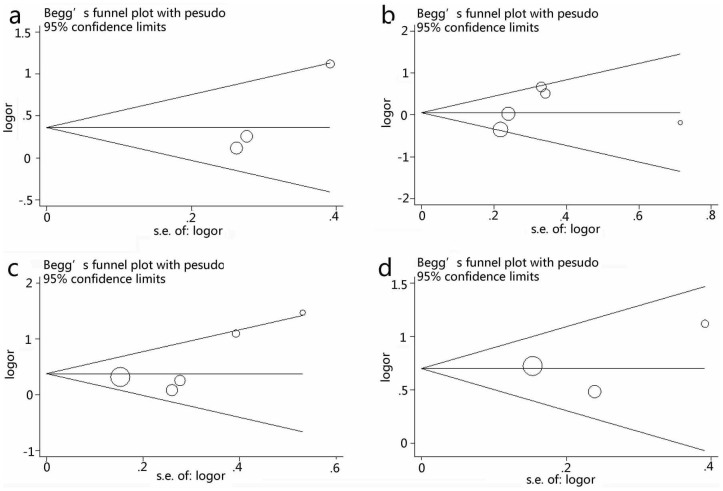
Both the Begg's test and Egger's regression test were performed to assess publication bias. The shapes of the funnel plots do not show obvious evidence of asymmetry (Figure 5). However, the P-value of Egger's test confirmed the existence of publication bias regarding the WNK4 C1155547T polymorphism and hypertension under allele contrast (P = 0.02). No statistical significance of publication bias was observed by Egger's test to G1156666A (P = 0.50), G1155942T (P = 0.18) and C6749T (P = 0.75) polymorphisms under allele contrast.
Figure 5. Begg's funnel plot of publication bias in the meta-analysis of the association of WNK4 Polymorphisms with hypertension risk under allele genetic model.
a: C1155547T and hypertension; b: G1156666A and hypertension; c: G1155942T and hypertension; d: C6749T and hypertension.
Sensitivity analysis
The sensitivity analyses were performed by sequential omitting individual studies to examine the influence of individual studies on the pooled ORs. The pooled ORs and corresponding 95% CIs were not significantly altered after excluding individual study in sequence (see Supplementary Fig. S1 online), suggesting that the overall results of our meta-analysis were stable and credible to some extent.
Cumulative meta-analysis
Cumulative meta-analysis of the relationship between WNK4 SNPs and hypertension were performed according to the year of publication. Regarding to C1155547T, as studies published in order, the statistical results had a trend to close to significant gradually and slightly. However, for G1156666A, G1155942T and C6749T polymorphisms, no obvious evolution rules were observed as records accumulated by year (see Supplementary Fig. S2 online).
Discussion
Ten eligible articles contained 16 independent case-control studies with 6089 hypertensive cases and 4881 normotensive controls were included in this meta- analysis. To the best of our knowledge, this was the first meta-analysis to date regarding the association of WNK4 gene polymorphisms with the risk of hypertension. The present study suggested a significant relationship between WNK4 G1155942T polymorphism and hypertension in the allelic genetic model and dominant genetic model. Moreover, WNK4 C6749T polymorphism was also found to be significantly associated with hypertension susceptibility under allele contrast, dominant genetic model and homozygous genetic model. Nevertheless, no significant association was identified between WNK4 gene C1155547T and G1156666A polymorphisms and hypertension risk. Although the statistical bias could not be avoided completely, this study indicated that WNK4 gene G1155942T and C6749T polymorphisms were associated with an increased risk of hypertension.
Human WNK4 gene is mapped to chromosome 17 with 19 exons and spanning 16 kb of genomic DNA12. It has suggested that WNK4 is expressed almost exclusively in the kidney, and specifically localizes to the distal convoluted tubule (DCT) and the cortical collecting duct (CCD), the segment of the distal nephron involved in regulating the ion homestasis11,36. Previous studies both in vivo and in vitro suggested that WNK4 modulated the balance between NaCl reabsorption and K+ secretion as a molecular switch via regulating the activities of the thiazide-sensitive Na-Cl cotransporter (NCC), the epithelia sodium channel (ENaC), the K+ channel (ROMK) and the paracellular Cl− pathways principally15,16,37,38,39. Loss-of-function mutations of WNK4 may cause increased NCC expression in the DCT and increase paracellular Cl− permeability in the distal tubule and reduce the surface expression of ROMK channels19,40,41. The overactivity of NCC resulted of Na+ retention in the DCT contributing to development of hypertension. Consistently, the results of this meta-analysis confirmed the association of WNK4 gene variation with the susceptibility of hypertension.
Wilson FH et al. firstly reported that mutations of WNK1 and WNK4 genes might be associated with PHA II, a rare autosomal dominant disease with the main performance of hypertension14. Since then, a series of animal experiments and epidemiologic studies concentrated on the association between WNK1 and WNK4 gene mutations and hypertension or blood pressure regulation. Studies by Putku et al.21 and Newhouse et al.23 separately demonstrated that polymorphisms in the WNK1 gene were related with BP variation and replicated the association in their meta-analysis as well. However, no meta-analysis examining the association of WNK4 gene mutations with the risk of hypertension existed. This study emerged under the circumstances in order to elucidate the role of WNK4 gene polymorphisms in development of hypertension. According to the limited quantities of relevant eligible studies, we only selected four polymorphisms in WNK4 gene (G1155942T, G1156666A, T1155547C, and C6749T) with at least 3 available relevant studies for further analyses. The mutations of WNK4 gene G1662A, K1169E, Pro556Thr and Q565E were also suggested to be associated with blood pressure42,43,44. Better designed case-control or cohort studies involved in these gene loci may be retrieved to update our study much more comprehensively.
Though we tried our best to control the potential bias from statistical aspect and estimated the association between WNK4 gene polymorphisms and hypertension as reliable as possible, some limitations of our study should be concerned. First, the included eligible studies without of large enough sample size limited the statistical power to some extent. Moreover, the quantities of each studied gene polymorphism were quite small and no sufficient data could be included for further analysis when we attempted to perform subgroup analyses. More studies with lager sample size are needed in the future to better interpret our results. Second, although the comprehensive literature search was performed in both English and Chinese database without language restriction, however, the P-value of Egger's test indicated the existence of publication bias for C1155547T polymorphism in association with hypertension. Third, the pathogenesis of hypertension is considered to be quite complicated involving gene-gene and gene-environment interactions. In this meta-analysis we tried to collect all the potentially relevant information such as BMI, gender distribution of the studied population, age and the level of serum lipid, however, we failed to assess the effect of gene-gene and gene-environment interactions. It has been reported that WNK kinases are upstream activators of ste20-related pralinealanine-rich kinase (SPAK) and ORS1 and the WNK-SPAK/OSR1 signaling pathway is very important in regulation of ion cotransporters and blood pressure45. The polymorphism of STK39 gene encoding SPAK protein has been confirmed to be associated with hypertension by a meta-analysis recently46. Moreover, some other relevant gene polymorphisms including the calcium/calmodulin kinase IV (CaMK4) and G-protein-coupled receptor kinases (GRKs) are also demonstrated to related to regulation of blood pressure47,48.Together with intake of dietary salt, age, sex, many gene polymorphisms can play an influence on the regulation of blood pressure.
In conclusion, the current meta-analysis suggested that the G1155942T and C6749T polymorphisms of WNK4 gene may increase the susceptibility of hypertension. Whereas, biological evidence was absent to support the association between WNK4 C115547T and G1156666A polymorphisms and risk of hypertension. Hypertensive disease is one of the major causes of mortality and global burden of disease, timely recognition and effective intervention of hypertension can help control the hospital mortality of cardiovascular disease49,50. The results of our meta-analysis indicate that WNK4 might be a potential target for hypertension therapeutic intervention. However, due to the given limitations above, better designed studies with lager sample size and well-matched controls considering gene-gene and gene-environment interactions are needed in the future.
Methods
Strategy for literature search
A comprehensive literature search for relevant articles without language restrictions was performed through the following electronic database: PubMed, MEDLINE, Web of Science, China National Knowledge Infrastructure (CNKI), Wanfang Data and China Biology Medicine (CBM). To identify all possible studies as complete as possible, various keywords as “with-no-lysine kinase”, “WNK”, “variant”, “polymorphism”, “blood pressure”, “high blood pressure” and “hypertension” were used. We also manually searched the reference lists of the included studies to find additional eligible studies. The literature search was finally performed on March 20, 2014.
Inclusion criteria
The studies were included in this meta-analysis if they comply with the following inclusion criteria: 1) evaluated the WNK4 (G1155942T, G1156666A, T1155547C, and C6749T) polymorphisms and hypertension; 2) used case-control or cohort design; 3) provided sufficient data of sample size, genotype distribution and allele frequencies or other information such as ORs with 95% CIs for statistical analysis; 4) diagnosis of hypertension patients based on the criteria of systolic blood pressure ≥ 140 mmHg or diastolic blood pressure ≥ 90 mmHg. If multi-studies reported the same or reduplicative data, we selected the latest published or the one with the largest sample size.
Data extraction
Two authors (X. G. Guo and J Ding) extracted the following information from each qualified studies independently: first author's name, publication year, ethnicity of study population, study design, genotyping methods, source of the controls, sample size of cases and controls, the distribution of genotypes in cases and controls, and diagnostic criteria for hypertensive cases and normotensive controls. We also collected the baseline information of each study population, such as age, gender, body mass index (BMI), mean blood pressure of cases and controls, and serum concentration of total cholesterol (TC) and triglyceride (TG). If the inconsistent evaluations are encountered, the two authors reached a consensus by discussion or intervention of a third author.
Statistical analysis
ORs and corresponding 95% CIs were used to evaluate the possible association between WNK4 polymorphisms and hypertension. The heterogeneity among different studies was assessed by chi-square-based Q-tests51. And the value of I2 was used to quantify the effect of heterogeneity. If the P-value of the Q-test >0.10 or I2 <50%, we chose a fixed-effects model by Mantel-Haenszel method for the absence of significant heterogeneity. Otherwise, a random-effects model with the method of Dersimonian & Laird was used52,53.
We also performed sensitivity analysis by omitting an individual study each time to check whether any of these estimates can bias the overall estimate. Furthermore, cumulative meta-analysis was performed to estimate the influence on the subsequent studies by the first published study, and identified the evolution of pooled estimates as time goes by54.
Finally, publication bias was assessed by both Begg's test55 and Egger's regression test56. P < 0.05 was considered the existence of statistically significant publication bias. The HWE of controls was calculated using Pearson χ2 -test. The genotypes and allele frequencies of controls were considered in HWE if P > 0.05. All statistical analyses were performed using STATA 11.0 (StataCorp LP, College Station, Texas, USA).
Author Contributions
Conceived and designed the experiments: G.X.G., D.J. Performed the experiments: X.T.M., J.W.Q., Y.X. Analyzed the data: S.Y.P., Z.F.R., Contributed reagents/materials/analysis tools: Z.J.H., Z.L.R., Wrote the paper: D.J., X.H., G.X.G.
Supplementary Material
Supporting Information
Acknowledgments
This study was supported by National Natural Science Foundation of China (Grant No. 81470370) and the Zhejiang Provincial Natural Science Foundation of China (Grant No.LY12H02006) and the Health Bureau of Zhejiang Province of China, (Grant No. 2011RCB016).
References
- Mancia G. et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the Task Force for the Management of Arterial Hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). Eur Heart J 34, 2159–2219 (2013). [DOI] [PubMed] [Google Scholar]
- Kearney P. M. et al. Global burden of hypertension: analysis of worldwide data. Lancet 365, 217–223 (2005). [DOI] [PubMed] [Google Scholar]
- Santulli G. Epidemiology of Cardiovascular Disease in the 21st Century: updates numbers and updated facts. J. Cardiovasc. Dis. 1, 1–2 (2013). [Google Scholar]
- Lim S. S. et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2224–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanira M. O. & Al Balushi K. A. Genetic variations related to hypertension: a review. J Hum Hypertens 19, 7–19 (2005). [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy K. M. The genetics of essential hypertension. Br J Clin Pharmacol 51, 5–11 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl L. K. Salt and hypertension. Am J Clin Nutr 25, 231–244 (1972). [DOI] [PubMed] [Google Scholar]
- Khaw K. T. & Barrett-Connor E. The association between blood pressure, age, and dietary sodium and potassium: a population study. Circulation 77, 53–61 (1988). [DOI] [PubMed] [Google Scholar]
- Morris R. C. Jr, Schmidlin O., Frassetto L. A. & Sebastian A. A Relationship and interaction between sodium and potassium. J Am Coll Nutr 25, 262S–270S (2006). [DOI] [PubMed] [Google Scholar]
- Xu B. et al. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275, 16795–16801 (2000). [DOI] [PubMed] [Google Scholar]
- Wilson F. H. et al. Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 (2001). [DOI] [PubMed] [Google Scholar]
- Verissimo F. & Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 20, 5562–5569 (2001). [DOI] [PubMed] [Google Scholar]
- Choate K. A., Kahle K. T., Wilson F. H., Nelson-Williams C. & Lifton R. P. WNK1, a kinase mutated in inherited hypertension with hyperkalemia, localizes to diverse Cl- -transporting epithelia. Proc Natl Acad Sci U S A 100, 663–668 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson F. H. et al. Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 (2001). [DOI] [PubMed] [Google Scholar]
- Yang C. L., Angell J., Mitchell R. & Ellison D. H. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest 111, 1039–1045 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle K. T. et al. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet 35, 372–376 (2003). [DOI] [PubMed] [Google Scholar]
- Yang S. S. et al. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4 (D561A/+) knockin mouse model. Cell Metab 5, 331–344 (2007). [DOI] [PubMed] [Google Scholar]
- Lalioti M. D. et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet 38, 1124–1132 (2006). [DOI] [PubMed] [Google Scholar]
- Gamba G. Role of WNK kinases in regulating tubular salt and potassium transport and in the development of hypertension. Am J Physiol Renal Physiol 288, F245–252 (2005). [DOI] [PubMed] [Google Scholar]
- Lazrak A., Liu Z. & Huang C. L. Antagonistic regulation of ROMK by long and kidney-specific WNK1 isoforms. Proc Natl Acad Sci U S A 103, 1615–1620 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putku M. et al. Novel polymorphic AluYb8 insertion in the WNK1 gene is associated with blood pressure variation in Europeans. Hum Mutat 32, 806–814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhouse S. J. et al. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum Mol Genet 14, 1805–1814 (2005). [DOI] [PubMed] [Google Scholar]
- Newhouse S. et al. Polymorphisms in the WNK1 gene are associated with blood pressure variation and urinary potassium excretion. PLoS One 4, e5003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin M. D. et al. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation 112, 3423–3429 (2005). [DOI] [PubMed] [Google Scholar]
- Erlich P. M. et al. Genetic variants of WNK4 in whites and African Americans with hypertension. Hypertension 41, 1191–1195 (2003). [DOI] [PubMed] [Google Scholar]
- Speirs H. J. & Morris B. J. WNK4 intron 10 polymorphism is not associated with hypertension. Hypertension 43, 766–768 (2004). [DOI] [PubMed] [Google Scholar]
- Mendes A. I. et al. A WNK4 gene variant relates to osteoporosis and not to hypertension in the Portuguese population. Mol Genet Metab 102, 465–469 (2011). [DOI] [PubMed] [Google Scholar]
- Sun Z. J. et al. Association of Ala589Ser polymorphism of WNK4 gene with essential hypertension in a high-risk Chinese population. J Physiol Sci 59, 81–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. et al. Hypertension associated polymorphisms in WNK1/WNK4 are not associated with hydrochlorothiazide response. Clin Biochem 44, 1045–1049 (2011). [DOI] [PubMed] [Google Scholar]
- Cao F. F. et al. Study on the association between genetic polymorphism on WNK4 genes and essential hypertension among Kazakhs ethnic population, in Xinjiang. Zhonghua Liu Xing Bing Xue Za Zhi 31, 375–378 (2010). [PubMed] [Google Scholar]
- Cao F. F. et al. Association of the C1155547T polymorphism in WNK4 gene with essential hypertension in Xinjiang Kazakhs. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 27, 546–549 (2010). [DOI] [PubMed] [Google Scholar]
- Lu M. et al. WNK4 polymorphisms and essential hypertension in the Uyghur population. Clin Exp Hypertens 31, 179–185 (2009). [DOI] [PubMed] [Google Scholar]
- Wang, F. Relationship between C6749T polymorphism in WNK4 gene and essential hypertension in Xinjiang Kazakhs population. Master's Degree Thesis, Shandong university, Shandong, China (2008) (in Chinese). [Google Scholar]
- Lv J. Y. The research on the expression of WNK genes. Ph.D. Thesis, China Medical University, Beijing (2003) (in Chinese). [Google Scholar]
- Zhang L. P. et al. Association between WNK4 polymorphism and essential hypertension in Kazak population in China. Chinese Journal of Geriatrics, 27, 548–551 (2008) (in Chinese). [Google Scholar]
- Ohno M. et al. Immunolocalization of WNK4 in mouse kidney. Histochem Cell Biol 136, 25–35 (2011). [DOI] [PubMed] [Google Scholar]
- Kahle K. T. et al. WNK4 regulates apical and basolateral Cl- flux in extrarenal epithelia. Proc Natl Acad Sci U S A 101, 2064–2069 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. S. et al. Regulation of apical localization of the thiazide-sensitive NaCl cotransporter by WNK4 in polarized epithelial cells. Biochem Biophys Res Commun 330, 410–414 (2005). [DOI] [PubMed] [Google Scholar]
- Ring A. M. et al. WNK4 regulates activity of the epithelial Na+ channel in vitro and in vivo. Proc Natl Acad Sci U S A 104, 4020–4024 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle K. T., Wilson F. H. & Lifton R. P. Regulation of diverse ion transport pathways by WNK4 kinase: a novel molecular switch. Trends Endocrinol Metab 16, 98–103 (2005). [DOI] [PubMed] [Google Scholar]
- Flatman P. W. Cotransporters, WNKs and hypertension: an update. Curr Opin Nephrol Hypertens 17, 186–192 (2008). [DOI] [PubMed] [Google Scholar]
- Kamide K. et al. Three novel missense mutations of WNK4, a kinase mutated in inherited hypertension, in Japanese hypertensives: implication of clinical phenotypes. Am J Hypertens 17, 446–449 (2004). [DOI] [PubMed] [Google Scholar]
- Mayan H. et al. Hypercalciuria in familial hyperkalemia and hypertension accompanies hyperkalemia and precedes hypertension: description of a large family with the Q565E WNK4 mutation. J Clin Endocrinol Metab 89, 4025–4030 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. Identification of a novel WNK4 mutation in Chinese patients with pseudohypoaldosteronism type II. Nephron Physiol 118, 53–61 (2011). [DOI] [PubMed] [Google Scholar]
- Richardson C. & Alessi D. R. The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121, 3293–3304 (2008). [DOI] [PubMed] [Google Scholar]
- Xi B. et al. STK39 polymorphism is associated with essential hypertension: a systematic review and meta-analysis. PLoS One 8, e59584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G. et al. CaMK4 Gene Deletion Induces Hypertension. J Am Heart Assoc 1, e1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G., Trimarco B. & Iaccarino G. G-protein-coupled receptor kinase 2 and hypertension: molecular insights and pathophysiological mechanisms. High Blood Press Cardiovasc Prev 20, 5–12 (2013). [DOI] [PubMed] [Google Scholar]
- Liu C. & Xi B. Pooled analyses of the associations of polymorphisms in the GRK4 and EMILIN1 genes with hypertension risk. Int J Med Sci 9, 274–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G. Coronary heart disease risk factors and mortality. JAMA 307, 1137–1138 (2012). [DOI] [PubMed] [Google Scholar]
- Higgins J. P. & Thompson S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–1558 (2002). [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran W. G. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics 24, 295–313 (1968). [PubMed] [Google Scholar]
- Ioannidis J. P. & Trikalinos T. A. Early extreme contradictory estimates may appear in published research: the Proteus phenomenon in molecular genetics research and randomized trials. J Clin Epidemiol 58, 543–549 (2005). [DOI] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey, Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information