Abstract
Background and Aims
Although xyloglucans are ubiquitous in land plants, they are less abundant in Poales species than in eudicotyledons. Poales cell walls contain higher levels of β-1,3/1,4 mixed-linked glucans and arabinoxylans than xyloglucans. Despite the relatively low level of xyloglucans in Poales, the xyloglucan endotransglucosylase/hydrolase (XTH) gene family in rice (Oryza sativa) is comparable in size to that of the eudicotyledon Arabidopsis thaliana. This raises the question of whether xyloglucan is a substrate for rice XTH gene products, whose enzyme activity remains largely uncharacterized.
Methods
This study focused on OsXTH19 (which belongs to Group IIIA of the XTH family and is specifically expressed in growing tissues of rice shoots), and two other XTHs, OsXTH11 (Group I/II) and OsXTH20 (Group IIIA), for reference, and measurements were made of the enzymatic activities of three recombinant rice XTHs, i.e. OsXTH11, OsXTH20 and OsXTH19.
Key Results
All three OsXTH gene products have xyloglucan endohydrolase (XEH, EC 3·2·1·151) activity, and OsXTH11 has both XEH and xyloglucan endotransglycosylase (XET, EC 2·4·1207) activities. However, these proteins had neither hydrolase nor transglucosylase activity when glucuronoarabinoxylan or mixed-linkage glucan was used as the substrate. These results are consistent with histological observations demonstrating that pOsXTH19::GUS is expressed specifically in the vicinity of tissues where xyloglucan immunoreactivity is present. Transgenic rice lines over-expressing OsXTH19 (harbouring a Cauliflower Mosaic Virus 35S promoter::OsXTH19 cDNA construct) or with suppressed OsXTH19 expression (harbouring a pOsXTH19 RNAi construct) did not show dramatic phenotypic changes, suggesting functional redundancy and collaboration among XTH family members, as was observed in A. thaliana.
Conclusions
OsXTH20 and OsXTH19 act as hydrolases exclusively on xyloglucan, while OsXTH11 exhibits both hydrolase and XET activities exclusively on xyloglucans. Phenotypic analysis of transgenic lines with altered expression of OsXTH19 suggests that OsXTH19 and related XTH(s) play redundant roles in rice growth.
Keywords: Poales, Oryza sativa, cell wall, xyloglucan endotransglucosylase, xyloglucan endohydrolase, XTH
INTRODUCTION
Recent phylogenetic studies suggest that xyloglucan is unique to and ubiquitous among land plants (reviewed by Popper, 2008; Popper and Tuohy, 2010; Popper et al., 2011; Fangel et al., 2012). In angiosperms other than commelinoid monocotyledons, xyloglucan is the major structural component of the cell-wall matrix in several cell types at different developmental stages, including cell plates formed in dividing cells (Moore and Staehelin, 1988) and primary walls in growing cells (Hayashi, 1989). Xyloglucan is also considered to play a role in fully differentiated secondary walls (Bourquin et al., 2002). By contrast, the xxt1 xxt2 double mutant resulted in lack of detectable xyloglucan content with concomitant changes in the mechanical properties of the cell wall, but showed a rather subtle phenotype restricted to certain tissues. These results challenge conventional models of the plant primary cell wall, and the function of xyloglucan and xyloglucan endotransglucosylase/hydrolases (XTHs) in controlling the growth and development in Arabidopsis remains controversial (Cavalier et al., 2008).
The most widely adopted structural models for the primary cell walls in eudicotyledonous plants envisage xyloglucan directly or indirectly crosslinking cellulose microfibrils, forming the load-bearing framework of type I cell walls (Carpita, 1996; Vincken et al., 1997; Cosgrove, 2005; Wang et al., 2012). Given the suggested importance of xyloglucan in maintaining the load-bearing framework of the cell wall, it is of considerable interest and importance to understand the molecular basis by which xyloglucans are integrated into the cell-wall framework and modified during growth and differentiation in individual cell types.
The key enzymes responsible for xyloglucan metabolism in muro are XTHs. These enzymes may have xyloglucan endohydrolase (XEH) and/or xyloglucan endotransglucosylase (XET) activities, which mediate splitting and reconnection of the xyloglucan crosslinks in the cell wall. Due to their potential enzymatic actions, XTHs are thought to play a pivotal role in the construction, remodelling and disassembly of the xyloglucan/cellulose framework in type I cell walls during cell growth and differentiation (Nishitani, 1997; Rose et al., 2002; Becnel et al., 2006; Nishitani and Vissenberg, 2007).
XTHs are a large family of enzymes. Thirty-three and 41 open reading frames encoding XTH-like proteins have been identified in Arabidopsis thaliana and poplar, respectively (Yokoyama and Nishitani, 2000, 2001; Geisler-Lee et al., 2006; Eklöf and Brumer, 2010; Del Bem and Vincentz, 2010). Given the ubiquitousness of xyloglucans and the tissue-specific expression of individual XTH genes, it appears that xyloglucan dynamics as mediated by XTHs participate in a wide range of physiological processes in eudicotyledonous plants.
The cell walls of Poales species, such as rice (Oryza sativa), are referred to as type II cell walls, and the amounts and structural features of the major hemicellulosic polysaccharides differ dramatically from those in type I cell walls (Carpita and Gibeaut, 1993; Vogel, 2008; Hsieh and Harris, 2009). In type II cell walls, xyloglucan is not a major component, and its size and branching patterns are different from those in type I cell walls (Carpita, 1996; Hsieh and Harris, 2009). The most characteristic structural feature of xyloglucans in type II cell walls is having XXGG-type and XXGGG-type building units (Kato et al., 1982; Kato and Matsuda, 1985; Vincken et al., 1997; Gibeaut et al., 2005). The occurrence of fucosylated xyloglucans, which are abundantly found in type I cell walls, is only restricted to specific tissues, such as sieve tubes (Brennan and Harris, 2011). The predominant glycans that crosslink the cellulose microfibrils are, instead, thought to be glucuronoarabinoxylan (GAX) and β1,3/1,4-mixed-linkage glucan (MLG). Consequently, xyloglucans have not been considered important components of type II walls, even though correlations between xyloglucan metabolism and growth parameters have been reported in monocotyledonous plants (Inouhe et al., 1984; Pritchard et al., 1993; Gibeaut et al., 2005).
Given the quantitative and structural differences of xyloglucan in type I and II cell walls, it is tempting to hypothesize that the XTH gene families evolved differently and that there would be fewer XTH genes in plants with type II cell walls than in eudicotyledonous plants such as A. thaliana. However, this hypothesis is not supported by the facts, as there are as many as 29 XTH (OsXTH) genes in the rice genome, which is similar to the number of arabidopsis XTH (AtXTH) genes (Yokoyama et al., 2004; Yokoyama and Nishitani, 2004).
XTH family members are categorized into two groups, Group I/II and III, based on the structural features of the individual members (Yokoyama et al., 2004, 2010). Group III consists of two distinct clades designated Group III-A and Group III-B. On the basis of crystallography data, members of Group III-A, to which OsXTH19 belongs, are predicted to have hydrolase activity instead of XET activity (Baumann et al., 2007; Eklöf and Brumer, 2010). Note that most of the OsXTH family members show distinct tissue-specific expression patterns. For example, OsXTH19, which belongs to Group III-A, is specifically expressed in the elongating zones of various organs, suggesting that this protein has an important function in the regulation of cell expansion in rice (Uozu et al., 2000; Jan et al., 2004; Yokoyama et al., 2004). The rate of tissue growth has also been shown to correlate with XTH mRNA abundance and XET activity in other grasses, such as barley (Hordeum vulgare L.) (Smith et al., 1996) and maize (Zea mays L.) (Pritchard et al., 1993; Wu et al., 1994; Palmer and Davies, 1996; Genovesi et al., 2008). Thus, the XTH family may play important roles even in commelinoid monocotyledons.
Additionally, there are many members of the XTH family in the non-vascular land plant Physcomitrella patens (Yokoyama et al., 2010), in which the structural features of xyloglucan differ from those in angiosperms (Pena et al., 2008). Furthermore, a possible role for XTH in mediating heteotransglycosylation between MLG and xyloglucans has been proposed in barley (Hrmova et al., 2007) and in Equisetum (Mohler et al., 2013).
Accordingly, whether xyloglucan is actually the principal substrate of XTHs in type II cell walls remains an intriguing but unanswered question (Yokoyama et al., 2004, 2010; Hrmova et al., 2007; Mohler et al., 2013). To address this question, we investigated the enzymatic activities of OsXTH19 and OsXTH20 as representatives of Group III-A XTHs, and OsXTH11 in Group I/II XTH as a reference, and examined whether xyloglucan is the principal substrate for these OsXTHs. To address the in planta function of XTH, we further examined the action of XTH using transgenic rice lines in which OsXTH19 was over-expressed or suppressed.
MATERIAL AND METHODS
Plant material
Rice (Oryza sativa ‘Nipponbare’) plants were grown in individual cylindrical plastic pots containing approximately 100 mL of soil. The pots were placed in a container filled with tap water fortified with fertilizer (HYPONeX; Hyponex Japan Corp. Ltd, Osaka, Japan). Plants were grown in growth chambers (model LH220S; Nippon Medical & Chemical Instruments Co., Ltd, Osaka, Japan) at 28 °C under a 15-h light (at 150 µmol−2 s−1)/9-h dark cycle for vegetative growth and then switched to a 9-h light/15-h dark cycle.
Construction of expression vectors
The cDNA of each of the three XTH proteins (OsXTH19, Os03g0108300; OsXTH20, Os10g0545500; and OsXTH11, Os06g0696400) was amplified without its predicted secretion signal sequence using the following forward and reverse PCR-primer sets: for OsXTH19, 5′-TGCCTCGAGAAAAGAGCGCAGCCATCGCCG-3′ and 5′-TGCTCTAGACTAGCACTCGGGGTA-3′; for OsXTH20, 5′-CCACTCGAGAAAAGACCTTCTCCTGGCTAC-3′ and 5′-GACTCTAGACATCAGCACTCGGGGTAGAA-3′; and for OsXTH11, 5′-CCACTCGAGAAAAGAAACTTCTTCCAGGAC-3′ and 5′-GCTTCTAGAGATCTCAACGGAGCTTGCACT-3′. The cDNAs were ligated into pBluescript II SK+ vector (Stratagene, La Jolla, CA, USA). The XhoI/XbaI fragment from this plasmid was cloned into the pPICZaA vector in-frame with the alpha factor secretion signal sequence for production and secretion in the Pichia pastoris expression system (Invitrogen/Molecular Probes, Carlsbad, CA, USA). The pPICZaA-OsXTH expression vector was linearized with BstXI and introduced into P. pastoris strain X-33 according to the manufacturer's protocol. Multiple-copy integrants were selected on YPDS plates (yeast/peptone/dextrose/sorbitol regeneration medium) containing zeocin at elevated concentrations up to 1 g L−1. Selected colonies were screened for the presence of the expression construct in the yeast genome by yeast colony PCR.
Recombinant protein production and purification
For OsXTH19, positive transformants were selected using specific antibodies against peptide sequences specific to OsXTH19, and positive transformants for OsXTH20 and OsXTH11 were selected using a polyclonal antibody raised against full-length recombinant Vigna angularis VaXTH1 protein derived from azuki bean (Yokoyama and Nishitani, 2001; Nakamura et al., 2003).
The recombinant OsXTH protein was expressed essentially as described in the Invitrogen Pichia Expression Kit manual (Invitrogen/Molecular Probes, Eugene, OR, USA). This culture was grown overnight at 28 °C. Induction was performed in 2-L flasks containing 500 mL of culture at 28 °C for 4 d with the addition of methanol to a final concentration of 1 % every 24 h. At the end of the induction period, yeast cells were discarded by centrifugation (1000 g for 5 min), and proteins were recovered from the medium by precipitation with 60 % saturated ammonium sulphate and centrifugation (7500 ×g for 30 min). Pellets were redissolved in 20 mm triethanolamine, pH 7·5, for OsXTH19 and OsXTH20, or in 50 mm MOPS-NaOH, pH 7·0, for OsXTH11. The resuspended proteins were dialysed four times against the same buffer to obtain the ammonium sulphate precipitate fraction. The ammonium sulphate precipitate fractions of OsXTH19 and OsXTH20 were subjected to successive anion-exchange chromatography using HiTrap Q FF (5 mL; GE Healthcare, Menlo Park, CA, USA), followed by RESOURCE Q (6 mL; GE Healthcare) equilibrated with 20 mm triethanolamine, pH 7·5, and eluted with a linear gradient of 0–0·5 m NaCl in the same buffer. The ammonium sulphate precipitate fraction of OsXTH11 was subjected to successive cation-exchange chromatography using HiTrap SP FF (5 mL; GE Healthcare) and RESOURCE S (6 mL; GE Healthcare) equilibrated with MOPS-NaOH, pH 7·0. The resulting XTH protein fractions were further purified by size exclusion chromatography on Superdex 75 10/300 GL (GE Healthcare) using 50 mm sodium acetate, pH 5·5, as the eluent, yielding highly purified recombinant XTH.
Hydrolase assay
Hydrolytic activity was measured using 50 ng of recombinant OsXTH protein, 10 mg of tamarind xyloglucan, barley β-1,3/1,4-glucan or maize glucuronoarabinoxylan (Nishitani and Tominaga, 1992) in 50 µL of 50 mm sodium acetate, pH 4·0–7·0. For the control experiments, OsXTH that had been heat-denatured at 100 °C for 5 min was added. After incubation at 30 °C for 1 h, the molecular weight distribution profile of the polysaccharide substrate was monitored by size exclusion chromatography on a TSK-GEL G3000PWXL and TSK-GEL G5000PWXL (Tosoh Corp., Kanagawa, Japan) connected in series using a high-performance liquid chromtography (HPLC) system (D500; Dionex Corp., Sunnyvale, CA, USA) equipped with a pulsed amperometry detector as described by Nishitani and Tominaga (1992). From the chromatogram, mass-average molecular weight was calculated according to Nishitani and Masuda (1981) using a calibration curve derived from standard pullulans (molecular mass = 380, 48 and 5·8 kDa) purchased from Showa Denko, Tokyo, Japan. One unit of hydrolase activity was defined as that which caused a decrease in the mass-average molecular weight by half in 1 min under the incubation conditions employed.
XET assay
XET activity was measured using 50 ng of recombinant OsXTH, 10 µg of the donor substrate and 50 pmol of the acceptor substrate in 50 µL of 25 mm sodium acetate, pH 4·0–5·5. Tamarind xyloglucan, barley MLG and maize GAX were used as the donor substrates. XXXG, XLLG and GGGG were coupled with 2-aminopyridine to obtain respective 2-aminopyridyl xyoglucan oligomers, and were used as the acceptor substrates (Nishitani and Tominaga, 1992). After 1 h of incubation at 30 °C, the reaction mixture was subjected to size exclusion chromatography, as described above, using a Dionex D500 HPLC system equipped with both a pulsed amperometry detector and a fluorescence detector (Nishitani, 1992; Nishitani and Tominaga, 1992). Endotransglycosylation activity was determined by measuring the peak area of enzyme product in the fluorescence chromatogram, which was then converted to the molar amount of 2-amimopyridyl oligosaccharide using a calibration curve.
Plasmid constructs
The 3 kb promoter region of OsXTH19 was amplified from rice (O. sativa ‘Nipponbare’) genomic DNA using the primer set 5′-CTCAAGCTTTACATTCTGCTTAATTGAAGGAGATC-3′ and 5′-CTCGTCGACCGATAATCGTCCTTAATTTCTAGCTG-3′. The amplified DNA fragment was TA-cloned into the pBluescript II SK+ plasmid. The HindIII/SalI fragment from this vector was cloned upstream of the β-glucuronidase (GUS) gene into the respective restriction sites of the binary vector pGPTV-HPT. The resulting construct was named pOsXTH19::GUS.
An OsXTH19 RNAi construct was designed in which transcription of a hairpin OsXTH19 mRNA fragment was driven by the 3 kb OsXTH19 promoter. Two fragments of the 3′-untranscribed region of OsXTH19 were amplified by PCR using the following forward and reverse PCR-primer sets: for the first fragment, 5′-CTCTCTAGACACGTATCTACTATTACCC-3′ and 5′-TTCGAGCTCACACAGCTGCACACCTCAA-3′; for the second fragment, 5′-TTCGGATCCACACAGCTGCACACCTCAA-3′ and 5′-CGCTCTAGAGGTCCATGGTCTACTACTA-3′.
The amplified fragments were TA-cloned into the pBluescript II SK+ plasmid. The second fragment was cloned into the plasmid at the downstream end of the first fragment in an anti-sense orientation. The OsXTH19 RNAi construct that was cloned in pBluescript II SK+ was excised with SmaI and SacI, and the resulting fragment was cloned into the SmaI/SacI restriction site of the pOsXTH19::GUS binary vector to replace the GUS structural gene with the OsXTH19 RNAi construct.
An OsXTH19 over-expression construct was designed in which transcription of OsXTH19 mRNA was driven by the cauliflower mosaic virus (CaMV) 35S promoter. The cDNA of OsXTH19 and the nopaline synthase terminator (Tnos) were amplified using the following forward and reverse PCR-primer sets: for OsXTH19, 5′-GGATCTAGAATGGAGCAGAAGCCACC-3′ and 5′-GTTGGATCCCTAGCACTCGGGGTAC-3′; for Tnos, 5′-GGTGGATCCGAATTTCCCGGATCGTTCAAA-3′ and 5′-AGTGAATTCCCGATCTAGTAACATA-3′.
The amplified fragments were TA-cloned into the pBluescript II SK+ plasmid. The Tnos fragment was cloned into the plasmid at the downstream end of the OsXTH19 cDNA. The OsXTH19 cDNA–Tnos construct that was cloned in pBluescript II SK+ was excised with XbaI and EcoRI and then cloned into the XbaI/EcoRI restriction site of the binary vector pBI121 (Clontech Laboratories, Inc., Palo Alto, CA, USA) to replace the GUS structural gene with the OsXTH19 cDNA–Tnos construct. The CaMV35S promoter–OsXTH19 cDNA–Tnos construct that was cloned in pBI121 was excised with HindIII and EcoRI and was cloned into the HindIII/EcoRI restriction site of the binary vector pGPTV-HPT.
Production of rice transformants
The constructs were used to transform Agrobacterium tumefaciens strain C58C1. Transformation of rice calli was carried out as reported previously (Toki et al., 2006). The hygromycin-resistant rice plants were subsequently acclimated in Murashige and Skoog medium without hormones for 4 weeks and transplanted to soil in pots. T2 generation plants (homozygous lines) were used for all experiments in this study.
RNA extraction and real-time RT-PCR
Seven-day-old seedlings of wild-type and transgenic plants were frozen in liquid nitrogen. RNA was extracted from the plant materials using the conventional SDS-phenol protocol. Oligo DNA primer sets for the OsXTH19 genes were prepared and used as previously described (Yokoyama et al., 2004). Quantitative one-step real-time RT-PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in an ABI Prism TM 5700 Sequence Detection System (Applied Biosystems) as described by Yokoyama and Nishitani (2001).
Histochemical GUS activity assay
For histochemical analysis of GUS expression, seedlings were incubated in 0·1 m sodium phosphate, pH 7, containing 0·1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid cyclohexylammonium at 37 °C for 2–20 h, followed by washing in 70 % ethanol as described by Jefferson et al. (1987). Stained seedlings were observed under a Leica MZ APO binocular microscope (Leica, Heerbrugg, Switzerland), and images were recorded using a Nikon DXM1200 digital camera (Nikon, Tokyo, Japan).
Immunofluorescence
The basal part of the elongating second internode of each shoot was cut into 70-mm cross-sections using a Vibratome 3000 Deluxe (Vibratome, St Louis, MO, USA) and immediately fixed in 4 % paraformaldehyde in phosphate-buffered saline. For ultrasensitive immunodetection, the tyramide signal amplification method was used based on the peroxidation of Alexa Fluor 488 tyramide according to the manufacturer's protocol (Tyramide Signal Amplification kits, Invitrogen/Molecular Probes). The samples were reacted with LM15 (PlantProbes, Leeds, UK; Marcus et al., 2008) diluted 1 : 10 (w/v) as the primary antibody and Alexa Fluor 488 goat anti-mouse highly cross-adsorbed secondary antibodies. Photographs were taken using appropriate filters for fluorescein isothiocyanate fitted to an epifluorescence microscope (DMRXP; Leica).
RESULTS
Heterologous expression and purification of three recombinant OsXTHs
We focused on OsXTH19 because we previously showed a clear correlation between its transcript abundance and shoot cell expansion (Yokoyama et al., 2004). OsXTH19 belongs to the Group IIIA subfamily of XTHs. In arabidopsis, AtXTH31 (Zhu et al., 2012) and AtXTH32 (Kaewthai et al., 2013), which belong to the Group IIIA subfamily, have been experimentally demonstrated. To allow comparison of the enzymatic characteristics of OsXTH19 with those of the other OsXTHs, we also examined OsXTH20, which belongs to Group IIIA and is the most closely related protein to OsXTH19 and OsXTH11, which is a member of Group I/II XTHs and used as a reference for the Group IIIA subfamily. We expressed the recombinant proteins in the methylotrophic yeast Pichia pastoris using the pPICZaA system. Transformants that actively secreted recombinant enzyme were selected using specific antibodies, and the recombinant protein in each of the culture media was purified using a three-step HPLC procedure to obtain highly purified preparations of recombinant OsXTH19, OsXTH20 and OsXTH 11. Each of the three protein preparations appeared as a single band using SDS–polyacrylamide gel electrophoresis, with a relative molecular mass of approximately 33, 36 and 31 kDa, respectively (Supplementary Data Fig. S1). Thus, the three recombinant OsXTHs were not post-translationally degraded during or after heterologous expression in P. pastoris.
Enzymatic properties of the three OsXTHs
The three recombinant OsXTHs were tested for their XET activity using a high-molecular-weight xyloglucan as a donor substrate and 2-aminopyridyl xyloglucan oligosaccharides as acceptor substrates. XET activity was measured by monitoring the generation of fluorescently labelled high-molecular-weight xyloglucan. OsXTH11 produced a high-molecular-weight fluorescent xyloglucan when 2-aminopyridyl-XLLG and -XXXG was used as the acceptor substrate but not when 2-aminopyridyl-GGGG was used (Fig. 1; for nomenclature of xyloglucan-derived oligosaccharides, see Fry et al., 1993). In other words, OsXTH11 mediated transfer of the xyloglucan segment to the xyloglucan oligosaccharide and therefore exhibited XET activity (Table 1). As xylosyl residues in rice xyloglucans are less often substituted by galactosyl or fucosyl residues (Kato and Matsuda, 1985; Hsieh and Harris, 2009), it appears that OsXTH11 prefers 2-aminopyridyl-XXXG as the acceptor substrate over 2-aminopyridyl-XLLG (Table 1). OsXTH11 showed a slightly acidic pH optimum and a rapid loss of activity when the pH was below 5 (Fig. 1). A similar pH dependency of XET activity has been reported for other XTHs (Nishitani and Tominaga, 1991, 1992; Fry et al., 1992; Campbell and Braam, 1999; Henriksson et al., 2003; Kallas et al., 2005; Maris et al., 2009). By contrast, neither OsXTH19 nor OsXTH20 mediated detectable transfer of the donor xyloglucan to either 2-aminopyridyl-XXXG or -XLLG. Thus, of the three OsXTHs, only OsXTH11 has XET activity (Table 1).
Fig. 1.
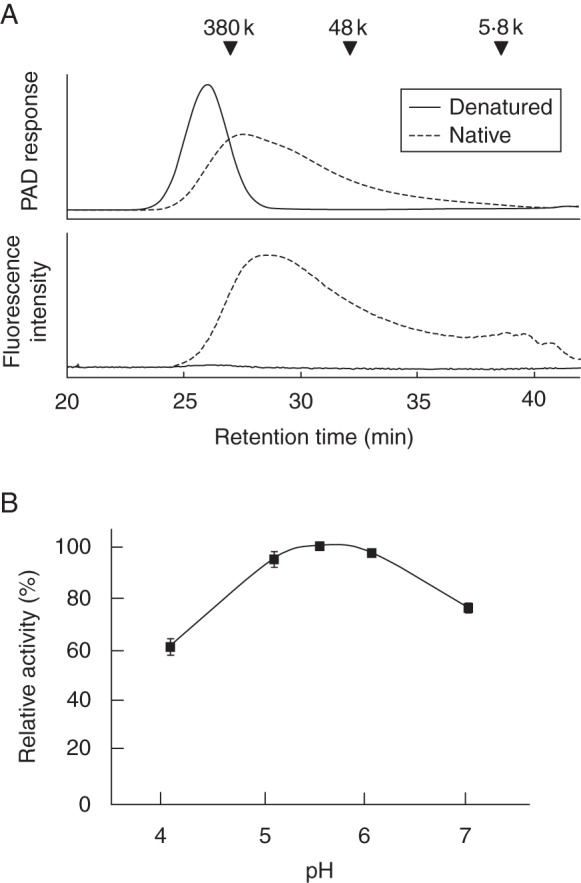
(A) Size exclusion chromatography showing XET activity between high-molecular-weight xyloglucan and 2-amimopyridyl xyloglucan oligosaccharide (XXXG-PA). A mixture of tamarind xyloglucan (donor substrate) and 2-amimopyridyl xyloglucan oligosaccharide XXXG-PA (acceptor substrate) was incubated for 1 h at 30 °C with native or denatured recombinant OsXTH11 protein (as indicated in the key). After the reaction, the reaction mixture was separated by size exclusion chromatography, and the elution profile of the high-molecular-weight fluorescently labelled product was measured with a fluorescence detector. PAD, pulsed amperometric detector. (B) pH dependency of OsXTH11-mediated XET activity.
Table 1.
Donor and acceptor substrate specificities for XET activity of purified OsXTH11, 19 and 20 proteins
| XET activity [pmol min−1 (μg−1 protein)] |
||||
|---|---|---|---|---|
| Donor substrate | Acceptor substrate | OsXTH11 | OsXTH19 | OsXTH20 |
| Tamarind xyloglucan | XXXG-PA | 6·88 ± 0·10* | 0·06 ± 0·11 | 0·03 ± 0·08 |
| XLLG-PA | 2·64 ± 0·13* | −0·21 ± 0·01 | 0·00 ± 0·00 | |
| GGGG-PA | 0·02 ± 0·03 | 0·04 ± 0·07 | −0·01 ± 0·02 | |
| Barley β-1,3/1,4-d-glucan | XXXG-PA | −0·12 ± 0·15 | −0·01 ± 0·01 | 0·03 ± 0·04 |
| GGGG-PA | −0·03 ± 0·04 | 0·00 ± 0·00 | −0·03 ± 0·04 | |
| Maize glucuronoarabinoxylan | XXXG-PA | 0·07 ± 0·09 | 0·01 ± 0·02 | −0·04 ± 0·06 |
| GGGG-PA | 0·01 ± 0·14 | 0·02 ± 0·03 | −0·16 ± 0·21 | |
Values are means ± s.d.
We next examined the XEH activity of the three recombinant OsXTHs using unlabelled high-molecular-weight xyloglucan. All three OsXTHs reduced the mass-average molecular weight of the xyloglucans (Fig. 2), although the hydrolytic activity varied among XTHs, indicating that, more or less, all have XEH activity. OsXTH19 exhibited the highest hydrolytic activity, and OsXTH20 was the weakest of the three (Fig. 2, Table 1). Heat-inactivated OsXTHs did not cause any detectable changes in the molecular weight profile of the xyloglucans (data not shown).
Fig. 2.
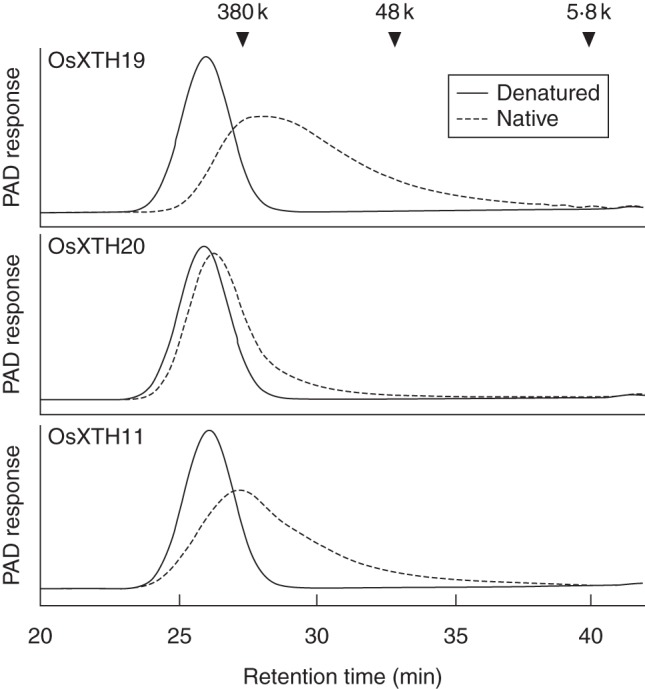
Changes in the distribution of molecular masses of high-molecular-weight xyloglucans during incubation with recombinant OsXTH19, OsXTH20 and OsXTH11. Tamarind xyloglucan was incubated for 1 h at 30 °C with native or denatured recombinant OsXTH19, OsXTH20 or OsXTH11 (as indicated). After the reaction, the reaction mixture was separated by size exclusion chromatography, and the elution profile of xyloglucan was analysed with a pulsed amperometric detector (PAD) using a Dionex D500 chromatograph.
Substrate specificity of the three OsXTHs
Hrmova et al. (2007) report that some grass XTHs may use a range of donor and acceptor substrates other than xyloglucan and may mediate hetero-transglucosylation between xyloglucan and MLG (Fincher, 2009). By contrast, Mohler et al. (2013) reported that barley cell-wall preparations do not exhibit MLG:xyloglucan endotransglucosylase activity, despite the fact that they contain both of its substrates and high XET activity, and the biological role of XTH in Poales species remains controversial.
In the present study, we focused on OsXTH19, which is implicated in shoot growth in rice, and examined the ability of the three OsXTHs to transfer other hemicellulosic polysaccharides to 2-aminopyridyl-XXXG or -GGGG (Table 1). None of the three OsXTHs exhibited significant hetero-transglycosylase activity when barley MLG, or maize GAX, was used as a donor substrate under standard conditions (Table 1). Similarly, none of the three OsXTHs exhibited detectable hydrolase activity when barley MLG, or maize GAX, was used as a substrate (Table 2).
Table 2.
Substrate specificity for hydrolase activity of purified OsXTH11, 19 and 20 proteins
| Substrate | Hydrolase activity [milli-units (μg−1 protein)] |
||
|---|---|---|---|
| OsXTH11 | OsXTH19 | OsXTH20 | |
| Tamarind xyloglucan | 129 ± 12 | 298 ± 3 | 121 ± 21 |
| Barley β-1,3/1,4-d-glucan | 5 ± 4 | 1 ± 3 | 9 ± 8 |
| Maize glucuronoarabinoxylan | 10 ± 8 | –1 ± 3 | –7 ± 6 |
Values are means ± s.d. (n = 3).
Localization of OsXTH19 and xyloglucan
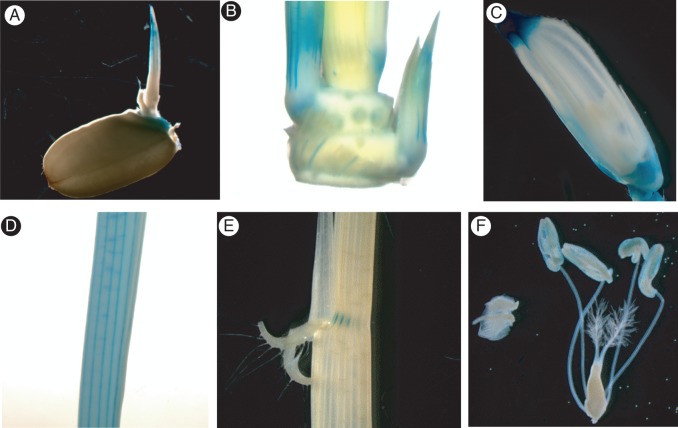
To assess the tissue-specific expression of OsXTH19, we generated transgenic rice lines expressing the β-glucuronidase (GUS) gene fused to the promoter of OsXTH19 (pOsXTH19::GUS). Expression of pOsXTH19::GUS was prominent in the elongating zones of both leaves and internodes of rice plants (Figs 3A and 4). After full elongation of the stem, GUS staining was undetectable. A more detailed analysis of transverse sections of the elongating region of the internode revealed that pOsXTH19::GUS was mainly expressed in the large vascular bundles, particularly in cells surrounding the protoxylem (Fig. 3Aii, iii). In transverse sections of mature stems, expression of this reporter gene was absent (data not shown). We also found that pOsXTH19::GUS was expressed in a variety of tissues, such as endosperm caps during germination and in the tillers and collars during vegetative growth. After the transition to the reproductive phase of growth, GUS activity was high at the tops and bottoms of young florets, whereas it was quite low at the anthers (Fig. 4).
Fig. 3.
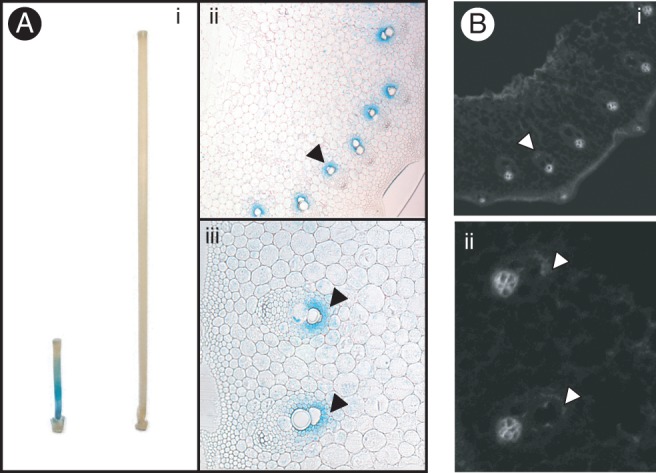
(A)Histochemical analysis of OsXTH19 expression in the second internodes of transgenic rice plants carrying pOsXTH19::GUS. (i) GUS staining in an elongating internode (left) and a fully expanded internode (right). (ii) GUS localization in transverse sections of an elongating internode. (iii) High-magnification view of vascular tissues of the transverse section shown in (ii). GUS expression is indicated by arrowhead. Bars: 1 cm in (i). (B) Immunohistochemical staining of xyloglucan associated with the stems of wild-type rice plants. (i) Immunofluorescent staining of 70-mm-thick transverse sections taken from an elongating second internode. Sections were stained with antibodies against xyloglucan (LM15). (ii) High-magnification view of vascular tissues of the transverse section shown in (i). Anti-xyloglucan staining, which overlaps with GUS localization, is indicated by arrowheads. PX, protoxylem.
Fig. 4.
Histochemical localization of OsXTH19 expression using pOsXTH19::GUS: (A) 3-d-old seedling, (B) tillers, (C) flower, (D) young leaf, (E) lamina joint and (F) pistil and stamens.
To confirm that xyloglucan is present in tissues where pOsXTH19::GUS is expressed, we examined the tissues using an immunofluorescence assay with monoclonal antibody LM15, which specifically recognizes the xylosylated epitope of xyloglucan (Marcus et al., 2008). In the large vascular bundles, there was weak but clear fluorescence in cells surrounding the protoxylem, where OsXTH19 was moderately but stably expressed (Fig. 3B). There was also a prominent signal for the xyloglucan epitope in other tissues, including the phloem cell walls, although pOsXTH19::GUS was not expressed in this tissue.
Function of OsXTH19 in rice plants
To assess the effects of loss of function of the OsXTH19 on rice growth and development in planta, we generated ten transgenic rice lines expressing an OsXTH19 RNAi construct. The RNAi construct was designed to express two copies of a 3′ untranslated region fragment of OsXTH19 mRNA connected inversely under the control of the OsXTH19 promoter. We chose three homozygous transgenic RNAi lines (R3, R4 and R10) for further characterization. The level of OsXTH19 mRNA was slightly but significantly reduced in two transgenic lines, R3 and R10, compared with that of the wild type, whereas a significant reduction was not observed for line R4 (Fig. 5).
Fig. 5.
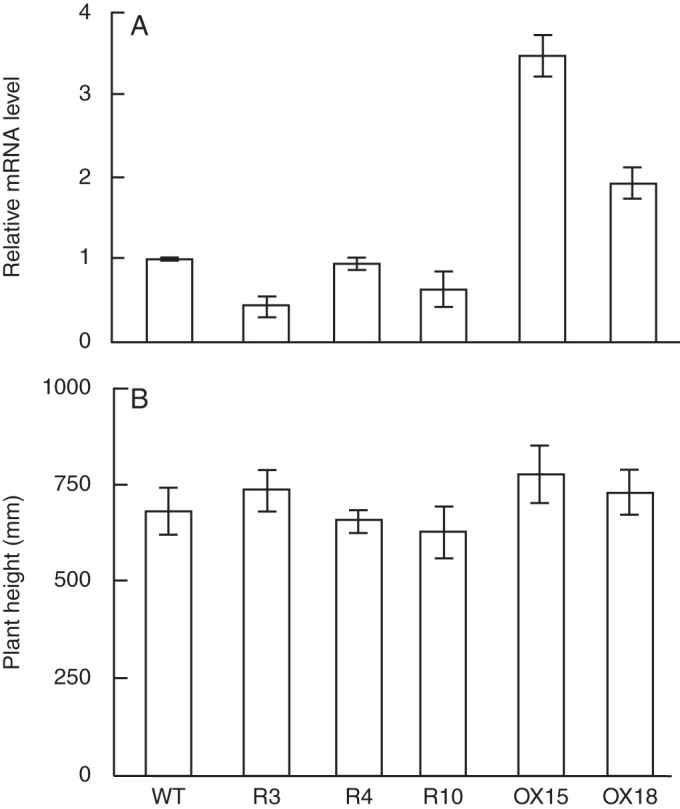
Analysis of OsXTH19 RNAi-expressing and OsXTH19-over-expressing transgenic plants. (A) Expression of OsXTH19 in 7-d-old rice seedling. OsXTH19 transcript levels were examined by real-time RT-PCR in wild-type (WT), OsXTH19 RNAi (lines R3, R4 and R10), and OsXTH19-over-expressing (OX15 and OX18) transgenic plants. Relative transcript levels of OsXTH19 compared with those for the WT plant are shown. Results represent the means ± s.d. of triplicate biological samples. (B) Comparison of the heights of WT, OsXTH19 RNAi and OsXTH19-over-expressing transgenic plants grown for 28 d under normal growth conditions. Results represent the means ± s.d. of eight plants for each line.
To examine the effects of ectopic over-expression of OsXTH19, we generated ten independent transgenic lines in which transcription of OsXTH19 mRNA was driven by the CaMV35S promoter. We selected two homozygous T2 lines, OX15 and OX18, which expressed relatively high levels of the mRNA for further characterization of growth parameters (Fig. 5). At the macroscopic level, neither down-regulation by RNAi nor over-expression by the CaMV35S promoter affected growth parameters, including plant height (Fig. 5, Supplementary Data Fig. S2), although morphometric analysis of these transgenic lines showed statistically significant changes in certain growth parameters, such as leaf length (line R3), number of tillers (lines OX15, OX18 and R3) and length of the second internode (line OX15) (Table 3).
Table 3.
Morphometric analysis of wild-type and transgenic plants with OsXTH19 gene expression modified
| Wild-type | OsXTH19 over-expression lines |
OsXTH anti-sense lines |
|||
|---|---|---|---|---|---|
| OX15 | OX18 | R3 | R4 | R10 | |
| Leaf length (mm) | |||||
| 342·5 ± 18·7 | 389·9 ± 52·3 | 316·0 ± 67·6* | 322·0 ± 48·6 | 321·1 ± 43·5* | 381·4 ± 36·8 |
| No. of tillers | |||||
| 5·3 ± 0·9 | 3·3 ± 0·9* | 7·8 ± 1·7* | 3·8 ± 1·4* | 4·4 ± 1·4 | 5·3 ± 2·1 |
| Stem length (mm) | |||||
| Internode I | |||||
| 225·1 ± 24·2 | 191·6 ± 43·9 | 225·5 ± 31·5 | 206·0 ± 18·6 | 197·9 ± 48·1 | 224·1 ± 19·3 |
| Internode II | |||||
| 108·0 ± 15·3 | 81·3 ± 19·2* | 103·0 ± 27·2 | 98·4 ± 14·5 | 95·9 ± 21·7 | 114·4 ± 12·2 |
| Stem diameter (mm) | |||||
| Internode I | |||||
| 1·1 ± 0·1 | 1·3 ± 0·2 | 1·2 ± 0·1 | 1·1 ± 0·1 | 1·0 ± 0·2 | 1·1 ± 0·1 |
| Internode II | |||||
| 2·1 ± 0·2 | 2·5 ± 0·4 | 2·2 ± 0·3 | 2·2 ± 0·2 | 1·9 ± 0·3 | 2·2 ± 0·2 |
Data are means ± s.d. (n = 8 plants). Asterisks indicate values of wild-type plants that are significantly different between wild-type and transgenic plants as determined by Student's t test (P < 0·05).
DISCUSSION
All three OsXTHs examined in the present study had a strict preference for xyloglucan as a substrate. OsXTH19 and OsXTH20 had only XEH activity, as was predicted by crystallography analysis (Baumann et al., 2007; Eklöf and Brumer, 2010). By contrast, OsXTH11 had not only XET activity but also XEH activity, despite being categorized as a member of Group I/II. Contamination with hydrolytic activity from the yeast culture can be excluded, as we used a highly purified recombinant protein preparation. Thus, this study demonstrates that the principal substrate for these OsXTHs is xyloglucan.
To our knowledge, Group I/II XTHs capable of mediating both XET and XEH activities have not been previously predicted (Baumann et al., 2007) or reported. Therefore, we propose that OsXTH11 is a new type of Group I/II XTH that exhibits dual enzymatic activity. Whether this type of bifunctional Group I/II XTH is restricted to commelinoid monocots or is distributed more widely in eudicotyledons with type I walls is currently unknown, which raises the question of how the Group I/II subfamily of XTHs evolved during the diversification of angiosperms. Further studies are needed to determine the structural basis for this dual function of OsXTH11.
The current study shows that xyloglucan functions as a substrate for the three rice XTHs, OsXTH11, 19 and 20, suggesting the possibility that xyloglucan is the prime candidate of the substrate for XTHs in rice. However, why the XTH gene family in rice is at least as large as that in A. thaliana, despite the much lower level of xyloglucan in rice, remains unanswered. It is possible that xyloglucans are restricted to certain types of tissues in commelinoid monocotyledons and that, although quantitatively less abundant on a whole-plant basis, xyloglucan is a major glycan in specific tissues and plays an important role in controlling cell-wall properties. Indeed, our immunofluorescence results support the idea of cell type-specific localization of xyloglucan.
Several questions remain, including whether xyloglucan is truly a substrate of all XTHs in commelinoid monocots. Moreover, if xyloglucan is not a substrate for some XTHs, what is the genuine substrate for these XTHs? As grass cell walls contain high levels of GAX and MLG, and these glycans may function as load-bearing molecules in type II cell walls (Carpita, 1996; Cosgrove, 2005; Vogel, 2008), one can speculate that some XTH proteins in grasses may be involved in the modification of GAX or MLG. This idea is supported by a report by Hrmova et al. (2007), who found that certain XTHs purified from barley have very low but measureable transglycosylation activity when MLG is used as the donor substrate. Fry et al. (2008) reported that horsetail (Equisetum species), a pteridophyte, contains endotransglucosylase activity that can mediate transglycosylation between MLG and xyloglucan oligosaccharides, implying that land plants have enzymes capable of catalysing hetero-transglycosylation (Fry et al., 2008).
The results of the current study show that OsXTH11, OsXTH19 and OsXTH20 have very weak, if any, activity on non-XG polysaccharides, and that xyloglucan : MLG hetero-transglycosylation activity was about 0·3 % of the xyloglucan : xyloglucan homo-transglycosylation activity in the case of OsXTH11. Therefore, it may be expected that the main function of these XTHs is to work on xyloglucans. Mohler et al. (2013) did not detect hetero-transglycosylation activity between xyloglucan and MLG in barley, despite the fact that barley contains both of its substrates and high XET activity. Thus, although the results of the present study do not exclude the possibility that other XTHs in Poales species can mediate still unknown enzymatic activity, a biological role for transglycosylation activity for polysaccharides other than xyloglucan has not been demonstrated in rice. Thus, the biological implications of hetero-transglycosylation in Poales, if any, remain unclear.
What is the function of xyloglucan and XTHs in controlling the growth and development of rice? Neither the OsXTH19 RNAi lines nor the OsXTH19 over-expression lines showed gross morphological changes. This, however, is not surprising, as similar outcomes have been reported in RNAi lines or knockout mutants for several arabidopsis XTH genes (Nishitani, 2002; Osato et al., 2006).
One possible reason for this apparent lack of phenotype in XTH transgenic lines is functional redundancy within the rice XTH family, as was demonstrated in the arabidopsis XTH family. This notion is supported by the results from Jan et al. (2004), who showed that XTH transcripts in rice are down-regulated in a non-specific manner when the rice ectopically expresses an OsXTH RNAi construct under control of the CaMV35S promoter. In these RNAi lines, shoot growth was moderately reduced, suggesting a role for the XTH family in rice growth. The apparent lack of a phenotype in our transgenic lines may also have been due to the fact that the phenotype may have been too subtle to be noticed by a rough survey of growth parameters and morphological observation due to tissue- and growth-stage-specific expression.
Zhu et al. (2012) reported that a single knockout line of Group III-A XTHs, xth31, showed a subtle but significant reduction in root elongation and more aluminium resistant than the wild type. By contrast, Kaewthai et al. (2013) showed that double knockout lines of arabidopsis Group III-A XTHs, AtXTH31 and AtXTH32, which completely lack xyloglucan hydrolase activity, do not exhibit significant growth or developmental phenotypes, suggesting that the morphological effects due to the loss of function of xyloglucan-specific hydrolases are either subtle or may be compensated for by other mechanisms.
This consideration can be applied to the role of OsXTH19 in rice growth and development as a xyloglucan-specific hydrolase. Further detailed analysis of transgenic rice lines under various growth conditions are needed to elucidate the primary function of individual members of the OsXTH family.
The phylogeny of xyloglucan and the XTH gene family in plants indicates that the xyloglucan/XTH system emerged and diversified during the evolution of embryophytes (Popper and Tuohy, 2010; Yokoyama et al., 2010; Fangel et al., 2012). The marked divergence in this system between commelinoid monocots and eudicotyledons may reflect parallel evolution of structural features of xyloglucan and enzymatic properties of the XTH family of proteins. To help clarify the evolution of the xyloglucan/XTH system, it will be important to better understand XTH-mediated remodelling of the cellulose/xyloglucan framework in embryophytes. In the current study, we showed that at least three of the rice XTHs apparently remodel xyloglucan. A further challenge will be to determine the enzymatic functions of other OsXTHs, such as OsXTH11 and OsXTH19, and how they regulate the growth of plants.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Area ‘Plant cell wall as information-processing system’ (grant numbers 24114001 and 24114005) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Professor Harry Brumer III of the University of British Columbia and Dr Kiyohiko Igarashi of The University of Tokyo for helpful discussions and critical reading of the manuscript.
LITERATURE CITED
- Baumann MJ, Eklöf JM, Michel G, et al. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. The Plant Cell. 2007;19:1947–1963. doi: 10.1105/tpc.107.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Molecular Biology. 2006;61:451–467. doi: 10.1007/s11103-006-0021-z. [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, et al. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. The Plant Cell. 2002;14:3073–3088. doi: 10.1105/tpc.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M, Harris PJ. Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Molecular Plant. 2011;4:144–156. doi: 10.1093/mp/ssq067. [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. The Plant Journal. 1999;18:371–382. doi: 10.1046/j.1365-313x.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC. The structure and biogenesis of the cell walls of grasses. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 2008;20:1519–1537. doi: 10.1105/tpc.108.059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Del Bem LEV, Vincentz M. Evolution of xyloglucan-related genes in green plants. BMC Evolutionary Biology. 2010;10(341) doi: 10.1186/1471-2148-10-341. doi:10.1186/1471-2148-10-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiology. 2010;153:456–466. doi: 10.1104/pp.110.156844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangel JU, Ulvskov P, Knox JP, et al. Cell wall evolution and diversity. Frontiers in Plant Science. 2012;3:152. doi: 10.3389/fpls.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiology. 2009;149:27–37. doi: 10.1104/pp.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosing enzyme activity from plants. Biochemical Journal. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiologia Plantarum. 1993;89:1–3. [Google Scholar]
- Fry SC, Mohler KE, Nesselrode BHWA, Frankova L. Mixed-linkage β-glucan:xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. The Plant Journal. 2008;55:240–252. doi: 10.1111/j.1365-313X.2008.03504.x. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, et al. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiology. 2006;140:946–962. doi: 10.1104/pp.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesi V, Fornalé S, Fry SC, et al. ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. Journal of Experimental Botany. 2008;59:875–889. doi: 10.1093/jxb/ern013. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Pauly M, Bacic A, Fincher GB. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta. 2005;221:729–738. doi: 10.1007/s00425-005-1481-0. [DOI] [PubMed] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annual Review of Plant Physiology Plant Molecular Biology. 1989;40:139–168. [Google Scholar]
- Henriksson H, Denman SE, Campuzano IDG, et al. N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochemical Journal. 2003;375:61–73. doi: 10.1042/BJ20030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Farkas V, Lahnstein J, Fincher GB. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates and (1,3;1,4)-β-d-glucans. Journal of Biological Chemistry. 2007;282:12951–12962. doi: 10.1074/jbc.M611487200. [DOI] [PubMed] [Google Scholar]
- Hsieh YS, Harris PJ. Xyloglucans of monocotyledons have diverse structures. Molecular Plant. 2009;2:943–965. doi: 10.1093/mp/ssp061. [DOI] [PubMed] [Google Scholar]
- Inouhe M, Yamamoto R, Masuda Y. Auxin-induced changes in the molecular weight distribution of cell wall xyloglucans in Avena coleoptiles. Plant and Cell Physiology. 1984;25:1341–1351. [Google Scholar]
- Jan A, Yang G, Nakamura H, et al. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiology. 2004;136:3670–3681. doi: 10.1104/pp.104.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewthai N, Gendre D, Eklöf JM, et al. Group III-A XTH genes of Arabidopsis encode predominant xyloglucan endohydrolases that are dispensable for normal growth. Plant Physiology. 2013;161:440–454. doi: 10.1104/pp.112.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas AM, Piens K, Denman SE, et al. Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula×tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochemical Journal. 2005;390:105–113. doi: 10.1042/BJ20041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Matsuda K. Xyloglucan in the cell walls of suspension-cultured rice cells. Plant and Cell Physiology. 1985;26:437–445. [Google Scholar]
- Kato Y, Ito S, Iki K, Matsuda K. Xyloglucan and β-d-glucan in cell walls of rice seedlings. Plant and Cell Physiology. 1982;23:351–364. [Google Scholar]
- Marcus SE, Verhertbruggen Y, Hervé Ordaz-Ortiz JJ, et al. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biology. 2008;8(60) doi: 10.1186/1471-2229-8-60. doi:10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen J-P, Vissenberg K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. Journal of Experimental Botany. 2009;60:3959–3972. doi: 10.1093/jxb/erp229. [DOI] [PubMed] [Google Scholar]
- Mohler KE, Simmons TJ, Fry SC. Mixed-linkage glucan: xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytologist. 2013;197:111–122. doi: 10.1111/j.1469-8137.2012.04371.x. [DOI] [PubMed] [Google Scholar]
- Moore PJ, Staehelin LA. Immunogold localization of the cell-wall matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.: implication for secretory pathways. Planta. 1988;178:353–366. doi: 10.1007/BF00634471. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokoyama T, Tomita E, Nishitani K. Two azuki bean XTH genes, VaXTH1 and VaXTH2, with similar tissue-specific expression profiles, are differently regulated by auxin. Plant and Cell Physiology. 2003;44:16–24. doi: 10.1093/pcp/pcg002. [DOI] [PubMed] [Google Scholar]
- Nishitani K. A novel method for detection of endo-xyloglucan transferase. Plant and Cell Physiology. 1992;33:1159–1164. [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell wall. International Review of Cytology. 1997;173:157–206. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Nishitani K. Genome-based approach to study the mechanism by which cell-wall type is defined and constructed by means of collaborative actions of wall-related enzymes. Journal of Plant Research. 2002;115:303–307. doi: 10.1007/s10265-002-0032-z. [DOI] [PubMed] [Google Scholar]
- Nishitani K, Masuda Y. Auxin-induced changes in the cell wall structure: changes in the sugar compositions, intrinsic viscosity and molecular weight distributions of matrix polysaccharides of the epicotyl cell wall of Vigna angularis. Physiologia Plantarum. 1981;52:485–494. [Google Scholar]
- Nishitani K, Tominaga R. In vitro molecular weight increase in xyloglucans by an apoplastic enzyme preparation from epicotyls of Vigna angularis. Physiologia Plantarum. 1991;82:490–497. [Google Scholar]
- Nishitani K, Tominaga R. Endoxyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. Journal of Biological Chemistry. 1992;267:21058–21064. [PubMed] [Google Scholar]
- Nishitani K, Vissenberg K. Roles of the XTH protein family in the expanding cell. Plant Cell Monographs. 2007;6:89–116. [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research. 2006;119:153–162. doi: 10.1007/s10265-006-0262-6. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Davies WJ. An analysis of relative elemental growth rate, epidermal cell size and xyloglucan endotransglycosylase activity through the growing region of ageing maize leaves. Journal of Experimental Botany. 1996;47:339–347. [Google Scholar]
- Pena MJ, Darvill AG, Eberhard S, York WS, O'Neill MA. Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology. 2008;18:891–904. doi: 10.1093/glycob/cwn078. [DOI] [PubMed] [Google Scholar]
- Popper ZA. Evolution and diversity of green plant cell walls. Current Opinion in Plant Biology. 2008;11:286–292. doi: 10.1016/j.pbi.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Tuohy MG. Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiology. 2010;153:373–383. doi: 10.1104/pp.110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, et al. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual Review of Plant Biology. 2011;62:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Hetehrington PRE, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. Journal of Experimental Botany. 1993;44:1281–1289. [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Smith RC, Matthews PR, Schunmann PHD, Chandler PM. The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in Himalay barley (Hordeum vulgare L) Journal of Experimental Botany. 1996;47:1395–1404. [Google Scholar]
- Toki S, Hara N, Ono K. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. The Plant Journal. 2006;47:961–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M. Characterization of XET-related genes of rice. Plant Physiology. 2000;122:853–860. doi: 10.1104/pp.122.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken J-P, York WS, Beldman G, Voragen AGJ. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiology. 1997;114:9–13. doi: 10.1104/pp.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. Unique aspects of the grass cell wall. Current Opinion in Plant Biology. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. Root growth maintenance at low water potentials. Plant Physiology. 1994;106:607–615. doi: 10.1104/pp.106.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zabotina O, Hong M. Pectin–cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry. 2012;51:9846–9856. doi: 10.1021/bi3015532. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. Functional diversity of xyloglucan-related proteins and its implications in the cell wall dynamics in plants. Plant Biology. 2000;2:598–604. [Google Scholar]
- Yokoyama R, Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant and Cell Physiology. 2004;45:1111–1121. doi: 10.1093/pcp/pch151. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiology. 2004;134:1088–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Uwagaki Y, Sasaki H, et al. Biological implications of the occurrence of 32 members of XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. The Plant Journal. 2010;64:658–659. doi: 10.1111/j.1365-313X.2010.04351.x. [DOI] [PubMed] [Google Scholar]
- Zhu XF, Shi YZ, Lei GJ, et al. XTH31, encoding an in-vitro XEH/XET-active enzyme, controls Al sensitivity by modulating in-vivo XET action, cell wall xyloglucan content and Al binding capacity in Arabidopsis. Plant Cell. 2012;24:4731–4747. doi: 10.1105/tpc.112.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.