Abstract
Most cases of cystic fibrosis (CF) are caused by mutations that block the biosynthetic maturation of the CF gene product, the CF transmembrane conductance regulator (CFTR) chloride channel. CFTR-processing mutants fail to escape the endoplasmic reticulum and are rapidly degraded. Current efforts to induce the maturation of CFTR mutants target components of the biosynthetic pathway (e.g., chaperones) rather than CFTR per se. Such methods are inherently nonspecific. Here we show that the most common CF-causing mutant (ΔF508-CFTR) can form mature, functional chloride channels that reach the cell surface when coexpressed with several other CFTR-processing mutants or with amino fragments of the wild-type CFTR protein. This transcomplementation effect required a specific match between the region flanking the disease-causing mutation and the complementing fragment; e.g., amino fragments complemented ΔF508-CFTR but not H1085R (a carboxy-processing mutant), whereas a carboxy fragment complemented H1085R but not ΔF508-CFTR. Transcomplementing fragments did not affect CFTR interactions with Hsc70, a chaperone previously implicated in CFTR biosynthesis. Instead, they may promote CFTR maturation by blocking nonproductive interactions between domains within the same or neighboring CFTR polypeptides that prevent normal processing. These findings indicate that it may be possible to develop CF therapies (e.g., mini-cDNA constructs for gene therapy) that are tailored to specific disease-causing mutants of CFTR.
Cystic fibrosis (CF) transmembrane conductance regulator (CFTR) is a complex polytopic membrane protein with several large cytosolic domains and two cytoplasmically oriented tails (see Fig. 1 A). Each of these domains is a site for CF-causing mutations that disrupt biosynthetic processing. The most common mutation is the ΔF508 mutation, which localizes to the first nucleotide-binding domain (NBD1) and is present on ≈90% of all CF chromosomes. Little or no ΔF508-CFTR protein escapes the endoplasmic reticulum (ER) under normal conditions; instead, this mutant is rapidly degraded by the proteasome (1). How the ΔF508 mutation and other CF-causing mutations block CFTR exit from the ER is unknown. Possible mechanisms include the exposure of ER retention signals [i.e., amino acid sequences that promote ER retention (2)], the occlusion of ER exit codes (3), or aggregation caused by local misfolding.
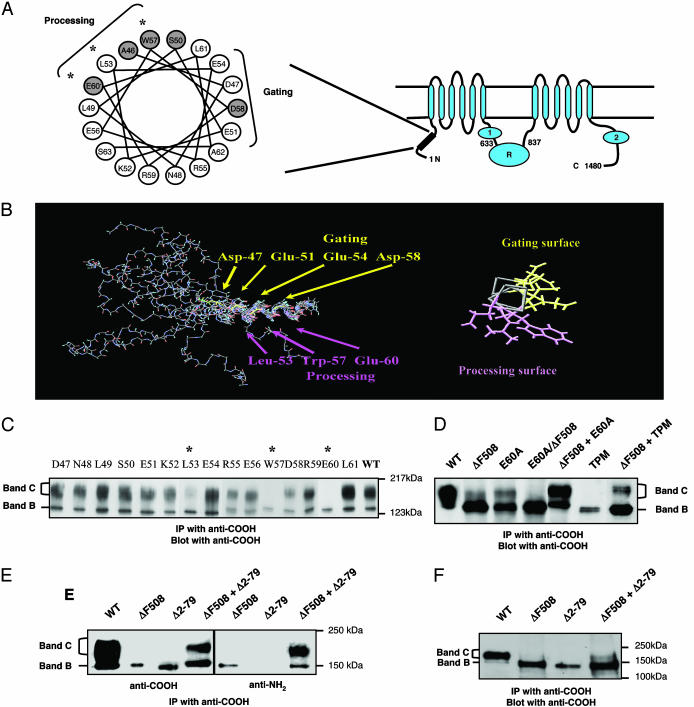
Fig. 1.
Identification of N-tail processing mutants that transcomplement ΔF508-CFTR. (A) CFTR topology showing the two cytoplasmic tails NBD1 and NBD2 and the regulatory (R) domain. (Left) Helical wheel representation of amino acids 46–63 in the N-tail. Amino acids that are sites for CF mutations (www.genet.sickkids.on.ca) are shaded. (B) NMR structure of a synthetic N-tail peptide (amino acids 30–63) depicting an α-helix from residues 47–62. The end-on representation illustrates two distinct surfaces, one implicated in channel gating (18) and the other in CFTR processing (present findings). (C) Single alanine-substituted mutants of CFTR expressed in COS-7 cells. Asterisks indicate processing mutants for which band B (the core-glycosylated form of CFTR) is predominant. CFTR protein was detected 24 h after transfection as described in Methods. (D) Coexpression of ΔF508-CFTR with N-tail processing mutants increased the amount of mature protein (band C). A CFTR polypeptide that harbored two mutations in cis (E60A/ΔF508) exhibited no band C formation. (E) Transcomplementation of ΔF508-CFTR by coexpression with a deletion mutant that lacks the N-tail Δ2–79-CFTR. (F) Coexpression of ΔF508-CFTR and Δ2–79-CFTR led to a modest appearance of band C in human embryonic kidney cells, HEK293T. All results are representative of three to five experiments.
A method that reliably and specifically induces the processing of CFTR mutants in patients does not exist. (We use the term “processing” to indicate the appearance of the mature, complex glycosylated protein.) Several nonspecific maneuvers that affect chaperone expression and/or CFTR-chaperone interactions can promote ΔF508-CFTR processing in tissue culture cells (e.g., growth at low temperature and high concentrations of chemical chaperones, such as glycerol, or calcium pump inhibitors) (4–7). Whether these maneuvers can be developed into therapeutically useful strategies is unknown. Gene therapy is a possible alternative, but current efforts to introduce the wild-type gene into CF cells are hampered by a number of factors (reviewed in ref. 8), including the relatively large size of the CFTR DNA, which limits the choice of suitable vectors.
We reasoned that it may be possible to use a transcomplementation strategy to induce the processing of mutants, such as ΔF508-CFTR, by coexpressing them with other mutants or with specific polypeptide fragments of the wild-type CFTR. The rationale for these experiments was based on the fact that there exists multiple interactions between domains within each CFTR polypeptide (9, 10) and perhaps between neighboring CFTR polypeptides. [Although controversial, there have been recent reports that CFTR polypeptides may form dimers under certain conditions (11).] Polypeptide fragments that associate with ΔF508-CFTR protein conceivably could occlude ER retention signals or provide ER exit codes in trans that promote protein export to the Golgi (2, 3). A strategy that targets CFTR per se should be more specific than methods that target chaperones. The identification of peptide fragments able to rescue the processing of CF mutants would have value as prospective gene therapy constructs. Here we report that we could overcome the processing defect of ΔF508-CFTR in vitro by coexpressing this mutant with specific fragments of the wild-type polypeptide.
Methods
DNA Constructs. All point mutations were introduced into a 1-kb fragment (from the start codon to the BspMII site) of the human CFTR coding region by the QuickChange Site-Directed Mutagenesis Kit (Stratagene). This fragment was sequenced completely to verify each mutation and then ligated into pcDNA3 vector (Invitrogen) containing the rest of the CFTR coding region. CFTR fragments (1–633 and 837-1,480) were generated by PCR and introduced into pcDNA3 after digestion with Asp-718/XhoI. The 1–633 fragment was tagged at the carboxy terminus with V5 epitope by introducing this fragment into the pcDNA3.1/V5-His vector (Invitrogen).
Cell Culture, Transfections, and Immunoprecipitations (IPs). COS-7 cells and HEK293T cells were cultured in DMEM media (GIBCO) supplemented with 10% FBS (Sigma) and penicillin/streptomycin (GIBCO). The COS-7 cells were infected/transfected by using recombinant vaccinia virus as described (12) and analyzed 48 h after transfection unless stated otherwise. HEK293T cells were transiently transfected by using FuGENE6 (Roche) following the manufacturer's recommendations and analyzed for CFTR expression 6 days after transfection. Plasmid DNA amounts used for COS-7 and HEK293T cells were 2 μg for full-length CFTR (wild type or mutants) and 1 μg for fragment 1–633 or 837-1,480 per 35-mm dish. Cells were lysed in 0.2% Triton X-100 in PBS (COS-7) or 1% Triton X-100 in PBS (HEK293T) and a mixture of protease inhibitors (Roche). CFTR was subjected to IP by using 1 μg of carboxy-terminal CFTR monoclonal antibody (R & D Systems, 24–1) or V5 antibody (Invitrogen) cross-linked to A/G agarose beads (Santa Cruz Biotechnology). After incubating the lysates with cross-linked antibody, the beads were pelleted and washed three times in PBS containing 0.2% Triton X-100, and the samples were processed for Western blotting by using the carboxy-terminal CFTR antibody, a polyclonal NBD1-R CFTR antibody (a gift from J. Collawn, University of Alabama, Birmingham), a monoclonal NH2-terminal CFTR antibody (Chemicon), a monoclonal Hsc70 antibody, or a polyclonal Hsp70 antibody (Santa Cruz Biotechnology) as indicated in the figures.
The glycosylation status of transcomplemented ΔF508-CFTR protein was assessed by treating ΔF508-CFTR immunoprecipitates prepared from lysates of transfected COS-7 cells with N-glycosydase F (2 units per sample; Boehringer Mannheim, which is now Roche Molecular Biochemicals) or endoglycosidase H (10 milliunits per sample; Sigma) for 90 mins by following manufacturers' protocols. The samples were then analyzed by immunoblotting.
Patch Clamp Experiments. CFTR channels were analyzed in inside-out membrane patches excised from COS-7 cells 24 h after transfection. Bath and pipette solution contained (in mM) 140 N-methyl-d-glucamine, 10 TES, 3 MgCl2, and 1 EGTA (pH adjusted to 7.3 with HCl). Pipettes were pulled from Corning 8161 glass-to-tip resistances of 4–8 mOhm. CFTR channels were activated with 200 units/ml protein kinase A catalytic subunit (Promega) and 1.5 mM Mg-ATP. CFTR channels were identified on the basis of their single-channel conductances (6–8 pS), linear current–voltage behavior, and voltage-dependent block by glibenclamide (250 μM) (13).
SPQ [6-Methoxy-N-(3-sulfopropyl)quinolinium] Assay. Transport activity in COS-7 cells transfected with CFTR constructs was assayed as the increase in fluorescence of an intracellular dye, SPQ, that is quenched by halides. Briefly, cells were grown on coated glass coverslips (Vectabond reagent, Vector Laboratories) in DMEM media supplemented with serum. Cells were loaded by hypotonic shock at 37°C for 15 min in Opti-MEM/water (1/1) containing 10 mM SPQ, washed, and then mounted in a perfusion chamber for fluorescence measurements. Fluorescence of single cells was measured with a Zeiss inverted microscope, a PTI (Lawrenceville, NJ) imaging system, and a Hamamatsu (Tokyo) camera. Excitation was at 340 nm, and emission was >410 nm. All functional studies were at 23°C. At the beginning of the experiments, cells were bathed in a quenching buffer (NaI) and, after establishment of a stable baseline, switched to a halide-free (NO3) dequenching buffer at 200 s and to an activating mixture (10 μM forskolin/100 μM 8-(4-chlorophenylthio)-cAMP/100 μM IBMX) in the same NO3 buffer at 500 s. Fluorescence was normalized to the baseline (quenched) value (average fluorescence from 100–200 s), with increases presented as percent increase in fluorescence over baseline. The buffers used in the SPQ assay were NaI buffer [130 mM NaI/5 mM KNO3/2.5 mM Ca(NO3)2/2.5 mM Mg(NO3)2/10 mM d-glucose/10 mM N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic) acid (Hepes, pH 7.30)] and NaNO3 buffer (identical to NaI buffer except that 130 mM NaNO3 replaced NaI).
Structural Analysis of N-tail Peptide. Far UV CD spectra were obtained on a peptide fragment of CFTR (amino acids 30–63) by using a spectropolarimeter (model 62DS, Aviv Associates, Lakewood, NJ). A dramatic increase in helical content was demonstrated up to 20% 2,2,2-trifluoroethanol (TFE). NMR spectra (Bruker DRX400) also demonstrated a similar change in chemical shifts and NOESY contacts with TFE. Two-dimensional NMR spectra (COSY, NOESY, and total correlation spectroscopy) of the peptide (4 mM) in a 25:75 (vol/vol) TFE:100 mM NaOAc, pH 5, solution were obtained at 20°C. The peptide was modeled with x-plor (14) by using nuclear Overhauser effect and J-coupling constant constraints obtained from NMR by using a standard distance geometry-simulated annealing protocol (15). A family of structures was obtained (10 shown in Fig. 1B), each of which was consistent with residues Asp-47 through Glu-60 being present in an α-helix.
Results
Identification of Amino-Terminal Processing Mutants That Transcomplement ΔF508-CFTR. To test whether neighboring CFTR molecules could associate with ΔF508-CFTR within the ER and promote its export, we coexpressed the ΔF508-CFTR mutant with processing mutants that harbor mutations in the aminoterminal cytoplasmic tail (N-tail) (Fig. 1). We used N-tail mutants for these initial complementation experiments because this tail is essential for CFTR protein processing (16, 17) and is the site for multiple CF-causing mutations (Fig. 1 A). In pilot experiments, we identified a helical region within the N-tail (amino acids 46–62) that plays a significant role in CFTR processing and channel gating (18) (Figs. 1 B and C). This region, which is predicted to form an α-helix by secondary structure algorithms, was confirmed to adopt a helical structure by 2D NMR analysis of a synthetic N-tail peptide in solution (Fig. 1B). In a mutational analysis of this helix, we observed that a cluster of residues (L53, W57, and E60) that maps to one surface of the helix is essential for CFTR processing (Fig. 1C). CFTR traffic from the ER to the Golgi can be monitored by SDS/PAGE analysis in which the immature ER form and the mature postGolgi form migrate at ≈150 kDa (band B) and 170–190 kDa (band C), respectively (Fig. 1C). Unlike wild-type CFTR, these N-tail processing mutants exhibit primarily the immature, band B form of the protein when expressed in COS-7 cells (Fig. 1). Replacing all three of these residues with alanine [triple processing mutant (TPM)], resulted in a complete loss of the mature, band C form of the protein (Fig. 1D), whereas mutating the other residues along this helix had no effect.
Figs. 1 D and E show that the steady-state amount of mature ΔF508-CFTR protein can be enhanced substantially by coexpressing this mutant with the N-tail processing mutants (e.g., E60A-CFTR) in COS-7 cells by using a strong viral expression system (vaccinia virus). This effect requires that the mutations be present on different polypeptides (i.e., in trans). No cis complementation was observed when the E60 mutation and the ΔF508 mutation were introduced into the same polypeptide (Fig. 1D). The most striking result (and easiest to interpret) was obtained when we coexpressed the ΔF508 mutant with a processing mutant that lacks the entire N-tail (Δ2–79-CFTR) (Fig. 1E). A robust band C signal could be obtained by using an N-tail antibody that recognizes only ΔF508-CFTR; thus, a portion of the mature protein that was detected under these conditions must represent ΔF508-CFTR protein. Qualitatively similar results were obtained when we coexpressed ΔF508-CFTR and Δ2–79-CFTR in HEK-293T cells by using a standard lipid transfection method (Fig. 1F), although the transcomplementation signal obtained in this expression system was not as strong. Because the Δ2–79-CFTR mutant is blocked for ER export, even under conditions that promote the processing of the slightly “leaky” ΔF508 mutant (e.g., growth at low temperature; results not shown), Δ2–79-CFTR presumably interacts initially with ΔF508-CFTR within this compartment.
ΔF508-CFTR Can Be Transcomplemented by Amino Fragments of the Wild-Type Protein. We next determined whether smaller fragments of the wild-type protein could transcomplement CFTR-processing mutants, such as ΔF508-CFTR. We observed no effect of expressing the N-tail alone (amino acids 1–79) on Δ2–79-CFTR processing in COS-7 cells or of expressing NBD1 alone (amino acids 433–634) on ΔF508-CFTR processing (results not shown). These negative results could be caused by the poor expression of these soluble fragments in COS-7 cells (data not shown). Conversely, a larger amino fragment that includes the N-tail, the first membrane-spanning domain and NBD1 (1–633) was able to transcomplement the ΔF508 mutant (Fig. 2 A; see also quantitation in Fig. 4F). This fragment could also rescue the processing of N-tail mutants (Δ2–79-CFTR and TPM-CFTR) in vaccinia-infected COS-7 cells and in HEK-293T cells analyzed 6 days after cotransfection with a lipid reagent, FuGENE6 (Figs. 2 B and C and data not shown). Similar results were obtained for HEK-293T cells 2 days after transfection with another lipid reagent (Lipofectamine 2000, Invitrogen) that exhibited higher transfection efficiency in these cells (data not shown). In vaccinia-infected COS-7 cells, mature ΔF508-CFTR protein could be detected biochemically by IP within 48 h of cotransfection with 1–633, whereas expression of the 1–633 fragment could be detected by immunoblotting in the lysate 24 h after transfection. (Note that mature ΔF508-CFTR could not be detected by immunoblotting alone at either time point; thus, the fragment was expressed at considerably higher levels than the full-length protein). Functional channels could be detected at the cell surface within 24 h of cotransfection by using more sensitive transport assays, as discussed below.
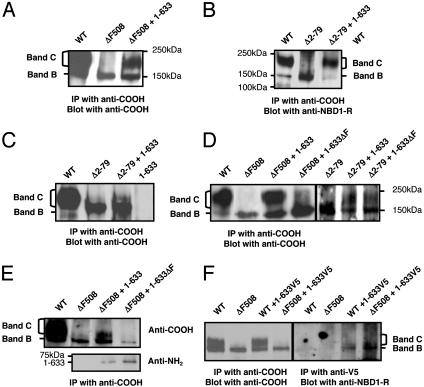
Fig. 2.
Transcomplementation of ΔF508-CFTR and N-tail processing mutants by an amino fragment (amino acids 1–633) of wild-type CFTR. (A and B) Coexpression of 1–633 with ΔF508-CFTR (A) or with Δ2–79-CFTR (B) in vacciniainfected COS-7 cells resulted in the appearance of band C. (C) Fragment 1–633 induced the modest appearance of band C when coexpressed with Δ2–79-CFTR in HEK-293T cells. (D) Introduction of the ΔF508 mutation in the 1–633 fragment (1–633ΔF) eliminated transcomplementation of ΔF508-CFTR in COS-7 cells but not transcomplementation of Δ2–79-CFTR. (E) The 1–633 fragment coimmunoprecipitates with ΔF508-CFTR. (F) CoIP of the band B forms of wild-type CFTR and ΔF508-CFTR with V5-tagged-1–633 from COS-7 lysates 24 h after transfection. All results are representative of 3–10 experiments.
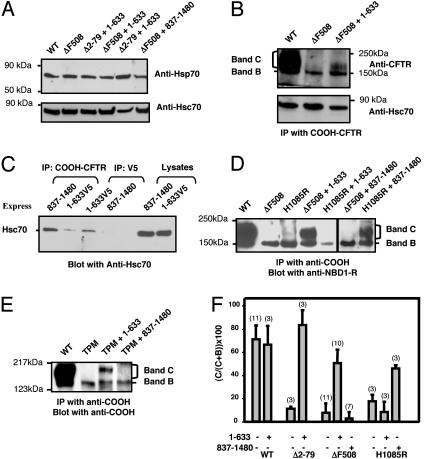
Fig. 4.
Transcomplementation is not a general consequence of sequestering Hsc70 or Hsp70 but requires a specific match between the CFTR mutation and the complementing fragment. (A) Steady-state amounts of Hsp70 and Hsc70 in COS-7 cells transfected with the indicated constructs. (B) CoIP of Hsc70 with ΔF508-CFTR expressed in the presence or absence of fragment 1–633. (C) Hcs70 coimmunoprecipitated with either an amino fragment (1–633V5) or a carboxy fragment (837–1,480) of CFTR. (D) ΔF508-CFTR was not transcomplemented by a carboxy fragment (837–1,480), whereas H1085R-CFTR, a cytosolic-loop-4-processing mutant that associates with severe disease, was complemented by fragment 837–1,480 but not by fragment 1–633. (E) Fragment 837-1,480 also had no effect on the processing of the TPM. (F) Quantitative analysis of the correction of CFTR mutant processing by the indicated constructs. Blots were analyzed by densitometry by using the scion program. In each case, the band C signal was normalized to the total band B plus band C signal. Numbers of experiments are shown in parentheses. Data are means ± SEMs.
As an initial test of the specificity of this effect, we determined whether the full-length ΔF508-CFTR mutant could be transcomplemented by an amino-terminal fragment in which we introduced the same ΔF508 mutation (1–633ΔF). This mutant fragment was unable to rescue ΔF508-CFTR processing, even when its expression in COS-7 cells was similar to that of the wild-type fragment (Figs. 2 D and E). Conversely, 1–633ΔF could transcomplement Δ2–79-CFTR (Fig. 2D Right). Interestingly, both the wild-type and mutant fragments physically associated with full-length ΔF508-CFTR protein as detected by coIP (Fig. 2E). In reciprocal coIP experiments performed with an epitope (V5)-tagged 1–633 construct that also promotes ΔF508-CFTR processing (ΔF508-CFTR band C could be detected 2 days after cotransfection with 1–633V5, data not shown), we observed a physical association between this fragment and the band B forms of both wild-type CFTR and ΔF508-CFTR 24 h after transfection (Fig. 2F). The CFTR fragment 1–633V5 did not apparently interact with CFTR band C, given that no band C for either wild-type CFTR or ΔF508-CFTR was detected in the 1–633V5 immunoprecipitates, although >70% of the total wild-type CFTR protein was present in the lysate as band C. These results imply that these interactions are restricted to the ER (i.e., transient) and indicate that transcomplementation correlates with a physical interaction between the complementing fragment and the ΔF508 mutant. However, the fact that the 1–633ΔF construct can physically associate with ΔF508-CFTR but cannot rescue processing implies that the gross physical interaction that is detectable by coIP probably involves multiple points of contact in addition to those that may confer complementation.
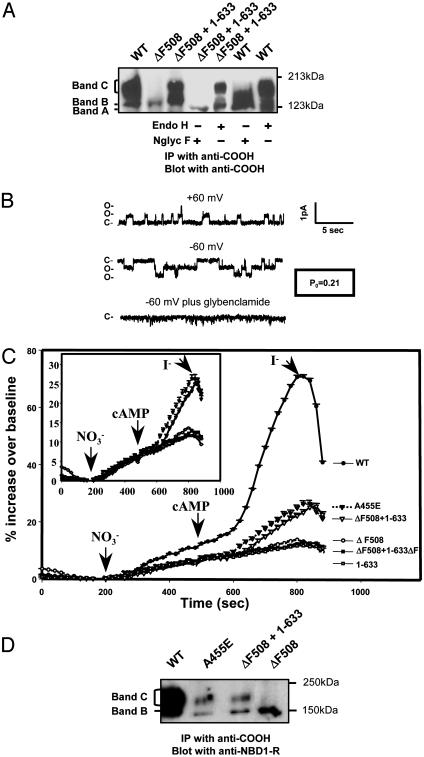
Transcomplemented ΔF508-CFTR Is a Functional Chloride Channel. Fig. 3 shows that the transcomplemented ΔF508 mutant is fully glycosylated and reaches the plasma membrane as a functional ion channel. The band C form of the transcomplemented mutant could not be deglycosylated by endoglycosidase H, as expected for fully mature CFTR protein (19) (Fig. 3A). In patch clamp experiments CFTR channels could be detected in inside-out patches excised from COS-7 cells 24 h after transfection with ΔF508-CFTR and 1–633 together (Fig. 3B; 5 of 26 patches had active channels that totaled to 17 channels). No channels were detected for cells transfected with ΔF508-CFTR alone (16 patches) or 1–633 alone (13 patches). The detected channels had the appropriate single-channel conductance (6–8 pS) and linear current–voltage behavior for CFTR channels and could be blocked by the CFTR inhibitor, glibenclamide (11) (Fig. 3B). By using a macroscopic halide transport assay, we observed that COS-7 cells that were cotransfected with ΔF508-CFTR and 1–633 exhibited a cAMP-dependent increase in halide permeability, a hallmark of wild-type CFTR expression (18, 20) (Fig. 3C). No such response was observed for cells that were transfected with ΔF508-CFTR alone, 1–633 alone, or ΔF508-CFTR plus 1–633ΔF.
Fig. 3.
Rescued ΔF508-CFTR is fully glycosylated and exhibits chloride channel activity at the cell surface. (A) Transcomplemented ΔF508-CFTR was digested by N-glycosidase F (Nglyc F) but not by endoglycosidase H (endo H). Band A represents the unglycosylated protein. (B) CFTR channel activity in inside-out membrane patch excised from a COS-7 cell 24 h after cotransfection with ΔF508-CFTR and fragment 1–633. This patch had two detectable channels with linear current–voltage behavior, single-channel conductances of 6–8 pS, and sensitivity to 250 μM glibenclamide, all features of CFTR channels. Po was estimated from a 5-min record by using pclamp 8 software (Axon Instruments), assuming two active channels in the patch. (C) Rescued ΔF508-CFTR exhibits macroscopic transport activity comparable to that of A455E-CFTR, a partial processing mutant associated with mild disease. Macroscopic halide permeability was monitored for COS-7 cells 24 h after transfection by using a fluorescence-based assay (SPQ assay) and conditions identical to those described in Methods (18). Each data point represents the mean change in fluorescence for 9–42 responding cells (i.e., cells that exhibit an increase in fluorescence in response to a cAMP mixture) from at least three different coverslips. Error bars (SEMs) are smaller than the symbols. (Inset) Enlarged view of the permeability responses of cells transfected with A455E (▾) or cotransfected with fragment 1–633 and ΔF508-CFTR (▿) to the cAMP mixture. (D) Biochemical analysis of the band C forms of transcomplemented ΔF508-CFTR and A455E-CFTR in transfected COS-7 cells. Cells were analyzed for protein expression 48 h after transfection as described in Methods.
To gauge the potential physiologic significance of this transcomplementation effect, we compared the transport activity and the amount of mature ΔF508-CFTR protein that could be induced by transcomplementation to that observed for a partial processing mutant that associates with mild CF (A455E) (21). Although the transport activity and the amount of mature protein for the rescued ΔF508-CFTR was considerably less than that observed for wild-type CFTR, both parameters were similar to that detected for the A455E mutant (Fig. 3 C and D). The A455E mutant associates with a mild phenotype presumably because it retains partial function [it has been reported that only 5–10% of “normal” CFTR activity is required to prevent severe disease (22, 23)]. The results of this comparison indicate that it may be feasible to achieve physiologically meaningful levels of correction by using this transcomplementation strategy.
Transcomplementation Requires Specific Interactions Between the Complementing Fragment and the Processing Mutant. We addressed the mechanism of transcomplementation by testing two alternative possibilities: (i) that complementing fragments nonspecifically promote the processing of CFTR mutants by sequestering chaperones that otherwise prevent ER exit or (ii) that complementation involves specific interactions between the complementing fragment and the CFTR processing mutant. To begin to address the first possibility we analyzed CFTR interactions with Hsp70 and Hsc70, two chaperones previously implicated in CFTR processing and in the block of ΔF508-CFTR exit from the ER (24–26). We observed no effect of expressing CFTR fragments including 1–633 on the steady-state amounts of either Hsp70 or Hsc70 in COS-7 cells (Fig. 4A). (Hsp70 expression was detected presumably because of the use of the vaccinia virus expression system). In addition, although we could detect a physical interaction between ΔF508-CFTR and Hsc70 (but not Hsp70) in coIP experiments (Fig. 4B), we observed no consistent effect of expressing the amino fragment on this interaction. Perhaps more significantly, interactions between Hsc70 and several CFTR fragments (1–633 and 837-1,480) could be detected by coIP (Fig. 4C), but these interactions showed no correlation with the complementing activities of the fragments. Regarding the latter point, we addressed the issue of specificity by comparing the transcomplementation of the ΔF508-CFTR mutant to that of a severe processing mutant (H1085R-CFTR) for which the mutation resides in the carboxy-terminal half of the protein (27). Unlike for ΔF508-CFTR, the processing of H1085R-CFTR could not be rescued by coexpression with the 1–633 amino fragment (Figs. 4 D and F). Importantly, however, H1085R-CFTR could be transcomplemented by a carboxy-terminal fragment (837–1,480), which had no effect on the processing of ΔF508-CFTR or the N-tail processing mutants (Figs. 4 D, E, and F). Thus, transcomplementation requires a specific match between the complementing fragment and the CF mutation.
Discussion
The present findings indicate that it is possible to promote the processing of CFTR-processing mutants, including ΔF508-CFTR, by using a transcomplementation strategy. The observed specificity argues against a generic effect of complementing fragments on chaperone function. Instead, we favor the hypothesis that transcomplementation requires specific interactions between CFTR polypeptides and fragments probably within the ER. We cannot rule the possibility that some portion of the observed increase in steady-state ΔF508-CFTR protein levels is due to interactions that occur beyond the biosynthetic pathway (i.e., certain fragments might stabilize the very small amount of mature ΔF508-CFTR that continually escapes the ER). However, a primary effect within the ER is the most likely explanation, because (i) ΔF508-CFTR could be transcomplemented by a severe processing mutant (Δ2–79-CFTR) that is completely restricted to the ER under all conditions examined (including growing the cells at low temperature; data not shown) and (ii) because only the ER form of CFTR could be detected in 1–633V5 immunoprecipitates (Fig. 2). The implication of these data are that separate CFTR polypeptides and fragments can associate within the ER at least at high levels of expression. It should be emphasized, however, that the current findings do not speak to the controversial issue of whether wild-type CFTR polypeptides normally associate to form dimers (11, 28). For example, the conformation of the wild-type protein may be less permissive for homotypic interactions.
There are several potential mechanisms by which complementing fragments might promote the export of the ΔF508-CFTR mutant from the ER. Such fragments could occlude ER retention signals on the mutant polypeptide by analogy to the effects of coexpressing SUR1 with K-channel subunits on the trafficking of ATP-sensitive K channels to the cell surface (29). This model is favored by Owsianik et al. (30), who recently provided a limited amount of functional evidence that ΔF508-CFTR processing can be promoted by coexpression with CFTR fragments. However, it is also possible that complementing fragments promote ΔF508-CFTR processing by providing ER exit codes in trans (3) or by burying hydrophobic surfaces on the mutant CFTR protein that otherwise promote aggregation. The aggregation or polymerization of mutant proteins by “domain swapping” is a feature of many genetic diseases (31). To what extent such polymerization contributes to the ΔF508-CFTR defect is unclear, but because the aggregation of some mutant proteins can be blocked by synthetic peptides or protein fragments in vitro (32, 33), it seems plausible that complementing fragments could have similar effects in vivo. Complementing fragments might provide useful biochemical tools for identifying the surfaces of the CFTR polypeptide that are critical for ER export.
Peptide therapy or gene therapy with mini-cDNAs might be worth considering if complementing peptides can be further miniaturized and optimized for use in vivo. Indeed, based on the present findings, it appears possible to tailor rescuing peptides to specific CF-causing mutations. A strategy based on mini-cDNAs might overcome the packaging problems of certain gene therapy vectors (8).
Acknowledgments
We thank Lois Musgrove and Katie Davis for excellent technical assistance. We thank M. J. Welsh for the H1085R-CFTR construct. This work was supported by the Cystic Fibrosis Foundation (E.C.-B.) and the National Institutes of Health (K.L.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CF, cystic fibrosis; CFTR, CF transmembrane conductance regulator; IP, immunoprecipitation; ER, endoplasmic reticulum; NBD, nucleotide-binding domain; SPQ, 6-methoxy-N-(3-sulfopropyl)quinolinium; TPM, triple processing mutant.
References
- 1.Ward, C. L., Omura, S. & Kopito, R. R. (1995) Cell 83, 121–127. [DOI] [PubMed] [Google Scholar]
- 2.Chang, X. B., Cui, L., Hou, Y. X., Jensen, T. J., Aleksandrov, A. A., Mengos, A. & Riordan, J. R. (1999) Mol. Cell 4, 137–142. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura, N. & Balch, W. E. (1997) Science 277, 556–558. [DOI] [PubMed] [Google Scholar]
- 4.Denning, G. M., Anderson, M. P., Amara, J. F., Marshall, J., Smith, A. E. & Welsh, M. J. (1992) Nature 358, 761–764. [DOI] [PubMed] [Google Scholar]
- 5.Sato, S., Ward, C. L., Krouse, M. E., Wine, J. J. & Kopito, R. R. (1996) J. Biol. Chem. 271, 635–638. [DOI] [PubMed] [Google Scholar]
- 6.Egan, M. E., Glockner-Pagel, J., Ambrose, C., Cahill, P. A., Pappoe, L., Balamuth, N., Cho, E., Canny, S., Wagner, C. A., Geibel, J., et al. (2002) Nat. Med. 8, 485–492. [DOI] [PubMed] [Google Scholar]
- 7.Rubenstein, R. C., Egan, M. E. & Zeitlin, P. L. (1997) J. Clin. Invest. 100, 2457–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driskell, R. A. & Engelhardt, J. F. (2003) Annu. Rev. Physiol. 65, 585–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csanady, L., Chan, K. W., Seto-Young, D., Kopsco, D. C., Nairn, A. C. & Gadsby, D. C. (2000) J. Gen. Physiol. 116, 477–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostedgaard, L. S., Rich, D. P., DeBerg, L. G. & Welsh, M. J. (1997) Biochemistry 36, 1287–1294. [DOI] [PubMed] [Google Scholar]
- 11.Ramjeesingh, M., Kidd, J. F., Huan, L. J., Wang, Y. & Bear, C. E. (2003) Biochem. J. 374, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormet-Boyaka, E., Di, A., Chang, S. Y., Naren, A. P., Tousson, A., Nelson, D. J. & Kirk, K. L. (2002) Proc. Natl. Acad. Sci. USA 99, 12477–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson, D. C., Smith, S. S. & Mansoura, M. K. (1999) Physiol. Rev. 79, Suppl. 1, S47–S75. [DOI] [PubMed] [Google Scholar]
- 14.Wuthich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 15.Brunger, A. T. (1992) x-plor 3.1, A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven, CT).
- 16.Naren, A. P., Quick, M. W., Collawn, J. F., Nelson, D. J. & Kirk, K. L. (1998) Proc. Natl. Acad. Sci. USA 95, 10972–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prince, L. S., Peter, K., Hatton, S. R., Zaliauskiene, L., Cotlin, L. F., Clancy, J. P., Marchase, R. B. & Collawn, J. F. (1999) J. Biol. Chem. 274, 3602–3609. [DOI] [PubMed] [Google Scholar]
- 18.Naren, A. P., Cormet-Boyaka, E., Fu, J., Villain, M., Blalock, J. E., Quick, M. W. & Kirk, K. L. (1999) Science 286, 544–548. [DOI] [PubMed] [Google Scholar]
- 19.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R. & Smith, A. E. (1990) Cell 63, 827–834. [DOI] [PubMed] [Google Scholar]
- 20.Rich, D. P., Anderson, M. P., Gregory, R. J., Cheng, S. H., Paul, S., Jefferson, D. M., McCann, J. D., Klinger, K. W., Smith, A. E. & Welsh, M. J. (1990) Nature 347, 358–363. [DOI] [PubMed] [Google Scholar]
- 21.Sheppard, D. N., Ostedgaard, L. S., Winter, M. C. & Welsh, M. J. (1995) EMBO J. 14, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu, C. S., Trapnell, B. C., Curristin, S., Cutting, G. R. & Crystal, R. G. (1993) Nat. Genet. 3, 151–156. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, L. G., Olsen, J. C., Sarkadi, B., Moore, K. L., Swanstrom, R. & Boucher, R. C. (1992) Nat. Genet. 2, 21–25. [DOI] [PubMed] [Google Scholar]
- 24.Meacham, G. C., Lu, Z., King, S., Sorscher, E., Tousson, A. & Cyr, D. M. (1999) EMBO J. 18, 1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Y., Janich, S., Cohn, J. A. & Wilson, J. M. (1993) Proc. Natl. Acad. Sci. USA 90, 9480–9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubenstein, R. C. & Zeitlin, P. L. (2000) Am. J. Physiol. Cell Physiol. 278, C259–C267. [DOI] [PubMed] [Google Scholar]
- 27.Cotton, J. F., Ostedgaard, L. S., Carson, M. R. & Welsh, M. J. (1996) J. Biol. Chem. 271, 21279–21284. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J. H., Chang, X. B., Aleksandrov, A. A. & Riordan, J. R. (2002) J. Membr. Biol. 188, 55–71. [DOI] [PubMed] [Google Scholar]
- 29.Zerangue, N., Schwappach, B., Jan, Y. N. & Jan, L. Y. (1999) Neuron 22, 537–548. [DOI] [PubMed] [Google Scholar]
- 30.Owsianik, G., Cao, L. & Nilius, B. (2003) FEBS Lett. 554, 173–178. [DOI] [PubMed] [Google Scholar]
- 31.Horwich, A. (2002) J. Clin. Invest. 110, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahadeva, R., Dafforn, T. R., Carrell, R. W. & Lomas, D. A. (2002) J. Biol. Chem. 277, 6771–6774. [DOI] [PubMed] [Google Scholar]
- 33.Friedler, A., Hansson, L. O., Veprintsev, D. B., Freund, S. M., Rippin, T. M., Nikolova, P. V., Proctor, M. R., Rudiger, S. & Fersht, A. R. (2002) Proc. Natl. Acad. Sci. USA 99, 937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]