Abstract
Characteristic for cruciferous plants is their production of N- and S-containing indole phytoalexins with disease resistance and cancer-preventive properties, previously proposed to be synthesized from indole independently of tryptophan. We show that camalexin, the indole phytoalexin of Arabidopsis thaliana, is synthesized from tryptophan via indole-3-acetaldoxime (IAOx) in a reaction catalyzed by CYP79B2 and CYP79B3. Cyp79B2/cyp79B3 double knockout mutant is devoid of camalexin, as it is also devoid of indole glucosinolates [Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., Normanly, J., Chory, J. & Celenza, J. L. (2002) Genes Dev. 16, 3100–3112], and isotope-labeled IAOx is incorporated into camalexin. These results demonstrate that only CYP79B2 and CYP79B3 contribute significantly to the IAOx pool from which camalexin and indole glucosinolates are synthesized. Furthermore, production of camalexin in the sur1 mutant devoid of glucosinolates excludes the possibility that camalexin is derived from indole glucosinolates. CYP79B2 plays an important role in camalexin biosynthesis in that the transcript level of CYP79B2, but not CYP79B3, is increased upon induction of camalexin by silver nitrate as evidenced by microarray analysis and promoter–β-glucuronidase data. The structural similarity between cruciferous indole phytoalexins suggests that these compounds are biogenetically related and synthesized from tryptophan via IAOx by CYP79B homologues. The data show that IAOx is a key branching point between several secondary metabolic pathways as well as primary metabolism, where IAOx has been shown to play a critical role in IAA homeostasis.
Characteristic for cruciferous plants is the synthesis of a wide range of species-specific phytoalexins that structurally are sulfur-containing indole alkaloids (1) and the synthesis of glucosinolates (reviewed in ref. 2). Both groups of natural products are involved in plant defense and have cancer-preventive properties (3, 4). Very little is known about the biosynthetic pathway of the S-containing indole phytoalexins. Their similar structure with N- and S-containing side chains at C-3 of the indole ring suggests a biogenetic relationship (5). Camalexin (3-thiazol-2′-yl-indole) is produced in the model plant Arabidopsis thaliana (6). It is induced by a variety of microorganisms, e.g., Pseudomonas syringae (6) and Alternaria brassisicola (7), and by abiotic factors, such as AgNO3 (8). These findings make camalexin a good model compound for studying biosynthesis and regulation of cruciferous indole phytoalexins. In vivo feeding experiments where radiolabeled anthranilate and tryptophan were applied on leaves treated with AgNO3 led to the suggestion that tryptophan was not a precursor in camalexin biosynthesis because tryptophan was much less efficiently incorporated into camalexin compared with anthranilate (8, 9). The data were further supported by labeling studies performed in three tryptophan mutants (8), where reduced levels of camalexin accumulated in trp1–100 with an impaired anthranilate transferase but not in trp3–1 and trp2–1 mutants with mutations in the α and β subunits of tryptophan synthase, respectively. In vivo feeding experiments with radiolabeled anthranilate and indole showed that indole was most efficiently incorporated into camalexin, and that feeding of anthranilate was followed by a transient accumulation of indole (10). Accordingly, it was suggested that indole destined for camalexin is produced by a pathway that does not involve tryptophan synthase (10) but, e.g., a homolog of the α-subunit tryptophan synthase such as BX1 from maize that catalyzes the formation of indole in 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one biosynthesis (11).
Glucosinolates are derived from amino acids that are converted to the corresponding oximes by cytochromes P450 belonging to the CYP79 family (2). In Arabidopsis, CYP79B2 (12, 13) and CYP79B3 (12) catalyze the conversion of tryptophan into indole-3-acetaldoxime (IAOx) that is channeled by the oxime-metabolizing CYP83B1 into the biosynthetic pathway of indole glucosinolates (14, 15). Furthermore, IAOx has been shown to be a precursor of the phytohormone indole-3-acetic acid (IAA) as evidenced by enzyme studies with radiolabeled IAOx (16) and the high-auxin phenotype of knockout mutants in postoxime enzymes in glucosinolate biosynthesis, such as sur1 (17) and sur2 (14, 18). Based on structural similarity, it has been suggested that brassinin and possibly other cruciferous phytoalexins were derived from the indole glucosinolate glucobrassicin (5). However, in a recent in vivo feeding study in turnip (Brassica rapa) roots with deuterated IAOx and glucobrassicin, IAOx, and not glucobrassicin, was incorporated into brassinin and brassinin-derived phytoalexins, indicating that IAOx is an intermediate in brassinin biosynthesis (19).
In this manuscript, we show that camalexin is synthesized from tryptophan via IAOx by CYP79 homologues, and that it is not derived from indole glucosinolates. Furthermore, we provide data that indicate that CYP79B2 and CYP79B3 have differential functions in camalexin biosynthesis.
Methods
Plant Material. Cyp79B2, cyp79B3, and cyp79B2/cyp79B3 knockout mutants (20) originating from the Salk Institute collection were kindly provided by Y. Zhao (University of California at San Diego, La Jolla). Generation of 35S::CYP79B2 has been described (13). Seeds of sur1–3 were a kind gift from C. Bellini (Institut de la Recherche Agronomique, Versailles, France). All mutants and transgenic lines are in Col-0 background. Plants were grown in soil mixed with sand (3:1) in a walk-in growth chamber (HEMZ 20/240/S, Heraeus) at 12 h light, 80–100 μmol of photons per m2 per sec, 18°C, and 40% relative humidity.
Silver Nitrate Induction and Extraction of Camalexin. For camalexin induction, 4- and 6-week-old rosette leaves were sprayed with a thin film of 5 mM AgNO3/0.02% Silwet L-77. Until harvest, plants were covered with a transparent plastic hood to avoid desiccation. For each measurement, three leaves were cut at the petiole, weighed, and frozen in liquid nitrogen. Camalexin was extracted as described (6), with the following minor modifications. Extraction was performed twice for 30 min in 600 μl of MeOH/H2O (1:1) at 60°C. Samples were analyzed by reverse-phase HPLC (LiChroCART 250-4, RP-18, 5 μm, Merck; 1 ml·min–1 MeOH/H2O (3:2) for 1 min, followed by a 14-min linear gradient to 100% MeOH and then 3 min at 100% MeOH). The peak at 16.2 min was identified as camalexin by comparison with authentic standard with respect to retention time and UV spectrum (photodiode array detector 168, Beckman Instruments, Fullerton, CA) and quantified by using a Shimadzu F-10AXL fluorescence detector (318 nm excitation and 370 nm emission) and by UV absorption at 318 nm.
Synthesis of Labeled IAOx. IAOx was synthesized chemically from indole-3-acetaldehyde, [2′-14C]IAOx was synthesized from [3′-14C]tryptophan, and 6-f luoro-indole-3-acetaldoxime (6-F-IAOx) was synthesized from 20 mg 6-F-tryptophan, essentially as described (21). Rf values of 0.35 and 0.21 were determined for 6-F-indole-3-acetaldehyde and 6-F-IAOx, respectively, in TLC analysis (CHCl3/MeOH, 96:4). IAOx and 6-F-IAOx were characterized by NMR (Avance 400, Bruker, Billerica, MA, in MeOH-d4, 1H at 400.1 and 13C at 100.6 Mhz). δH values are relative to internal TMS and δC-values are based on δC(MeOH-d4) = 49.05. Assignments (Fig. 1) are based on 1D spectra (1H, 13C, and distortionless enhancement by polarization transfer) as well as 2D spectra (COSY, heteronuclear sequential quantum correlation, heteronuclear multiple bond correlation, and J-resolved 2D correlation). Some chemical shifts have been retrieved from the 2D spectra. The (E)- and (Z)-isomers were present in a ratio of ≈2:1 and ≈17:1 for IAOx and 6-F-IAOx, respectively.
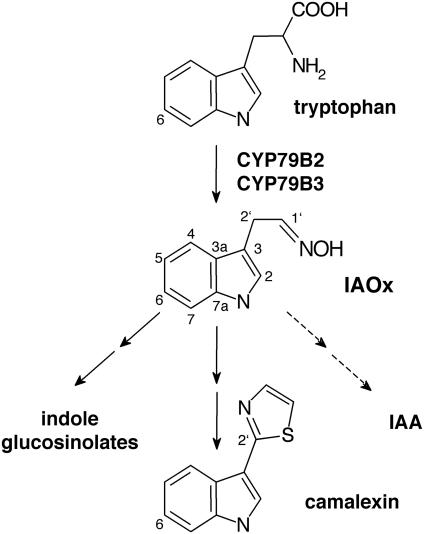
Fig. 1.
Schematic view of biosynthetic pathways of IAOx-derived indole compounds in Arabidopsis.
(E)-IAOx. 1H NMR: 7.10 (br s, H-2), 7.507 (dt, J = 7.9/1.0, H-4), 6.997 (m, J = 7.5 1.1, H-5), 7.094 (m, J = 7.6 1.2, H-6), 7.336 (dt, J = 8.2/1.0, H-7), 6.79 (t, J1′,2′ = 5.3, H-1′), 3.80 (dd, J = 5.3/0.9, H-2′). 13C NMR: 123.7 (C-2), 111.1 (C-3), 119.2 (C-4), 119.8 (C-5), 122.6 (C-6), 112.3 (C-7), 128.7 (C-3a), 138.3 (C-7a), 152.1 (C-1′), 22.3 (C-2′).
(Z)-IAOx. 1H NMR: 7.07 (br s, H-2), 7.54 (dt, J = 8.0/1.0, H-4), 6.994 (m, J = 7.5 1.2, H-5), 7.33 (dt, J = 8.2/0.9, H-7), 7.48 (t, J = 6.4, H-1′), 3.60 (dd, J = 6.4/0.9, H-2′). 13C NMR: 123.7 (C-2), 119.4 (C-4), 128.7 (C-3a), 151.4 (C-1′), 26.9 (C-2′).
(E)-6-F-IAOx. 1H NMR: 7.10 (t, J = 0.9, H-2), 7.46 (dd, J4,5 = 8.7 J4,F = 5.3, H-4), 6.79 (ddd, J4,5 = 8.7, J5,7 = 2.3, J5,F = 9.8, H-5), 7.03 (br dd, J5,7 = 2.3, J7,F = 10.0, H-7), 6.77 (t, J1′,2′ = 5.4, H-1′), 3.78 (dd, J1′,2′ = 5.4, J2,2′ = 0.9, H-2′). 13C NMR: C-2: 124.2 (C-2), 111.4 (C-3), 120.2 (JC,F = 10, C-4), 108.2 (JC,F = 25, C-5), 161.3 (JC,F≈220, C-6), 98.2 (JC,F = 26, C-7), 125.5 (C-3a), 138.2 (C-7a), 151.7 (C-1′), 22.2 (C-2′).
(Z)-6-F-IAOx. 1H NMR: 7.06 (t, J2,10 = 0.9, H-2), 7.47 (H-1′), 3.58 (dd, J1′,2′ = 6.4, J2,2′ = 0.9, C-2′). 13C NMR: 124.8 (C-2), 151.2 (C-1′), 27.0 (C-2′).
In Vivo Labeling Experiments. For each sample, fluoro- or 14C-labeled tracers of indole, tryptophan, and IAOx were incubated with three 6-week-old Arabidopsis rosette leaves for 16 h, starting 8 h after treatment with 5 mM AgNO3. For fluorolabeling, leaves were cut at their petiole and incubated in 150 μl of 0.5 mM fluorinated precursor with or without 0.5 or 2.5 mM unlabeled competitor in H2O/EtOH/DMSO (96.5/0.5/3). Extracts were analyzed on an Agilent (Santa Clara, CA) 1100 LC liquid chromatograph-MS coupled to a Bruker Esquire 3000Plus ion trap spectrometer (electrospray ionization in positive mode). A 2-mm ID XTerra C18 column (Waters) was used (0.2 ml/min, solvent A, H2O with 0.1% formic acid; solvent B, 80% aceto-nitrile with 0.1% formic acid; 0–2 min 10% B, 2–10 min 10–100% B, 10–20 min 100% B). Camalexin (Rt = 13.8 min) and 6-F-camalexin (Rt = 14.6 min) were quantified from the selected ion trace chromatogram of m/z 201 and m/z 219, respectively.
To confirm their identity, camalexin and 6-F-camalexin were purified by HPLC from an extract of 30 leaves labeled with F-IAOx, as described above, and analyzed by GC-MS with a HP5890 Series II gas chromatograph (Hewlett–Packard) coupled to a JEOL JMS-AX505W mass spectrometer (source conditions: electrospray ionization mode and 70 eV at 200°C) with head pressure at 100 kPa and splitless injection. An SGE column (BPX5, 25 m × 0.25 mm, 0.22-mm film thickness) was used. The oven temperature program was 80°C for 2 min, 80–200°Cat20°C/min, 200–300°Cat5°C/min, and 300°C for 30 min. The molecular ion was base peak for camalexin as well as 6-F-camalexin (at m/z 200 and 218), and Rt was 17.6 and 17.4 min, respectively.
For 14C-precursors, the labeling method was modified to avoid a strong unknown background signal comigrating with camalexin in the TLC analysis after feeding with [2′-14C]IAOx. The cuticula was removed with a razor blade, and 10 μl (1 nCi/μl; 1 Ci = 37 GBq) of indole (50 mCi/mmol), tryptophan (58.1 mCi/mmol), and IAOx (58.1 mCi/mmol), respectively, in H2O/EtOH (1:1) was spotted on each leaf in 1-μl droplets. After 16 h, extracts of the leaves were separated by TLC (CHCl3/MeOH, 9:1) and analyzed on a Storm 860 phosphorimager (Amersham Pharmacia). Radiolabeled bands comigrating with camalexin were analyzed. Background from parallel analysis of extracts of noninduced plants was subtracted.
RNA Isolation and Microarray Analysis. The custom-designed oligoarray used in this experiment contains oligonucleotide probes (50-mers) for 244 cytochromes P450, 107 glycosyltransferases, 35 genes from aromatic amino acid biosynthesis, 53 genes from secondary metabolism, 9 normalization genes, and 6 control genes. Probes were printed in duplicate, i.e., each gene was represented twice on each array. Each of the genes had been manually annotated (C. Kristensen and S. Bak, personal communication), and this information was used by MWG Biotec (Ebersberg, Germany) to design gene-specific 50-mer oligonucleotides (22). Synthesis of oligonucleotides and microarray fabrication was performed by MWG Biotec. Six-week-old plants were sprayed with 5 mM AgNO3/0.02% Silwet L-77 or 0.02% Silwet L-77 (control) and incubated for 16 h. Total RNA was isolated from eight leaves by using TRIzol reagent (Life Technologies, Rockville, MD). Twenty micrograms of each RNA sample (one control and one treated sample) was reverse transcribed, labeled with Cy3 and Cy5, and purified by using the RPN 5660 Cy Scribe cDNA Post Labeling Kit and the GFX purification kit (Amersham Pharmacia). Sample tubes containing Cy3- and Cy5-labeled cDNA were pooled and dried under vacuum to a final volume of 1 μl, and 15 μl of formamide hybridization buffer (MWG Biotec) was added. Before hybridization, the probe was left at 95°C for 2 min and then applied to the microarray under a coverslip. Slides were placed in hybridization chamber, and 10 μl of H2O was placed in each corner of the chamber before sealing. Slides were incubated for 15 h at 42°C, followed sequentially by washes for 5 min in 2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS, 0.5× SSC/0.1% SDS, and finally in 0.5× SSC. Slides were dried by centrifugation at 3,000 × g for 5 min.
Microarrays were scanned with a GMS 418 Array Scanner (Genetic MicroSystems, Woburn, MA). The two channels were normalized with respect to signal intensity by adjusting photo-multiplier and laser power settings such that the number of saturated spots was the same in the two channels. The microarray analysis, image analysis, and signal quantification were performed by using imagene 5.5 and genesight 3.5.2 (Biodiscovery, San Francisco). Background correction was performed, and spots showing a signal value lower than twice the background value were discarded. The experiment was performed twice, which result in four ratio values for each gene because one slide contains two replicate spots. Statistically significantly expressed genes were obtained by using sam statistical software (23).
Generation of CYP79B3p::β-glucuronidase (GUS) Plants and Infection with P. syringae. A 2,668-bp fragment, containing 2,413 bp of the promoter and 255 bp of the 5′ end of the coding region of CYP79B3, was PCR amplified from gDNA of Arabidopsis (Col-0) by using the primers 5′-tagtcagatctaccatgggtcctggcgggagaggatg-3′ introducing a BglII restriction site at the 3′ end and 5′-gctacactgcaggtccattagtattagtttgaggttggag-3′ introducing a PstI restriction site at the 5′ end. The fragment was digested with PstI and BglII and ligated into pCambia 3301. Arabidopsis (Col-0) transformants were selected with Basta. Generation of CYP79B2p::GUS plants and GUS staining has been described (13). Infection with P. syringae was performed at 24°C (24) by using a suspension of the strain DC3000/avrRps4 (25) at OD600 0.1 in 10 mM MgCl2.
Results
IAOx Is a Precursor of Camalexin. The role of IAOx as intermediate in camalexin biosynthesis was investigated by in vivo feeding of isotope-labeled indole, tryptophan, and IAOx to Arabidopsis leaves in which camalexin production was induced by spraying with AgNO3 (8). We used this treatment because it is highly reproducible and eliminates the contribution of microbial metabolic activity. Spraying with 0.02% Silwet L-77 alone did not result in camalexin formation (data not shown). When AgNO3-treated leaves were incubated with 6-F-indole, 6-F-tryptophan, and 6-F-IAOx all three compounds were shown to be precursors for 6-F-camalexin because 27 ± 2% (n = 2), 12 ± 1% (n = 2), and 60 ± 2% (n = 2), respectively, of total camalexin was fluorinated as calculated after liquid chromatography-MS analysis. Addition of an equal amount or a 5-fold molar excess of unlabeled IAOx in combination with 0.5 mM 6-F-IAOx yielded, respectively, 31% and 14% fluorinated camalexin, suggesting that 6-F-IAOx incorporation is not due to discrimination between hydrogen and fluoro substitution. Similarly, administration of 14C-labeled indole, tryptophan, and IAOx resulted in incorporation into camalexin of, respectively, 20 ± 7.0, 3.5 ± 1.2, and 8.9 ± 4.6 nCi/μg camalexin (n = 4).
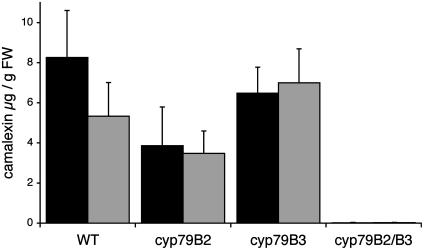
Camalexin Levels in cyp79B2/cyp79B3 and Other Knockout Mutants. We analyzed the production of camalexin in rosette leaves of 4- and 6-week-old plants of single knockout mutants of cyp79B2 and cyp79B3, and in cyp79B2/cyp79B3 double mutants (20), to investigate the origin of the IAOx involved in camalexin biosynthesis (Fig. 2). A sensitive HPLC method based on fluorescence for quantification of camalexin was used allowing detection of 1 pmol of camalexin. Twenty-four hours after AgNO3 treatment, 4- and 6-week-old WT plants synthesized, respectively, 8.3 ± 2.3 and 5.3 ± 1.7 μg of camalexin per g of fresh weight, whereas less than 1% of the WT level (18 ± 16 and 19 ± 15 ng/g, respectively) was detected in the cyp79B2/cyp79B3 double mutant. In the cyp79B2 single mutant, a tendency toward reduced camalexin production was observed in comparison with WT and cyp79B3 mutant (Fig. 2). Six-week-old 35S::CYP79B2 plants synthesized WT levels of camalexin (5.6 ± 2.1 μg/g, n = 6).
Fig. 2.
Induction of camalexin in leaves of Arabidopsis after treatment with AgNO3. Camalexin was measured in plant extracts from WT, cyp79B2, cyp79B3, and cyp79B2/cyp79B3 mutants 24 h after treatment with 5 mM AgNO3. Black bars are 4-week-old plants (n = 6), and gray bars are 6-week-old plants (n = 10).
We investigated camalexin concentrations in the “high-auxin” mutant sur1 that is completely devoid of glucosinolates (17). Four-week-old sur1 mutants grown on Murashige and Skoog medium synthesized 2.1 ± 0.7 μg of camalexin per g (n = 5). This finding demonstrates that camalexin is not derived from indole glucosinolates or its degradation products. To test whether the reduced camalexin production in sur1 was an effect of high auxin levels, WT seedlings were grown for 4 weeks on Murashige and Skoog medium containing 1 μM of the auxin 2,4-D. This treatment yielded a decrease in camalexin production (0.34 ± 0.05 μg/g, n = 4) compared with 3.6 ± 2.7 μg/g (n = 7) in seedlings grown on Murashige and Skoog medium without 2,4-D. This finding suggests that the high level of auxin in sur1 is inhibitory to camalexin production.
Camalexin levels in 6-week-old pad3 (phytoalexin deficient 3) plants (26) that are mutated in CYP71B15, which is suggested to be involved in camalexin biosynthesis, was ≈2% of WT level (0.13 ± 0.11 μg/g, n = 6), as previously shown (27). The presence of camalexin in induced cyp71B15/pad3 plants was confirmed by liquid chromatography-MS.
Induction of CYP79B2. Analysis of gene expression following treatment with AgNO3 was performed by probing a custom-designed oligoarray, which carries a selection of Arabidopsis genes including all cytochromes P450, with labeled RNA isolated 16 h after AgNO3 treatment, at which time point camalexin is synthesized at a high rate (8). Twenty-five genes were significantly (>2-fold) induced or repressed (Tables 1 and 2). Specifically, CYP79B2 was induced 3-fold, whereas no significant up-regulation of CYP79B3 was observed (Table 1). CYP71B15/PAD3, suggested to be involved in camalexin biosynthesis (26), was not up-regulated.
Table 1. Microarray data of genes significantly induced in Arabidopsis rosette leaves 16 h after spraying with AgNO3.
| Gene | Locus | Up-regulation, fold |
|---|---|---|
| AAD23027 | At2g24850 | 15.9 |
| CYP82G1 | At3g25180 | 10.7 |
| CYP71A18 | At1g11610 | 4.5 |
| CYP71A12 | At2g30750 | 3.5 |
| AAC31939 | At5g54140 | 3.4 |
| CYP79B2 | At4g39950 | 3.0 |
| CAB88998 | At3g44300 | 2.8 |
| CAA73905 | At1g51760 | 2.5 |
| CAA23026 | At4g23600 | 2.4 |
| CAB89000 | At3g44320 | 2.0 |
Four independent experiments were performed on an custom-designed oligoarray representing all Arabidopsis cytochromes P450 (CYP) and selected metabolic genes (see Methods).
Table 2. Microarray data of genes significantly repressed in Arabidopsis rosette leaves 16 h after spraying with AgNO3.
| Gene | Locus | Down-regulation, fold |
|---|---|---|
| CYP706A6 | At4g12320 | 2.0 |
| CYP77A4 | At5g04660 | 2.1 |
| CYP86A8 | At2g45970 | 2.1 |
| CYP71B7 | At1g13110 | 2.1 |
| CYP705A4 | At4g15380 | 2.1 |
| CYP709A1 | At5g38450 | 2.3 |
| CYP707A2 | At2g29090 | 2.5 |
| CYP706A2 | At4g22690 | 2.7 |
| CYP705A3 | At3g20960 | 2.7 |
| CYP71A14 | At5g24960 | 2.8 |
| CAB43691 | At4g32540 | 3.0 |
| CYP707A3 | At5g45340 | 3.0 |
| CYP75B1 | At5g07990 | 3.0 |
| CAB94971 | At3g55120 | 3.8 |
| AAF23561 | At5g13930 | 3.9 |
Four independent experiments were performed on an custom-designed oligoarray representing all Arabidopsis cytochromes P450 (CYP) and selected metabolic genes (see Methods).
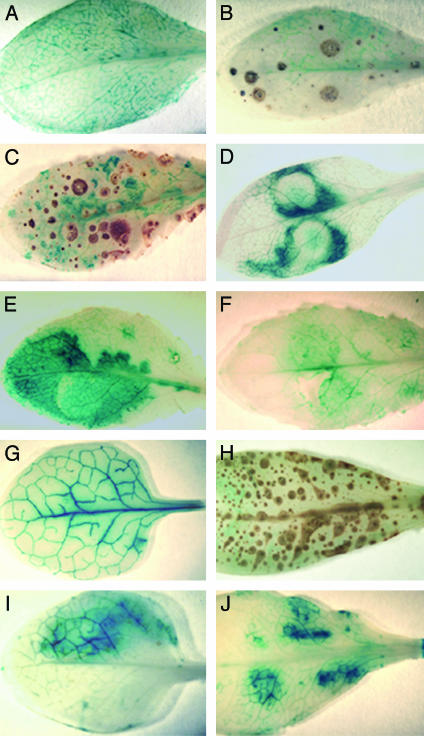
Tissue-specific induction of CYP79B2 was further investigated in 6-week-old CYP79B2p::GUS plants (13). As shown above, basal levels of CYP79B2 were expressed in untreated rosette leaves (Fig. 3A). After AgNO3 treatment, enhanced GUS expression was observed in tissue surrounding cells showing symptoms of hypersensitive response (Fig. 3 B and C). After infiltration of the plants with P. syringae DC3000/avrRps4, strong GUS staining was observed (Fig. 3 D and E), whereas control leaves infiltrated with 10 mM MgCl2 showed a weak, wounding-induced response (Fig. 3F). In comparison, CYP79B3p::GUS plants showed higher constitutive GUS expression (Fig. 3G), but no enhanced GUS expression was detected after AgNO3 treatment (Fig. 3H). CYP79B3 expression in plants infiltrated with P. syringae (Fig. 3I) was comparable with CYP79B3 expression in plants infiltrated with 10 mM MgCl2 (Fig. 3J), indicating that the observed expression reflects a wounding-induced response.
Fig. 3.
GUS expression in rosette leaves of 6-week-old CYP79B2p::GUS and CYP79B3p::GUS plants after AgNO3 and P. syringae treatment. CYP79B2p::GUS plants are shown untreated (A), 8 h after spraying with 5 mM AgNO3 (B), 16 h after spraying with 5 mM AgNO3 (C), 16 h after infiltration with P. syringae (D), 24 h after infiltration with P. syringae (E), and 24 h after infiltration with 10 mM MgCl2 (control) (F). CYP79B3p::GUS plants are shown untreated (G), 16 h after spraying with 5 mM AgNO3 (H), 24 h after infiltration with P. syringae (I), and 24 h after infiltration with 10 mM MgCl2 (control) (J). Weakly colored rings (D and E) are syringe marks.
Discussion
In this article we demonstrate that camalexin is synthesized from IAOx that is derived from tryptophan in a reaction catalyzed by CYP79B2 and CYP79B3. In addition, we suggest that CYP79B2 and CYP79B3 have differential functions, because CYP79B2 induction, specifically, is correlated with camalexin biosynthesis.
In Vivo Labeling of Camalexin. We show that IAOx is a precursor for camalexin as evidenced by incorporation in vivo of externally applied fluorinated and 14C-labeled IAOx into camalexin. Incorporation of radioactivity from [2′-14C]IAOx shows that C-2′ in the thiazolin group of camalexin is derived from IAOx, which excludes incorporation of radioactivity via degradation of IAOx to free indole. In parallel experiments, we confirm that indole is a precursor (10). Fluorinated and 14C-labeled tryptophan was incorporated with low efficiency. Low tryptophan incorporation has been observed in a study using 3H-labeled tryptophan and anthranilate (8, 9) and led to the suggestion that tryptophan was not a direct, or was at best a poor, precursor of camalexin. Generally, interpretation of in vivo labeling experiments is complicated by difference in the ability of compounds to be taken up and transported to the site of the biosynthetic machinery that might be localized to different compartments even for the same pathway. In support of tryptophan-independency in camalexin biosynthesis, it was mentioned that the tryptophan synthase mutants trp3–1 and trp2–1 are not impaired in camalexin biosynthesis (8). However, it was recently shown that trp3–1 is not tryptophan-limited, because it synthesizes elevated levels of tryptophan-derived glucobrassicin (28). Possibly, externally applied tryptophan is not efficiently transported to the compartment where camalexin is synthesized and/or it is consumed by other metabolic processes.
IAOx Is a Central Metabolic Branching Point in Arabidopsis. The absence of indole glucosinolates and camalexin in Arabidopsis cyp79B2/cyp79B3 double mutant shows that only CYP79B2 and CYP79B3 contribute significantly to the IAOx pool(s), from which camalexin and indole glucosinolates are synthesized. Accordingly, neither the biochemically characterized plasma membrane-bound peroxidase catalyzing IAOx production (29) nor the proposed IAOx-forming pathway via YUCCA (30) contribute significantly to biosynthesis of camalexin or indole glucosinolates.
It has been hypothesized that cruciferous indole phytoalexins are derived from indole glucosinolates (5). However, we observe induction of camalexin in the mutant sur1 that is completely blocked in its biosynthesis of glucosinolates (17). This finding excludes the possibility that camalexin is synthesized via indole glucosinolates. Interestingly, in vivo labeling of brassinin from Brassica rapa has shown more efficient incorporation of deuterated IAOx than deuterated indole glucosinolate into brassinin, suggesting that brassinin is not derived from the intact glucosinolate but directly from IAOx (19). Based on these observations and our data, we propose that all cruciferous indole phytoalexins with N- and S-containing side chains at the C-3 position are synthesized from tryptophan via IAOx in a reaction catalyzed by CYP79B homologs and that at least some of the indole phytoalexins including camalexin are not derived from indole glucosinolates.
Early enzymatic studies have identified IAOx as a precursor of IAA (16). A possible role of IAOx in IAA biosynthesis is further evidenced by the high-auxin phenotype of sur1 and sur2 that are blocked in postoxime enzymes in the glucosinolate pathway, resulting in channeling of IAOx into IAA (14, 17, 18). Expression of the IAOx-metabolizing CYP83B1 in glucosinolate biosynthesis was not significantly affected by treatment with AgNO3 in our microarray experiments (Tables 1 and 2). Because CYP83B1 has a very high affinity for IAOx (14), this observation suggests that the IAOx-metabolizing enzymes in camalexin and IAA biosynthesis compete with CYP83B1 for IAOx, unless independent pools exist. Interestingly, S-alkyl thiohydroximate intermediates that are produced in vitro by the oxime-metabolizing CYP83 enzymes in the glucosinolate pathway undergo internal cyclization to form a thiazoline ring with structural similarity to the thiazol ring in camalexin (15). CYP83A1 and CYP83B1 belong phylogenetically to the CYP71 family (31), as does the oxime-metabolizing enzyme in the biosynthesis of cyanogenic glucosides (32). This relationship suggests that the oxime-metabolizing enzyme in camalexin biosynthesis may belong to the CYP71 family. CYP71B15/PAD3 is implicated to be involved in camalexin biosynthesis.
In conclusion, IAOx represents an interesting metabolic branching point, at which the flux of IAOx into camalexin, indole glucosinolates, and IAA must be controlled in a well structured organization at the 3D level of the different metabolons.
Differential Function of CYP79B2 and CYP79B3 in Camalexin Biosynthesis. A difference in the expression pattern of CYP79B2 and CYP79B3 was observed in rosette leaves 16 h after AgN03 treatment. The transcript level of CYP79B2, and not that of CYP79B3, was increased, as is evident from our cytochrome P450 microarray data. Similarly, GUS staining of leaves after AgNO3 treatment showed induction of CYP79B2, but not of CYP79B3, and the induction was localized to areas surrounding spots that received AgNO3 spray (Fig. 3C). This finding suggests that the local induction of CYP79B2 in the leaf is much higher than observed in the array data where whole-leaf RNA was used. The GUS and array data are in agreement with the observed tendency to reduction of camalexin production in the cyp79B2 mutant compared with the cyp79B3 mutant (Fig. 2).
The effect of infection with camalexin-inducing pathogens on gene expression has been investigated in a number of microarray technology studies. We surveyed the available array data to evaluate our observation that CYP79B2 and CYP79B3 play different roles in camalexin biosynthesis. Upon Alternaria brassicola infection, CYP79B2 was induced 3-fold after 24 h and 4.7-fold after 48 h induction (33). By using the same pathogen in a different experimental setup, as high as 45-, 50-, and 71-fold induction of CYP79B2 was observed in WT plants 12, 24, and 36 h, respectively, after infection (34). Upon P. syringae pv. ES4326 infection, a 27- to 42-fold induction of CYP79B2 was observed after 30 h (35), in accordance with our GUS-staining data after infection with P. syringae DC3000/avrRps4. Under the various experimental conditions, no or minor CYP79B3 induction was detected. The strong induction of CYP79B2 by eukaryotic and prokaryotic pathogens and by the abiotic elicitor AgNO3 indicates that CYP79B2 plays a critical role in camalexin biosynthesis.
In CYP79B2 overexpressing plants, no elevated camalexin levels have been observed. Therefore, the rate-limiting step of camalexin biosynthesis after AgNO3 induction is downstream of IAOx. The transcript level of CYP71B15/PAD3 was shown to be strongly induced by both A. brassicola and P. syringae infection (34, 35) but not significantly increased 16 h after AgNO3 treatment in our microarray experiment, despite camalexin induction. A possible explanation is that CYP71B15/PAD3 induction is stronger after treatment with A. brassicola and P. syringae in comparison with AgNO3, and the constitutive CYP71B15/PAD3 level is sufficient for camalexin response, suggesting a yet unknown enzyme to be rate-limiting for camalexin biosynthesis.
In summary, the present data have made us reformulate the biosynthetic pathway of camalexin, because we demonstrate that camalexin is synthesized from tryptophan via IAOx by CYP79B homologues and that it is not derived from glucosinolates or their degradation products. This is an important step in the elucidation of the biosynthetic pathways of cruciferous indole phytoalexins for future metabolic engineering of plants with improved disease-resistance and cancer-prevention properties. The data show that IAOx is a key branching point between several secondary metabolic pathways as well as primary metabolism, where IAOx has been shown to play a critical role in IAA homeostasis.
Acknowledgments
We thank Dr. Catherine Bellini for providing sur1 seeds, Dr. Yunde Zhao for providing seeds of cyp79B2, cyp79B3, and cyp79B2/cyp79B3 mutants, originated from the SALK collection (Salk Institute Genomic Analysis Laboratory), Prof. Alfons Gierl for his continuous support, Charlotte Kristensen, Marc Morant, Dr. Mari-Anne Newman, and Dr. Bent Larsen Petersen for helpful suggestions, Katharina Lange and Thomas Rauhut for practical help, and Dr. Søren Bak for providing the P450 array. Funding for the SIGnAL-indexed insertion mutant collection was provided by the National Science Foundation. This work was supported by a European Molecular Biology Organization long-term fellowship (to E.G.) and Deutsche Forschungsgemeinschaft Grant GL346/1.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: 6-F-IAOx, 6-fluoro-indole-3-acetaldoxime; GUS, β-glucuronidase; IAA, indole-3-acetic acid; IAOx, indole-3-acetaldoxime.
References
- 1.Pedras, M. S. C., Okanga, F. I., Zaharia, I. L. & Khan, A. Q. (2000) Phytochemistry 53, 161–176. [DOI] [PubMed] [Google Scholar]
- 2.Glawischnig, E., Mikkelsen, M. D. & Halkier, B. A. (2003) in Sulphur in Plants, eds. Abrol, Y. & Ahmad, A. (Kluwer, Dordrecht, The Netherlands), pp. 145–162.
- 3.Mezencev, R., Mojzis, J., Pilatova, M. & Kutschy, P. (2003) Neoplasma (Bratisl.) 50, 239–245. [PubMed] [Google Scholar]
- 4.Zhang, Y., Talalay, P., Cho, C.-G. & Posner, G. H. (1992) Proc. Natl. Acad. Sci. USA 89, 2399–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanley, A. B., Parsley, K. R., Lewis, J. A. & Fenwick, G. R. (1990) J. Chem. Soc. Perkin Trans. 1 , 2273–2276.
- 6.Tsuji, J., Jackson, E. P., Gage, D. A., Hammerschmidt, R. & Somerville, S. C. (1992) Plant Physiol. 98, 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomma, B. P., Nelissen, I., Eggermont, K. & Broekaert, W. F. (1999) Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji, J., Zook, M., Hammerschmidt, R., Somerville, S. & Last, R. (1993) Physiol. Mol. Plant Pathol. 43, 221–229. [Google Scholar]
- 9.Zook, M. & Hammerschmidt, R. (1997) Plant Physiol. 113, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zook, M. (1998) Plant Physiol. 118, 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grün, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R. B., Briggs, S. P., et al. (1997) Science 277, 696–699. [DOI] [PubMed] [Google Scholar]
- 12.Hull, A. K., Vij, R. & Celenza, J. L. (2000) Proc. Natl. Acad. Sci. USA 97, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen, M. D., Hansen, C. H., Wittstock, U. & Halkier, B. A. (2000) J. Biol. Chem. 275, 33712–33717. [DOI] [PubMed] [Google Scholar]
- 14.Bak, S., Tax, F. E., Feldmann, K. A., Galbraith, D. W. & Feyereisen, R. (2001) Plant Cell 13, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, C. H., Du, L., Naur, P., Olsen, C. E., Axelsen, K. B., Hick, A. J., Pickett, J. A. & Halkier, B. A. (2001) J. Biol. Chem. 276, 24790–24796. [DOI] [PubMed] [Google Scholar]
- 16.Helminger, J., Rausch, T. & Hilgenberg, W. (1985) Phytochemistry 24, 2497–2502. [Google Scholar]
- 17.Mikkelsen, M. D., Naur, P. & Halkier, B. A. (2004) Plant J. 37, 770–777. [DOI] [PubMed] [Google Scholar]
- 18.Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G. & Bellini, C. (2000) Proc. Natl. Acad. Sci. USA 97, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedras, M. S. C., Montaut, S., Xu, Y., Khan, A. Q. & Loukaci, A. (2001) Chem. Commun., 1572–1573. [DOI] [PubMed]
- 20.Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., Normanly, J., Chory, J. & Celenza, J. L. (2002) Genes Dev. 16, 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, F., Rausch, T. & Hilgenberg, W. (1981) J. Labelled Compds. Radiopharm. 18, 1491–1495. [Google Scholar]
- 22.Kane, M. D., Jatkoe, T. A., Stumpf, C. R., Lu, J., Thomas, J. D. & Madore, S. J. (2000) Nucleic Acids Res. 28, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glazebrook, J. & Ausubel, F. M. (1994) Proc. Natl. Acad. Sci. USA 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinsch, M. & Staskawicz, B. (1996) Mol. Plant–Microbe Interact. 9, 55–61. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, N., Tootle, T. L. & Glazebrook, J. (1999) Plant Cell 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mert-Türk, F., Bennet, M. H., Mansfield, J. H. & Holub, E. B. (2003) Physiol. Mol. Plant Pathol. 62, 137–145. [Google Scholar]
- 28.Müller, A. & Weiler, E. W. (2000) Planta 211, 855–863. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig-Müller, J. & Hillgenberg, W. (1992) Plant Cell Physiol. 38, 1115–1125. [Google Scholar]
- 30.Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D. & Chory, J. (2001) Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
- 31.Paquette, S. M., Bak, S. & Feyereisen, R. (2000) DNA Cell Biol. 19, 307–317. [DOI] [PubMed] [Google Scholar]
- 32.Bak, S., Kahn, R. A., Møller, B. L. & Halkier, B. A. (1998) Plant Mol. Biol. 36, 393–405. [DOI] [PubMed] [Google Scholar]
- 33.Narusaka, Y., Narusaka, M., Seki, M., Ishida, J., Nakashima, M., Kamiya, A., Enju, A., Sakurai, T., Satoh, M., Kobayashi, M., et al. (2003) Plant Cell Physiol. 44, 377–387. [DOI] [PubMed] [Google Scholar]
- 34.Van Wees, S. C., Chang, H. S., Zhu, T. & Glazebrook, J. (2003) Plant Physiol. 132, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glazebrook, J., Chen, W., Estes, B., Chang, H. S., Nawrath, C., Metraux, J. P., Zhu, T. & Katagiri, F. (2003) Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]