Abstract
We have developed a simple method to measure RNA synthesis in real time. In this technique, transcription reactions are performed in the presence of molecular beacons that possess a 2′-O-methylribonucleotide backbone. These probes become fluorescent as they hybridize to nascent RNA during the course of synthesis. We found that molecular beacons synthesized from natural deoxyribonucleotides were not suitable, because they are copied by RNA polymerases, generating complementary product strands that bind to the molecular beacons, causing a conformational change that results in unwanted fluorescence. However, when the molecular beacons are synthesized from 2′-O-methylribonucleotides, they are not copied and fluorescence is strictly dependent upon transcription of the added template. Utilizing these modified molecular beacons, quantitative comparisons were made of the activity of a variety of RNA polymerases and the effect of an inhibitor of transcription was determined.
INTRODUCTION
In order to study the DNA-directed synthesis of RNA by RNA polymerases, it is important to develop methods that enable the sequence-specific detection of RNA products in real time. The ability to monitor RNA synthesis in real time will aid in understanding the mechanisms by which inhibitors, repressors and transcription factors modulate the activities of RNA polymerases. If these methods can be adapted to high-throughput formats, they will be useful for identifying new drugs that inhibit clinically relevant polymerases, such as the polymerases of bacteria and viruses. We have developed single-stranded oligonucleotide probes called ‘molecular beacons’ that are suitable for monitoring the synthesis of RNA in real time, because they undergo a conformational reorganization when they bind to single-stranded nucleic acids that renders them brightly fluorescent (1).
MATERIALS AND METHODS
DNA templates and molecular beacons
The artificial DNA templates used in the transcription reactions were generated by performing PCR using tailed primers that introduce a bacteriophage T3, bacteriophage T7 (2,3) or Bacillus subtilis sinP3 (4) promoter sequence at one end of the amplified DNA and a poly(A) sequence at the other end of the amplified DNA. The sequences of the coding strands of these templates are shown in Table 1. Sequences to the left of the dotted segments identify the forward primers and sequences to the right of the dotted segments identify sequences that are complementary to the reverse primers. DNA templates containing a segment of the B.subtilis sinR gene were derived from a plasmid containing the entire sinR gene, including its promoter; and a DNA template containing a segment of the rpoB gene was derived from Mycobacterium tuberculosis total genomic DNA. In order to generate large quantities of DNA templates, 500-µl PCRs were prepared, each containing 1 pmol DNA template, 200 nM forward primer, 200 nM reverse primer, 20 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Forster City, CA), 250 µM each dNTP, 4 mM MgCl2, 50 mM KCl and 10 mM Tris–HCl (pH 8.0). The thermal cycling program consisted of 10 min at 95°C to activate the DNA polymerase, followed by 35 cycles of 30 s at 95°C, 30 s at 50°C and 30 s at 72°C.
Table 1. DNA templates (coding strands) and molecular beacons used in transcription reactions.
| Templates | ||
| T7 promoter sinR poly(A) | ||
| T7-sinR-poly(A) | 5′-GACTAATACGACTCACTATAGGGAGGA|TTTGCTCGATGAGAAA……….AAACAATTTCGTGAATTT|(A)25-3′ | 167 bp |
| T3 promoter sinR poly(A) | ||
| T3-sinR-poly(A) | 5′-GACTTATTAACCCTCACTAAAGGGAAG|TTTGCTCGATGAGAAA……….AAACAATTTCGTGAATTT|(A)25-3′ | 167 bp |
| T7 promoter rpoB poly(A) | ||
| T7-rpoB-poly(A) | 5′-GACTAATACGACTCACTATAGGGAGGA|GGCCGGTGGTCGCCGCG……….GCCCGGCGGTCTGTCACGT|(A)25-3′ | 187 bp |
| T7 promoter sinR | ||
| T7-sinR | 5′-GACTAATACGACTCACTATAGGGAGGA|TTTGCTCGATGAGAAA……….AAACAATTTCGTGAATTT-3′ | 161 bp |
| sinP3 promoter sinR poly(A) | ||
| sinP3-sinR-poly(A) | 5′-GAAGTCATACCGTAAATCCTTTCTGAATGTGCTATAATATCACAAGG|TTTGCTCGATGAGAAA……….AAACAATTTCGTGAATTT|(A)25-3′ | 187 bp |
| Molecular beacons | ||
| MB-sinR-TET-2′-O-methyl | 5′-tetrachlorofluorescein-CGCU UUUCGAUACCCCGGAUGUCAU AGCG-dabcyl-3′ | |
| MB-poly(A)-FAM-2′-O-methyl | 5′-fluorescein-CGC UUUUUUUUUUUU GCG-dabcyl-3′ | |
| MB-poly(A)-FAM-DNA | 5′-fluorescein-CGCTC TTTTTTTTTTTTTTTTTTTT GAGCG-dabcyl-3′ |
Molecular beacons possessing either a deoxyribonucleotide backbone or a 2′-O-methylribonucleotide backbone were prepared via β-cyanoethyl phosphoramidite chemistry on an Applied Biosystems 394 DNA/RNA synthesizer. A detailed protocol for the synthesis, purification and characterization of molecular beacons is available on the Internet at http://www.molecular-beacons.org. The sequences of the molecular beacons are shown in Table 1 and in Supplementary Material, Table S1. Underlined sequences identify the nucleotides that form the hairpin stem of each molecular beacon.
Real-time monitoring of transcription
Each 20-µl reaction contained 1 pmol DNA template, 10 mM MgCl2, 10 mM DTT, 1 mM each rNTP, 500 nM molecular beacon, 40 mM Tris–HCl (pH 8.0) and specific amounts of RNA polymerase that varied from experiment to experiment. In order to ensure that all of the transcription reactions started simultaneously, the reaction mixtures were prepared on an ice-cold metal block. The reactions were carried out in an Applied Biosystems 7700 Prism spectrofluorometric thermal cycler that was programmed to incubate the reactions for 60 min at 37°C, while recording the fluorescence intensity of each differently colored molecular beacon every 15 s. In the case of reactions that require sequential addition of reagents, 150-µl reactions were carried out in a thermostated cuvette using a Quantamaster spectrofluorometer (Photon Technology International, Lawrenceville, NJ). Transcription products labeled with [α-32P]CTP were analyzed by denaturing gel electrophoresis, using 10% polyacrylamide gels containing 7 M urea.
RNA polymerases and RNase H
Bacteriophage T3 and bacteriophage T7 RNA polymerases were purchased from Roche Diagnostics (Indianapolis, IN), Escherichia coli RNA polymerase was purchased from Epicentre Technologies (Madison, WI) and Mycobacterium smegmatis RNA polymerase was purified as described previously (4). RNase H was obtained from Pharmacia (Piscataway, NJ).
RESULTS AND DISCUSSION
Problems using DNA molecular beacons
We utilized a model in vitro transcription system to study the use of molecular beacons for real-time monitoring of RNA synthesis. The DNA templates that we used contained promoters favored by a number of different bacteriophage and bacterial RNA polymerases and they served as templates for short RNAs that ranged from 134 to 160 nt in length. These templates were used to initiate transcription in the presence of molecular beacons that were complementary to the transcripts. The fluorescence intensity of the molecular beacons in each reaction was monitored during the course of synthesis.
Using molecular beacons containing deoxyribonucleotide backbones for our first set of experiments, we noticed that the fluorescence of the molecular beacons increased with time, even in the absence of DNA templates (Fig. 1A). This increase in fluorescence background could have been due either to degradation of the molecular beacons, to binding of the polymerase to the molecular beacons and subsequent opening of their stems or to unintended synthesis of RNAs complementary to the molecular beacons. To investigate the origins of background generation, we added T7 RNA polymerase to a solution of molecular beacons in the absence of ribonucleoside triphosphates and found that no increase in fluorescence took place (Fig. 1B), indicating that the presence of the T7 RNA polymerase alone does not cause a conformational change or degradation of the molecular beacons. However, when ribonucleoside triphosphates were added to the mixture, fluorescence rose dramatically until it approached a plateau. When RNase H, which degrades RNA in RNA:DNA hybrids, was then added to this mixture, fluorescence decreased, reaching a level close to the initial level of fluorescence (Fig. 1B). These observations suggest that generation of the background signal was due to the synthesis of RNAs complementary to the molecular beacons.
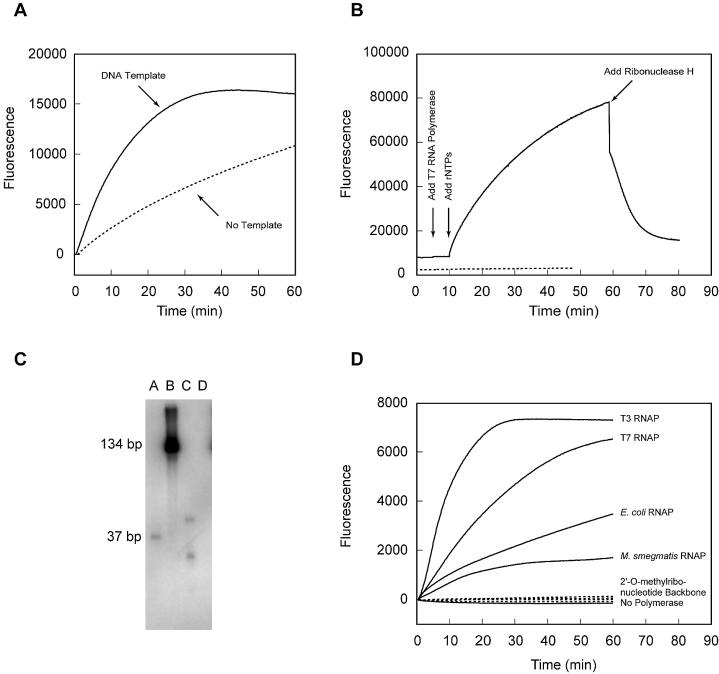
Figure 1.
Generation of background signals from molecular beacons containing a deoxyribonucleotide backbone during transcription reactions. (A) The fluorescence of transcription reactions containing T7 RNA polymerase and molecular beacon MB-poly(A)-FAM-DNA was monitored as a function of time. In the first reaction, DNA template T7-sinR-poly(A) was present (continuous line) and in the second reaction no template was present (dotted line). (B) The fluorescence of template-free reactions was monitored as the ingredients were added consecutively. The fluorescence of a reaction containing a molecular beacon [MB-poly(A)-FAM-DNA] that possessed a deoxyribonucleotide backbone (continuous line) was compared to the fluorescence from an otherwise identical reaction in which the molecular beacon [MB-poly(A)-FAM-2′-O-methyl] possessed a 2′-O-methylribonucleotide backbone (dotted line). (C) Denaturing gel electrophoresis analysis of the radioactively labeled products of template-free reactions. Lanes A and B contain size standards, lane C contains the products of a reaction containing MB-poly(A)-FAM-DNA (length 30 nt) and lane D contains the products of an otherwise identical reaction containing MB-poly(A)-FAM-2′-O-methyl (length 18 nt). (D) Template-free transcription reactions containing MB-poly(A)-FAM-DNA and four different RNA polymerases (RNAPs) (continuous lines) were compared to four otherwise identical reactions containing MB-poly(A)-FAM-2′-O-methyl (dotted lines).
The synthesis of complementary RNAs was confirmed by the incorporation of radioactive ribonucleotides during the reaction, followed by analysis of the products by denaturing gel electrophoresis and autoradiography. The results showed that RNAs of the same length as the molecular beacons were produced in the reaction (Fig. 1C). The same phenomenon was observed when RNA polymerases isolated from E.coli, M.smegmatis and bacteriophage T3 were used (Fig. 1D).
In order to explore whether this template-independent increase in fluorescence takes place with all molecular beacons containing a deoxyribonucleotide backbone, we carried out the reactions with 10 other molecular beacons containing a deoxyribonucleotide backbone (Supplementary Material, Table S1) and found that all of them exhibited the increase, although the rate of increase depended on their sequence (Supplementary Material, Fig. S1). The results of these experiments indicate that molecular beacons possessing deoxyribonucleotide backbones are transcribed by RNA polymerases in a promoter-independent manner. The transcripts then bind to the molecular beacons, which changes their conformation and results in a marked increase in background fluorescence. Promoter-independent synthesis of RNA from single-stranded DNA templates has been observed before (5). Our results explain why it has not been possible to reproduce the results of an earlier study in which molecular beacons possessing a deoxyribonucleotide backbone were used to monitor the progress of transcription reactions (6). That study was not able to reveal the presence of non- specific background signals, because template-free control transcriptions were not performed.
Use of 2′-O-methylribonucleotide molecular beacons
We reasoned that it might be possible to solve the background problem by using molecular beacons synthesized from unnatural nucleotides, which would not be recognized as templates by DNA-directed RNA polymerases. When molecular beacons synthesized from 2′-O-methylribonucleotides (in which the 2′-hydroxyl group of ribose is replaced by a 2′-O-methyl group) were used in template-free transcription reactions, we observed no increase in fluorescence (Figs 1B and 2). In addition, no radioactive RNA products could be detected in these reactions (Fig. 1C). Furthermore, no other RNA polymerases produced background signals (Fig. 1D) and eight other molecular beacons possessing 2′-O-methylribonucleotides (Supplementary Material, Table S1) that were tested with T7 RNA polymerase did not produce any background signals (Supplementary Material, Fig. S1). However, when a DNA template encoding an RNA complementary to the molecular beacon was included in the reaction, an increase in fluorescence was observed (Fig. 2, continuous line). Thus, in reactions containing molecular beacons possessing a 2′-O-methylribonucleotide backbone, the only fluorescence that occurs is due to the binding of the molecular beacons to the transcripts synthesized from the added template DNA. We therefore used these probes in all subsequent experiments.
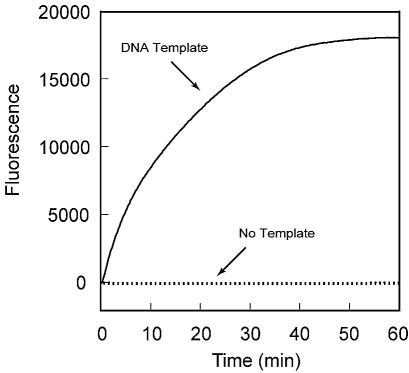
Figure 2.
Signal generation is entirely dependent upon transcription from an added DNA template when a molecular beacon possessing a 2′-O-methylribonucleotide backbone [MB-poly(A)-FAM-2′-O-methyl] is used as the probe. The fluorescence of a reaction initiated with a DNA template [T7-sinR-poly(A)] (continuous line) was compared with the fluorescence of an otherwise identical template-free reaction (dotted line).
In order to see whether the progress of relatively fast transcription reactions can be monitored with molecular beacons, we utilized the extremely efficient RNA polymerases from bacteriophages T3 and T7 and DNA templates possessing corresponding promoters (2,3). The concentration of the molecular beacons was chosen so that the probes would always be in excess of the amount of RNA produced. In these transcription reactions fluorescence rose rapidly at first and then asymptotically approached a plateau (Fig. 3). The same kinetic profile was obtained when we utilized the intercalating fluorescent dye SYBR green, which binds to transcripts as they are synthesized, indicating that molecular beacons faithfully monitor the rate of RNA synthesis. By systematically changing the amount of RNA polymerase, we demonstrated that the initial rate of increase in fluorescence intensity is linearly proportional to polymerase concentration (Fig. 3, insets). Similarly, we showed that the initial rate of increase in fluorescence intensity is proportional to the amount of added DNA template. Quantitative analysis indicated that multiple copies of RNA were produced for each DNA template molecule. These data indicate that fluorescence intensity at a given time reflects the amount of RNA produced by that time and the activities of different RNA polymerases can be quantitatively determined in real time.
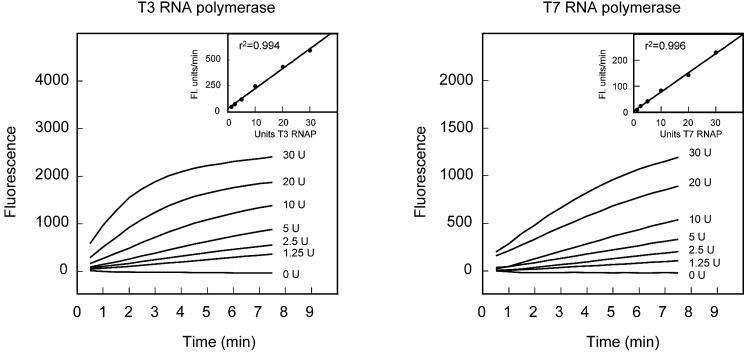
Figure 3.
Demonstration that the rate of RNA synthesis determined by the fluorescence of molecular beacons in real time is proportional to the concentration of added bacteriophage T3 or bacteriophage T7 RNA polymerase. The DNA template used to initiate each group of transcription reactions contained a promoter that was specific for the added RNA polymerase. The molecular beacon used to monitor the course of synthesis was specific for the 3′-terminal poly(A) sequence in the expected transcripts. The results shown in the insets demonstrate that the initial rate of transcription (measured during the first 3 min of each reaction) is linearly proportional to the concentration of the RNA polymerase (RNAP). Each reaction was initiated with 1 pmol template DNA, which was T3-sinR-poly(A) (for T3 RNA polymerase reactions) or T7-sinR-poly(A) (for T7 RNA polymerase reactions). The reactions were monitored with molecular beacon MB-poly(A)-FAM-2′-O-methyl.
Multiplex transcription monitoring
In order to monitor the synthesis of two different RNAs in the same reaction tube, we prepared two DNA templates possessing different coding sequences, but the same promoter sequence. The first DNA contained a promoter for T7 RNA polymerase and a segment of the M.tuberculosis rpoB gene that serves as a template for the synthesis of a transcript possessing a 3′-poly(A) sequence [T7-rpoB-poly(A)]. The second DNA template contained the same promoter for T7 RNA polymerase and a segment of the B.subtilis sinR gene (T7-sinR). In order to distinguish between the RNA products of these templates, two different molecular beacons were prepared. The first molecular beacon was specific for the poly(A) sequence in the rpoB transcript and was labeled with fluorescein [MB-poly(A)-FAM-2′-O-methyl] and the second molecular beacon was specific for a coding region of the sinR transcript and was labeled with tetrachlorofluorescein (MB-sinR-TET-2′-O-methyl). Three transcription reactions were carried out. Each reaction contained both molecular beacons (Fig. 4) and fluorescence was monitored in real time. When T7-rpoB-poly(A) was the only template present in the reaction, only fluorescence from the fluorescein-labeled molecular beacons increased. When T7-sinR was the only template present in the reaction, only fluorescence from the tetrachlorofluorescein-labeled molecular beacons increased. When both templates were present in the reaction, fluorescence from both molecular beacons increased with time. In order to make these measurements, the entire fluorescence emission spectrum from each reaction tube was recorded as a function of time. The complex fluorescence spectrum recorded at each time point was analyzed in real time by the computer program controlling the spectrofluorometric thermal cycler to determine the fluorescence intensity resulting from each of the molecular beacons that was present in the reaction. There was no significant spillover of signal from one fluorophore to another. These results indicate that synthesis of RNA from several templates and from different polymerases can be monitored simultaneously in the same reaction by utilizing a set of molecular beacons, each specific for a different transcript and each labeled with a differently colored fluorophore (7). The ability to monitor multiple reactions in the same tube will be useful for the inclusion of internal controls and for increasing the throughput of assays.
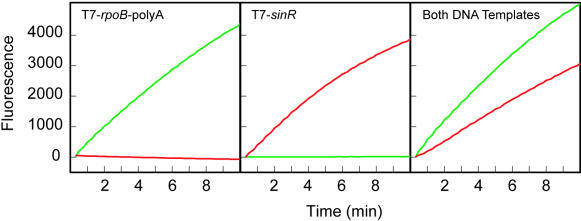
Figure 4.
Monitoring the synthesis of two different RNA transcripts in the same tube. Each reaction contained bacteriophage T7 RNA polymerase, fluorescein-labeled molecular beacon MB-poly(A)-FAM-2′-O-methyl and tetrachlorofluorescein-labeled molecular beacon MB-sinR-TET-2′-O-methyl. T7-rpoB-poly(A) DNA template was added to the first reaction (left), T7-sinR DNA template was added to the second reaction (middle) and both DNA templates were added to the third reaction (right). The fluorescence intensities of fluorescein (green lines) and tetrachlorofluorescein (red lines) were monitored simultaneously during each reaction. The results show that synthesis of rpoB poly(A) transcripts only elicits fluorescence from poly(A)-specific molecular beacons and synthesis of sinR transcripts only elicits fluorescence from sinR-specific molecular beacons.
Inhibition of transcription
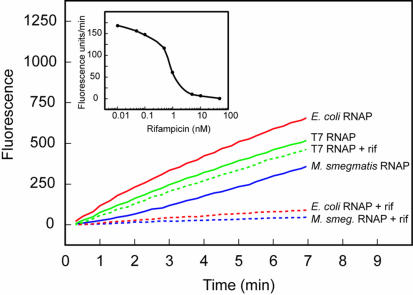
In order to demonstrate that molecular beacons possessing a 2′-O-methylribonucleotide backbone can be used to study the inhibition of RNA polymerization by specific inhibitors, we decided to use rifampicin, which is a potent inhibitor of bacterial RNA polymerases (8) but does not inhibit bacteriophage RNA polymerases (9). Exploiting our previous observation that the RNA polymerases from E.coli and M.smegmatis are able to initiate transcription from the B.subtilis sinP3 promoter (4), we prepared a DNA template containing a segment of the sinR gene that possessed this promoter [sinP3-sinR-poly(A)]. There was no rise in fluorescence when rifampicin was added to transcription reactions containing this DNA template, molecular beacons complementary to the transcripts and either E.coli RNA polymerase or M.smegmatis RNA polymerase. However, in the absence of rifampicin, a normal rise in fluorescence was observed (Fig. 5). Significantly, the presence of rifampicin had no effect on the increase in fluorescence that occurred during transcription reactions containing T7 RNA polymerase, demonstrating the ability of this technique to specifically measure inhibition by rifampicin. We were also able to determine the effective inhibitory concentration of rifampicin on the activity of E.coli RNA polymerase by monitoring the effect of different concentrations of rifampicin on the initial rate of increase in fluorescence (Fig. 5, inset). The results of these experiments demonstrate the ease with which it is possible to use this method to characterize new drugs that are designed to inhibit particular RNA polymerases.
Figure 5.
Use of molecular beacons to assess the inhibition of different RNA polymerases by rifampicin. Each transcription reaction contained either 1 U/µl bacteriophage T7 RNA polymerase, 0.05 U/µl E.coli RNA polymerase or 1 µl M.smegmatis RNA polymerase (4). Each reaction contained DNA templates possessing a promoter specific for the RNA polymerase (RNAP) that was present and molecular beacons that were specific for the resulting transcripts. The kinetic course of reactions carried out in the absence of rifampicin (continuous lines) was compared to the kinetic course of otherwise identical reactions carried out in the presence of 5 nM rifampicin (rif) (dotted lines). The results shown in the inset illustrate the relationship between the initial rate of transcription catalyzed by E.coli RNA polymerase and the logarithm of rifampicin concentration. T7-sinR-poly(A) was used as template in the T7 RNA polymerase reactions and sinP3-sinR-poly(A) was used as template in both the E.coli RNA polymerase reactions and the M.smegmatis RNA polymerase reactions. All reactions were monitored using molecular beacon MB-poly(A)-FAM-2′-O-methyl.
Applications
Transcription reactions containing molecular beacons should aid in the analysis of interactions that occur between transcription factors, RNA polymerases and regulatory elements that are present in DNA templates. Even though we monitored RNA synthesis catalyzed by DNA-directed RNA polymerases, these reactions can easily be adopted for monitoring the activity of replicases, reverse transcriptases and other enzymes that catalyze the synthesis of single-stranded nucleic acids. Since the time course of fluorogenic reactions can be monitored in a high-throughput format with commonly available instruments, the screening of potential drugs that inhibit the RNA polymerases and reverse transcriptases of pathogens will be more efficient. The advantage of this method, compared to conventional measurements of polymerase activities using intercalating dyes, is that the probes do not generate a signal from unrelated nucleic acids, aberrant polymerase activities can be distinguished from promoter-specific polymerization and transcription from different templates can be followed simultaneously in the same reaction.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Eugenie Dubnau for critically reading the manuscript. This work was supported by National Institutes of Health grants EB-00277 to F.R.K., GM-070357-01 to S.T. and AI-44856 to I.S.
REFERENCES
- 1.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Davanloo P., Rosenberg,A.H., Dunn,J.J. and Studier,F.W. (1984) Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 81, 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris C.E., Klement,J.F. and McAllister,W.T. (1986) Cloning and expression of the bacteriophage T3 RNA polymerase gene. Gene, 41, 193–200. [DOI] [PubMed] [Google Scholar]
- 4.Predich M., Doukhan,L., Nair,G. and Smith,I. (1995) Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol. Microbiol., 15, 355–366. [DOI] [PubMed] [Google Scholar]
- 5.Krupp G. (1989) Unusual promoter-independent transcription reactions with bacteriophage RNA polymerases. Nucleic Acids Res., 17, 3023–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Feldman,P. and Chung,T.D. (2002) Real-time monitoring of in vitro transcription using molecular beacons. Anal. Biochem., 300, 40–45. [DOI] [PubMed] [Google Scholar]
- 7.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann G., Honikel,K.O., Knusel,F. and Nuesch,J. (1967) The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta, 145, 843–844. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlin M. and Ring,J. (1973) Characterization of T7-specific ribonucleic acid polymerase. II. Inhibitors of the enzyme and their application to the study of the enzymatic reaction. J. Biol. Chem., 248, 2245–2250. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.