ABSTRACT
The emerging epidemic of drug resistance places the development of efficacious and safe antibiotics in the spotlight of current research. Here, we report the design of next-generation aminoglycosides. Discovery efforts were driven by rational synthesis focusing on 4′ alkylations of the aminoglycoside paromomycin, with the goal to alleviate the most severe and disabling side effect of aminoglycosides—irreversible hearing loss. Compounds were evaluated for target activity in in vitro ribosomal translation assays, antibacterial potency against selected pathogens, cytotoxicity against mammalian cells, and in vivo ototoxicity. The results of this study produced potent compounds with excellent selectivity at the ribosomal target, promising antibacterial activity, and little, if any, ototoxicity upon chronic administration. The favorable biocompatibility profile combined with the promising antibacterial activity emphasizes the potential of next-generation aminoglycosides in the treatment of infectious diseases without the risk of ototoxicity.
IMPORTANCE
The ever-widening epidemic of multidrug-resistant infectious diseases and the paucity of novel antibacterial agents emerging from modern screening platforms mandate the reinvestigation of established drugs with an emphasis on improved biocompatibility and overcoming resistance mechanisms. Here, we describe the preparation and evaluation of derivatives of the established aminoglycoside antibiotic paromomycin that effectively remove its biggest deficiency, ototoxicity, and overcome certain bacterial resistance mechanisms.
INTRODUCTION
The rise in levels of antibiotic resistance has resulted in an emerging global public health crisis (1). Thus, there is an urgent need not only to identify new drug targets and to develop new antibiotic compound classes (2) but also to improve available antibacterial agents. Synthetic modification of natural product scaffolds has been the most successful strategy in antibacterial drug discovery (3–5). This has led to the development of multiple generations of β-lactams, macrolides, and tetracycline antibiotics. Antibiotic scaffolds are modified to improve therapeutic use, e.g., by synthesizing derivatives less affected by resistance or by designing derivatives with improved biocompatibility.
With the development of new orally administered antibacterial compound classes, interest in improving the clinical efficacy of aminoglycosides declined in the 1970s. However, aminoglycoside antibiotics still represent one of the critically important classes of antimicrobial agents for human therapy (6) and continue to be used as broad-spectrum antibiotics whose targets include Gram-negative pathogens, methicillin-resistant Staphylococcus aureus (MRSA), multidrug-resistant Mycobacterium tuberculosis, and complex infectious diseases such as sepsis, complicated urinary tract infections, chronic obstructive pulmonary disease in hospitalized patients, and exacerbated cystic fibrosis (7, 8).
Aminoglycosides target bacterial protein synthesis by inducing codon misreading and by inhibiting translocation of the tRNA-mRNA complex (9–12). Aminoglycosides affect translation of the ribosome by direct interaction with decoding A-site rRNA. The molecular details of antibiotic-target interaction have been revealed by X-ray crystallography (13, 14) and mutagenesis studies (15, 16). Nucleotides 1408 and 1491 in the decoding A site of the small-subunit rRNA (Escherichia coli numbering used throughout) are critical for properly positioning ring I of the 2-deoxystreptamine aminoglycoside compounds (15–18). The phylogenetic variability of rRNA residues 1408 and 1491 provides the basis and limits for aminoglycoside selectivity (19–22; for a review, see reference 23).
The therapeutic use of aminoglycosides is compromised by significant toxicity, in particular ototoxicity, which is irreversible and results in hearing damage affecting 20% of patients following brief courses of treatment (24) and more than 90% of patients after long-term regimens (25). Ototoxicity of aminoglycoside antibiotics occurs in both a sporadic, dose-dependent fashion and in a genetically inherited fashion, the latter linked to mutations A1555G and C1494U in mitochondrial rRNA (26–28). Recent evidence points to a key role of mitochondrial dysfunction in aminoglycoside ototoxicity (29–31).
There is a renewed interest in aminoglycosides with a focus on overcoming resistance mechanisms (32–34; for reviews, see references 35, 36, and 37). Few, if any, systematic studies have been conducted to discover aminoglycoside structures with less ototoxicity. Previous studies led to the hypothesis that aminoglycoside ototoxicity is related to the drug's mechanism of action and linked to limited target selectivity, i.e., it stems from the drug’s action on the eukaryotic ribosome (29, 38). Following this line of research, we recently disclosed the discovery of a highly target-selective set of novel 4′,6′-O-acetals and 4′-O-aralkyl-ethers of the clinical aminoglycoside paromomycin (1), which are active against a range of bacterial pathogens (39). Despite these initial promising results at the drug target level, subsequent validation in vivo pointed to dose-related acute toxicity (unpublished data). Given our previous success in modifying the drug-target interaction, we were interested in exploring further novel 4′-aminoglycoside analogues with a view to combining target selectivity with ameliorated ototoxicity and antibacterial efficacy. Here, we report on the design and synthesis of 4′-O-(alkyl) substituted derivatives of paromomycin. The ribosomal target selectivity, antibacterial efficacy, and toxicity profile of these compounds, which are devoid of the acute toxicity exhibited by the previous aralkyl series, were examined in both in vitro and in vivo systems. Lead structures, exhibiting enhanced selectivity for the bacterial ribosome, potent antibacterial activity, and little, if any, ototoxicity in vivo were identified.
RESULTS
Chemical synthesis and target specificity.
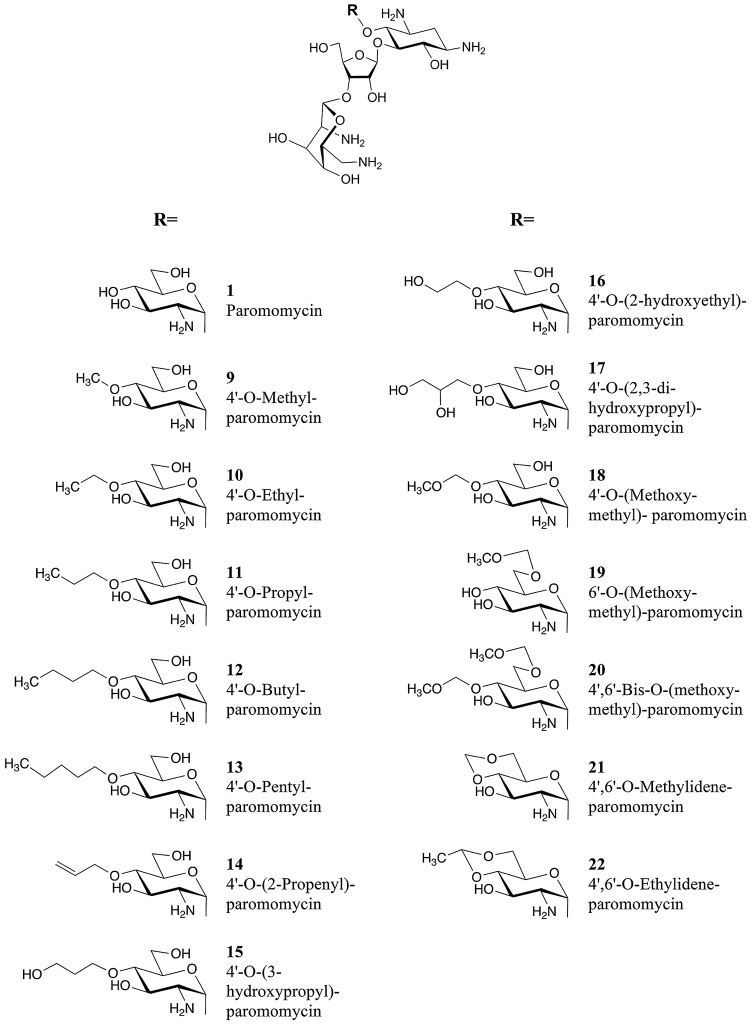
Ring I of paromomycin was modified by substitutions of the 4′-hydroxy group. We designed a series of aliphatic 4′-O-substituted compounds, compounds 9 to 22, derived from the known paromomycin derivative, compound 2, via the intermediates, compounds 23 to 27 (Fig. 1; see also the supplemental material). The derivatives were tested for target activity in cell-free ribosomal translation assays in comparison to the parental paromomycin, 4′-deoxy paromomycin (40), 4′-O-(3-phenylpropyl) paromomycin, and 4′,6′-O-(3-phenylpropylidene) paromomycin—the latter two compounds as representatives of the previous series of 4′-O-aralkyl ethers and 4′,6′-O-acetals (39). To assess target activity in cell-free translation assays, we used wild-type bacterial ribosomes and recombinant bacterial ribosomes with single point mutations in the drug binding pocket. Dose-response curves of aminoglycoside-dependent inhibition of luciferase synthesis were analyzed to define the drug concentrations required to inhibit in vitro synthesis of functional firefly luciferase to 50% (IC50s).
FIG 1 .
Chemical structures of compounds synthesized. The compounds are indicated by boldface numbers (e.g., compound 1 is shown as 1).
The point mutations introduced into the bacterial ribosomes were chosen to reflect the phylogenetically variable 16S rRNA residues in the drug binding pocket, i.e., nucleotides 1408 (adenine in bacterial/mitochondrial ribosomes and guanine in cytosolic ribosomes) and 1491 (guanine in bacterial ribosomes, adenine in cytosolic ribosomes, and cytosine in mitochondrial ribosomes [see Fig. S1 in the supplemental material]) in the decoding A site. Interaction of paromomycin with the ribosome is little affected by alteration of A1408, while mutational alterations of residue G1491 significantly decrease drug-target interaction. The interaction of the 4′-O-alkyl derivatives with the ribosomal drug binding site, however, largely involves both residues 1408 and 1491 (Table 1). This is illustrated by comparing the drug susceptibility of mutant ribosomes with that of wild-type ribosomes and by calculating the ratio of the IC50 for the mutant ribosomes to that for the wild-type ribosomes. For paromomycin, the ratio of the IC50 for the G1408 mutant ribosomes to the IC50 for the wild-type ribosomes is 13, and the ratio of the IC50 for the C1491 mutant ribosomes to the IC50 for the wild-type ribosomes is 520. For compound 11, the ratio of IC50s for the G1408 mutant ribosomes to the wild-type ribosomes is 80, and the ratio of IC50s for C1491 mutant ribosomes to the wild-type ribosomes is 2,000; for compound 10, the ratio of IC50s for the G1408 mutant ribosomes to the wild-type ribosomes is 100, and the ratio of IC50s for the C1491 mutant ribosomes to the wild-type ribosomes is 3,000.
TABLE 1 .
Interaction of 4′-O-alkyl derivatives with polymorphic residues 1408 and 1491 in the drug binding pocket
| Drug | IC50 (mg/liter) with mutant bacterial A sites in the ribosomea: |
|||
|---|---|---|---|---|
| Wild-type | G1491C | G1491A | A1408G | |
| Paromomycin | 0.02 ± 0.01 | 10.42 ± 2.86 | 0.57 ± 0.09 | 0.26 ± 0.04 |
| 4′ Deoxy paromomycin | 0.05 ± 0.01 | 28.6 ± 0.9 | 2.4 ± 0.1 | 0.89 ± 0.4 |
| 4′,6′-O-(3-Phenylpropylidene) paromomycin | 0.10 ± 0.03 | 124.1 ± 0.1 | 43.0 ± 11.3 | 3.8 ± 2.4 |
| 4′-O-(3-Phenylpropyl) paromomycin | 0.20 ± 0.07 | 200.73 ± 25.84 | 51.26 ± 15.72 | 14.52 ± 5.77 |
| Compound 13 | 0.40 ± 0.11 | 229.92 ± 32.26 | 62.13 ± 9.35 | 21.73 ± 5.04 |
| Compound 12 | 0.30 ± 0.03 | 348.38 ± 23.69 | 90.87 ± 16.44 | 24.33 ± 5.82 |
| Compound 11 | 0.14 ± 0.02 | 288.85 ± 148.66 | 45.54 ± 23.65 | 11.74 ± 4.94 |
| Compound 14 | 0.14 ± 0.04 | 268.65 ± 18.91 | 44.68 ± 3.15 | 12.34 ± 1.57 |
| Compound 10 | 0.08 ± 0.03 | 258.93 ± 31.67 | 40.77 ± 16.93 | 8.83 ± 3.10 |
| Compound 22 | 0.12 ± 0.08 | 180.47 ± 45.33 | 41.77 ± 7.39 | 5.42 ± 1.23 |
| Compound 9 | 0.22 ± 0.05 | 242.42 ± 6.20 | 68.99 ± 11.53 | 14.21 ± 6.74 |
| Compound 21 | 0.89 ± 0.24 | 522.20 ± 44.59 | 119.58 ± 49.65 | 32.57 ± 9.13 |
| Compound 16 | 0.05 ± 0.02 | 152.22 ± 51.28 | 5.99 ± 1.19 | 1.93 ± 0.38 |
| Compound 15 | 0.73 ± 0.19 | >700 | 143.69 ± 15.86 | 38.12 ± 5.73 |
| Compound 17 | 0.03 ± 0.01 | 67.35 ± 21.25 | 2.57 ± 0.48 | 0.87 ± 0.07 |
| Compound 18 | 0.20 ± 0.03 | 167.43 ± 11.30 | 11.45 ± 0.55 | 4.48 ± 0.32 |
| Compound 19 | 1.57 ± 0.42 | 585.91 ± 16.81 | 83.71 ± 18.04 | 5.47 ± 1.24 |
| Compound 20 | 10.87 ± 0.53 | >700 | 529.40 ± 76.03 | 80.44 ± 20.17 |
The interaction of 4′-O-alkyl derivatives with the bacterial A site of the drug binding pocket is measured by the drug concentrations (in milligrams per liter) required to inhibit in vitro synthesis of functional firefly luciferase to 50% (IC50s). The values are means ± standard deviations (SD) for experiments performed in triplicate.
Compared to a simple deletion of the 4′-hydroxy group as in 4′-deoxy paromomycin, introduction of a 4′-O-alkyl group provokes a substantial loss of activity against mutant 1491C, 1491A, and 1408G ribosomes. The optimum size for the 4′-O-alkyl group conferring little change in activity toward bacterial wild-type ribosomes but greatly affecting interaction with mutant 1491A, 1491C, and 1408G ribosomes is two or three carbon atoms. The effect of the 4′-O-alkyl substituent is in part offset by the inclusion of a hydroxyl group, as in the 2-hydroxy alkyl ether compounds (compounds 16 and 17). Shift of the methoxymethyl group from the 4′ position to the 6′ position causes a significant loss of inhibitory activity for wild-type bacterial ribosomes (e.g., compare compound 9 with compound 19). Addition of two methoxymethyl groups (one at 4′-O and the other at 6′-O) as in compound 20 totally disrupts antiribosomal activity.
Antibacterial activity.
To screen for antibacterial activity, we first determined the MIC values against Mycobacterium smegmatis (see Table S1 in the supplemental material). The most active compounds were selected and further tested against clinical isolates of Escherichia coli and Staphylococcus aureus. Compared to the parental paromomycin, the antibacterial activity of the 4′-O-alkylated aminoglycosides was mostly modest (Table 2). However, the ethyl ether (compound 10) and propyl ether (compound 11) showed promising antibacterial activity including clinical S. aureus isolates that are highly resistant toward paromomycin.
TABLE 2 .
MICs of clinical isolates
| Clinical isolate | MIC (mg/liter)a: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Paromomycin | c12 | c11 | c14 | c10 | c22 | c9 | c16 | c17 | c18 | |
| Staphylococcus aureus (MRSA) isolates | ||||||||||
| AG 038 | 4 | 8−16 | 8−16 | 8−16 | 8−16 | 32−64 | 32 | 32 | 16−32 | 16−32 |
| AG 039 | >256 | 16−32 | 16 | 16 | 16 | 64−128 | 32 | 32 | 16−32 | 64 |
| AG 042 | >256 | 16 | 8−16 | 16 | 8−16 | 32 | 32 | 16−32 | 16 | 32 |
| AG 044 | 4−8 | 16 | 8 | 8−16 | 8−16 | 32−64 | 32 | 32−64 | 32 | 16−32 |
| AG 045 | 4 | 16 | 8 | 8−16 | 8−16 | 16−32 | 32 | 16−32 | 16 | 32 |
| Escherichia coli isolates | ||||||||||
| AG 002 | 8−16 | 32−64 | 16−32 | 64 | 32 | 128 | ≥128 | 32 | 16−32 | 32−64 |
| AG 003 | 8−16 | 64 | 32 | 64 | 32 | 64−128 | ≥128 | 32 | 16 | 64−128 |
| AG 001 | 16−32 | 64 | 32 | 64 | 64 | >128 | >128 | 32−64 | 32−64 | 128 |
| AG 055 | 8−16 | 64 | 32 | 64 | 32 | 64 | >128 | 32−64 | 16−32 | 64−128 |
Compounds 9 to 22 are indicated by a lowercase c before the compound number (e.g., c12 stands for compound 12).
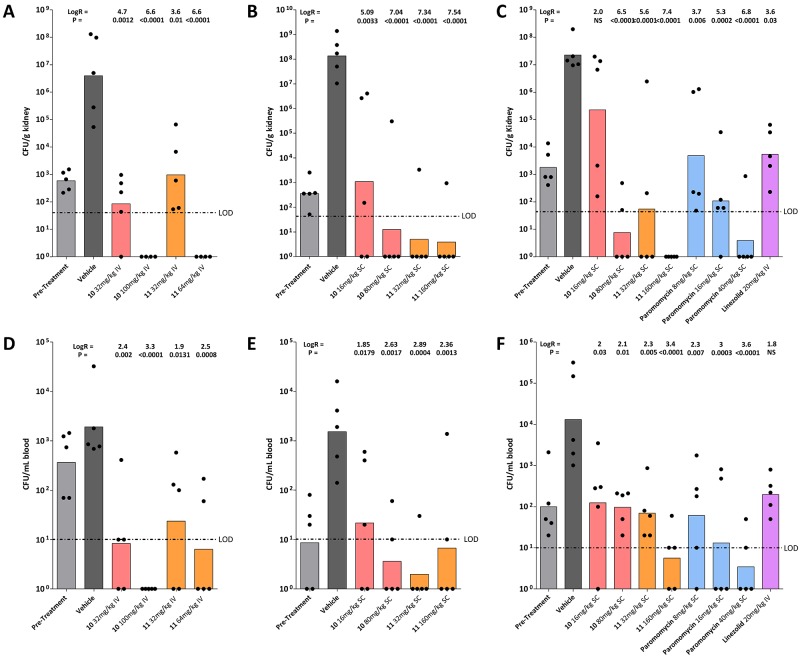
To determine whether the in vitro activity of the designed compound series translates into in vivo activity, we assessed the antibacterial activity of the ethyl ether (compound 10) and propyl ether (compound 11) in a neutropenic murine model of Staphylococcus aureus septicemia. Two different routes of drug application were initially investigated—intravenous and subcutaneous. In animals infected with methicillin-resistant S. aureus, treatment with compounds 10 and 11 via both routes effectively reduced the bacterial burden in blood and kidney in a dose-dependent manner (Fig. 2). For subcutaneous drug application, the efficacy of compounds 10 and 11 was compared to that of the parental paromomycin. Compared to the vehicle-treated mice, drug treatment reduced bacterial burden in the kidney between 2 and 7 log10 units and in blood between 2 and 3 log10 units in a dose-dependent manner. At the highest dose tested, compounds 10 and 11 and paromomycin reduced the bacterial burden in the kidney below detectable limits (Fig. 2C and F).
FIG 2 .
In vivo activity of aminoglycoside compounds and comparators in a murine septicemia model. Compounds were administered intravenously (IV) (A and D) or subcutaneously (SC) (B, C, E, and F). Compounds 10 and 11 are indicated by boldface numbers. (A to C) Bacterial burden in kidney (CFU/g tissue); (D to F) bacterial burden in blood (CFU/ml). The values for 1 h pretreatment and vehicle (25 h) are shown as controls. Each symbol represents the value for an individual animal (CFU). The bar represents the geometric mean of the group (5 mice per treatment group). The log reduction (LogR) compared to the vehicle control and the P value (nonparametric Kruskal-Wallis using pairwise comparisons [Conover-Inman]) are given above each group (NS, not significant). The limit of detection is indicated by the horizontal line labeled LOD. The different treatments are color coded as follows: light gray, pretreatment; dark gray, vehicle control; red, compound 10; orange, compound 11; blue, paromomycin; violet, linezolid.
Activity toward eukaryotic ribosomes and cell cytotoxicity.
The toxicity of aminoglycosides, at least in part, relates to limited selectivity, i.e., their activity toward the eukaryotic ribosome (20). The two characteristic features of aminoglycoside action on the eukaryotic ribosome relevant in this respect are translation inhibition and induction of misreading (23).
To assess target selectivity of the 4′-O-alkyl derivatives, i.e., their interaction with the various eukaryotic drug binding pockets, we made use of hybrid bacterial ribosomes carrying decoding A-site cassettes representing the various eukaryotic drug binding pockets. These ribosomes carry the cytoplasmic A site, mitochondrial A site, or mitochondrial mutant A1555G deafness A site (see Fig. S1 in the supplemental material) and have been shown to faithfully reflect the drug susceptibility of the corresponding eukaryotic ribosomes (21, 29, 30). Translation inhibition was assessed by determining the inhibition of protein synthesis in cell-free translation assays. Compared to paromomycin, the 4′-O-alkyl derivatives interact poorly with any of the eukaryotic drug binding pockets, including cytosolic hybrid ribosomes, mitochondrial hybrid ribosomes, and mitochondrial deafness hybrid ribosomes (Table 3).
TABLE 3 .
Interaction of 4′-O-alkyl derivatives with eukaryotic drug binding pockets
| Drug | IC50 (mg/liter) with eukaryotic A sites in the ribosomea: |
|||
|---|---|---|---|---|
| Mitochondrial wild-type | Mitochondrial mutant A1555G | Cytosolic wild-type | Rabbit reticulocytes | |
| Paromomycin | 50.66 ± 13.33 | 5.77 ± 2.32 | 9.89 ± 2.73 | 9.55 ± 4.94 |
| 4′ Deoxy paromomycin | 74.1 ± 12.4 | 24.2 ± 10.5 | 28.4 ± 13.8 | 17.3 ± 4.2 |
| 4′,6′-O-(3-Phenylpropylidene) paromomycin | 305.7 ± 74.1 | 126.9 ± 54.4 | 150.5 ± 28.0 | ND |
| 4′-O-(3-Phenylpropyl) paromomycin | 49.85 ± 15.39 | 105.68 ± 39.23 | 268.57 ± 54.97 | ND |
| Compound 13 | 109.02 ± 6.73 | 81.11 ± 8.77 | 92.32 ± 9.20 | 49.74 ± 3.48 |
| Compound 12 | 149.54 ± 38.62 | 148.36 ± 30.59 | 161.48 ± 13.93 | 49.83 ± 5.47 |
| Compound 11 | 194.88 ± 57.17 | 151.92 ± 31.44 | 165.52 ± 28.30 | 59.93 ± 30.10 |
| Compound 14 | 102.39 ± 9.99 | 90.88 ± 9.96 | 125.18 ± 3.07 | 49.98 ± 16.75 |
| Compound 10 | 96.82 ± 4.22 | 119.83 ± 45.62 | 94.65 ± 30.97 | 37.58 ± 14.14 |
| Compound 22 | 206.31 ± 112.31 | 98.00 ± 39.68 | 126.41 ± 71.71 | 37.40 ± 4.61 |
| Compound 9 | 106.58 ± 24.30 | 174.76 ± 46.48 | 128.22 ± 27.65 | 40.92 ± 8.39 |
| Compound 21 | 325.05 ± 123.72 | 252.74 ± 37.61 | 240.92 ± 69.84 | 130.03 ± 41.37 |
| Compound 16 | 87.49 ± 30.97 | 81.35 ± 17.06 | 74.36 ± 11.94 | 24.00 ± 5.10 |
| Compound 15 | >700 | >700 | >700 | 374.34 ± 117.07 |
| Compound 17 | 31.84 ± 14.21 | 24.32 ± 3.30 | 25.54 ± 2.30 | 8.81 ± 4.42 |
| Compound 18 | 68.24 ± 5.66 | 46.90 ± 4.07 | 89.83 ± 4.87 | 43.10 ± 1.88 |
| Compound 19 | 504.69 ± 107.23 | 288.80 ± 10.81 | 171.37 ± 22.52 | 91.19 ± 18.69 |
| Compound 20 | >700 | >700 | 482.63 ± 58.51 | 90.97 ± 8.00 |
The interaction of 4′-O-alkyl derivatives with the eukaryotic drug binding pocket is measured by the drug concentrations (in milligrams per liter) required to inhibit in vitro synthesis of functional firefly luciferase to 50% (IC50s). The values are means ± standard deviations (SD) for experiments performed in triplicate. ND, not determined.
To study aminoglycoside-induced misreading, we used a sensitive gain-of-function assay taking advantage of recombinant firefly luciferase constructs where amino acid 245 in the active site of firefly luciferase has been mutated from wild-type CAC (His) to the near-cognate codon CGC (Arg). The CAC-to-CGC mutation results in loss of enzymatic activity, and enzymatic activity can be restored by aminoglycoside-induced misreading of the near-cognate CGC codon (30). On the basis of aminoglycoside-induced misreading and drug-mediated inhibition of translocation, we calculated a ribosomal damage index (RDI) by relating inhibition of eukaryotic protein synthesis (IC50) and induction of ribosomal misreading (1/IC50 × misreading at IC50/misreading in the absence of the drug × 100). Compared to the parental scaffold paromomycin, compounds 22, 10, and 11 show a significantly lower ribosomal damage index for all three hybrid ribosomes—cytosolic hybrid, mitochondrial hybrid, and mitochondrial A1555G mutant hybrid (Table 4). To challenge the results of the hybrid ribosomes, we determined the RDI for native cytosolic eukaryotic ribosomes. Rabbit reticulocyte ribosomes were used, and the RDI values were compared to those of the cytosolic hybrid ribosomes. An excellent correlation (Pearson correlation coefficient, R = 0.998) corroborated our conclusion that the designed aminoglycosides are less toxic toward eukaryotic ribosomes.
TABLE 4 .
Ribosomal damage index
| Drug | Ribosomal damage indexa |
|||
|---|---|---|---|---|
| Mitochondrial wild-type | Mitochondrial mutant A1555G | Cytosolic wild-type | Rabbit ribosomes | |
| Paromomycin | 5.0 | 125.0 | 40.0 | 120.0 |
| Compound 22 | 0.4 | 2.5 | 2.5 | 10.5 |
| Compound 10 | 0.8 | 2.0 | 2.0 | 16.5 |
| Compound 11 | 1.5 | 1.5 | 3.0 | 14.0 |
The ribosomal damage index is calculated as follows: 1/IC50 × misreading at IC50/misreading in the absence of the drug × 100.
Aminoglycoside-mediated dysfunction of the eukaryotic ribosome may manifest as drug cytotoxicity in mammalian cells (21, 29, 41). The cytotoxicity of compounds 22, 10, and 11 was determined. Compared to Geneticin, an aminoglycoside known to be cytotoxic to mammalian cells, there was no detectable cytotoxicity of compound 22, 10, or 11 against mammalian NIH 3T3 cells (see Fig. S2 in the supplemental material).
We performed an extensive in vitro profiling screen of compounds 10 and 11 against 44 primary human molecular targets (42) to detect any relevant off-target pharmacological effects. With the possible exception of Lck kinase, no significant off-target effects were observed, including hERG potassium ion channel and G-protein-coupled receptors (see Table S2 in the supplemental material). In addition, the compounds show low plasma protein binding properties (the mean values for percent protein bound were 13% for compound 10 and 9% for compound 11).
In vivo ototoxicity.
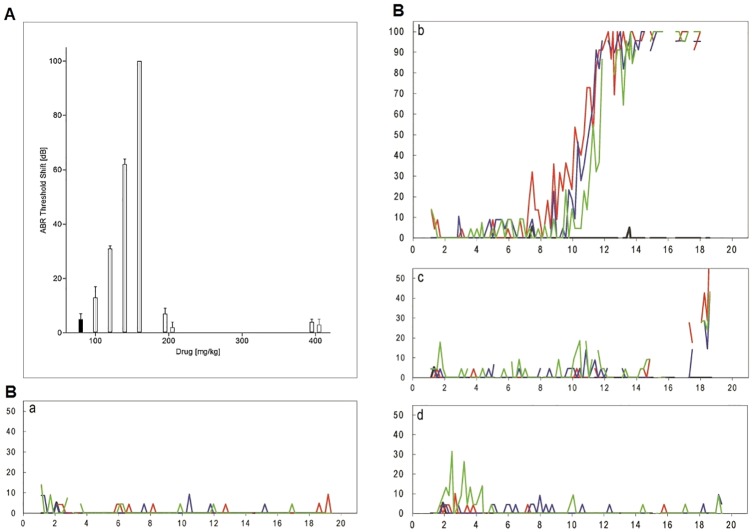
The guinea pig model of chronic ototoxicity is the reference for determination of aminoglycoside-induced hearing loss in vivo (24). We used this model to compare the ototoxicity of compounds 10 and 11 to that of the standard gentamicin. Notably, the acute toxicity that prevented such studies in the previous aralkyl series is absent from the present class of compounds. Guinea pigs were treated for 2 weeks with various doses of aminoglycosides, and ototoxicity was assessed by measuring hearing thresholds at 12 kHz and 32 kHz using auditory brain stem responses (ABR). Gentamicin-induced threshold shifts (i.e., loss of auditory function) exhibit a steep dose-response curve—little effect at 100 mg of gentamicin/kg of body weight but complete deafness at 160 mg gentamicin/kg. Experiments with 180 mg gentamicin/kg were discontinued after the premature death of two animals. In contrast, treatment with compounds 10 and 11 showed no effect on ABR thresholds, even at doses as high as 400 mg/kg (Fig. 3A). A modest but significant threshold shift at 32 kHz was observed for compound 10 at 400 mg/kg (see Table S3 in the supplemental material). In order to correlate the functional hearing data with histopathology, we quantified loss of outer hair cells in cochleae harvested after the ABR measurements. Loss of hair cells seen in cochlear surface preparations was in agreement with the functional results obtained from the ABR measurements (Fig. 3B). In animals treated with 140 mg gentamicin/kg, extensive loss of outer hair cells was seen at the base of the cochlea, consistent with the observed high-frequency hearing loss. In contrast, in animals treated with 400 mg of compound 10 or 11 per kg, little (compound 10) or no (compound 11) loss of outer hair cells was found.
FIG 3 .
Loss of auditory function and hair cells in vivo. (A) Effect of chronic aminoglycoside treatment in vivo on ABR at 12 kHz. The threshold shift is the difference in the auditory threshold before and 3 weeks after treatment, calculated for individual animals. The threshold shift is given in decibels (dB) and corresponds to a logarithmic scale, i.e., a 10-dB difference indicates a difference in energy of 1 log10 unit. The different treatments are indicated as follows: black bars, control; gray bars, gentamicin; white bars outlined in black, compound 10; white bars outlined in gray, compound 11. Data represent means plus standard errors of the means (SEM) (error bars) (n = 3 to 11 per drug concentration). (B) Quantitative evaluation of hair cell loss. Surface preparations of guinea pig cochlea were evaluated quantitatively by determining the presence or absence of hair cells along the entire length of the cochlea. Representative examples are shown. Red, green, and blue lines indicate outer hair cells from the first, second, and third row, respectively. y axis, missing hair cells (%); x axis, distance from apex (mm). (a) Control (no drug treatment). Typical low-level scattered loss of individual cells along the length of the cochlea was observed. (b) Gentamicin given at 140 mg/kg. A steep rise of hair cell loss from the middle cochlea to complete loss at the base was seen. (c) Compound 10 given at 400 mg/kg. There was some hair cell loss but only at the very base of the cochlea. (d) Compound 11 given at 400 mg/kg. There was no significant hair cell loss.
DISCUSSION
Here, we have approached the development of new lead antibacterial agents through the informed modification of an existing compound class and accomplished the synthesis of potent aminoglycoside compounds devoid of ototoxic side effects. Aminoglycoside antibiotics offer several features that make them an attractive target for further development: high efficacy, potent bactericidal activity, lack of drug-related allergy, and absence of life-threatening side effects related to disturbances of the human microbiome, e.g., postantibiotic enterocolitis (7). Thus, we followed the idea to substantially improve aminoglycoside antibiotics with their potent broad-spectrum activity against many clinically relevant resistant pathogens and to render them more broadly applicable by engineering out their biggest deficiency, ototoxicity.
Few systematic attempts have been undertaken to design more-biocompatible aminoglycosides. Our approach is based on the mechanistic hypothesis that the toxic side effects of aminoglycosides are related to limitations in ribosomal target selectivity (29–31, 38). On the basis of this hypothesis, we have recently disclosed a set of new aminoglycosides by optimizing the structure-activity relationship of the paromomycin scaffold toward increased target selectivity (39). Here, we report on further optimizing the C-4′-O-substituent toward improved biocompatibility by ameliorating toxicity. We demonstrate that the critical C-4′-alkoxy moieties identified confer substantial antibacterial activity in vitro and in vivo accompanied by little, if any, hearing loss or morphological cochlear damage. In addition, there was no detectable cytotoxicity against mammalian cells.
Compared to the parental paromomycin, introduction of a 4′-O-alkyl group—with an optimum size of two or three carbons—affords only a slight loss of activity toward wild-type bacterial ribosomes but offers a substantial increase in selectivity. The target selectivity profile of the novel 4′-alkoxy compounds compares favorably to that of the former 4′,6′-O-acetals and 4′-O-aralkyl ether derivatives. Modeling the drug-target interaction of the 4′ alkoxy compounds and the ribosomal A site on the basis of available crystal structures of 4′-O-acetals and 4′-O-ethers in complex with the small ribosomal subunit of Thermus thermophilus (39) suggests that the 4′ substituent is positioned in proximity to the backbone and ribose of A1492 and points out into the bulk solvent between A1492 and G1491 (see Fig. S3 in the supplemental material). At the ribosomal drug target level, the series of aliphatic 4′-O-alkylated 4,5-disubstituted 2-deoxystreptamines largely avoids interaction with the eukaryotic drug binding pockets in cytosolic and mitochondrial ribosomes (Table 3). Most likely, this reflects the susceptibility of the current compound series to base substitutions at polymorphic 16S rRNA residues 1408 and 1491 in the drug binding pocket, as revealed by the interaction with recombinant bacterial ribosomes carrying the corresponding point mutations (Table 1). Thus, compared to the parental paromomycin, the 4′-O-alkyl derivatives take additional advantage of the polymorphic rRNA residues to increase target selectivity. Alkylation of the 4′-OH of the parental paromomycin tends to decrease antibacterial efficacy. However, compared to the initially synthesized compounds, the MIC values of the subsequently synthesized compounds 10 and 11 demonstrate rather potent antibacterial activity (Table 2), indicating that there is opportunity to further enhance antibacterial activity of 4′-O-substituted compounds.
The determination of aminoglycoside ototoxicity requires the assessment of auditory function in vivo. Although the guinea pig model of chronic ototoxicity requires the synthesis of large amounts of compound, it is unequivocally considered the gold standard, which best reflects the pathophysiology involved in human aminoglycoside-induced ototoxicity (24). The minimal ototoxicity of the designed compounds, predicted on the basis of their in vitro ribosomal damage index, is clearly demonstrated in vivo. There was very little hearing loss induced by compound 10 or 11, even at excessive drug concentrations. This is in stark contrast to the well-characterized and commonly used aminoglycoside gentamicin, which caused significant auditory threshold shifts. Consistent with the functional results from ABR measurements, the in vivo treatment with the newly designed aminoglycosides resulted in little or no hair cell loss. In addition to ototoxicity, treatment with gentamicin also resulted in signs of general toxicity, most likely reflecting nephrotoxicity, i.e., significant weight loss at doses below 160 mg/kg, and death upon doses exceeding 160 mg/kg. In contrast, compounds 10 and 11 were well tolerated. All animals treated with compound 10 or 11 remained healthy during the complete course of the experiment. No drug-related death or weight loss was observed even at doses of 400 mg/kg body weight. Analysis of blood urea nitrogen and creatinine likewise did not reveal any nephrotoxic side effects of the compounds. Notably, the concentrations of drugs used in the animal model exceed those used in clinical medicine by an order of magnitude so as to obtain relatively high and reliable drug levels in serum, resulting in consistent ABR threshold shifts.
The rationale for choosing the 4′ position for modification was suggested by the target selectivity profile observed with the previously synthesized 4′,6′-O-acetals and 4′-O-aralkyl-ethers (39). Our data extend these findings and demonstrate a key role for 4′-substituents in determining drug toxicity. We have found that 4′-O-alkylation is a valuable structural element that significantly affects target selectivity and ototoxicity. In addition, extensive in vitro testing against mammalian enzymes and receptors showed that the 4′-alkoxy leads have little potential for off-target effects that result in adverse reactions. Our combined chemical synthesis, biochemical, and toxicology studies have identified 4′-alkoxy groups of paromomycin as a promising new pharmacophore that affects aminoglycoside ototoxicity and significantly improves biocompatibility. These efforts validate the potential to modify the neamine core of the aminoglycosides to obtain potent new sets of this compound class that are less compromised by adverse side effects. Together with strategies to increase the antibacterial efficacy of these compounds and their recalcitrance against resistance determinants (32, 33, 43, 44; for a review, see reference 45), this should result in a revival of this important class of antibacterials in combating the global emergence of antibiotic resistance.
MATERIALS AND METHODS
Antibiotic compounds.
Paromomycin was obtained from Sigma. Chemical synthesis procedures, including Schemes 1 to 4, are detailed in Text S1 in the supplemental material.
Bacterial strains.
Clinical isolates of Escherichia coli and Staphylococcus aureus were obtained from the Diagnostic Department, Institute of Medical Microbiology, University of Zurich. MIC values were determined by broth microdilution assays. Microtiter plates were incubated overnight for E. coli and S. aureus and 72 h for Mycobacterium smegmatis.
Recombinant microorganisms.
The construction of these strains derived from an M. smegmatis ΔrrnB strain with a single rRNA operon has been described previously (21, 28).
Purification of ribosomes and cell-free translation assays.
Ribosomes were purified from bacterial cell pellets, and purified 70S hybrid ribosomes were used in translation reaction mixtures as described previously (30). A typical translation reaction mixture with a total volume of 30 µl contained 0.25 µM 70S ribosomes, 4 µg firefly luciferase (F-luc) mRNA, 40% (vol/vol) M. smegmatis S100 extract, 200 µM amino acid mixture, 24 units of RiboLock (Thermo Scientific), and 0.4 mg ml−1 tRNAs, and energy was supplied by addition of 12 µl of commercial S30 Premix without amino acids (Promega). In addition to ribosomes, rabbit reticulocyte lysate (Promega) was used for in vitro translation. Following addition of serially diluted aminoglycosides, the reaction mixture was incubated at 37°C for 35 min and stopped on ice. Thirty-microliter samples of the reaction mixture were assayed for luciferase activities using the Dual-Luciferase reporter assay system (Promega). Luminescence was measured using a luminometer FLx800 (Bio-Tek Instruments).
Misreading was assessed in a gain-of-function assay as described previously (30). In brief, we introduced Arg245 (CGC, near-cognate codon) into the firefly luciferase protein to replace residue His245 (CAC codon). Arg245 F-luc mRNA and wild-type (wt) F-luc mRNA were used in in vitro translation reaction mixtures, and renilla luciferase (R-luc) mRNA was used as internal control. We quantified misreading by calculating mutant firefly/renilla luciferase activity compared with wild-type firefly/renilla luciferase activity.
Cytotoxicity assays.
For cytotoxicity assays, mouse embryonic fibroblast (NIH 3T3) cells were grown in 96-well plates (4,000 cells/well) in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% glutamine at 37°C and 5% CO2. Following overnight incubation, serial dilutions of compound were added, and the cells were incubated for an additional 72 h. Cell viability was measured using alamarBlue fluorimetric assay (Life Technologies) according to the manufacturer’s instructions. Fluorescence was measured using an FLx800 plate reader (Bio-Tek Instruments). Cell viability was calculated as the ratio between the numbers of living cells in cultures grown in the presence of the tested compounds and those in cultures grown under identical conditions without the tested compound. The LC25 values, i.e., the drug concentration at which 25% of the cells in culture are nonviable, were calculated from fitting concentration-response curves to the data of at least three independent experiments using PRISM 5 software.
In vivo infection experiments.
Animal experiments were performed under United Kingdom Home Office Licenses with clearance by the ethical review committee at the University of Manchester. Male mice aged 7 to 8 weeks old were used in this study. Mice were supplied by Charles River United Kingdom and were specific pathogen free. The strain of mouse used was Hsd:ICR (CD-1), a well-characterized outbred strain. The weight of the mice at the start of the experiment was 22 to 25 g. Mice were housed in sterile individual ventilated cages with free access to sterile food and water and were exposed to 12-h light/dark cycles with dawn/dusk phases. There were five mice in each group.
Mice were rendered temporarily neutropenic by immunosuppression with cyclophosphamide at 200 mg/kg 4 days before infection and 150 mg/kg 1 day before infection by intraperitoneal injection. The immunosuppression regimen leads to neutropenia starting 24 h after the administration of cyclophosphamide, and neutropenia continues throughout the study. For in vivo infection, a methicillin-resistant strain of Staphylococcus aureus, clinical isolate MRSA AG041, was used. Twenty-four hours after the second round of immunosuppression, mice were infected with S. aureus MRSA AG041 by intravenous injection into the lateral tail vein using ~1 × 107 CFU/mouse. This strain had the following MIC values—4.0 mg/liter for paromomycin, 8 mg/liter for compound 10, 16 mg/liter for compound 11, and 1.0 mg/liter for linezolid. Test compounds and comparators were reconstituted and diluted in 0.9% saline. Dosing solutions were prepared immediately prior to administration of the first dose and stored at 4°C between treatments. Antibacterial treatment was initiated 1 h postinfection; linezolid at a dose of 20 mg/kg was used as a positive control and was always given by intravenous bolus injection. All drugs were administered 1, 9, and 17 h postinfection.
At 1 h (pretreatment group) or 24 h postinfection, blood samples were collected by cardiac puncture under isoflurane anesthesia, and mice were humanely euthanized using pentobarbitone overdose. Both kidneys were removed and homogenized in 2 ml ice-cold sterile phosphate-buffered saline (PBS). Kidney homogenates were quantitatively cultured onto mannitol salt agar (MSA) and incubated at 37°C for 24 h before being counted. Individual blood samples were quantitatively cultured onto cysteine lactose electrolyte-deficient (CLED) agar and incubated at 37°C for 24 h before being counted. Data were analyzed, using StatsDirect software (version 2.7.8), using the nonparametric Kruskal-Wallis test (pairwise comparisons, Conover-Inman).
In vivo ototoxicity.
Male Hartley guinea pigs of initially about 200 g body weight (Charles River Breeding Laboratories) had free access to water and food and were acclimated for 1 week prior to experiments. Drugs were administered once daily subcutaneously for 14 days at dosages indicated in the figure legends; saline injections of the same volume served as controls. Auditory function was measured as auditory brain stem response (ABR) at 12 and 32 kHz under anesthesia with an intraperitoneal injection of xylazine (7 mg/kg body weight), ketamine (65 mg/kg), and acepromazine (2 mg/kg). Each animal had its threshold determined before and 3 weeks after the end of drug treatment.
Following the final ABR, cochleae were harvested and dissected into segments from the apical, middle, and basal turns. These segments were permeabilized in 3% Triton X-100 for 30 min at room temperature (RT), washed three times with PBS, and incubated with rhodamine phalloidin (1:100) at RT for 1 h. Hair cells were counted on a Leitz Orthoplan microscope whose right objective had a 0.19-mm scale imposed on the field. Beginning at the apex, consecutive 0.19-mm fields were evaluated by observers in a blind manner (unaware of the experimental conditions). Hair cell counts were compared to a normative database (KHRI Cytocochleogram, version 3.0.6; Kresge Hearing Research Institute).
SUPPLEMENTAL MATERIAL
Supplemental methods Download
Small-subunit rRNA secondary structures Download
Compound cytotoxicity Download
Overview of compound 10 bound to the bacterial A site Download
MICs (mg/liter) of Mycobacterium smegmatis
Toxicity and secondary pharmacology profiling of compounds 10 and 11
ABR threshold shifts at 32 kHz
ACKNOWLEDGMENTS
We thank Tanja Matt and Tanja Janusic for help with the ribosomal assays and MIC determinations, Kim Wearne for help with the ototoxicity assessment, and Susanna Salas for typing the manuscript.
This study was supported by the University of Zurich (E. C. Böttger), and grants R01 DC003685 and P30 DC005188 from the National Institute on Deafness and Other Communication Disorders, National Institutes of Health (J. Schacht).
E. C. Böttger, A. Vasella, J. Schacht, and D. Crich designed the study; S. Salian, S. Silva, T. Kato, and D. Perez-Fernandez performed chemical syntheses and recorded nuclear magnetic resonance (NMR) and mass spectrometry (MS) spectra; B. Bernet, T. Kato, and D. Crich analyzed NMR spectra and stereochemical aspects; S. Duscha, H. Boukari, D. Shcherbakov, and R. Akbergenov prepared ribosomes and conducted the in vitro ribosomal assays and MIC determinations; A. Kendall, S. Vaddi, and P. Thommes performed the in vivo experiments. All authors analyzed and discussed the data. E. C. Böttger, A. Vasella, J. Schacht, and D. Crich checked the data and wrote the paper with input and contribution from all authors.
E. C. Böttger and A. Vasella are named inventors on patents filed by the University of Zurich and pertaining to this work. We declare that we have no conflicts of interest.
Footnotes
Citation Duscha S, Boukari H, Shcherbakov D, Salian S, Silva S, Kendall A, Kato T, Akbergenov R, Perez-Fernandez D, Bernet B, Vaddi S, Thommes P, Schacht J, Crich D, Vasella A, Böttger EC. 2014. Identification and evaluation of improved 4′-O-(alkyl) 4,5-disubstituted 2-deoxystreptamines as next-generation aminoglycoside antibiotics. mBio 5(5):e01827-14. doi:10.1128/mBio.01827-14.
REFERENCES
- 1. French GL. 2010. The continuing crisis in antibiotic resistance. Int. J. Antimicrob. Agents 36(Suppl 3):S3–S7. 10.1016/S0924-8579(10)70003-0 [DOI] [PubMed] [Google Scholar]
- 2. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40. 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- 3. Wright GD, Sutherland AD. 2007. New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 13:260–267. 10.1016/j.molmed.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 4. Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093. 10.1126/science.1176667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newman DJ, Cragg GM. 2007. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70:461–477. 10.1021/np068054v [DOI] [PubMed] [Google Scholar]
- 6. World Health Organization 2007. Critically important antimicrobials for human medicine. Categorization for the development of risk management strategies to contain antimicrobial resistance due to non-human antimicrobial use. Report of the Second WHO Expert Meeting, Copenhagen, 29 to 31 May 2007. World Health Organization, Geneva, Switzerland: http://www.who.int/foodborne_disease/resistance/antimicrobials_human.pdf [Google Scholar]
- 7. Chambers HF. 1996. Chemotherapy of microbial diseases, p 1103–1121 In Hardman JG, Limbird LE. (ed), Goodmann & Gilman’s the pharmaceutical basis of therapeutics, 10th ed. The McGraw-Hill Companies, New York, NY. [Google Scholar]
- 8. Hanberger H, Edlund C, Furebring M, Giske CG, Melhus A, Nilsson LE, Petersson J, Sjölin J, Ternhag A, Werner M, Eliasson E, Swedish, Reference Group for Antibiotics (SRGA) 2013. Rational use of aminoglycosides—review and recommendations by the Swedish Reference Group for Antibiotics (SRGA). Scand. J. Infect. Dis. 45:161–175. 10.3109/00365548.2012.747694 [DOI] [PubMed] [Google Scholar]
- 9. Davies J, Gorini L, Davis BD. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93–106 [PubMed] [Google Scholar]
- 10. Cabañas MJ, Vázquez D, Modolell J. 1978. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 83:991–997. 10.1016/0006-291X(78)91493-6 [DOI] [PubMed] [Google Scholar]
- 11. Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. 2004. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 343:1183–1194. 10.1016/j.jmb.2004.08.097 [DOI] [PubMed] [Google Scholar]
- 12. Feldman MB, Terry DS, Altman RB, Blanchard SC. 2010. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat. Chem. Biol. 6:244. 10.1038/nchembio0310-244a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. 10.1038/35030019 [DOI] [PubMed] [Google Scholar]
- 14. François B, Russell RJ, Murray JB, Aboul-ela F, Masquida B, Vicens Q, Westhof E. 2005. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 33:5677–5690. 10.1093/nar/gki862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfister P, Hobbie S, Vicens Q, Böttger EC, Westhof E. 2003. The molecular basis for A-site mutations conferring aminoglycoside resistance: relationship between ribosomal susceptibility and X-ray crystal structures. Chembiochem 4:1078–1088. 10.1002/cbic.200300657 [DOI] [PubMed] [Google Scholar]
- 16. Pfister P, Hobbie S, Brull C, Corti N, Vasella A, Westhof E, Böttger EC. 2005. Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6'OH and 6'NH3+ aminoglycosides. J. Mol. Biol. 346:467–475. 10.1016/j.jmb.2004.11.073 [DOI] [PubMed] [Google Scholar]
- 17. Hobbie SN, Pfister P, Brüll C, Westhof E, Böttger EC. 2005. Analysis of the contribution of individual substituents in 4,6-aminoglycoside-ribosome interaction. Antimicrob. Agents Chemother. 49:5112–5118. 10.1128/AAC.49.12.5112-5118.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hobbie SN, Pfister P, Bruell C, Sander P, François B, Westhof E, Böttger EC. 2006. Binding of neomycin-class aminoglycoside antibiotics to mutant ribosomes with alterations in the A site of 16S rRNA. Antimicrob. Agents Chemother. 50:1489–1496. 10.1128/AAC.50.4.1489-1496.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Recht MI, Douthwaite S, Puglisi JD. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133–3138. 10.1093/emboj/18.11.3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. 2001. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2:318–323. 10.1093/embo-reports/kve062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hobbie SN, Kalapala SK, Akshay S, Bruell C, Schmidt S, Dabow S, Vasella A, Sander P, Böttger EC. 2007. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 35:6086–6093. 10.1093/nar/gkm658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan-Minogue H, Bedwell DM. 2008. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA 14:148–157. 10.1261/rna.805208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matt T, Akbergenov R, Shcherbakov D, Böttger EC. 2010. The ribosomal A-site: decoding, drug target, and disease. Isr. J. Chem. 50:60–70. 10.1002/ijch.201000003 [DOI] [Google Scholar]
- 24. Forge A, Schacht J. 2000. Aminoglycoside antibiotics. Audiol. Neurootol. 5:3–22. 10.1159/000013861 [DOI] [PubMed] [Google Scholar]
- 25. Duggal P, Sarkar M. 2007. Audiologic monitoring of multi-drug resistant tuberculosis patients on aminoglycoside treatment with long term follow-up. BMC Ear Nose Throat Disord. 7:5. 10.1186/1472-6815-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N. 1993. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4:289–294. 10.1038/ng0793-289 [DOI] [PubMed] [Google Scholar]
- 27. Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. 2004. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 74:139–152. 10.1086/381133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobbie SN, Bruell CM, Akshay S, Kalapala SK, Shcherbakov D, Böttger EC. 2008. Mitochondrial deafness alleles confer misreading of the genetic code. Proc. Natl. Acad. Sci. U. S. A. 105:3244–3249. 10.1073/pnas.0707265105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hobbie SN, Akshay S, Kalapala SK, Bruell CM, Shcherbakov D, Böttger EC. 2008. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc. Natl. Acad. Sci. U. S. A. 105:20888–20893. 10.1073/pnas.0811258106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matt T, Ng CL, Lang K, Sha SH, Akbergenov R, Shcherbakov D, Meyer M, Duscha S, Xie J, Dubbaka SR, Perez-Fernandez D, Vasella A, Ramakrishnan V, Schacht J, Böttger EC. 2012. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc. Natl. Acad. Sci. U. S. A. 109:10984–10989. 10.1073/pnas.1204073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shulman E, Belakhov V, Wei G, Kendall A, Meyron-Holtz EG, Ben-Shachar D, Schacht J, Baasov T. 2014. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: a strategy for the treatment of genetic diseases. J. Biol. Chem. 289:2318–2330. 10.1074/jbc.M113.533588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob. Agents Chemother. 54:4636–4642. 10.1128/AAC.00572-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao F, Zhao Q, Blount KF, Han Q, Tor Y, Hermann T. 2005. Molecular recognition of RNA by neomycin and a restricted neomycin derivative. Angew. Chem. Int. Ed. Engl. 44:5329–5334. 10.1002/anie.200500903 [DOI] [PubMed] [Google Scholar]
- 34. Hanessian S, Giguere A, Grzyb J, Maianti JP, Saavedra OM, Aggen JB, Linsell MS, Goldblum AA, Hildebrandt DJ, Kane TR, Dozzo P, Gliedt MJ, Matias RD, Feeney LA, Armstrong ES. 2011. Toward overcoming Staphylococcus aureus aminoglycoside resistance mechanisms with a functionally designed neomycin analogue. ACS Med. Chem. Lett. 2:924–928. 10.1021/ml200202y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450. 10.1128/CMR.16.3.430-450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477–498. 10.1021/cr0301088 [DOI] [PubMed] [Google Scholar]
- 37. Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. 2010. The future of aminoglycosides: the end or renaissance? Chembiochem 11:880–902. 10.1002/cbic.200900779 [DOI] [PubMed] [Google Scholar]
- 38. Francis SP, Katz J, Fanning KD, Harris KA, Nicholas BD, Lacy M, Pagana J, Agris PF, Shin JB. 2013. A novel role of cytosolic protein synthesis inhibition in aminoglycoside ototoxicity. J. Neurosci. 33:3079–3093. 10.1523/JNEUROSCI.3430-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez-Fernandez D, Shcherbakov D, Matt T, Leong NC, Kudyba I, Duscha S, Boukari H, Patak R, Dubbaka SR, Lang K, Meyer M, Akbergenov R, Freihofer P, Vaddi S, Thommes P, Ramakrishnan V, Vasella A, Böttger EC. 2014. 4'-O-substitutions determine selectivity of aminoglycoside antibiotics. Nat. Commun. 5:3112. 10.1038/ncomms4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pathak R, Perez-Fernandez D, Nandurdikar R, Kalapala SK, Böttger EC, Vasella A. 2008. Synthesis and evaluation of paromomycin derivatives modified at C(4'). Helv. Chim. Acta 91:1533–1552. 10.1002/hlca.200890167 [DOI] [Google Scholar]
- 41. Kandasamy J, Atia-Glikin D, Shulman E, Shapira K, Shavit M, Belakhov V, Baasov T. 2012. Increased selectivity toward cytoplasmic versus mitochondrial ribosome confers improved efficiency of synthetic aminoglycosides in fixing damaged genes: a strategy for treatment of genetic diseases caused by nonsense mutations. J. Med. Chem. 55:10630–10643. 10.1021/jm3012992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S. 2012. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 11:909–922. 10.1038/nrd3845 [DOI] [PubMed] [Google Scholar]
- 43. Sucheck SJ, Wong AL, Koeller KM, Boehr DD, Draker K, Sears P, Wright GD, Wong CH. 2000. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J. Am. Chem. Soc. 122:5230–5231. 10.1021/ja000575w [DOI] [Google Scholar]
- 44. Bastida A, Hidalgo A, Chiara JL, Torrado M, Corzana F, Pérez-Cañadillas JM, Groves P, Garcia-Junceda E, Gonzalez C, Jimenez-Barbero J, Asensio JL. 2006. Exploring the use of conformationally locked aminoglycosides as a new strategy to overcome bacterial resistance. J. Am. Chem. Soc. 128:100–116. 10.1021/ja0543144 [DOI] [PubMed] [Google Scholar]
- 45. Wright GD, Berghuis AM, Mobashery S. 1998. Aminoglycoside antibiotics—structure, function and resistance, p 27–69. In Rosen B, Mobashery S. (ed), Resolving the antibiotic paradox. Springer Verlag, New York, NY. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods Download
Small-subunit rRNA secondary structures Download
Compound cytotoxicity Download
Overview of compound 10 bound to the bacterial A site Download
MICs (mg/liter) of Mycobacterium smegmatis
Toxicity and secondary pharmacology profiling of compounds 10 and 11
ABR threshold shifts at 32 kHz