Abstract
Cys2His2 zinc finger proteins make up the largest class of transcription factors encoded in the genomes of higher eukaryotes. Recent studies of the Ikaros transcription factor demonstrated that this zinc finger protein undergoes cell cycle-dependent changes in association with DNA that seem to be due to phosphorylation of Thr or Ser residues in the linker regions connecting adjacent zinc finger domains. The high degree of conservation of this linker sequence within the Cys2His2 superfamily suggested a common mechanism for the cell cycle-dependent modulation of DNA-binding affinity throughout this large class of transcription factors. The effects of linker phosphorylation on DNA-binding affinity were investigated through a direct comparison of the DNA-binding properties of four synthetic zinc finger proteins produced by native chemical ligation. The four proteins, comprising three zinc finger domains joined by two consensus Thr-Gly-Glu-Lys-Pro linkers, correspond to all four possible combinations of linker Thr phosphorylation states. Fluorescence-based DNA-binding studies of a specific DNA-binding site revealed that phosphorylation of a single linker reduced binding affinity ≈40-fold, whereas phosphorylation of both linkers reduced binding affinity 130-fold. These results with purified components demonstrate that linker phosphorylation does, indeed, produce a significant reduction in DNA-binding affinity and support a model wherein a single cell cycle-dependent Ser/Thr kinase could simultaneously inactivate a large number of zinc finger transcription factors.
The Cys2His2 zinc finger proteins represent one of the largest classes of proteins found in the genomes of all eukaryotes that have been sequenced to date (1, 2). These proteins typically have multidomain architectures in which individual zinc-binding domains approximating the consensus sequence (Tyr,Phe)-X-Cys-X2,4-Cys-X3-Phe-X5-Leu-X2-His-X3-5-His are joined into arrays of two or more by relatively short linker sequences. As this protein class initially came to be characterized, it was observed that the linker sequence connecting adjacent zinc finger domains tends to be remarkably highly conserved. This sequence typically deviates only slightly from the consensus His-Thr-Gly-Glu-Lys-Pro-(Tyr,Phe)-X-Cys. Indeed, the conservation of this sequence allowed cDNAs encoding many zinc finger proteins to be identified before genomic sequencing efforts (3). Although structural studies (4-7) and mutagenesis experiments (8-12) have suggested a role for the linker in DNA binding, it has been noted that this contribution is relatively minor and probably insufficient to explain the high degree of evolutionary conservation of the linker sequence (10).
Recently, an attractive hypothesis explaining linker sequence conservation was proposed based on the observation that the Cys2His2 zinc finger protein Ikaros was excluded from mitotic chromosomes (13). Ikaros protein isolated from mitotic cell extracts was found to be phosphorylated and seemed to have reduced DNA-binding affinity. The sites of phosphorylation were localized to residues within the linkers connecting the protein's four zinc finger domains. The linkers in murine Ikaros have the sequences Thr-Gly-Glu-Arg-Pro, Ser-Gly-Glu-Lys-Pro, and Ser-Val-Gly-Lys-Pro, approximating the Thr-Gly-Glu-Lys-Pro consensus sequence (phosphorylation residues are in italics). Replacement of the Thr and Ser residues in these sequences with Ala resulted in a loss-of-protein phosphorylation, suggesting that these residues are the sites of modification. Based on these observations, it was proposed that the linker sequence acts as the substrate for an unidentified cellular kinase capable of phosphorylating Cys2His2 zinc finger proteins in a cell cycle-dependent manner. It has long been known that transcription ceases upon entry into mitosis and that this phenomenon is due, in part, to the mitotic inactivation of gene-specific transcription factors (14). The proposal that a cell cycle-dependent kinase can phosphorylate the Thr-Gly-Glu-Lys-Pro linker sequence provides a plausible explanation for the linker's high degree of evolutionary conservation while at the same time offering a mechanism for the mitotic inactivation of a large number of sequence-specific transcription factors.
Because the previous report was limited to semipurified components from whole cell extracts, it was impossible for the authors to evaluate the effects of linker phosphorylation on DNA-binding affinity in a quantitative manner. To test the hypothesis that phosphorylation of linker sequences is responsible for a marked decrease in DNA-binding affinity and to provide quantitative information regarding the impact of the addition of each phosphoryl group, we compared the DNA-binding properties of a prototypical three-zinc finger protein in its four possible states of linker phosphorylation. The production of these proteins was made possible through the use of native chemical ligation (15) to join synthetic peptide fragments incorporating phosphothreonine in the linker sequence. DNA-binding studies revealed that phosphorylation of the Thr residue in a single linker produced a reduction in DNA-binding affinity of ≈40-fold (at pH 7.5, 125 mM NaCl), whereas phosphorylation of both linkers produced a 130-fold reduction. These results support the hypothesis that a cellular kinase capable of phosphorylating Thr (or Ser) residues in the conserved linker in Cys2His2 zinc finger proteins would be able to inactivate a large number of sequence-specific transcription factors in a coordinated fashion.
Materials and Methods
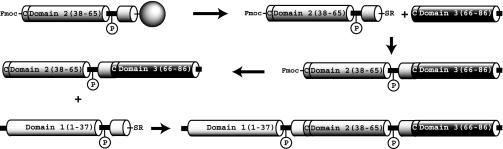
Construction of Zinc Finger Proteins. The sequence of the zinc finger protein QNK-QNK-RHR is given in Fig. 1. Proteins corresponding to all four phosphorylation states of QNK-QNK-RHR were synthesized by means of the strategy outlined in Fig. 2. All peptides were made by fluorenylmethoxycarbonyl (Fmoc)-based solid phase peptide synthesis on a Milligen 9050 peptide synthesizer. A peptide comprising residues 38-65 (corresponding to the middle domain) was first synthesized on a sulfamylbutyryl AM resin (Novabiochem) to yield a C-terminal thioester as described (16). To prevent an intramolecular ligation reaction, the peptide was liberated from the resin without prior removal of the Fmoc group from the N-terminal cysteine residue. After side chain deprotection in Reagent R (95% trifluoroacetic acid/5% thioanisole/3% ethanedithiol/2% anisole), the Fmoc-protected peptide thioester was precipitated with ether and purified by reversed-phase HPLC. This peptide was prepared twice, once incorporating phosphothreonine into the Thr-Gly-Glu-Lys-Pro linker sequence by using an N-Fmoc-protected derivative [Fmoc-Thr(PO(OBzl)OH)-OH, Bachem]. A peptide comprising residues 66-86 (corresponding to the C-terminal domain) was synthesized on a PEG-PS resin (Applied Biosystems). After removal of the N-terminal Fmoc group, the peptide was cleaved from the resin and deprotected in Reagent R. The peptide was then ether precipitated and HPLC purified as before. About 20 μmol of the purified C-terminal domain peptide were added to ≈10 μmol of the middle domain thioester (either unmodified or phosphorylated) in 5 ml of ligation buffer (2 M guanidine-HCl/100 mM sodium phosphate, pH 7.5/2 mM 2-mercaptoethanesulfonic acid), and the reactions were stirred overnight at room temperature. The pH of the solutions was then raised to 13 with the addition of NaOH for 5 min to remove the N-terminal Fmoc group. After neutralization with HCl, the solutions were clarified by centrifugation, and the ligated peptides corresponding to residues 38-86 were isolated by HPLC. Peptide thioesters were then prepared from these resins and purified as before. Two micromoles of each thioester were then added to 2 μmol of each of the previously prepared two-domain peptides, and the segments were allowed to ligate overnight in 1 ml of ligation buffer. The full-length three-domain proteins were purified by HPLC and dried under anaerobic conditions. After resuspension in water, protein concentrations were determined spectrophotometrically (based on absorption at 276 nm), and the four proteins were diluted to a concentration of 10 μmol in a 20 mM Tris, pH 7.5/125 mM NaCl/100 μM ZnCl2 solution.
Fig. 1.
The amino acid sequence of the three-zinc finger protein QNK-QNK-RHR (referred to as QNK) showing the numbering scheme. Metal-binding residues are shown in bold, direct DNA contact residues are underlined, and the Thr residues that are the sites of phosphorylation and shaded. The positions of the linkages formed by peptide ligation are indicated by dashed lines.
Fig. 2.
The synthetic scheme for the preparation of QNK-QNK-RHR in its fully phosphorylated form (referred to as QNKp123). The zinc finger domains are shown as cylinders, and the linkers are shown as thick lines.
Cobalt Binding Titrations. About 60 nmol of each purified protein was titrated with CoCl2 in 200 mM Hepes/50 mM NaCl, pH 7.0, buffer. Cobalt binding was evaluated by monitoring absorbance at 650 nM on a Perkin-Elmer Lambda 9 spectrophotometer. Titration data were fit by using kaliedagraph (Synergy Software, Reading, PA) to yield dissociation constants for the protein-metal complexes. Under these experimental conditions, it was not possible to accurately determine dissociation constants <1 × 10-7 M. Consequently, reported Kd values represent upper limits.
DNA-Binding Assay. All oligonucleotides were ordered from Operon Technologies (Alameda, CA). A fluorescein-labeled DNA probe was prepared by annealing oligonucleotides with the sequences 5′-CGATGCTTGAGGCAGAACCTGATCA-3′ and 5′-TGATCAGGXTCTGCCTCAAGCATCG-3′, where X is fluorescein-dT (protein-binding site is in italics). Three milliliters of a 5 nM solution of this probe in 20 mM Tris, pH 7.5/125 mM NaCl/10 μg/ml BSA were added to a 4.5-ml acrylic fluorescence cuvette (Fisher) fitted with a micro stir bar. The fluorescence anisotropy was then determined at 25°C on a SPEX Fluorolog-3 spectrofluorometer in the T format with the excitation monochromator set at 490 nm and the emission monochromator at 515 nm. Subsequent anisotropy measurements were taken after the stepwise addition of purified zinc finger protein, allowing 5 min at room temperature before each measurement. All titrations were carried out in triplicate. No significant changes in fluorescence intensity were observed over the course of the titrations. Plots of the change in anisotropy as a function of added protein concentration were fit by using kaleidagraph to yield dissociation constants for the protein-DNA complexes. To rule out the possibility that binding was being influenced by the fluorophore on the labeled DNA probe, competition experiments were conducted to determine dissociation constants for the four proteins in complex with an unlabeled oligonucleotide binding site (data not shown). In all cases, Kd values determined for the labeled and unlabeled sites were within experimental error.
Alkaline Phosphatase Treatment. A 100-μmol solution of each protein was prepared in 20 mM Tris, pH 8.0, with 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 500 μM ZnCl2. Two microliters of calf intestinal phosphatase (20 units, New England Biolabs) were added to 50 μl of each protein solution, and the reactions were heated at 37°C overnight. These solutions were then diluted 10-fold into 20 mM Tris, pH 7.5, with 125 mM NaCl and used fluorescence anisotropy-based DNA-binding assays.
Results
Synthesis of Phosphoproteins. The four proteins under investigation are variants of a designed zinc finger protein termed QNK-QNK-RHR (Fig. 1). This protein consists of three Cys2His2 zinc fingers joined by a pair of Thr-Gly-Glu-Lys-Pro linkers. The individual zinc finger domains are based on the consensus zinc finger peptide CP-1, representing an average zinc finger sequence (17). Peptides and proteins based on this consensus sequence have been extensively characterized and found to closely mimic their natural counterparts in terms of both structure (18) and DNA-recognition properties (19). It is believed, therefore, that the results obtained with our consensus sequence-based prototypical proteins almost certainly will apply to most natural zinc finger proteins.
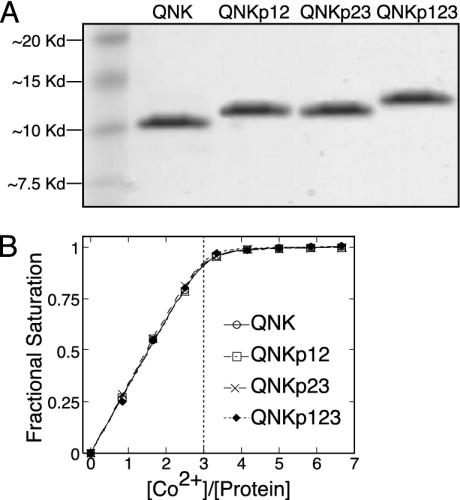
The synthetic proteins were constructed by native chemical ligation by using the strategy outlined in Fig. 2. This approach is similar to that used by Beligere and Dawson (20) for the synthesis of the zinc finger region from the natural zinc finger protein Zif268. The present protocol, however, has been modified to be suitable for automated Fmoc-based peptide synthesis. This synthetic strategy allowed the construction of three-zinc finger domain proteins incorporating phosphothreonine in the first position of the Thr-Gly-Glu-Lys-Pro linker. Thus, with the synthesis of four variant proteins, it was possible to characterize all possible phosphorylation states of the three-finger protein. The protein referred to as QNK is unphosphorylated QNK-QNK-RHR. QNKp12 incorporates phosphothreonine into the linker between fingers 1 and 2 (the N-terminal two fingers), QNKp23 is phosphorylated on the linker between fingers 2 and 3 (the C-terminal two fingers), and QNKp123 is phosphorylated on both linkers. The purity and molecular masses of the four synthetic proteins were verified by SDS/PAGE and matrix-assisted laser desorption ionization mass spectrometry (Fig. 3A). Cobalt binding studies (Fig. 3B) revealed that each protein bound three equivalents of metal, consistent with the successful synthesis of three-zinc finger domain proteins. Cobalt titration data were fit to yield an upper-limit of 1 × 10-7 M for the dissociation constant of all four protein-cobalt complexes. This value is consistent with previous investigations of CP-1-based peptides (17). As zinc finger proteins can be expected to bind Zn(II) with three to four orders of magnitude greater affinity than they bind Co(II) (21), it can be assumed that all four synthetic proteins would be saturated with zinc under physiological conditions.
Fig. 3.
Characterization of the synthetic zinc finger proteins. (A) SDS/PAGE gel showing the four purified proteins. Molecular masses (m + 1) of 9,780.4 daltons (9,778.5 calculated) (QNK), 9,860.6 daltons (9,858.5 calculated) (QNKp12), 9,860.9 daltons (QNKp23) (9,858.5 calculated), and 9,941.3 daltons (9,938.5 calculated) (QNKp123) were determined by matrix-assisted laser desorption ionization mass spectrometry. (B) Cobalt titrations to determine metal binding affinity. All four proteins bound three equivalents of cobalt each, and titration data were fit to yield dissociation constants lower than 1 × 10-7 M for all metal binding sites.
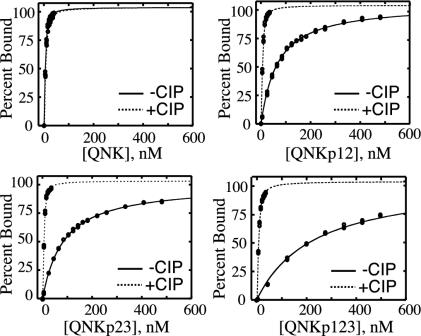
DNA-Binding Activities of the Phosphoproteins. A fluorescence anisotropy-based DNA binding assay was used to evaluate the affinities of the four proteins for DNA (Fig. 4). The QNK-QNK-RHR protein, named for the DNA contacting amino acids in positions -1, 3, and 6 of its three zinc finger helices, binds with high affinity to the DNA site 5′-GA/GGGAAGAA-3′ (22). The protein is known to be highly tolerant of substitutions in the fifth and eighth positions of its binding site so that a fluorescein-labeled, double-stranded DNA probe containing the sequence 5′-GAGGCAGAA-3′ was suitable for the characterization of DNA binding. The results of these binding studies are shown in Fig. 3. The unphosphorylated QNK protein was found to bind this sequence with a Kd value of 1.7 ± 0.2 nM. In contrast, QNKp12 bound the DNA probe with a Kd value of 62 ± 6 nM, a 36-± 9-fold reduction in affinity. QNKp23 bound the DNA site with a Kd value of 78 ± 9 nM, a 46-± 11-fold loss in affinity over the unphosphorylated protein. The doubly phosphorylated QNKp123 bound to the DNA with still lower affinity, binding to the fluorescent probe with a Kd value of 221 ± 8 nM, a 130-± 23-fold reduction in affinity relative to QNK.
Fig. 4.
DNA binding by QNK-QNK-RHR in its four phosphorylation states. The figure shows the percentage of the oligonucleotide probe bound (deduced from changes in fluorescence anisotropy) as a function of protein concentration, comparing the results before (-CIP) and subsequent to (+CIP) treatment with calf intestinal phosphatase.
DNA-binding activity was readily restored to the phosphorylated proteins by treatment with alkaline phosphatase (Fig. 3). After overnight phosphatase treatment, the four proteins bound to the DNA probe with comparable dissociation constants of 1.7 ± 0.2 nM (QNK), 2.1 ± 0.3 nM (QNKp12), 1.5 ± 0.1 nM (QNKp23), and 2.7 ± 0.2 (QNKp123). These results indicate that the loss in DNA-binding activity in the phosphorylated proteins is reversible and is a direct consequence of the phosphoryl groups attached to the linker threonine residues.
Discussion
It was observed over 40 years ago that the passage of eukaryotic cells into mitosis is accompanied by a pause in transcription until cell division is complete (23). There seem to be several underlying causes for this phenomenon, including limited access to the DNA template upon chromatin condensation, the production of general transcription repressor proteins, and the inactivation of transcription machinery. The latter mechanism has both a general and a gene-specific component. A general block to transcription is afforded by the inhibition of RNA polymerase II through phosphorylation of transcription factors TFIID (24) and TFIIH (25) and of the polymerase itself (26). It has also been observed that several components of the SWI/SNF chromatin remodeling complex are phosphorylated upon entry into mitosis and excluded from mitotic chromosomes (27). In addition to these general mechanisms for inhibiting transcription, several gene-specific transcription factors are known to be inactivated upon entry into mitosis. The oncoproteins Myc and Myb were found to be hyperphosphorylated at mitosis (27), as were several members of the POU family of homeodomain proteins (27-30). These site-specific DNA-binding proteins all exhibited reduced DNA-binding activity in mitotic cell extracts. It is unclear why these gene-specific transcription factors are inactivated upon entry into mitosis, but it has been proposed that this type of inhibition is necessary for cell division and the proper reestablishment of expression patterns upon completion of mitosis (13).
The existence of a common mechanism for the mitotic inactivation of Cys2His2 zinc finger transcription factors is an attractive hypothesis given the abundance of this type of protein encoded in eukaryotic proteomes. The simplest model for such a system would be the direct inhibition of DNA binding through phosphorylation of a conserved site within all zinc finger domains by a cell cycle-dependent kinase. This model places strict restrictions on the site of phosphorylation within the zinc finger region. The site must be (i) conserved between proteins and, therefore, not directly involved in site-specific DNA recognition, (ii) accessible to the kinase active site when bound to DNA, and (iii) able to interfere with DNA binding when phosphorylated. The Thr-Gly-Glu-Lys-Pro linker sequence appears to meet these criteria. It is highly conserved between zinc finger proteins but is not part of the DNA-recognition helix responsible for conferring sequence specificity. As such, the linker sequence can remain constant even as members of the family evolve to have novel DNA recognition properties. The accessibility of the site to a cellular kinase is difficult to investigate until the kinase in question is identified. The existing structures of zinc finger proteins in complex with DNA, however, reveal that the linker forms an extended structure that is reasonably distant from the DNA. The threonine side chain that is proposed to be the target for phosphorylation is on the outer surface of this loop and exposed to solvent. As such, this residue is positioned such that it would be accessible to regulatory factors even when bound to DNA. Also consistent with this proposal is our finding that calf-intestinal phosphatase was able to remove the phosphate groups from our phosphorylated proteins. This result indicates that, at least in the absence of DNA, the zinc finger structure does not interfere substantially with enzymatic activity at the linker Thr residue. Moreover, this result lends support to the proposal that a cellular phosphatase could reactivate DNA binding and reestablish transcription patterns following mitosis.
Our results demonstrate conclusively that phosphorylation of the linker Thr residues does, indeed, lead to a substantial decrease in DNA-binding affinity for a prototypical zinc finger-based DNA-binding protein. The biophysical explanation for the decrease in DNA-binding affinity remains to be established. Examination of the structure of the unphosphorylated protein bound to DNA (18) reveals several features: (i) there is sufficient space for a phosphoryl group to be introduced without the introduction of unfavorable steric clashes with either the protein backbone or the DNA; (ii) if no structure changes take place, the phosphoryl group would lie ≈10 Å from two DNA backbone phosphate groups; and (iii) the introduced phosphoryl group could interact with side chains within the linker or the zinc finger preceding it. Based on these features, several mechanisms may contribute to the decrease in DNA binding. Direct electrostatic interactions between the introduced phosphoryl group and the negatively charged DNA backbone could be responsible; an interaction between singly negatively charged groups separated by 10 Å in water corresponds to 0.4 kcal/mol (1 cal = 4.18 J), a value expected to be reduced further by the presence of counterions. The interaction of a single phosphate on the protein with two DNA backbone phosphates would be expected to decrease the affinity by approximately a factor of 4. This value is well below the 40-fold reduction in binding affinity that we observed with the singly phosphorylated proteins, suggesting that additional factors are at work. It is also possible that linker phosphorylation inhibits DNA binding by disrupting structural elements within the protein itself. It has been observed that the linker region becomes structured upon DNA binding, and this structure has been proposed to contribute to the relative positioning of individual zinc finger domains (4-6). In particular, the conserved threonine seems to acts as part of a C-cap, which terminates the DNA-recognition helix of the preceding zinc finger domain (7). Phosphorylation of the Thr side chain might disrupt these contacts and destabilize linker structures appropriate for positioning adjacent zinc finger domains along the DNA. Alternatively, the phosphorylation might stabilize difference conformations of the linker that are well suited for DNA binding. A potential explanation that phosphorylation leads to zinc release and, hence, destabilization of the zinc finger structure is ruled out by our observation that all four proteins exhibit comparable affinities for metal ions as demonstrated by direct cobalt(II)-binding studies.
In summary, we have used solid-phase peptide synthesis techniques to produce a set of zinc finger proteins that have precisely incorporated phosphoryl groups and have used these proteins to correlate phosphorylation state with DNA-binding affinity. Through the direct demonstration that linker phosphorylation does, in fact, impair DNA binding, we provide support for the existence of a cellular kinase/phosphatase pair capable of inactivating/reactivating Cys2His2 zinc finger proteins in a cell cycle-dependent manner. This hypothesis provides considerable insight into the previously enigmatic observation that the linker between zinc finger domains is one of the most conserved features among members of this large and important family of proteins.
Acknowledgments
We thank Dr. Gregory J. Gatto, Jr., for useful discussions and comments on the manuscript. This work was supported by National Institute of General Medical Sciences Grant GM046257.
Abbreviation: Fmoc, fluorenylmethoxycarbonyl.
References
- 1.Rubin, G. M., Yandell, M. D., Wortman, J. R., Gabor Miklos, G. L., Nelson, C. R., Hariharan, I. K., Fortini, M. E., Li, P. W., Apweiler, R., Fleischmann, W., et al. (2000) Science 287, 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke, N. D. & Berg, J. M. (1998) Science 282, 2018-2022. [DOI] [PubMed] [Google Scholar]
- 3.Becker, K. G., Nagle, J. W., Canning, R. D., Biddison, W. E., Ozato, K. & Drew, P. D. (1995) Hum. Mol. Genet. 4, 685-691. [DOI] [PubMed] [Google Scholar]
- 4.Hyre, D. E. & Klevit, R. E. (1998) J. Mol. Biol. 279, 929-943. [DOI] [PubMed] [Google Scholar]
- 5.Wuttke, D. S., Foster, M. P., Case, D. A., Gottesfeld, J. M. & Wright, P. E. (1997) J. Mol. Biol. 273, 183-206. [DOI] [PubMed] [Google Scholar]
- 6.Bowers, P. M., Schaufler, L. E. & Klevit, R. E. (1999) Nat. Struct. Biol. 6, 478-485. [DOI] [PubMed] [Google Scholar]
- 7.Laity, J. H., Dyson, H. J. & Wright, P. E. (2000) J. Mol. Biol. 295, 719-727. [DOI] [PubMed] [Google Scholar]
- 8.Thukral, S. K., Morrison, M. L. & Young, E. T. (1991) Proc. Natl. Acad. Sci. USA 88, 9188-9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, K. R., Zhang, P., Liao, X., McBryant, S. J., Wright, P. E. & Gottesfeld, J. M. (1994) J. Mol. Biol. 244, 23-35. [DOI] [PubMed] [Google Scholar]
- 10.Choo, Y. & Klug, A. (1993) Nucleic Acids Res. 21, 3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagaoka, M., Nomura, W., Shiraishi, Y. & Sugiura, Y. (2001) Biochem. Biophys. Res. Commun. 282, 1001-1007. [DOI] [PubMed] [Google Scholar]
- 12.Ryan, R. F. & Darby, M. K. (1998) Nucleic Acids Res. 26, 703-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dovat, S., Ronni, T., Russell, D., Ferrini, R., Cobb, B. S. & Smale, S. T. (2002) Genes Dev. 16, 2985-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesfeld, J. M. & Forbes, D. J. (1997) Trends Biochem. Sci. 22, 197-202. [DOI] [PubMed] [Google Scholar]
- 15.Dawson, P. E. & Kent, S. B. (2000) Annu. Rev. Biochem. 69, 923-960. [DOI] [PubMed] [Google Scholar]
- 16.Shin, Y., Winans, K. A., Backes, B. J., Kent, S. B., Ellman, J. A. & Bertozzi, C. R. (1999) J. Am. Chem. Soc. 121, 11684-11689. [Google Scholar]
- 17.Krizek, B. A., Amann, B. T., Kilfoil, V. J., Merkle, D. L. & Berg, J. M. (1991) J. Am. Chem. Soc. 113, 4518-4523. [Google Scholar]
- 18.Kim, C. A. & Berg, J. M. (1996) Nat. Struct. Biol. 3, 940-945. [DOI] [PubMed] [Google Scholar]
- 19.Shi, Y. & Berg, J. M. (1995) Chem. Biol. 2, 83-89. [DOI] [PubMed] [Google Scholar]
- 20.Beligere, G. S. & Dawson, P. E. (1999) Biopolymers 51, 363-369. [DOI] [PubMed] [Google Scholar]
- 21.Berg, J. M. & Merkle, D. L. (1989) J. Am. Chem. Soc. 111, 3759-3761. [Google Scholar]
- 22.Desjarlais, J. R. & Berg, J. M. (1994) Proc. Natl. Acad. Sci. USA 91, 11099-11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prescott, D. M. & Bender, M. A. (1962) Exp. Cell Res. 26, 260-268. [DOI] [PubMed] [Google Scholar]
- 24.Segil, N., Guermah, M., Hoffmann, A., Roeder, R. G. & Heintz, N. (1996) Genes Dev. 10, 2389-2400. [DOI] [PubMed] [Google Scholar]
- 25.Long, J. J., Leresche, A., Kriwacki, R. W. & Gottesfeld, J. M. (1998) Mol. Cell. Biol. 18, 1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellier, S., Dubois, M. F., Nishida, E., Almouzni, G. & Bensaude, O. (1997) Mol. Cell. Biol. 17, 1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sif, S., Stukenberg, P. T., Kirschner, M. W. & Kingston, R. E. (1998) Genes Dev. 12, 2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luscher, B. & Eisenman, R. N. (1992) J. Cell Biol. 118, 775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segil, N., Roberts, S. B. & Heintz, N. (1991) Science 254, 1814-1816. [DOI] [PubMed] [Google Scholar]
- 30.Caelles, C., Hennemann, H. & Karin, M. (1995) Mol. Cell. Biol. 15, 6694-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]