Abstract
Retrotransposons are mobile DNA elements present throughout eukaryotic genomes that can cause mutations and genome rearrangements when they replicate through reverse transcription. Increased expression and/or mobility of retrotransposons has been correlated with aging in yeast, Caenorhabditis elegans, Drosophila melanogaster, and mammals. The many copies of retrotransposons in humans and various model organisms complicate further pursuit of this relationship. The Saccharomyces cerevisiae Ty1 retrotransposon was introduced into a strain of S. paradoxus that completely lacks retrotransposons to compare chronological lifespans (CLSs) of yeast strains with zero, low, or high Ty1 copy number. Yeast chronological lifespan reflects the progressive loss of cell viability in a nondividing state. Chronological lifespans for the strains were not different in rich medium, but were extended in high Ty1 copy-number strains in synthetic medium and in rich medium containing a low dose of hydroxyurea (HU), an agent that depletes deoxynucleoside triphosphates. Lifespan extension was not strongly correlated with Ty1 mobility or mutation rates for a representative gene. Buffering deoxynucleoside triphosphate levels with threonine supplementation did not substantially affect this lifespan extension, and no substantial differences in cell cycle arrest in the nondividing cells were observed. Lifespan extension was correlated with reduced reactive oxygen species during early stationary phase in high Ty1 copy strains, and antioxidant treatment allowed the zero Ty1 copy strain to live as long as high Ty1 copy-number strains in rich medium with hydroxyurea. This exceptional yeast system has identified an unexpected longevity-promoting role for retrotransposons that may yield novel insights into mechanisms regulating lifespan.
Keywords: retrotransposons, aging, chronological lifespan, yeast, reactive oxygen species
RETROTRANSPOSONS are mobile DNA elements that replicate through reverse transcription of an RNA intermediate and are known to be capable of promoting genome instability (Belancio et al. 2009). Retrotransposon sequences can comprise up to 30–80% of eukaryotic genomes, and cells commonly inhibit retrotransposon expression and mobility through use of repressive chromatin marks and/or post-transcriptional silencing mechanisms (Slotkin and Martienssen 2007). Changes in chromatin and genome instability are observed with aging, and recent work is demonstrating that the regulation and mobility of retrotransposons also changes during aging (Moskalev et al. 2013; Wood and Helfand 2013). Increased expression of retrotransposons with age has been observed in gonads of Caenorhabditis elegans, brains of Drosophila melanogaster, somatic tissues in mice, normal human cells grown ex vivo, and in yeast mother cells (Wang et al. 2011; Dennis et al. 2012; De Cecco et al. 2013a,b; Li et al. 2013; Hu et al. 2014). Increased mobility of retrotransposons has been detected at late stages of Saccharomyces cerevisiae chronological lifespan (CLS) and in brains of aged D. melanogaster (Maxwell et al. 2011; Li et al. 2013). Inhibition of elevated Alu retrotransposon expression during senescence of normal human cells grown ex vivo reversed senescence phenotypes (Wang et al. 2011). Exploring the relationship between retrotransposons and aging might provide additional insights into the aging process.
Retrotransposons have been hypothesized to provide an evolutionary advantage through increased genetic variation at the cost of reduced lifespan due to genome instability (St Laurent et al. 2010). Complementary DNA (cDNA) generated by reverse transcription of retrotransposon RNA is integrated into the genome during retrotransposition, producing genetic variation and potentially mutating genes (Beauregard et al. 2008). Nearly 100 disease-causing alleles resulting from retrotransposon insertions in humans have been characterized (Hancks and Kazazian 2012). Retrotransposons can produce DNA double-strand breaks, even in the absence of successful retrotransposition (Gasior et al. 2006). Some retrotransposons, such as the mammalian L1 element and yeast Ty1 element, are frequently present at sites of chromosome rearrangements and can produce retrotransposed copies of gene transcripts (Derr et al. 1991; Esnault et al. 2000; Dunham et al. 2002; Gilbert et al. 2002; Umezu et al. 2002; Abeysinghe et al. 2003; Maxwell and Curcio 2007; Robberecht et al. 2013). Furthermore, mammalian L1 elements and yeast Ty1 elements are both activated by increased reactive oxygen species (ROS) and DNA damage (Rockwood et al. 2004; Beauregard et al. 2008; Stoycheva et al. 2010; Giorgi et al. 2011), which are stresses associated with aging (Burhans and Weinberger 2012; Kirkwood and Kowald 2012). However, the potential role of retrotransposons and the genome instability they cause in aging has not yet been well investigated.
The Saccharomyces research model offers a unique opportunity to address the potential contribution of retrotransposons to aging. S. cerevisiae has five families of long terminal repeat (LTR) retrotransposons, Ty1–Ty5, and Ty1 is an abundant and active retrotransposon in this yeast (Beauregard et al. 2008). LTR retrotransposons are also referred to as extrachromosomally primed (EP) retrotransposons, since they code for a protein that forms a cytoplasmic virus-like particle in which the retrotransposon RNA is packaged and reverse transcribed by an element-encoded reverse transcriptase prior to integration into a genomic site (Beauregard et al. 2008). S. paradoxus is a very closely related species to S. cerevisiae, and a strain of S. paradoxus has been reported to completely lack sequences from the coding regions of Ty retrotransposons (Moore et al. 2004). This is an exceptional system, since the multicopy nature of retrotransposons typically prevents the generation of populations of a species that do not have retrotransposons. Gains and losses of Ty1 elements and regulation by a copy-number control mechanism have been examined following insertion of defined numbers of genomic Ty1 elements into this Ty-less S. paradoxus strain (Garfinkel et al. 2005). Copy-number control results in reduced Ty1 mobility when many copies of Ty1 are present in the genome, but high Ty1 mobility when only one or a few Ty1 elements are present in the genome. Similarities in regulation by cellular stresses and influences on genome instability between Ty1 and mammalian retrotransposons make Ty1 a good model element for investigating the contribution of retrotransposons to a complex phenotype, such as aging (Beauregard et al. 2008; Stoycheva et al. 2010; Giorgi et al. 2011).
Two aging models have been used in yeast to obtain information relevant to aging in diverse species. Yeast replicative lifespan is the number of times a mother cell can divide to produce a daughter cell, while chronological lifespan is the length of time that cells remain viable in a nondividing state in nutrient-depleted medium (Longo et al. 2012). Yeast lifespan can be altered by the activity of evolutionarily conserved growth signaling pathways, such as the TOR kinase pathway, mitochondrial function, proteasome function, sirtuin gene function, levels of autophagy, dietary restriction, and oxidative stress, among other processes and gene functions (Kaeberlein 2010; Longo et al. 2012). Many of these processes and homologous pathways also regulate aging in other eukaryotes (Kaeberlein 2010; Longo et al. 2012). Facile genetic approaches and short lifespan have made yeast an efficient system for identifying and characterizing these aging-associated gene/pathway functions.
As associations between retrotransposons and aging continue to be identified, it becomes important to find approaches to test for any direct influence of retrotransposons on aging. We used the Ty-less S. paradoxus strain to determine whether the presence and mobility of Ty1 elements in the genome would reduce yeast chronological lifespan by comparing strains with zero, 1–3, or ∼20 genomic copies of Ty1. Lifespan data did not support this hypothesis, but surprisingly, we found that strains with high Ty1 copy number had extended lifespan when cells were grown either in synthetic medium or in rich medium in the presence of a low dose of the ribonucleotide reductase inhibitor hydroxyurea (HU). Hydroxyurea can stimulate Ty1 mobility and causes DNA replication stress, and replication stress has been correlated with reduced yeast chronological lifespan (Curcio et al. 2007; Weinberger et al. 2007, 2013). However, lifespan extension in high Ty1 copy-number strains did not appear to be due to high retrotransposition levels or substantial differences in DNA replication stress. Rather, lifespan extension was correlated with changes in ROS accumulation. These results identify a novel role for retrotransposons in promoting lifespan that may uncover additional associations between retrotransposons and ROS production or signaling.
Materials and Methods
Yeast strains, media, and plasmids
Standard rich (YPD) and synthetic (SC) media were used for growing yeast strains (Amberg et al. 2005). All strains were derivatives of a Ty-less strain of S. paradoxus, DG1768 (MATα, his3-∆200hisG, ura3, kindly provided by D. Garfinkel) (Garfinkel et al. 2003). A galactose-inducible plasmid copy of Ty1 containing a his3AI retrotransposition indicator gene (Curcio and Garfinkel 1991) was used to obtain derivatives of DG1768 with a single genomic copy of a Ty1his3AI element, as described previously (Maxwell et al. 2011). The high copy URA3-marked plasmid pGTy1H3CLA containing an unmarked Ty1 element under the control of a galactose-inducible promoter (Garfinkel et al. 1988) was used to introduce unmarked copies of Ty1 into the genome of strain DG1768. Ura+ transformants harboring pGTy1H3CLA were grown on SC + 2% glucose medium lacking uracil, and selected Ura+ transformants were then grown in liquid SC + 2% glucose medium lacking uracil at 30° overnight. Aliquots of these cultures were diluted and spread onto solid SC medium lacking uracil + 2% galactose to induce Ty1 retrotransposition, and were then grown at 20° or 30° to vary the level of retrotransposition (Ty1 mobility is restricted at 30°) (Paquin and Williamson 1984; Lawler et al. 2002). Single colonies obtained after induction were grown on YPD medium to allow for loss of pGTy1H3CLA and analyzed for Ty1 copy number by qPCR using a Roche LightCycler 480. Genomic DNA was extracted by glass bead disruption in phenol/chloroform and ethanol precipitation, followed by further purification using a Wizard SV gel and PCR Clean-up system (Promega). Equal concentrations of DNA from these strains were amplified with primers to the TYA1 region of Ty1 and primers to a single-copy gene, ACT1. The cycle threshold (Ct) values for the TYA1 PCR were normalized to the ACT1 Ct values for each strain, and approximate copy number was then determined by comparing these normalized values to the normalized TYA1 Ct for a single copy Ty1his3AI control strain. Strains were grouped as low copy (1–3 genomic Ty1 elements) or high copy (∼20 genomic Ty1 elements). A pGTy1H3kanMX plasmid was constructed by first amplifying the kanMX marker present in the S. cerevisiae MATα deletion collection (Thermo Scientific Open Biosystems) with PCR primers that added sequences corresponding to Ty1-H3 positions 5516–5566 and 5567–5616 to either end of kanMX (Ty1-H3 GenBank accession M18706.1). This PCR product was then cotransformed into strain DG1768 with pGTy1H3CLA linearized at the ClaI site present between the 3′ end of TYB1 and the 3′ LTR. Cells in which the Ty1-kanMX PCR product was used to repair the plasmid were selected on YPD medium containing 200 µg/ml G418 sulfate and verified by PCR. A high-copy URA3-marked control plasmid lacking Ty1 sequences was generated by cloning a BamHI–EcoRI fragment of the PCR-amplified kanMX gene into pRS426 digested with BamHI and EcoRI. Strains with pGTy1H3kanMX or pRS426-kanMX were grown using YPD medium containing 200 µg/ml G418 sulfate to maintain each plasmid.
Southern blotting to detect Ty1 insertions
Approximately 1–2 µg of genomic DNA prepared from cells grown at 30° by glass bead disruption in phenol/chloroform and ethanol precipitation was digested with PvuII or SphI restriction enzymes and used for Southern blotting as described previously (Maxwell et al. 2004). A radioactive probe was prepared from a PCR product corresponding to positions 4221–4842 of Ty1-H3 (GenBank accession M18706.1) by random priming using the Ambion DECAprime II kit (Life Technologies), and signal was visualized using a Typhoon Trio+ imager (GE Healthcare).
Growth rate experiments
Cells from fresh streaks were inoculated into 5 ml YPD broth with or without 30 mM hydroxyurea at an initial density of 1 × 105 cells/ml to determine cell-doubling times. Triplicate cultures were grown for each strain for each trial on a culture tube rotator at 20°. Cell densities were determined at several time points between 7 and 33 hr of growth by counting cells on a hemocytometer to measure growth rate from late lag phase to midexponential phase. Exponential function trend lines fit to plots of cell densities compared to hours of growth were used to calculate doubling times in hours.
Chronological lifespan determination
Cells from fresh streaks were inoculated at an initial density of 5 × 103 cells/ml in triplicate 25 ml cultures in 125-ml Erlenmeyer flasks and aged at 20° (or 30° for some trials) with shaking. For SC medium experiments, the same initial density of cells was inoculated into triplicate 4-ml cultures in culture tubes and aged at 20° on a tube rotator. Media used for lifespan experiments included YPD with or without 30 mM hydroxyurea, 0.8 mg/ml threonine, or 5 mM or 10 mM reduced glutathione, as well as SC + 2% glucose medium with or without 30 mM hydroxyurea. At early stationary phase and at regular time points thereafter, aliquots of cells were removed to assess viability and colony-forming units per milliliter (CFU/ml). Viability was measured by mixing diluted cells with two volumes of 0.4% Trypan blue in phosphate-buffered saline (PBS), incubating for 45 min at room temperature, and determining the fraction of unstained cells to total cells for populations of ∼200 cells by microscopy. CFU/ml were determined by spreading aliquots of diluted cells onto YPD agar. Trend lines were fit to graphs of viability vs. days of incubation and used to calculate days to 50 and 10% viability as measures of median and maximal lifespan, respectively.
Ty1 integration and retromobility frequency
A previously described semiquantitative assay to measure Ty1 integration upstream of 5S rRNA genes was modified in two ways to compare Ty1 retrotransposition levels in different yeast strains (Maxwell et al. 2011). First, primers to the S. paradoxus LEU2 gene were used to control for PCR efficiency. Second, the total signal intensity of Ty1 integration PCR products for each template minus the background intensity obtained from a zero Ty1 copy template was determined using a ChemiDoc XRS+ imager and Image Lab version 4.0 (Bio-Rad) and normalized to the signal intensity of the control LEU2 PCR product to obtain a semiquantitative relative measure of Ty integration. In some trials, retrotransposition was inhibited by growing cells in the presence of 200 µg/ml phosphonoformic acid to strongly inhibit Ty1 reverse transcriptase without noticeably altering cell growth (Lee et al. 2000). Ty1 retromobility was measured in strains with a chromosomal Ty1his3AI element by determining the frequency of His+ prototroph formation (Curcio and Garfinkel 1991). Seven replicate cultures initiated at a density of 5 × 103 cells/ml were grown to near saturation at 20° for each strain/trial, aliquots of diluted cells were spread onto YPD medium to determine CFU/ml, and appropriate volumes of cells were spread onto SC + 2% glucose medium lacking histidine. Mobility frequencies were calculated as the number of His+ prototrophs divided by the number of CFU in the same volume of culture. Median values for three independent trials were averaged.
Mutation rate and frequency measurements
Fluctuation tests were performed to obtain rates of loss-of-function mutations in the CAN1 gene that produce resistance to canavanine, and rates were calculated using the online calculator FALCOR (Foster 2006; Hall et al. 2009). Ten replicate 1-ml cultures in YPD or YPD with 30 mM hydroxyurea inoculated at initial densities of 1 × 103 cells/ml were grown to late log/early stationary phase at 20°. Aliquots of culture were diluted in water and spread onto YPD agar to determine CFU/ml and the remainder of each culture was pelleted and resuspended in water to spread onto SC + 2% glucose medium lacking arginine and containing 60 µg/ml canavanine. Plates were incubated for up to 4 days at 30°. Mutation frequencies were measured in a similar manner using three or seven replicate cultures inoculated at initial densities of 5 × 103 cells/ml grown in YPD or YPD with varying concentrations of hydroxyurea. Mutation frequencies were calculated as the number of mutant colonies obtained divided by the number of CFU spread on selective medium, and median values from independent trials were averaged.
Analysis of reactive oxygen species levels and DNA content
Strains were grown in flasks at 20° (or 30° for some trials) as described for chronological lifespan experiments and sampled after 3–7 days (1–3 days at 30°) of growth to determine levels of ROS. Methods to measure ROS using dihydroethidium (DHE) or dihydrorhodamine 123 (DHR) were based on published protocols (Mesquita et al. 2010; Weinberger et al. 2010). Briefly, ∼1 × 107 cells were washed in 1× PBS buffer twice, resuspended in 1× PBS, and incubated with a 50 µM final concentration of either DHR or DHE for 2 hr at room temperature (in the dark with rocking). After incubation, cells were washed twice in 1× PBS buffer before analysis on a BD LSR II flow cytometer using excitation at 488 nm and a 530/30 nm (long pass 505) emission filter for DHR or excitation at 515 nm and a 610/20 nm (long pass 600) emission filter for DHE. The median fluorescence intensity for each sample was determined using FlowJo version X.0.7 software (TreeStar) and normalized based on the median fluorescence intensity of the corresponding unstained sample. Similar results were obtained analyzing geometric means. Reported values are averages of median values for three or more trials. Analysis of DNA content in cells was performed as described previously (Weinberger et al. 2007), except that Propidium Iodide ReadyProbes Reagent (PI, Life Technologies) was used instead of SYBR Green. Cells were incubated with two drops per milliliter of the PI reagent for 15 min prior to analysis by flow cytometry using excitation at 515 nm and a 610/20 nm (long pass 600) emission filter.
Statistical analysis
Mean values were compared for significant differences using unpaired, two-tailed t-tests assuming equal or unequal variance, depending on the variance in particular datasets. Levels of significance are indicated in figures and the text.
Results
We used a plasmid to introduce copies of a Ty1 retrotransposon into the genome of a Ty-less lab strain of S. paradoxus (Garfinkel et al. 2003; Moore et al. 2004) to address the impact of retrotransposons on yeast chronological lifespan. Conditions to induce Ty1 mobility were varied to obtain strains with different Ty1 copy numbers, and the approximate Ty1 copy number in clonal populations was determined through quantitative PCR. We grouped the strains with 1–3 genomic Ty1 insertions as low-copy strains and the strains with ∼20 genomic Ty1 insertions as high-copy strains, since Ty1 mobility is restricted through a copy-number control mechanism (Garfinkel et al. 2003), and the initial expectation was that Ty1 mobility would be correlated with any observed influence on chronological lifespan. Four different low-copy strains and four different high-copy strains were used for this work to reduce the possibility that lifespan changes would simply be due to a particular Ty1 insertion or pattern of Ty1 insertions that directly altered expression or function of a gene regulating lifespan. To confirm the presence of independent insertions, genomic DNA from these strains was digested separately with two restriction enzymes to produce fragments containing sequences from the 3′ end of Ty1 and flanking genomic DNA. A Southern blot of these samples hybridized with a probe to the 3′ end of Ty1 demonstrated that different fragment sizes were present in different low- or high-copy strains with no distinct fragments common to all low or all high Ty1 copy strains (Figure 1). Low-copy strains had one to three fragments, and high-copy strains had >10 distinct fragments. This indicates that each low or high Ty1 copy strain harbors insertions at different genomic sites, so a consistent change in lifespan for all low Ty1 copy strains or for all high Ty1 copy strains would likely be a general phenotype of a low- or high-copy strain.
Figure 1.
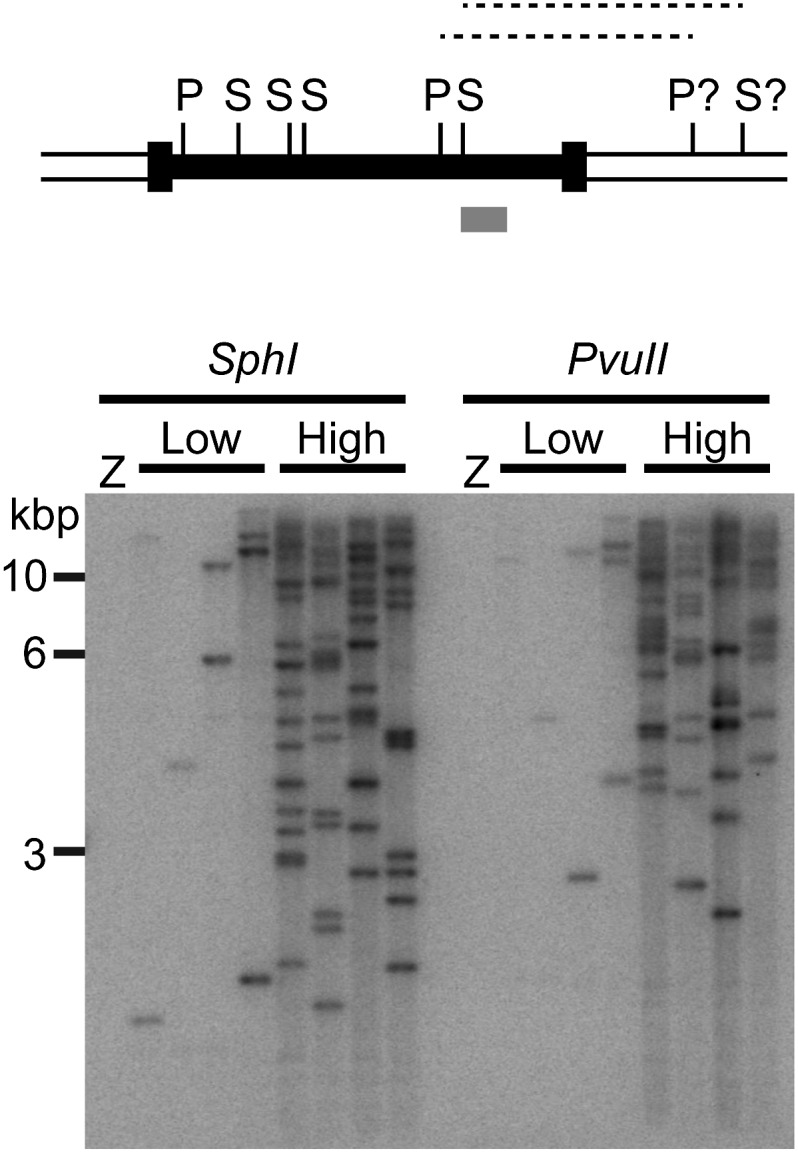
Individual low and high Ty1 copy strains have distinct Ty1 insertions. The diagram at the top depicts a Ty1 element as a solid box bounded by narrow taller boxes (long terminal repeats) and flanked by genomic DNA sequences (double lines). Relative positions of sites for the restriction enzymes PvuII (P) and SphI (S) are indicated and the question marks indicate that the distance to the nearest flanking site is variable for each insertion. The lightly shaded box shows the relative position of the probe and dotted lines indicate the restriction fragments detected by the probe. Below the diagram is a Southern blot of genomic DNA digested with PvuII or SphI from the zero copy, “Z,” four low copy, “low,” and four high copy, “high,” Ty1 strains probed for 3′ Ty1-flanking DNA restriction fragments. Positions of selected size standards in kilobase pairs (kbp), are indicated to the left of the blot image.
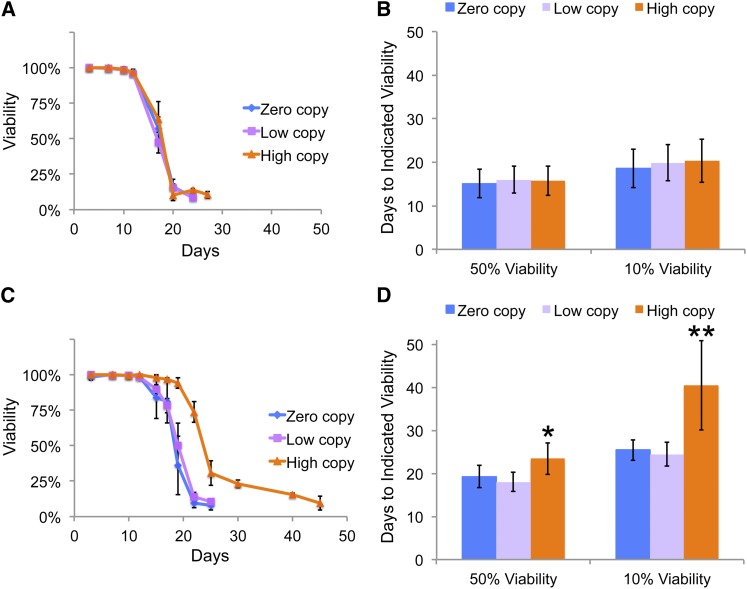
Strains were grown and aged in rich medium (YPD) at 20° to determine CLS so that Ty1 would be able to actively retrotranspose (Paquin and Williamson 1984; Lawler et al. 2002). Note that growth at 20° can substantially lengthen CLS compared to standard growth at 30° (Maxwell et al. 2011). The number of days required for populations to reach 50 and 10% viability were calculated as representations of median (50%) and maximum (10%) lifespans to facilitate comparisons between multiple strains and replicate trials. The mean days to 50 and 10% viability for the zero Ty1 copy parent strain were 15.1 and 18.6, respectively, and there was no significant difference in lifespan between the zero, low, or high Ty1 copy-number strains in YPD (Figure 2, A and B).
Figure 2.
High Ty1 copy strains have extended lifespan in the presence of hydroxyurea. (A) Representative chronological lifespan experiment in YPD medium using triplicate cultures of the zero Ty1 copy parent strain, one low Ty1 copy strain, and one high Ty1 copy strain. (B) Mean ± SD of median chronological lifespan (days to 50% viability) and maximum chronological lifespan (days to 10% viability) in YPD medium for the zero Ty1 copy strain (blue columns), four low Ty1 copy strains (light purple columns), and four high Ty1 copy strains (orange columns) from 5 to 9 independent trials. (C) Representative chronological lifespan experiment in YPD + 30 mM hydroxyurea using triplicate cultures of the zero Ty1 copy strain, one low Ty1 copy strain, and one high Ty1 copy strain. (D) Mean ± SD of median chronological lifespan (days to 50% viability) and maximum chronological lifespan (days to 10% viability) in YPD + 30 mM hydroxyurea medium for the zero Ty1 copy parent strain (blue columns), four low Ty1 copy strains (light purple columns), and four high Ty1 copy strains (orange columns) from 9 to 12 independent trials. Significant differences are indicated with asterisks: *P < 0.05, **P < 0.01.
We considered that it might be necessary to stress cells to detect an influence of Ty1 on lifespan, since various stresses induce retromobility and a number of stresses can also influence lifespan (Slotkin and Martienssen 2007; Beauregard et al. 2008; Haigis and Yankner 2010). Increased DNA replication stress has been correlated with decreased yeast CLS (Weinberger et al. 2007, 2013), so we repeated CLS experiments in YPD medium with a low concentration (30 mM) of the ribonucleotide reductase inhibitor HU. This concentration of HU led to a 30–40% increase in the doubling time of all strains (Table 1), but did not decrease viability when cells first reached stationary phase. Unexpectedly, chronic exposure to HU significantly increased the median lifespan of the zero Ty1 copy strain by 28% to 19.3 days (P < 0.05) and the maximum lifespan by 37% to 25.4 days (P < 0.01, Figure 2, C and D). Significant increases were also observed for the median lifespan of the low Ty1 copy strains (P < 0.05) and both the median and maximum lifespans of the high Ty1 copy strains (P < 0.01, Figure 2D). Since growth signaling is known to influence yeast lifespan (Longo et al. 2012), the slower growth of these strains in HU could be at least partly responsible for this lifespan extension. There was no significant difference in CLS between zero Ty1 copy and low Ty1 copy strains in HU. Surprisingly, median and maximum lifespans of high Ty1 copy strains were significantly extended by 22 and 59%, respectively, in YPD medium with HU compared to the zero-copy strain (Figure 2D). Cell doubling times of the high Ty1 copy strains were not significantly different from the parent zero-copy strain in YPD medium with HU (Table 1), indicating that a difference in growth rate is unlikely to be the reason for the lifespan extension.
Table 1. High Ty1 copy strains have a slightly slower growth rate in rich medium than the zero Ty1 copy strain.
| Strain (Ty1 copy number) | Trials | Doubling time in YPDa | Trials | Doubling time in YPD + 30 mM hydroxyureaa |
|---|---|---|---|---|
| Zero | 6 | 3.8 ± 0.2 | 7 | 5.2 ± 0.3 |
| Low | 6 | 3.9 ± 0.2 | 6 | 5.0 ± 0.3 |
| High | 8 | 4.0 ± 0.2* | 8 | 5.5 ± 0.4 |
Triplicate cultures were grown at 20° for each trial, and a total of four low Ty1 copy and four high Ty1 copy strains were tested. *P < 0.05 vs. zero Ty1 copy strain in same medium.
Mean ± SD in hours.
Increased lifespan of high Ty1 copy strains grown in medium with HU is not correlated with Ty1 retrotransposition
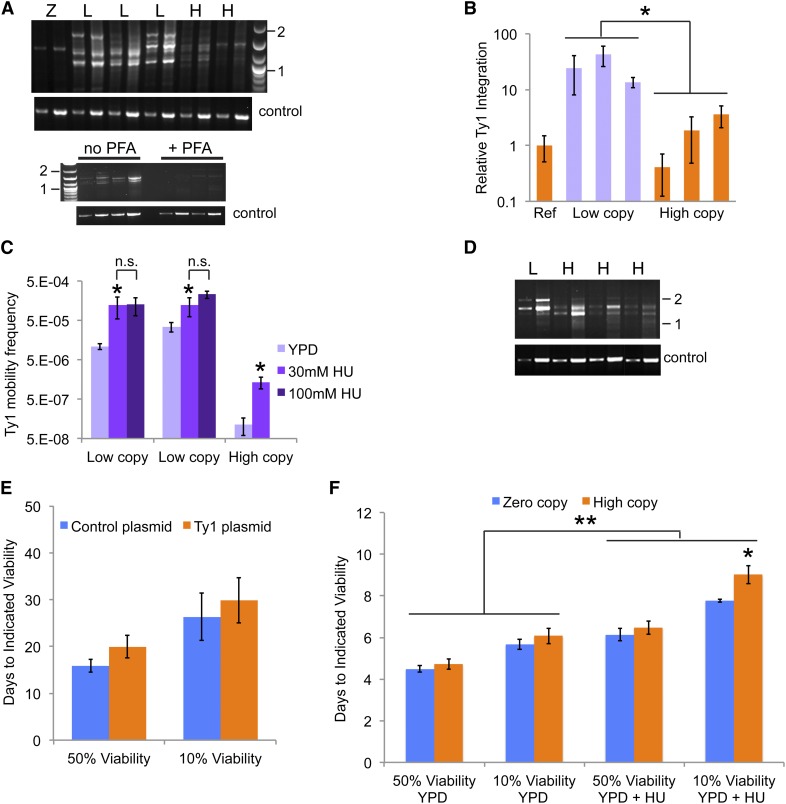
Since Ty1 mobility did not appear to negatively influence CLS, relative Ty1 retrotransposition levels were examined to confirm that low Ty1 copy strains had higher levels of retrotransposition and high Ty1 copy strains had lower levels of retrotransposition, due to restriction of retrotransposition by a copy-number control mechanism (Garfinkel et al. 2003). We assayed for Ty1 integration events upstream of 5S rRNA genes by PCR (Figure 3A), since Ty1 elements are known to frequently integrate upstream of genes transcribed by RNA polymerase III (Devine and Boeke 1996). The expected PCR result is a ladder of products representing different sites of Ty1 insertions relative to a 5S rRNA gene in different subpopulations of cells in a given culture. The yield of PCR products was greatly reduced using DNA templates prepared from cells grown in the presence of phosphonoformic acid, a reverse transcriptase inhibitor, demonstrating that the products resulted from new retrotransposition events (Figure 3A). Relative Ty1 integration levels were compared by normalizing the total signal intensities of all integration products for each template to the signal intensity of a PCR product for a single-copy gene and then using that normalized value for one high Ty1 copy strain to normalize all other integration PCR values. Low Ty1 copy strains had ∼10- to 40-fold higher Ty1 integration levels using this assay than the reference high Ty1 copy strain, and integration levels in other high-copy strains ranged from 0.5- to 4-fold the value of the reference strain (Figure 3B). These results indicate that copy number control is occurring in these strains.
Figure 3.
Ty1 mobility is elevated in low Ty1 copy strains and in the presence of hydroxyurea. (A) Representative pairs of ethidium bromide-stained agarose gel images showing Ty1 integration PCR products (top panel of each pair) obtained after 31 and 34 cycles from the same template or single-copy gene control product obtained after 17 and 19 or 20 and 22 cycles from the same template (top “control” and bottom “control” images, respectively). Results in the upper pair of images are for the zero Ty1 copy parent strain, “Z,” three low Ty1 copy strains, “L,” and two high Ty1 copy strains, “H.” Results in the lower pair of images are for two low Ty1 copy strains grown in the absence, “no PFA,” or presence, “+ PFA,” of 200 µg/ml phosphonoformic acid to inhibit Ty1 reverse transcriptase. The positions of 1 kbp and 2 kbp markers are indicated (“1” and “2”). (B) A comparison of the relative yield of Ty1 integration PCR products from two to three independent trials using a reference high Ty1 copy strain, “Ref,” three low Ty1 copy strains, “low copy,” and three additional high Ty1 copy strains, “high copy.” Columns indicate mean ± SD. *P < 0.05 for the means of low vs. high Ty1 copy strains. (C) Frequencies of forming His+ prototrophs as a measure of Ty1 mobility in two low Ty1 copy, “low copy,” and one high Ty1 copy, “high copy,” strains harboring a chromosomal Ty1his3AI element grown in the absence, “YPD,” or presence of two different concentrations of hydroxyurea, “30 mM HU” and “100 mM HU.” Columns indicate mean ± SD for three trials per strain per condition, *P < 0.05 and n.s., no significant difference. (D) Representative results of Ty1 integration PCR following growth of cells in YPD medium with 30 mM HU, as described for A, and control product was obtained after 20 and 22 cycles of PCR. (E) The median and maximum lifespans for zero-copy Ty1 strains harboring a control plasmid (blue columns) or a Ty1 plasmid (orange columns) grown in YPD + 30 mM hydroxyurea + 200 µg/ml G418 are shown as the mean ± SD of three trials. (F) Median, “50% viability,” and maximum, “10% viability,” lifespans for zero Ty1 copy (blue columns) and high Ty1 copy (orange columns) strains grown at 30° in YPD medium without or with 30 mM hydroxyurea, “YPD” or “YPD + HU,” respectively. Data are mean ± SD from three trials for the zero-copy strain and four trials for the high-copy strains. *P < 0.05 for maximum lifespans of zero compared to high Ty1 copy strains in YPD + HU, and **P < 0.01 for lifespans in YPD compared to those in YPD + HU.
The influence of HU on Ty1 retromobility in low- and high-copy strains was compared using the his3AI retromobility indicator gene (Curcio and Garfinkel 1991). Ty1 retromobility frequencies in two low-copy strains harboring only a single genomic Ty1his3AI element and a high Ty1 copy strain with a single genomic Ty1his3AI were measured as the frequencies with which cells became His+ prototrophs. HU was reported to activate Ty1 retromobility in a dose-dependent manner in a S. cerevisiae lab strain harboring Ty1his3AI (Curcio et al. 2007), which would be comparable to the high Ty1 copy strain with Ty1his3AI in the current study. Treatment with 30 mM HU increased Ty1his3AI retromobility ∼4- or 11-fold in the two low-copy strains and ∼12-fold in the high Ty1 copy strain (Figure 3C). Increasing the HU concentration to 100 mM led to a marginal, but not significant, further increase in mobility for the low-copy strains. The substantially higher mobility in the single-copy strains compared to the high Ty1 copy strain again verified Ty1 regulation through copy number control (Figure 3C). Furthermore, integration upstream of 5S rRNA genes was still observed with HU treatment, indicating that Ty1 insertion preferences were not grossly altered in response to HU (Figure 3D). These results indicate that lifespan extension in high Ty1 copy strains in the presence of HU occurs in the context of moderate levels of Ty1 mobility, and higher levels of retromobility in low Ty1 copy strains do not negatively influence CLS in rich medium with or without HU.
A high-copy plasmid with a Ty1 element under the control of a galactose-inducible promoter was introduced into the zero Ty1 copy strain to test whether the presence of many Ty1 DNA sequences in the nucleus of cells would somehow extend lifespan in the presence of HU. Strains with the Ty1 plasmid or a control plasmid were grown and aged in rich medium with HU and an antibiotic, G418, to maintain the plasmids. Glucose medium was used to repress the galactose-inducible promoter and thereby minimize expression of the Ty1 element. PCR with DNA extracts from early and late time points during aging confirmed the presence of the plasmid throughout aging (data not shown). No significant differences in median or maximum lifespan were identified between strains with the control plasmid or the Ty1 plasmid (Figure 3E). Therefore, the presence of Ty1 sequences on a high-copy plasmid is not enough to reproduce the phenotype of high Ty1 copy strains grown in YPD with HU.
Ty1 mobility is inhibited at 30°, as previously noted, which is largely due to reduced Ty1 protein levels and reduced proteolytic processing of Ty1 Gag–Pol fusion protein by Ty1 protease (Lawler et al. 2002). The lack of Gag–Pol processing inhibits reverse transcription, but Ty1 transcription is not altered at high temperature (Lawler et al. 2002). Earlier work also demonstrated that Ty1 transcripts are abundant in cells grown at 30° (Curcio et al. 1990). We therefore aged zero and high Ty1 copy strains at 30° in the presence and absence of HU to determine whether Ty1 protein expression and processing were required for lifespan extension. Incubation at 30° significantly decreased lifespan for all strains compared to incubation at 20° (P < 0.005), as expected (Figure 2, B and D; Figure 3F). No significant differences in lifespan between strains were noted during aging in YPD medium alone, and lifespans of all strains were significantly extended when HU was added to the medium (P < 0.001, Figure 3F). High Ty1 copy strains no longer had extended median lifespan relative to the zero-copy strain when incubated at 30° in YPD with HU (Figure 3F). Maximum lifespan was still significantly extended, but the relative extension was significantly reduced from 59% at 20° to 16% at 30° (P < 0.01, Figure 2D; Figure 3F). These results indicate that the presence and/or functions of Ty1 proteins may contribute to lifespan extension in HU.
Extension of lifespan in high Ty1 copy strains does not appear to result from substantial differences in DNA replication stress between zero and high Ty1 copy strains
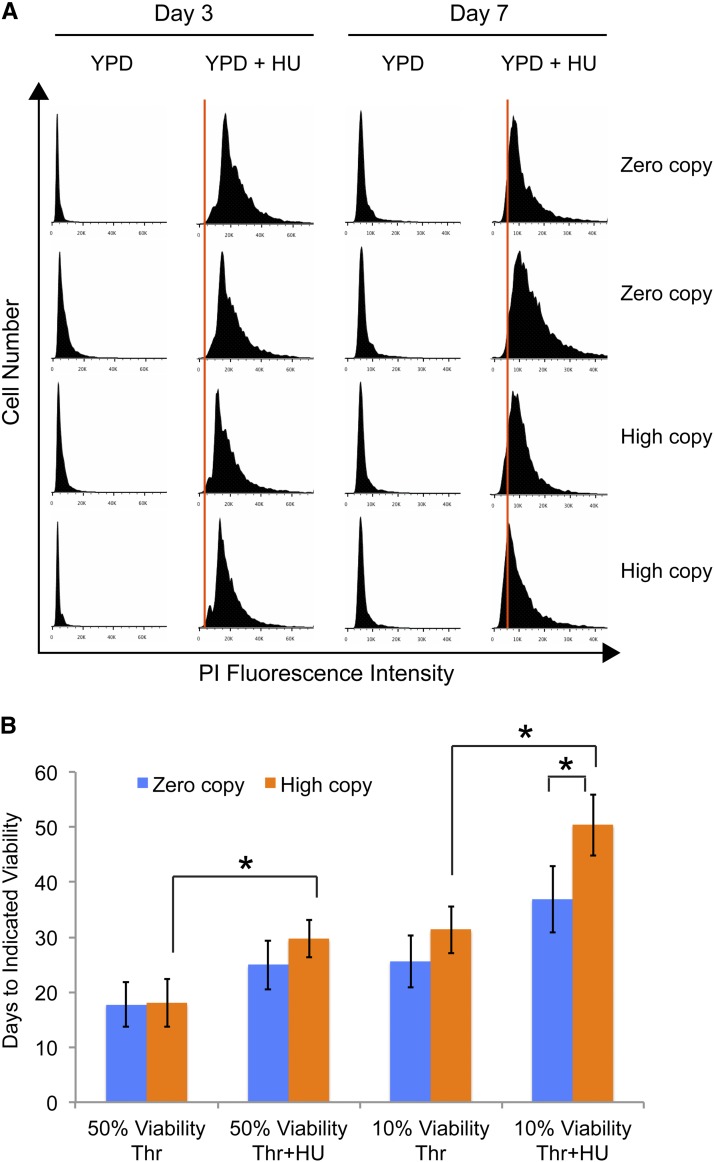
A negative correlation has previously been reported between treatments and mutations that alter yeast CLS and levels of DNA replication stress (Weinberger et al. 2007). This negative correlation was established in part by examining the ability of yeast cells to correctly arrest in G1 stage of the cell cycle when they reach stationary phase through DNA content analysis (Weinberger et al. 2007). The possibility that high Ty1 copy strains lived longer in HU medium due to improved arrest in G1 during the transition to stationary phase was examined through use of a fluorescent DNA stain and flow cytometry. Nearly all cells for both zero and high Ty1 copy strains had G1 DNA content when grown in YPD medium for 3 days, and there was little change in this profile at day 7 (Figure 4A). Growth in YPD with 30 mM HU caused a pronounced increase in the fluorescence intensities of cell populations at day 3, indicative of many cells in S and G2 stages, but this effect was diminished by day 7 as cells began to return to a G1 DNA content (Figure 4A). DNA content profiles for the zero and high Ty1 copy strains were very similar in the presence of HU at both time points (Figure 4A), and we did not observe consistent or substantial differences in profiles in multiple experiments. Therefore, lifespan extension in high Ty1 copy strains does not seem to be due to substantially improved arrest in G1 during stationary phase in the presence of HU.
Figure 4.
Differences in replication stress are unlikely to fully explain the lifespan extension in high Ty1 copy strains. (A) Representative histograms from flow cytometry using propidium iodide (PI) to determine DNA content of two replicates of the parent zero Ty1 copy strain, “zero copy,” and two different high Ty1 copy strains, “high copy.” Cells were analyzed after 3, “day 3,” or 7, “day 7,” days of growth in rich medium without or with 30 mM hydroxyurea, “YPD” or “YPD + HU,” respectively. Each panel for day 3 has the same x-axis scale, and each panel for day 7 has the same x-axis scale. An orange vertical line on the “YPD + HU” panels indicates the average position of the peak signal from the corresponding set of “YPD” graphs to facilitate comparisons. (B) Mean ± SD of median, “50% viability,” and maximum, “10% viability,” lifespans for the zero Ty1 copy strain (blue columns) and three high Ty1 copy strains (orange columns) grown in rich medium with an excess of threonine and without or with 30 mM hydroxyurea, “Thr” or “Thr + HU,” respectively. Data are from three trials, and *P < 0.05 shows comparison of high Ty1 copy strains grown without or with HU and maximum lifespans of zero and high Ty1 copy strains in Thr + HU.
We further investigated the potential relationship between HU-induced DNA replication stress and lifespan extension by high Ty1 copy number by growing and aging cells in the presence of excess threonine. S. cerevisiae cells deficient for threonine biosynthesis genes, such as THR1, show increased sensitivity to HU, which can be suppressed by growing cells with an excess of threonine (Hartman 2007). A metabolic pathway involving threonine has been proposed to buffer deoxynucleoside triphosphate (dNTP) levels in the presence of HU or when other means are used to inhibit ribonucleotide reductase activity (Hartman 2007). Also, thr1∆ mutants have been reported to have decreased CLS, which can be suppressed by overexpression of the ribonucleotide reductase subunit gene RNR1, and excess threonine can suppress the shortened CLS of cells experiencing replication stress due to high glucose (Weinberger et al. 2013). Strains were grown and aged in rich medium with four times the normal concentration of threonine to determine whether buffering dNTP levels would result in more similar lifespans for zero and high Ty1 copy strains in the presence of HU. The maximum lifespan of high Ty1 copy strains was still significantly longer by ∼37% compared to the parent strain in the presence of both excess threonine and HU, which was not significantly different from the lifespan extension originally observed in rich medium and HU without threonine (Figure 2D; Figure 4B). The difference in median lifespan between zero and high Ty1 copy strains in medium with threonine and HU was not significant, however. Two additional aspects of these data are noteworthy. First, HU still extended lifespan of all strains in the presence of excess threonine (compare Thr vs. Thr + HU in Figure 4B), but the increase was not quite significant for the zero-copy strain. Second, the median and maximum CLS of both zero and high Ty1 copy strains were significantly longer in YPD with HU and excess threonine than they were in YPD with only HU (P < 0.05 for zero copy and P < 0.01 for high copy; compare Figure 2D with Figure 4B). This indicates that depletion of dNTPs may limit CLS of these strains in HU, even though HU has an overall positive influence on CLS. Since the maximum CLS of high Ty1 copy strains was still longer than the parent strain despite threonine supplementation, differential response to DNA replication stress may not be the primary reason for the increased longevity.
High Ty1 copy strains have a very moderate reduction in mutation frequency when grown in medium with HU
Mutations and chromosome rearrangements accumulate during yeast chronological aging (Madia et al. 2007), so we examined young cell populations to determine whether reduced mutation rates early in lifespan could potentially contribute to lifespan extension in high Ty1 copy strains. CAN1 encodes a permease that transports arginine and the toxic arginine-analog canavanine into yeast cells, and loss-of-function mutations in CAN1 provide canavanine resistance (Whelan et al. 1979). Modest increases or decreases in CAN1 mutation rates per cell generation were observed for individual low Ty1 copy and high Ty1 copy strains compared to the zero-copy strain when grown in YPD medium, but the 95% confidence intervals were largely overlapping (Figure 5A). CAN1 mutation frequencies (fraction of cells in the population with a mutation) were significantly increased by growth in YPD with 15 mM HU compared to YPD only for both zero and high Ty1 copy strains, though the additional increases in mutation frequency with each additional increase in HU concentration were not quite significant in most cases (Figure 5B). High-copy Ty1 strains showed a moderately diminished mutation frequency response to HU compared to the zero-copy strain, particularly in 30 mM HU (Figure 5B). However, substantial differences in mutation rates for the zero-copy and four high Ty1 copy strains in YPD with 30 mM HU were not observed (Figure 5C). There were marginal decreases in rate for three high-copy strains and a modest increase in mutation rate for the fourth compared to the zero-copy strains, but the 95% confidence intervals were again largely overlapping. Differences in mutation rates therefore do not appear to be responsible for the lifespan extension in the high Ty1 copy strains.
Figure 5.
High Ty1 copy strains have a modest reduction in mutation frequency in the presence of hydroxyurea. (A) Rate per cell generation of forming canavanine-resistant mutants for two trials of the zero-copy Ty1 strain (blue columns), four low Ty1 copy strains (light purple columns), and four high Ty1 copy strains (orange columns) in YPD medium determined using the FALCOR calculator (Hall et al. 2009). Error bars indicate 95% confidence intervals. (B) Frequency of forming canavanine-resistant mutants following growth in rich medium without or with the indicated concentrations of hydroxyurea for zero (blue columns) or four high Ty1 copy (orange columns) strains. Data represent the mean ± SD from four or five trials, and **P < 0.01 shows comparison of zero to high Ty1 copy strains in 30 mM HU, or for indicated comparisons. (C) Mutation rates as for A for two trials of the zero-copy strain and four high Ty1 copy strains using YPD medium with 30 mM HU.
Differences in ROS accumulation play a role in lifespan extension of high Ty1 copy strains
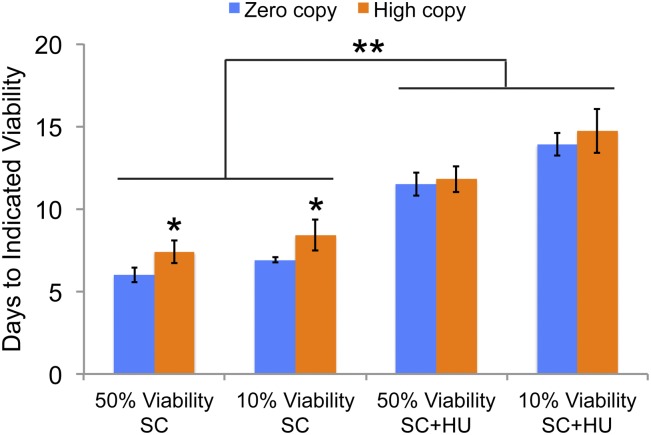
Yeast CLS is substantially reduced in synthetic medium compared to rich medium, which is at least partly due to acetic acid accumulation and is associated with increased replication stress (Weinberger et al. 2007; Burtner et al. 2009). We observed that these S. paradoxus strains had very short lifespans in synthetic medium with 2% glucose (SC), especially considering that their growth was at 20°, rather than the more typical 30° used for most published CLS experiments. In SC medium, high Ty1 copy strains had significantly increased median and maximum lifespans compared to the zero-copy strain (7.4 vs. 6.0 days and 8.4 vs. 6.9 days, respectively, Figure 6). All strains had a significantly longer lifespan when 30 mM HU was added to the SC medium, but there was no difference between zero and high Ty1 copy strains in this medium (Figure 6). The difference in results using YPD with HU and SC with HU could indicate that acetic acid accumulation, increased DNA replication stress, or other stresses/cellular changes associated with aging in SC medium may mask the ability of HU treatment to further extend lifespan of the high Ty1 copy strains.
Figure 6.
High Ty1 copy strains have moderately extended lifespan in synthetic medium without hydroxyurea. Median, “50% viability,” and maximum, “10% viability,” lifespans for zero Ty1 copy (blue columns) and high Ty1 copy (orange columns) strains grown in synthetic medium without or with 30 mM hydroxyurea, “SC” or “SC + HU,” respectively. Data are the mean ± SD from three to five trials. *P < 0.05 for lifespans of zero compared to high Ty1 copy strains in SC, and **P < 0.01 for lifespans in SC compared to those in SC + HU.
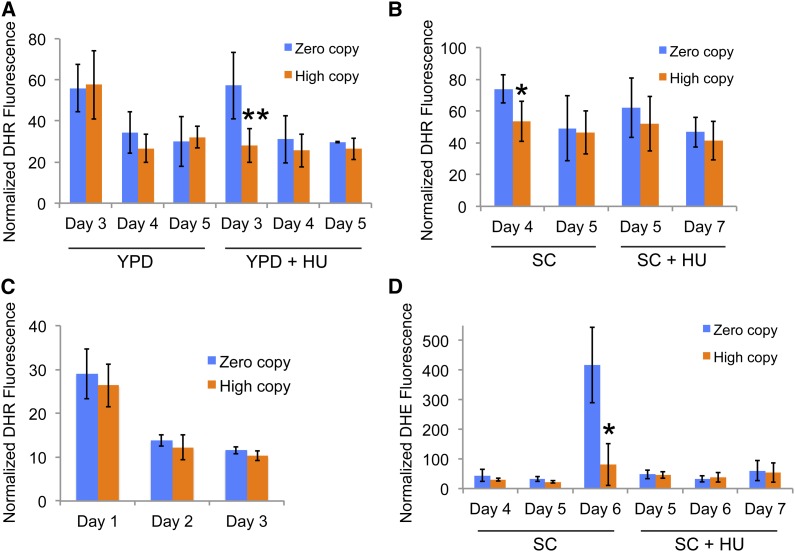
Since changes in ROS have been associated with yeast CLS (Fabrizio et al. 2003; Weinberger et al. 2010; Longo et al. 2012), we measured ROS levels in the zero and high Ty1 copy strains. No significant differences in levels of peroxides and other ROS detected by the fluorescent reagent DHR and flow cytometry were noted between zero and high Ty1 copy strains grown in YPD during the transition to stationary phase and early time points during aging. In contrast, high Ty1 copy strains had a significant reduction in ROS detected by DHR compared to the zero-copy strains at day 3 when 30 mM HU was present in the YPD medium (Figure 7A). This difference diminished and was no longer significant at later time points. Cells grown in SC medium were sampled at comparable points during early stationary phase and early aging, but sampling was on different days to compensate for slightly slower growth in SC medium compared to YPD and a further reduction in growth rate in SC medium with HU (data not shown). High-copy Ty1 strains again had a significant reduction in ROS detected by DHR compared to the zero-copy strain on the initial day of sampling in SC medium alone (Figure 7B), but no significant differences were seen for either day of sampling of cells grown in SC with HU. No differences in ROS levels measured with DHR were noted between zero and high Ty1 copy strains grown at 30° in YPD medium with HU, consistent with the diminished lifespan extension observed with incubation at 30° (Figure 7C). Sampling was done on different days in the latter case to account for faster growth at 30°. We also observed a significant increase in fluorescence with DHE, which detects superoxide anions, in the zero Ty1 copy strain as populations neared their median lifespan in SC medium alone that was significantly diminished in the high Ty1 copy strains at the same time point (Figure 7D). The results of these ROS experiments show a correlation between media conditions in which high Ty1 copy strains have extended longevity and significant differences in ROS levels compared to the zero Ty1 copy strain.
Figure 7.
Reduced ROS levels in high Ty1 copy strains at early time points may contribute to lifespan extension. (A) Mean of normalized dihydrorhodamine (DHR), fluorescence obtained from at least four trials of zero Ty1 copy (blue columns) and high Ty1 copy (orange columns) strains after the indicated number of days in rich medium without or with 30 mM hydroxyurea, “YPD” or “YPD + HU,” respectively. Error bars indicate standard deviation, and **P < 0.01 compares zero and high Ty1 copy strains at the indicated day. (B) As for A for three trials using synthetic medium without or with 30 mM hydroxyurea, “SC” or “SC + HU,” respectively. *P < 0.05, comparing zero and high Ty1 copy strains. (C) As for A for five trials using YPD with 30 mM hydroxyurea and incubation at 30°. (D) Mean of normalized dihydroethidium (DHE), fluorescence obtained from three trials of zero Ty1 copy (blue columns) and high Ty1 copy (orange columns) strains in synthetic medium without or with 30 mM hydroxyurea, “SC” or “SC + HU,” respectively. *P < 0.05, comparing zero and high Ty1 copy strains.
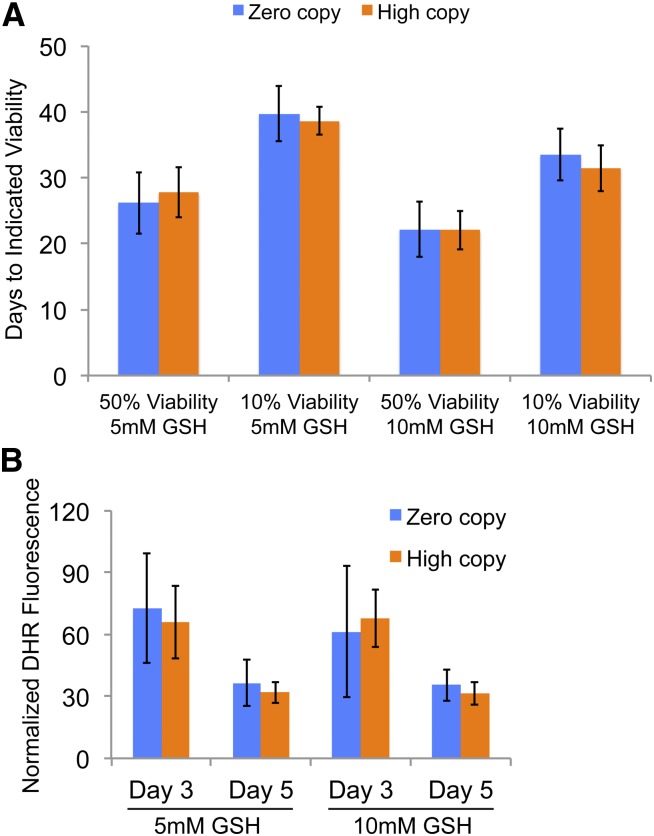
Zero and high Ty1 copy strains were grown and aged in the presence of reduced glutathione as an antioxidant to further explore the connection between ROS levels and extension of lifespan in high Ty1 copy strains. The median and maximum CLS of the zero Ty1 copy strain were virtually the same as for the high Ty1 copy strains in YPD medium with 30 mM HU and either 5 or 10 mM glutathione (Figure 8A). The moderate reduction in CLS of all strains when 10 mM glutathione was used indicates that excessive levels of glutathione may have negative effects on cells. The median and maximum CLS of the zero-copy strain in YPD with HU and 5 mM glutathione were not significantly different from the median and maximum CLS of the high Ty1 copy strains in YPD with HU only (Figure 2D; Figure 8A). This demonstrates that addition of glutathione to the medium increased the lifespan of the zero-copy strain so that it was equivalent to that of the high Ty1 copy strains in HU. Treatment with 5 mM glutathione led to a modest increase in median CLS of the high Ty1 copy strains (P < 0.05), but did not increase their maximum CLS in YPD with HU (Figure 2D; Figure 8A). This limited influence of glutathione is consistent with longevity-promoting changes in ROS accumulation occurring in the high-copy strains without exposure to an antioxidant. These data from glutathione exposure are consistent with changes in ROS contributing to the lifespan extension of high-copy Ty1 strains grown in YPD with HU. An overall comparison of the relative lifespan of high Ty1 copy strains compared to the zero-copy parent strain in glutathione-containing medium and all other media conditions tested is shown in Table 2.
Figure 8.
Lifespan of the zero Ty1 copy strain increases to that of high Ty1 copy strains in hydroxyurea with antioxidant treatment. (A) Median, “50% viability,” and maximum, “10% viability,” lifespans for zero Ty1 copy and high Ty1 copy strains (blue and orange columns, respectively) grown in YPD medium with 30 mM hydroxyurea and the indicated concentration of glutathione, “5 mM GSH” or “10 mM GSH.” Data represent means ± SDs of three to five trials. (B) Mean of normalized dihydrorhodamine (DHR), fluorescence obtained from at least three trials of zero Ty1 copy and high Ty1 copy (blue and orange columns, respectively) strains after the indicated number of days in rich medium with 30 mM hydroxyurea and the indicated concentration of glutathione, “5 mM GSH” or “10 mM GSH.” Error bars indicate standard deviation.
Table 2. Mean lifespan of high Ty1 copy strains as a percentage of mean lifespan of the zero Ty1 copy strain for each medium or growth condition.
| Medium/condition | Median lifespana | Maximum lifespanb |
|---|---|---|
| YPD | 104 | 109 |
| YPD + HU | 122* | 159** |
| YPD 30° | 106 | 107 |
| YPD 30° + HU | 106 | 116**c |
| YPD + threonine | 101 | 119 |
| YPD + threonine + HU | 122 | 137* |
| SC | 123* | 122* |
| SC + HU | 103 | 106 |
| YPD + 5 mM glutathione + HU | 106 | 97.2 |
| YPD + 10 mM glutathione + HU | 99.5 | 93.7 |
*P < 0.05 vs. zero Ty1 copy strain; **P < 0.01 vs. zero Ty1 copy strain.
Days to 50% viability.
Days to 10% viability.
Significantly reduced compared to high Ty1 copy strains in YPD + HU at 20° (P < 0.01).
DHR fluorescence was compared in zero and high Ty1 copy strains exposed to both HU and reduced glutathione to test whether ROS levels would be similar, since these strains had similar CLS. No significant differences were noted in ROS levels between these strains when grown and aged in YPD with HU and either 5 or 10 mM glutathione for 3 or 5 days (Figure 8B). This is in contrast to the results for YPD with HU and no glutathione (Figure 7A). These results further support the observation that conditions that increase CLS of the high Ty1 copy strains relative to the zero-copy strain are associated with changes in ROS between these strains, while conditions that do not increase CLS of the high Ty1 copy strains are associated with similar ROS levels between high and zero Ty1 copy strains.
Discussion
This work identifies an unanticipated positive contribution of yeast Ty1 elements to chronological lifespan through use of an exceptional yeast model system for studying retrotransposons. Much of the current focus on the possible contribution of retrotransposons to aging concerns the potential for these elements to negatively influence lifespan by promoting genome instability (St Laurent et al. 2010; Sedivy et al. 2013). We did not observe a negative influence of retrotransposition on lifespan of nondividing yeast cells, but rather, strains with many Ty1 copies and low retrotransposition levels survived longer in stationary phase when grown in rich medium with a low dose of the ribonucleotide reductase inhibitor HU or in synthetic medium. Lifespan extension was correlated with reduced ROS during early stationary phase, and antioxidant treatment allowed the strain with no Ty1 elements to live as long as the high Ty1 copy strains in rich medium with HU. The presence of many chromosomal Ty1 retrotransposons may indirectly or directly lead to cellular changes that affect ROS production and/or scavenging and promote chronological lifespan. The potential contribution of retrotransposons to aging could therefore be complex and should be investigated from a broad perspective.
The increased lifespan of all the S. paradoxus strains due to treatment with a low dose of HU is in contrast to a recent report of decreased CLS in S. cerevisiae strains treated with the same dose of HU (Weinberger et al. 2013). This discrepancy could be due to differences in methods, but we note that there are certain consistencies between our results and those of the previous report. First, S. cerevisiae cells exposed to 30 mM HU have substantially decreased reproductive potential (ability to form colonies on fresh medium) in stationary phase but no reduction in terminal cell density (Weinberger et al. 2013). We also observed no decrease in terminal cell density and a moderately lower density of colony forming units (lower reproductive potential) at the start of stationary phase with HU treatment (data not shown). Second, many cells exposed to HU in the previous study and in our work were in S or G2 stage of the cell cycle during stationary phase (Weinberger et al. 2013). Third, both our study and the previous study found that addition of excess threonine to buffer dNTP levels could extend lifespan of cells aging in the presence of HU (Hartman 2007; Weinberger et al. 2013). However, we found no loss of cell viability due to HU by direct staining of cells and longer maintenance of cell viability and reproductive potential in HU medium. An important difference between the studies is that we grew cells from a low initial density (5 × 103 cells/ml) to stationary phase, while the previous study used cultures inoculated at a higher initial density (5 × 107 cells/ml), so cells in our experiments spent more time actively growing in the presence of HU (Weinberger et al. 2013). We propose that a longevity-promoting effect resulting from exposure to a low dose of HU during exponential growth in our experiments overshadows a negative effect of HU on CLS due to replication stress during stationary phase.
Extended lifespan of high Ty1 copy strains was correlated with decreased ROS levels during early stationary phase. Changes in mitochondrial function and ROS levels during active growth that lead to decreased ROS levels during early stationary phase underlie yeast CLS extension due to deletion of the TOR1 gene (Bonawitz et al. 2007; Pan et al. 2011). TOR genes in diverse organisms encode kinases responsive to nutrient conditions and growth signaling that regulate processes such as translation, ribosome biogenesis, and autophagy to influence cell growth, metabolism, and aging (Bjedov and Partridge 2011). Functional mitochondria are required for increased CLS of yeast tor1∆ mutants (Bonawitz et al. 2007). These mutants exhibit elevated mitochondrial membrane potential and mitochondrial superoxide during active growth, but reduced mitochondrial membrane potential, superoxide, and ROS detected by DHR in early stationary phase (Pan et al. 2011). Overexpression of the mitochondrial superoxide dismutase, SOD2, prevents lifespan extension in tor1∆ mutants. Exposure to a low dose of the superoxide-generating reagent menadione during exponential growth of wild-type cells causes lifespan, ROS, and mitochondrial changes similar to those of tor1∆ mutants (Pan et al. 2011). These observations indicate that ROS changes during active growth and early stationary phase are important for mediating lifespan extension. The ROS changes we observed in high Ty1 copy strains could reflect similar changes in mitochondrial function or ROS production/scavenging that contribute to lifespan extension in these S. paradoxus strains. How the presence, expression, and/or mobility of Ty1 elements could indirectly or directly produce such changes is a topic that we are actively investigating.
Our findings do not rule out potential negative effects of retrotransposons on lifespan in other contexts or aging models, including replicative aging of actively dividing yeast mother cells (Kaeberlein 2010). Retrotransposon expression increases with age in gonads of C. elegans, brain tissue of D. melanogaster, normal human cells maintained ex vivo, multiple mouse tissues, and yeast mother cells (Wang et al. 2011; Dennis et al. 2012; De Cecco et al. 2013a,b; Li et al. 2013; Hu et al. 2014). Increased mobility of retrotransposons occurs at late time points during yeast chronological lifespan and in brains of aged D. melanogaster (Maxwell et al. 2011; Li et al. 2013). Increased copy numbers of retrotransposons in human cells aged ex vivo, somatic tissues from aged mice, and during yeast replicative aging have also been interpreted as signs of increased retrotransposition (De Cecco et al. 2013a,b; Hu et al. 2014). These correlations have led to proposals that retrotransposons might promote the aging process and/or genome instability during aging. The S. paradoxus Ty-less strain background has allowed us to carefully address such a potential role during chronological aging of haploid yeast strains in certain media conditions. Whether Ty1 elements could negatively influence CLS in the context of other lifespan altering mutations or treatments, and whether they have a negative influence on the lifespan of dividing cells will require further study.
Additionally, mutation rate of a representative gene was not increased due to elevated retrotransposition in our haploid strains in young populations, in contrast with previous work showing that retrotransposition was correlated with loss of heterozygosity and chromosome loss during chronological aging of diploid S. cerevisiae strains (Maxwell et al. 2011). The difference in these results could be due to the use of different cell types (haploids vs. diploids), cells of different ages, or to the different measures of genome instability. While high levels of retrotransposition did not negatively influence CLS, it remains possible that some forms of genome instability are elevated in haploid cells during chronological aging due to retrotransposition. Furthermore, while mutations and chromosome rearrangements accumulate during yeast chronological aging (Madia et al. 2007), additional research is needed to understand how this relates to the progressive loss of viability during stationary phase.
This work also provides a new perspective on the relationship between ROS and retrotransposons. Prior work identified increased mobility of yeast Ty1 and mammalian L1 elements in response to oxidative stress and that activation of Ty1 elements by the DNA-damaging agent MMS requires mitochondrial function and ROS production (Stoycheva et al. 2010; Giorgi et al. 2011). Regulation of retrotransposon expression and mobility by numerous stresses could have consequences for cell function and survival in stressful conditions (Capy et al. 2000; Slotkin and Martienssen 2007). However, our findings show that retrotransposons can indirectly or directly alter ROS levels in certain contexts, indicating that these elements might influence cellular responses to stress. Dispersed copies of retrotransposons that are subject to chromatin modifications and that initiate transcripts that can read through into neighboring genes may alter gene expression patterns (Slotkin and Martienssen 2007), which could affect cellular stress responses. Chimeric transcripts containing retrotransposon sequences and sequences of other genes are observed in human cells, including tumor cells, and can regulate gene expression (Belancio et al. 2010; Cruickshanks et al. 2013). Chromatin marks on mammalian short-interspersed nuclear element (SINE) retrotransposons can change in response to stress (Hunter et al. 2012), and RNA from these elements can directly interact with RNA polymerase to influence gene expression in response to heat shock (Ponicsan et al. 2010). Yeast Ty1 elements can alter transcription of neighboring genes and establish stress-responsive patterns of expression on genes (Lesage and Todeschini 2005; Servant et al. 2008). However, the results from growth at 30° are consistent with the expression and activity of Ty1 proteins directly or indirectly influencing lifespan and ROS, since substantial Ty1 transcription occurs at 30° (Curcio et al. 1990; Lawler et al. 2002). Also, the different patterns of Ty1 insertions in the high Ty1 copy strains indicate that it is unlikely that the same gene or genes would have altered expression due to neighboring Ty1 sequences in all the high-copy strains. Ty1 Gag protein forms the Ty1 virus-like particle and Ty1 Pol protein has protease, reverse transcriptase, and integrase activities (Beauregard et al. 2008). One or more of these activities might indirectly alter stress-response pathways or, in the case of reverse transcriptase or integrase, influence DNA metabolism. An alternative possibility is that Ty1 protein expression/function (or moderate levels of Ty1 mobility) is needed in combination with transcriptional effects on gene expression for lifespan extension.
Overall, a longevity-promoting role for retrotransposons in yeast chronological lifespan in certain contexts provides a new point of view for investigating the relationship between these elements and aging. The advantages of the S. paradoxus Ty-less model offer a unique opportunity to directly test for additional roles of retrotransposons during aging. Continued work in this system will likely produce findings relevant to aging and transposable elements in many species, considering common aspects of regulation and impacts of retrotransposons in diverse species, as well as fundamental similarities between aging in budding yeast and other eukaryotes.
Acknowledgments
The authors thank David Garfinkel for generously providing the Ty-less S. paradoxus parent strain and members of the Maxwell lab for helpful comments. We also thank Meredith Giblin, Alison Kenny, and Andrew Peifer for technical assistance. This work was supported in part by grant R00AG031911 from the National Institute on Aging to P.H.M. Content of this article does not necessarily represent official views of the National Institutes of Health.
Footnotes
Communicating editor: J. Schimenti
Literature Cited
- Abeysinghe S. S., Chuzhanova N., Krawczak M., Ball E. V., Cooper D. N., 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 22: 229–244 [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N., 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2005 Edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Beauregard A., Curcio M. J., Belfort M., 2008. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42: 587–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V. P., Deininger P. L., Roy-Engel A. M., 2009. LINE dancing in the human genome: transposable elements and disease. Genome Med 1: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V. P., Roy-Engel A. M., Deininger P. L., 2010. All y’all need to know ’bout retroelements in cancer. Semin. Cancer Biol. 20: 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I., Partridge L., 2011. A longer and healthier life with TOR down-regulation: genetics and drugs. Biochem. Soc. Trans. 39: 460–465 [DOI] [PubMed] [Google Scholar]
- Bonawitz N. D., Chatenay-Lapointe M., Pan Y., Shadel G. S., 2007. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Weinberger M., 2012. DNA damageDNA damage and DNA replication stressDNA replication stress in yeast models of aging. Subcell. Biochem. 57: 187–206 [DOI] [PubMed] [Google Scholar]
- Burtner C. R., Murakami C. J., Kennedy B. K., Kaeberlein M., 2009. A molecular mechanism of chronological aging in yeast. Cell Cycle 8: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P., Gasperi G., Biémont C., Bazin C., 2000. Stress and transposable elements: Co-evolution or useful parasites? Heredity (Edinb) 85(Pt 2): 101–106 [DOI] [PubMed] [Google Scholar]
- Cruickshanks H. A., Vafadar-Isfahani N., Dunican D. S., Lee A., Sproul D., et al. , 2013. Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res. 41: 68557–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Hedge A. M., Boeke J. D., Garfinkel D. J., 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220: 213–221 [DOI] [PubMed] [Google Scholar]
- Curcio M. J., Kenny A. E., Moore S., Garfinkel D. J., Weintraub M., et al. , 2007. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol. Cell. Biol. 27: 8874–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Criscione S. W., Peckham E. J., Hillenmeyer S., Hamm E. A., et al. , 2013a Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Criscione S. W., Peterson A. L., Neretti N., Sedivy J. M., et al. , 2013b Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany, N.Y. Online) 5: 867–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis S., Sheth U., Feldman J. L., English K. A., Priess J. R., 2012. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8: e1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr L. K., Strathern J. N., Garfinkel D. J., 1991. RNA-mediated recombination in S. cerevisiae. Cell 67: 355–364 [DOI] [PubMed] [Google Scholar]
- Devine S. E., Boeke J. D., 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633 [DOI] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T., 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367 [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Liou L. L., Moy V. N., Diaspro A., Valentine J. S., et al. , 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409: 195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Mastrangelo M. F., Sanders N. J., Shafer B. K., Strathern J. N., 1988. Transposon tagging using Ty elements in yeast. Genetics 120: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Nyswaner K., Wang J., Cho J. Y., 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165: 83–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Nyswaner K. M., Stefanisko K. M., Chang C., Moore S. P., 2005. Ty1 copy number dynamics in Saccharomyces. Genetics 169: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S. L., Wakeman T. P., Xu B., Deininger P. L., 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., Moran J. V., 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325 [DOI] [PubMed] [Google Scholar]
- Giorgi G., Marcantonio P., Del Re B., 2011. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 346: 383–391 [DOI] [PubMed] [Google Scholar]
- Haigis M. C., Yankner B. A., 2010. The aging stress response. Mol. Cell 40: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. M., Ma C. X., Liang P., Singh K. K., 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H., 2012. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J. L., 2007. Buffering of deoxyribonucleotide pool homeostasis by threonine metabolism. Proc. Natl. Acad. Sci. USA 104: 11700–11705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Chen K., Xia Z., Chavez M., Pal S., et al. , 2014. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 28: 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R. G., Murakami G., Dewell S., Seligsohn M., Baker M. E., et al. , 2012. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc. Natl. Acad. Sci. USA 109: 17657–17662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., 2010. Lessons on longevity from budding yeast. Nature 464: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood T. B., Kowald A., 2012. The free-radical theory of ageing–older, wiser and still alive: modelling positional effects of the primary targets of ROS reveals new support. BioEssays 34: 692–700 [DOI] [PubMed] [Google Scholar]
- Lawler J. F., Haeusser D. P., Dull A., Boeke J. D., Keeney J. B., 2002. Ty1 defect in proteolysis at high temperature. J. Virol. 76: 4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. S., Bi L., Garfinkel D. J., Bailis A. M., 2000. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol. Cell. Biol. 20: 2436–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P., Todeschini A. L., 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110: 70–90 [DOI] [PubMed] [Google Scholar]
- Li W., Prazak L., Chatterjee N., Grüninger S., Krug L., et al. , 2013. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 16: 529–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D., Shadel G. S., Kaeberlein M., Kennedy B., 2012. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 16: 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F., Gattazzo C., Fabrizio P., Longo V. D., 2007. A simple model system for age-dependent DNA damage and cancer. Mech. Ageing Dev. 128: 45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Curcio M. J., 2007. Retrosequence formation restructures the yeast genome. Genes Dev. 21: 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Coombes C., Kenny A. E., Lawler J. F., Boeke J. D., et al. , 2004. Ty1 mobilizes subtelomeric Y’ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 24: 9887–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Burhans W. C., Curcio M. J., 2011. Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. USA 108: 20376–20381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A., Weinberger M., Silva A., Sampaio-Marques B., Almeida B., et al. , 2010. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc. Natl. Acad. Sci. USA 107: 15123–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. P., Liti G., Stefanisko K. M., Nyswaner K. M., Chang C., et al. , 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21: 649–660 [DOI] [PubMed] [Google Scholar]
- Moskalev A. A., Shaposhnikov M. V., Plyusnina E. N., Zhavoronkov A., Budovsky A., et al. , 2013. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res. Rev. 12: 661–684 [DOI] [PubMed] [Google Scholar]
- Pan Y., Schroeder E. A., Ocampo A., Barrientos A., Shadel G. S., 2011. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 13: 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C. E., Williamson V. M., 1984. Temperature effects on the rate of ty transposition. Science 226: 53–55 [DOI] [PubMed] [Google Scholar]
- Ponicsan S. L., Kugel J. F., Goodrich J. A., 2010. Genomic gems: SINE RNAs regulate mRNA production. Curr. Opin. Genet. Dev. 20: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht C., Voet T., Esteki M. Z., Nowakowska B. A., Vermeesch J. R., 2013. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 23: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwood L., Felix K., Janz S., 2004. Elevated presence of retrotransposons at sites of DNA double strand break repair in mouse models of metabolic oxidative stress and MYC-induced lymphoma. Mutat. Res. 548: 117–125 [DOI] [PubMed] [Google Scholar]
- Sedivy J. M., Kreiling J. A., Neretti N., De Cecco M., Criscione S. W., et al. , 2013. Death by transposition: The enemy within? BioEssays 35: 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G., Pennetier C., Lesage P., 2008. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol. Cell. Biol. 28: 5543–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R., 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285 [DOI] [PubMed] [Google Scholar]
- St Laurent G., IIIHammell N., McCaffrey T., 2010. A LINE-1 component to human aging: Do LINE elements exact a longevity cost for evolutionary advantage? Mech. Ageing Dev. 131: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoycheva T., Pesheva M., Venkov P., 2010. The role of reactive oxygen species in the induction of Ty1 retrotransposition in Saccharomyces cerevisiae. Yeast 27: 259–267 [DOI] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Geesman G. J., Hostikka S. L., Atallah M., Blackwell B., et al. , 2011. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 10: 3016–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M., Feng L., Paul A., Smith D. L., Hontz R. D., et al. , 2007. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS ONE 2: e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M., Mesquita A., Caroll T., Marks L., Yang H., et al. , 2010. Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence. Aging (Albany, N.Y. Online) 2: 709–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M., Sampaio-Marques B., Ludovico P., Burhans W. C., 2013. DNA replication stress-induced loss of reproductive capacity in S. cerevisiae and its inhibition by caloric restriction. Cell Cycle 12: 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan W. L., Gocke E., Manney T. R., 1979. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics 91: 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. G., Helfand S. L., 2013. Chromatin structure and transposable elements in organismal aging. Front. Genet. 4: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]