Abstract
In multicellular organisms, genetic programs guide cells to adopt cell fates as tissues are formed during development, maintained in adults, and repaired after injury. Here we explore how a small molecule in the environment can switch a genetic program from one fate to another. Wild-type Caenorhabditis elegans XX adult hermaphrodites make oocytes continuously, but certain mutant XX adults make sperm instead in an otherwise hermaphrodite soma. Thus, puf-8; lip-1 XX adults make only sperm, but they can be switched from sperm to oocyte production by treatment with a small-molecule MEK inhibitor. To ask whether this chemical reprogramming is common, we tested six XX sperm-only mutants, but found only one other capable of cell fate switching, fbf-1; lip-1. Therefore, reprogramming competence relies on genotype, with only certain mutants capable of responding to the MEK inhibitor with a cell fate change. To gain insight into the molecular basis of competence for chemical reprogramming, we compared polyadenylated transcriptomes of competent and noncompetent XX sperm-only mutants in the absence of the MEK inhibitor and hence in the absence of cell fate reprogramming. Despite their cellular production of sperm, competent mutants were enriched for oogenic messenger RNAs relative to mutants lacking competence for chemical reprogramming. In addition, competent mutants expressed the oocyte-specific protein RME-2, whereas those lacking competence did not. Therefore, mutants competent for reprogramming possess an intersexual molecular profile at both RNA and protein levels. We suggest that this intersexual molecular signature is diagnostic of an intermediate network state that poises the germline tissue for changing its cellular fate in response to environmental cues.
Keywords: sex determination, germline, PUF protein, MAPK signaling, network states
CELLS in multicellular organisms acquire specific fates, either during development as tissues are generated or during adulthood as tissues are renewed or repaired. A classical view holds that during development, a cell follows a specific genetic program to determine its differentiated fate, but more recent studies reveal that cells can be induced to switch from one genetic program to another. Such “reprogramming” of a cell fate program can occur in vivo in cancer (Friedl and Alexander 2011; Byun and Gardner 2013), wound healing (Wong et al. 2013), and regeneration (Xin et al. 2013; Ziv et al. 2013). Moreover, reprogramming can be stimulated by introduction of transcription factors (Takahashi and Yamanaka 2006) or small molecules (Morgan et al. 2010; Zhu et al. 2010; Li et al. 2014). Here we explore the effect of genotype on the capacity for a particular case of chemically induced cell fate reprogramming within an organism.
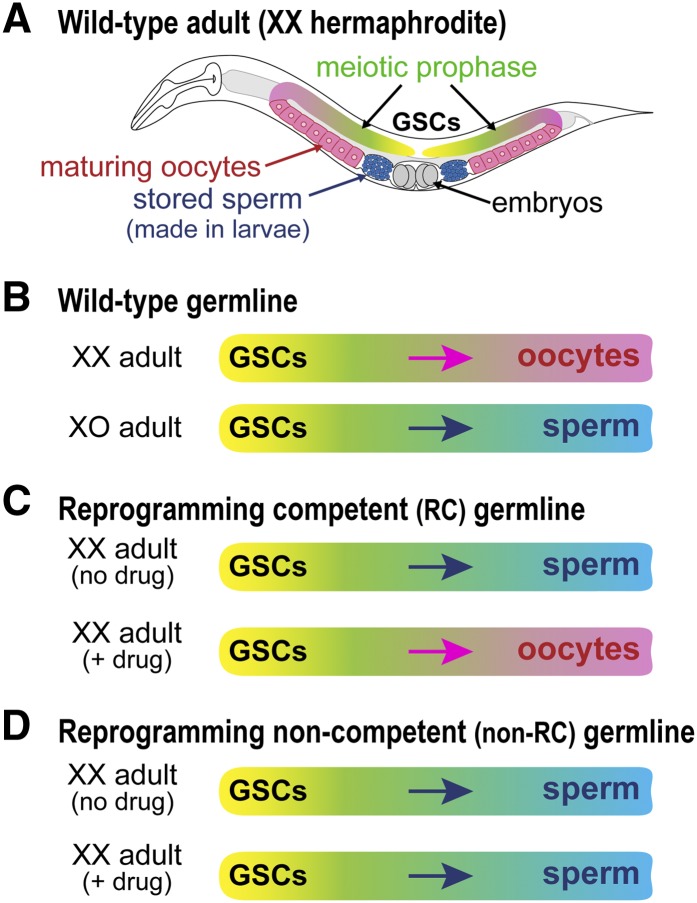
The Caenorhabditis elegans germline provides a tractable model for analyses of cell fate reprogramming. The adult germline is an elongate tissue with ∼2000 cells, including germline stem cells (GSCs) (Figure 1A, yellow). GSCs continuously generate daughter cells that enter the meiotic cell cycle (Figure 1A, green) and then differentiate as sperm (Figure 1A, blue) or oocytes (Figure 1A, pink). Wild-type adults generate only one type of gamete: XX adult hermaphrodites make only oocytes, and XO adult males make only sperm (Figure 1B). However, wild-type adults of either sex can be induced to reprogram their germline sex to produce the opposite gamete. Using either temperature-sensitive mutants or RNA interference (RNAi) in key germline sex determination genes, XX oogenic adults can be triggered to make sperm (Barton et al. 1987; Otori et al. 2006), and conversely XO spermatogenic adults can be triggered to make oocytes (Barton and Kimble 1990; Schedin et al. 1994; Chen et al. 2000).
Figure 1.
Chemical reprogramming of germline sex in C. elegans adults. (A) XX hermaphrodite adult, with germline tissue (color) housed in two U-shaped gonadal arms. Germline stem cells (GSCs), yellow; meiotic prophase, green; sperm, blue; oocytes, pink. XX hermaphrodites make sperm transiently as larvae and store them for fertilization of oocytes in adults. (B) Wild-type XX adult germlines make oocytes only (top), whereas XO adult germlines make sperm only (bottom). (C) Reprogramming competent XX adult germlines make sperm only in the absence of U0126, an inhibitor of MEK kinase (“no drug”) (top), but are reprogrammed to produce oocytes in the presence of U0126 (“+ drug”) (bottom) (Morgan et al. 2010). (D) Reprogramming noncompetent XX adult germlines make only sperm with or without drug treatment.
Reprogramming of germline sex using a small molecule supplied in the environment was recently achieved in C. elegans XX “sperm-only” mutant adults—animals with a masculinized germline in an otherwise hermaphrodite (essentially female) soma (Morgan et al. 2010) (Figure 1C). In this context, we use the term reprogramming to mean a switch from the genetic program driving production of sperm to that driving production of oocytes. This chemical reprogramming takes advantage of the discovery that MPK-1, the C. elegans homolog of extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) (Lackner et al. 1994), normally promotes the sperm fate at the expense of oogenesis (Lee et al. 2007b). Chemical MEK inhibitors (e.g., U0126) can induce oogenesis in XX puf-8; lip-1 sperm-only adults (Figure 1C) (Morgan et al. 2010). This reprogramming is efficient and fast: virtually all adults (>99%) switch from sperm to oocyte production within 24 hr (Morgan et al. 2010). Moreover, U0126 reduces ERK/MAPK activity within 15 min and abolishes expression of a sperm fate regulator, FOG-1, within 2 hr (Morgan et al. 2013). Importantly, the reprogrammed animals are self-fertile, which means that sperm made in the absence of reprogramming and oocytes made in the presence of reprogramming are both functional. The high penetrance and speed of this case of chemical cell fate reprogramming makes it particularly tractable.
The capacity of puf-8; lip-1 adults to respond to the MEK inhibitor by switching gamete sex fits the classical definition of developmental “competence,” which is defined as the ability to respond in a specific manner to a given stimulus (Gilbert 1997, p. 628). How might competence for chemical reprogramming be conferred in XX puf-8; lip-1 mutants? PUF-8 belongs to the Pumilio and FBF (PUF) family of RNA-binding proteins, which bind regulatory elements in the 3′-untranslated regions of target messenger RNAs (mRNAs) (Wickens et al. 2002). PUF proteins are best known for repressing their targets, either by decreasing mRNA stability or by inhibiting translation (Wickens et al. 2002; Friend et al. 2012). Indeed, PUF-8 associates with at least 160 germline mRNAs and represses most tested (Mainpal et al. 2011). Therefore, PUF-8 removal is predicted to relieve target mRNAs from repression. In puf-8 single mutants, XX adults are typically oogenic as in wild type, but XX adults with a masculinized sperm-only germline occur at low penetrance (<3%), suggesting a mild defect in the genetic network regulating gamete sex (Bachorik and Kimble 2005). This low penetrance masculinization is dramatically enhanced by loss of lip-1 activity: XX puf-8; lip-1 double mutants have a fully penetrant sperm-only phenotype (Morgan et al. 2010). LIP-1 is a homolog of dual-specificity phosphatases (Camps et al. 2000) and an upstream inhibitor of MPK-1 (Berset et al. 2001). Therefore, LIP-1 removal results in hyperactivation of ERK/MAPK, although it has no sexual fate defect of its own (Hajnal and Berset 2002). A simple explanation of why puf-8; lip-1 mutants are competent for reprogramming is that the MEK inhibitor reverses the lip-1 enhancer phenotype.
Here we investigate the genetic and molecular basis of this example of chemical reprogramming. Among a battery of XX sperm-only adults, we identified one more mutant (fbf-1; lip-1) that was competent for chemical reprogramming and others that were not competent. In contrast to puf-8; lip-1 and fbf-1; lip-1 double mutants, puf-8fbf-1; lip-1 triple mutants are not competent for reprogramming. Therefore, reprogramming competence relies on the presence of one of the two PUF proteins, PUF-8 or FBF-1. In the absence of the MEK inhibitor, RNA-Seq revealed an intermediate transcriptional signature of sperm- and oocyte-specific mRNAs in both puf-8; lip-1 and fbf-1; lip-1 reprogramming competent mutants, but not in mutants lacking competence. Moreover, sperm- and oocyte-specific proteins were expressed in reprogramming competent germlines, but not in those lacking competence. Therefore, competence for chemical reprogramming correlates with an intersexual molecular profile at both the RNA and protein levels. In the presence of drug, RNA-Seq identified no significant differences during the process of reprogramming, but it did reveal a dramatic change once overt differentiation had begun. We present a model for how the puf-8; lip-1 and fbf-1; lip-1 genotypes confer competence for chemical reprogramming and suggest that an intersexual molecular profile is diagnostic of an intermediate state in the gamete specification network.
Materials and Methods
C. elegans strains and assay for reprogramming competence
Strains used were JK4662, puf-8(q725)/ mIn1[mIs14dpy-10(e128)] II; lip-1(zh15) IV; JK3198, fbf-1(ok91)/ mIn1[mIs14dpy-10(e128)] II; lip-1(zh15) IV; JK3228: fbf-1(ok91) puf-8(q725)/ mIn1[mIs14dpy-10(e128)] II; JK4612, fem-3(q20ts,gf) IV; JK4888, mog-1(q151) III/ hT2[qIs48](I;III); and JK5021, puf-8(q725) fbf-1(ok91)/ mIn1[mIs14dpy-10(e128)] II; lip-1(zh15) IV. Bristol isolate N2 was used as wild type.
All strains were maintained at 20° except JK4612, which was maintained at 15° but shifted to 25° for reprogramming assays. Worms were synchronized as L1s and grown to adulthood (24 hr past mid-L4 larval stage), using standard methods. To test for U0126 reprogramming competence, we treated XX adults for 24 hr with either DMSO or 100 µM U0126 and then scored embryo production over the next 2 days as readout of oocyte production. Mutants competent for reprogramming made functional oocytes and therefore made embryos.
Generation of biological replicates and RNA extraction
To obtain transcriptomes of adults either competent or not competent for reprogramming, three biological replicates were collected for each relevant genotype, with 50–100 young adults per replicate. Because XX sperm-only adults are self-sterile, homozygotes were picked from balanced strains, all raised at 20° on OP50-seeded petri plates. Worms were washed, pelleted, placed in TRIzol (Invitrogen, Carlsbad, CA), and flash frozen in liquid nitrogen. RNA was extracted using the manufacturer’s protocol (Invitrogen, Carlsbad, CA). RNA was further purified over an RNeasy Micro column (QIAGEN, Valencia, CA) and eluted in DEPC-treated water. RNA purity and integrity were assessed using the RNA PicoChip assay (Agilent, Santa Clara, CA).
To obtain transcriptomes during the process of reprogramming, animals were treated with U0126 or DMSO for specific time intervals and two biological replicates of each condition and time point were collected and processed as described above.
Whole-genome libraries and Illumina-platform sequencing
DNA libraries were prepared from 1 μg of total RNA, using the mRNA Sequencing Sample Preparation Kit (Roche, Indianapolis, IN), according to manufacturer’s protocol. Subsequently, cDNA was purified using the E-Gel SizeSelect 2% Agarose gel (Invitrogen). Purified cDNA was enriched through 18 PCR cycles and then run over a ZymoGen Clean & Concentrator Column (Zymo Research). Libraries were sequenced on the Illumina GAIIx system (Roche).
Short read alignment
Reads were mapped using TopHat version 1.4.0 (Trapnell et al. 2009), with a custom build of the C. elegans poly(A)+ transcriptome (WS220) as the primary reference sequence and the C. elegans genome (WS220) as a secondary reference. TopHat options used were -m 2, -G, -g 1.
Differential expression analysis
To test for significant differences in mRNA abundances we used DESeq (Anders and Huber 2010). mRNA counts were determined from alignment files, using scripts that came with the DESeq package and both WormBase 220 and modENCODE gene models. We performed pairwise tests of differential expression between untreated mutants that were either reprogramming competent or not competent. We also tested for significant differences in mRNA abundances over a time course of reprogramming, using the generalized linear model function in DESeq with time and treatment type as main factors. To test for differential exon expression we used DEXSeq (version 1.2.0) (Anders et al. 2012). For all tests, we used a false discovery rate (FDR) cutoff of <0.01 and a minimum fold change requirement of twofold and excluded transcripts with <10 read counts.
Bioinformatic analyses
The DAVID database (Huang et al. 2009) was used to identify enriched GO categories. The significance threshold was set at FDR < 0.005. Significant terms were calculated separately for up- and downregulated mRNAs, with the entire genome used as the reference set.
modENCODE sequencing data for >100 conditions were obtained (www.modencode.org) (Gerstein et al. 2010). Transcription factor targets for all modENCODE-derived experiments were identified using the filter that a binding site had to be within 1 kb upstream of a transcription start site. Hypergeometric distribution testing for significant overlaps was performed using custom scripts.
Variance-stabilized count data from DESeq were used as input for all clustering and heat maps. Gene expression data were clustered and visualized with Cluster 3.0 and Java TreeView (Eisen et al. 1998). Principal component analysis was performed in R, using scaled and normalized expression data for the 485 mRNAs that were differentially expressed between reprogramming competent and noncompetent animals (FDR < 0.01). Fourteen samples were used, comprising 12 sperm-only replicates from both reprogramming competent and noncompetent animals, 1 oocyte-only replicate from wild-type XX hermaphrodites, and 1 sperm-only replicate from wild-type XO males. Principal components 1 and 2 (PC1 and PC2) captured almost all of the variations in gene expression and so we graphed PC1 and PC2 on a two-dimensional plot.
Motif analysis was performed first with MEME (Bailey et al. 2006) and second with custom scripts to identify PUF-8 and FBF consensus binding sites within C. elegans 3′-UTR sequences from BioMart (www.biomart.org).
Antibody staining
Antibody staining followed standard protocols (Lee et al. 2006). Primary antibodies were anti-RME-2 (used at a 1:500 dilution), anti-SP56 (used at a 1:100 dilution), anti-OMA-2 (1:500), anti-SPE-44 (1:200), and anti-GLD-1 (1:100). Affinity-purified secondary antibodies were animal Cy3-labeled donkey anti-rabbit IgG and FITC-labeled donkey anti-mouse IgG (Jackson Immunoresearch Laboratories, both used at a 1:500 dilution). Compound fluorescent images were taken with a Zeiss Axioscope.
RT–quantitative PCR validation
RT–quantitative PCR (RT-qPCR) was used to validate RNA-Seq data. Two hundred nanograms of mRNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen), with oligo(dT) priming. qPCR was carried out using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) and TaqMan Universal PCR Master Mix (Applied Biosystems) in a 7500 Fast Real-Time PCR System (Applied Biosystems). rps-25 was used as the endogenous control for normalization.
Data availability
Raw reads and mRNA expression data are available at the Gene Expression Omnibus under accession no. GSE54030.
Results
XX sperm-only adults differ in reprogramming competence
To ask whether competence for U0126 gamete sex reprogramming is a common feature of XX sperm-only adults, we tested a battery of mutants, all with a fully penetrant phenotype of germline masculinization: puf-8fbf-1 (Bachorik and Kimble 2005), fem-3(gf) (Barton et al. 1987), mog-1 (Graham and Kimble 1993), fbf-1; lip-1 (this work), and puf-8fbf-1; lip-1 (this work). As a positive control, we also assayed puf-8; lip-1, which was previously identified as competent for chemical reprogramming (Morgan et al. 2010). Synchronized adults of each genotype were treated for 24 hr with either 100 µM U0126 or the DMSO vehicle, or they were not treated and kept on OP50-seeded plates; all adults were then assayed for production of embryos, a hallmark of the switch from making functional sperm to functional oocytes (see Materials and Methods). Only one additional reprogramming competent genotype was found, fbf-1; lip-1; the others were not competent for chemical reprogramming, as assayed by embryo production and DIC inspection for oogenesis (Table 1 and Figure 1D). Therefore, only certain genotypes are competent for this chemical reprogramming, a conclusion limited by the caveat that our protocol uses a hypomorphic level of MEK inhibition.
Table 1. Tests of XX sperm-only mutants for reprogramming competence.
| Genotypea | Treatment | % making oocytesb | nc |
|---|---|---|---|
| Wild type | None | 100 | NA |
| lip-1 | None | 100 | NA |
| puf-8 | None | 97 | NA |
| fbf-1 | None | 99 | NA |
| puf-8; lip-1 | None | 0 (± 0) | 135 |
| DMSO | 0 (± 0) | 85 | |
| U0126 | 97 (± 6.3) | 121 | |
| fbf-1; lip-1 | None | 0 (± 0) | 39 |
| DMSO | 0 (± 0) | 29 | |
| U0126 | 69.7 (± 4.8) | 131 | |
| puf-8 fbf-1 | None | 0 (± 0) | 609 |
| DMSO | 0 (± 0) | 641 | |
| U0126 | 0 (± 0) | 658 | |
| puf-8 fbf-1; lip-1 | None | 0 (± 0) | 70 |
| DMSO | 0 (± 0) | 190 | |
| U0126 | 0 (± 0) | 42 | |
| fem-3(gf) | None | 0 (± 0) | 193 |
| DMSO | 0 (± 0) | 659 | |
| U0126 | 0 (± 0) | 1080 | |
| mog-1 | None | 0 (± 0) | 159 |
| DMSO | 0 (± 0) | 321 | |
| U0126 | 0 (± 0) | 284 |
Alleles: puf-8(q725), fbf-1(ok91), lip-1(zh15), fem-3(q20), and mog-1(q151).
Standard deviation from mean is in parentheses.
n, number animals scored. NA, not applicable if numbers cited are from literature, with references as follows: wild type, Brenner (1974); lip-1, Hajnal and Berset (2002); puf-8, Bachorik and Kimble (2005); and fbf-1, Crittenden et al. (2002).
Both genotypes competent for chemical reprogramming harbored the lip-1 mutation plus a mutation in either of two PUF protein-encoding genes, puf-8 or fbf-1. However, puf-8fbf-1 double mutants and puf-8fbf-1; lip-1 triple mutants could not be chemically reprogrammed. Therefore, either puf-8 or fbf-1 is required for reprogramming competence, and puf-8fbf-1 double mutants could not be further enhanced by lip-1 or suppressed by U0126. We suggest that removal of either puf-8 or fbf-1 sensitizes the gamete fate regulatory network to perturbations in ERK status, but that removal of both genes abolishes that sensitization (see Discussion). Identification of mutants with distinct capacities for chemical reprogramming allowed us to investigate their transcriptomes for differences that correlate with reprogramming competence in the absence of drug and differences that occur during reprogramming in the presence of drug.
puf-8; lip-1 and puf-8 fbf-1; lip-1 animals have distinct transcriptomes
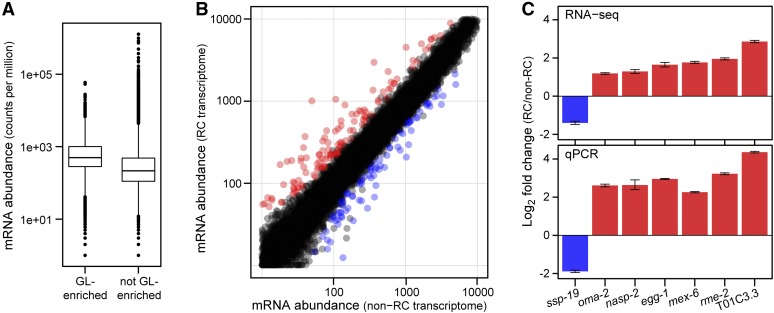
The identification of genotypically distinct XX adults that either were or were not capable of chemical reprogramming allowed us to investigate the molecular basis of competence for U0126 reprograming. To that end, we first analyzed the polyadenylated transcriptomes of puf-8; lip-1 reprogramming competent and puf-8fbf-1; lip-1 reprogramming noncompetent adults in the absence of MEK inhibition and hence in the absence of reprogramming (see Materials and Methods). These two mutants were phenotypically indistinguishable: both were XX adults with a fully penetrant masculinized germline. Their genotypes differed only at the fbf-1 locus so this analysis identified FBF-1-dependent transcriptome changes. For each of three replicates per genotype, 33 million reads on average mapped to unique locations in the C. elegans genome (Supporting Information, Table S1). We detected 16,313 mRNAs overall, corresponding to 77% of annotated C. elegans genes. These RNAs included >98% of germline-enriched RNAs from each of two prior studies: 3090 of 3143 mRNAs (Reinke et al. 2004) and 1049 of 1063 mRNAs (Wang et al. 2009). Moreover, germline-enriched mRNAs were typically more abundant in our data set than non-germline-enriched mRNAs and had a broad range of expression (Figure 2A). The puf-8; lip-1 and puf-8fbf-1; lip-1 transcriptional profiles were compared using DESeq (see Materials and Methods). A total of 219 mRNAs were differentially expressed (FDR < 0.01; twofold cutoff), with 135 transcripts more abundant in puf-8; lip-1 animals and 84 transcripts less abundant in puf-8; lip-1 animals (Figure 2B; File S1). Differential expression was validated using qPCR (100%; n = 7) (Figure 2C). By Gene Ontology (GO) analysis, the 135 mRNAs increased in puf-8; lip-1 were enriched for the terms sexual reproduction and gamete generation (Table S2), whereas the 84 mRNAs reduced in puf-8; lip-1 were not significantly enriched for any terms. We conclude that puf-8; lip-1 and puf-8fbf-1; lip-1 animals have distinct transcriptomes despite their common cellular morphology and gamete fate specification.
Figure 2.
Differential expression between puf-8; lip-1 reprogramming competent and puf-8 fbf-1; lip-1 reprogramming noncompetent transcriptomes. (A) Boxplots of “germline-enriched” (left) and “not germline-enriched” (right) mRNAs found in puf-8; lip-1 plus puf-8 fbf-1; lip-1 transcriptomes. Each boxplot shows the average counts per million for all mRNAs, normalized by library size. Sorting into the two categories was done following the classification of Reinke et al. (2004). (B) Differentially expressed mRNAs in puf-8; lip-1 (reprogramming competent, “RC”) vs. puf-8 fbf-1; lip-1 (noncompetent, “non-RC”) transcriptomes. mRNA abundance is shown as normalized counts per million. A total of 219 mRNAs were differentially expressed in RC vs. non-RC (FDR < 0.01 with twofold change or more), with 135 RC-increased mRNAs (red) and 84 RC-reduced mRNAs (blue). mRNAs not differentially expressed are black. (C) RNA-Seq fold changes (top) compared to qPCR fold changes (bottom). In both, puf-8; lip-1 mRNA levels were normalized to levels in puf-8 fbf-1; lip-1. Error bars represent standard deviation from the mean.
Germline and sex specificity of mRNAs associated with reprogramming competence
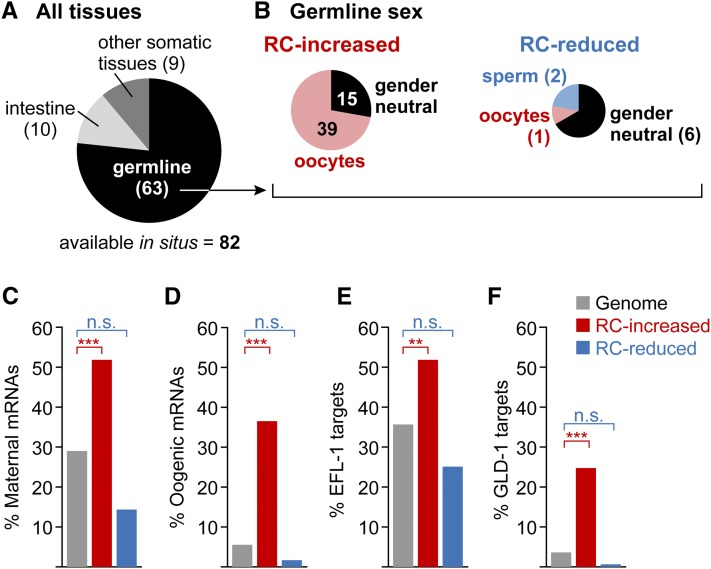
We took advantage of available databases to explore tissue expression of the 219 mRNAs differentially expressed (either increased or reduced) in puf-8; lip-1 reprogramming competent animals compared to puf-8fbf-1; lip-1 animals not competent for reprogramming. Specifically, we used the publicly available Nematode Expression Pattern DataBase (NEXTDB: nematode.lab.nig.ac.jp) as well as several genome-wide expression data sets (Baugh et al. 2003; Reinke et al. 2004; Wang et al. 2009; Gerstein et al. 2010). Eighty-two of 219 of the differentially expressed mRNAs were represented in NEXTDB, with 77% (63/82) expressed in the germline (Figure 3A). Moreover, these changed mRNAs had significant overlap with germline-enriched transcripts identified in prior studies (Reinke et al. 2004; Wang et al. 2009) (Table S3). Therefore, the differentially expressed mRNAs are enriched for germline mRNAs.
Figure 3.
puf-8; lip-1 increased mRNAs are enriched for oogenic mRNAs. (A) Tissue expression of mRNAs differentially expressed between reprogramming competent puf-8; lip-1 and noncompetent puf-8 fbf-1; lip-1 animals. The publicly available Nematode Expression Pattern DataBase in situ hybridization database (NEXTDB: nematode.lab.nig.ac.jp/) contained hybridizations for 82 of the 219 mRNAs differentially expressed between reprogramming competent and noncompetent animals; these mRNAs were scored for expression in germline, intestine, and other somatic tissues (pharynx, hypodermis, body wall muscle, vulva, and neuron). (B) Sex-specific germline expression of mRNAs differentially expressed between reprogramming competent and noncompetent animals. The 63 germline-expressed mRNAs from A were first separated as being either at higher levels in the reprogramming competent animals (“RC-increased”) or at lower levels in the reprogramming competent animals (“RC-reduced”); mRNAs in these two groups were then scored for expression during oogenesis (pink), spermatogenesis (blue), or both (gender neutral, in black). (C–F) mRNAs in distinct data sets were compared to RC-increased (red), RC-reduced (blue), and the genome (gray). (C) Maternally expressed mRNAs (Baugh et al. 2003). (D) Oogenesis mRNAs (Reinke et al. 2004). (E) EFL-1 targets (Gerstein et al. 2010). (F) GLD-1 targets (Jungkamp et al. 2011). n.s., not significant; **FDR < 1E-6; ***FDR < 1E-9.
We next queried the differentially expressed mRNAs for a bias in sex specificity. For those germline mRNAs represented in NEXTDB, most mRNAs increased in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were expressed in oocytes or developing oocytes and not in sperm, while most mRNAs reduced in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were expressed in both spermatogenic and oogenic germlines (Figure 3B). We also compared these increased and reduced mRNAs to genomic data sets of maternal mRNAs (Baugh et al. 2003) and oogenic mRNAs (Reinke et al. 2004). The 135 mRNAs increased in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were significantly enriched for maternal and oogenic mRNAs (hypergeometric test, FDR < 1E-9) (Figure 3, C and D). By contrast, the 84 mRNAs reduced in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were not enriched for oogenic mRNAs from either list (Figure 3, C and D). Finally, we compared our lists of increased and decreased mRNAs with a recent data set of mRNAs enriched in dissected oogenic gonads (Ortiz et al. 2014) and found a similar significant enrichment for the increased mRNAs (53%, 72/134; hypergeometric distribution, FDR < 1E-24), but not for the decreased mRNAs. We conclude that puf-8; lip-1 animals express high levels of oogenic mRNAs relative to puf-8fbf-1; lip-1 animals.
puf-8; lip-1 differentially expressed mRNAs are enriched for targets of certain germline sex regulators
To further characterize the differences between puf-8; lip-1 and puf-8fbf-1; lip-1 transcriptomes we asked whether the differentially expressed mRNAs were enriched for targets of germline sex regulators. Specifically, we queried published targets of (1) a transcription factor promoting female development in all tissues [TRA-1 (Berkseth et al. 2013)], (2) a transcription factor promoting oogenesis [EFL-1 (Kudron et al. 2013)], (3) several oocyte fate-promoting translational regulators [FBF (Kershner and Kimble 2010), GLD-2/RNP-8 (Kim et al. 2010), GLD-1 (Jungkamp et al. 2011), and PUF-8 (Mainpal et al. 2011)], and (4) a transcription factor promoting spermatogenesis [SPE-44 (Kulkarni et al. 2012)]. Additionally, we queried >70 genome-wide transcription factor target lists from the modENCODE consortium (Gerstein et al. 2010).
We found significant enrichment for targets of four germline sex regulators (FDR ≤ 1E-6). mRNAs that increased in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were enriched for targets of both EFL-1/E2F (Figure 3E) and GLD-1 (Figure 3F), whereas mRNAs reduced in puf-8; lip-1 vs. puf-8fbf-1; lip-1 were underenriched for those targets (Figure 3, E and F). We also found enrichment for FBF-1 and PUF-8 targets in mRNAs increased in puf-8; lip-1 vs. puf-8fbf-1; lip-1 but not in mRNAs reduced in puf-8; lip-1 vs. puf-8fbf-1; lip-1 (see below). No other enrichments were found. The association of mRNAs increased in puf-8; lip-1 vs. puf-8fbf-1; lip-1 with EFL-1 and GLD-1 targets is consistent with the enrichment of oocyte mRNAs among the mRNAs increased in reprogramming competent mutants.
Oocyte mRNAs are also enriched in fbf-1; lip-1, the second reprogramming competent mutant
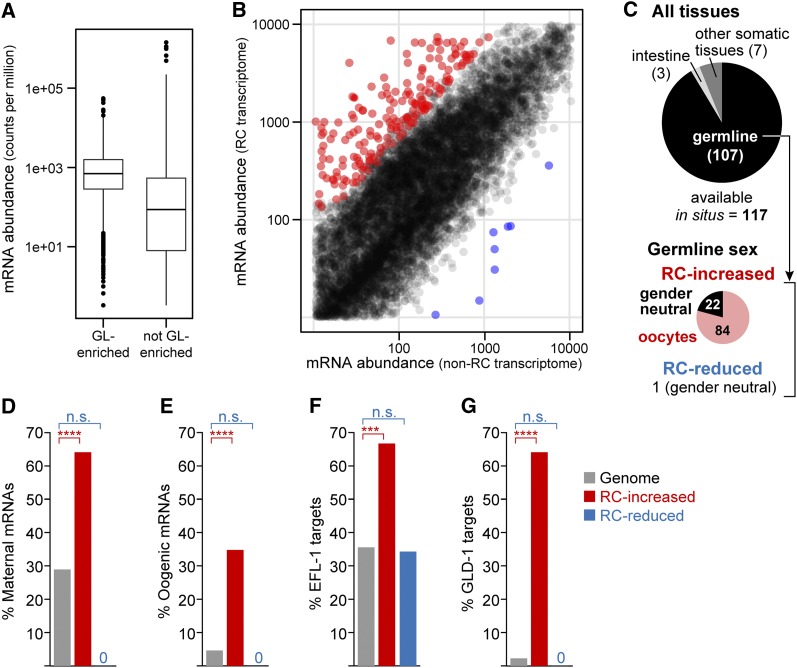
To ask whether enrichment for oocyte mRNAs was a consistent feature of reprogramming competent XX sperm-only adults, we compared the transcriptomes of the second reprogramming competent mutant identified in this study, fbf-1; lip-1, and the same reprogramming noncompetent mutant used earlier, puf-8fbf-1; lip-1. This distinct reprogramming competent mutant adult is also fully penetrant for a masculinized germline—it makes only sperm and no oocytes in the absence of drug (Table 1). This second comparison identifies PUF-8-dependent gene expression changes. For each of three replicates, 43 million reads on average mapped to unique locations in the C. elegans genome (Table S1). We detected 16,350 mRNAs or 77% of annotated C. elegans genes. Germline mRNAs were again higher in abundance in these transcriptomes than other mRNAs, and the range of abundances was broad (Figure 4A). Pairwise comparison of fbf-1; lip-1 to puf-8fbf-1; lip-1 transcriptomes identified 350 differentially expressed mRNAs (DESeq, FDR < 0.01) with a twofold or greater change in either direction (File S2). These 350 mRNAs were significantly enriched for germline mRNAs (Table S4). Of these 350 mRNAs, 335 were higher in fbf-1; lip-1 than in puf-8fbf-1; lip-1 and only 15 were lower (Figure 4B). Exon usage analyses revealed no distinct isoforms between the two sets. The mRNAs increased in fbf-1; lip-1 vs. puf-8fbf-1; lip-1 were enriched for genes involved in gamete generation, sexual reproduction, and oogenesis, among other GO terms (Table S5). The 15 mRNAs reduced in fbf-1; lip-1 vs. puf-8fbf-1; lip-1 were not enriched for any GO terms.
Figure 4.
mRNAs more abundant in fbf-1; lip-1 than in puf-8 fbf-1; lip-1 are enriched for oogenic mRNAs. (A) Boxplots of “germline-enriched” (left) and “not germline-enriched” (right) mRNAs found in fbf-1; lip-1 plus puf-8 fbf-1; lip-1 transcriptomes. Each boxplot shows the average counts per million for all mRNAs, normalized by library size. Sorting into the two categories was done following the classification of Reinke et al. (2004). (B) Differentially expressed mRNAs in fbf-1; lip-1 (reprogramming competent, “RC”) vs. puf-8 fbf-1; lip-1 (noncompetent, “non-RC”) transcriptomes. A total of 350 mRNAs differed between RC and non-RC transcriptomes (FDR < 0.01 with twofold change or more), with 335 increased mRNAs (red) and 15 reduced mRNAs (blue). All other mRNAs are shown in black. (C) Tissue expression of mRNAs differentially expressed between reprogramming competent and noncompetent animals. The NEXTDB database contained hybridizations for 117 of the 350 differentially expressed mRNAs; these were scored for expression in germline, intestine, and other somatic tissues (pharynx, hypodermis, body wall muscle, vulva, and neuron) (top pie chart). Germline-expressed mRNAs were separated into RC-increased and RC-reduced lists and scored for expression in oogenesis (pink), spermatogenesis (blue), or both (gender neutral, black). Bottom pie chart shows breakdown for mRNAs at higher levels in the reprogramming competent strain (“RC-increased”); the single germline mRNA that was reduced in the reprogramming competent strain (“RC-reduced”) was gender neutral. (D–G) Representation of mRNAs in distinct data sets to 335 RC-increased mRNAs (red), 15 RC-reduced mRNAs (blue), and the genome (gray). (D) Maternally expressed mRNAs (Baugh et al. 2003). (E) Oogenesis mRNAs (Reinke et al. 2004). (F) Regulatory targets of EFL-1 (Gerstein et al. 2010). (G) Regulatory targets of GLD-1 (Jungkamp et al. 2011). n.s., not significant; ***FDR < 1E-9; ****FDR < 1E-15.
As with the puf-8; lip-1 mutant, the 335 mRNAs increased in fbf-1; lip-1 vs. puf-8fbf-1; lip-1 were also enriched for mRNAs related to oogenesis. According to NEXTDB the 335 mRNAs were significantly enriched for oogenic transcripts (Figure 4C), maternal mRNAs (Figure 4D), oogenic mRNAs (Figure 4E), EFL-1 targets (Figure 4F) and GLD-1 mRNA targets (Figure 4G) (FDR ≤ 1E-6). Additionally, of these 335 mRNAs, 255 were also enriched in oogenic gonads compared to spermatogenic gonads (Ortiz et al. 2014) (FDR < 1E-24). We conclude that the fbf-1; lip-1 transcriptome, like the puf-8; lip-1 transcriptome, is enriched for mRNAs linked to oogenesis, even though both mutants make only sperm.
Similarities of puf-8; lip-1 and fbf-1; lip-1 transcriptomes
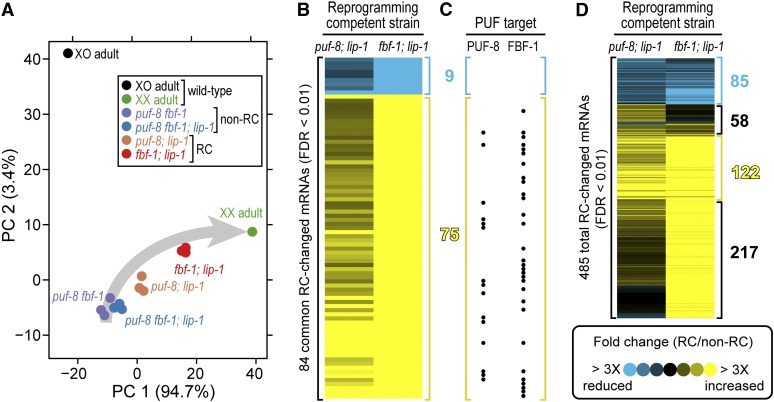
We performed principal component analysis (PCA) to seek a pattern in the transcriptional expression profiles. PCA was performed on RNA-Seq data from individual replicates of the four XX mutants with a fully masculinized germline [(1) puf-8; lip-1, (2) fbf-1; lip-1, (3) puf-8fbf-1; lip-1, and (4) puf-8fbf-1] plus wild-type XX oogenic adult hermaphrodites and wild-type XO adult males (Materials and Methods). All sequencing data were mapped and quantified as described in Materials and Methods (Table S1). The first principal component (PC1) accounted for 94.7% of the variation in the expression data and spread samples along a trajectory corresponding to germline sexuality (Figure 5A; see Materials and Methods). Thus according to PC1, the transcriptomes of two XX sperm-only mutants (puf-8fbf-1; lip-1 and puf-8fbf-1) were closest to XO spermatogenic males in that both had negative coefficients, while the transcriptomes of two other XX sperm-only mutants (puf-8; lip-1 and fbf-1; lip-1) were intermediate between wild-type XO spermatogenic males and wild-type XX oogenic adults (Figure 5A). The second principal component (PC2) accounted for 3.4% of the variability in the data and separated the wild-type XO male sample from all of the XX hermaphrodite samples (Figure 5A). We conclude that the transcriptomes of the two reprogramming competent puf-8; lip-1 and fbf-1; lip-1 mutants occupy intermediate states in the expression landscape.
Figure 5.
Similarities between puf-8; lip-1 and fbf-1; lip-1 transcriptomes. (A) Principal component analysis. Each point represents a biological replicate of the following: wild-type XO adult males (black), wild-type XX adult oogenic hermaphrodites (green), the reprogramming competent mutants (puf-8; lip-1 in orange, fbf-1; lip-1 in red), and the noncompetent mutants (puf-8 fbf-1 in purple, puf-8 fbf-1; lip-1 in blue). PC, principal component. Gray arrow shows a deduced trajectory of germline sexuality in XX animals from most spermatogenic (start of arrow) to overtly oogenic (arrowhead). PCA was carried out on the pooled 485 total RC-changed mRNAs in one or both reprogramming competent mutants. (B and D) Each heat map shows mRNA abundances in the puf-8; lip-1 reprogramming competent mutant (left column) and the fbf-1; lip-1 reprogramming competent mutant (right column) normalized to mRNA abundances in the puf-8 fbf-1; lip-1 noncompetent mutant; each row represents an mRNA whose abundance is the average of three biological replicates. Groups were visually identified in hierarchically clustered data. (B) Heat map of the 84 mRNAs identified as differentially expressed in both reprogramming competent mutants vs. the noncompetent control (FDR < 0.01 with twofold change or more in both lists). (C) Dots correspond to mRNAs in B that are PUF-8 targets (left) and FBF-1 targets (right). (D) Heat map of all 485 mRNAs differentially expressed from either reprogramming competent mutant vs. the noncompetent control (FDR < 0.01 with twofold change or more).
We next compared the mRNAs differentially expressed in each of the reprogramming competent mutants (puf-8; lip-1 and fbf-1; lip-1) relative to their noncompetent counterpart (puf-8fbf-1; lip-1). The 219 mRNAs differentially expressed in puf-8; lip-1 vs. puf-8fbf-1; lip-1 and the 350 mRNAs differentially expressed in fbf-1; lip-1 vs. puf-8fbf-1; lip-1 shared 84 mRNAs (File S3) (FDR < 0.01 with twofold change), including 75 elevated in both mutants (Figure 5B, yellow), 9 reduced in both (Figure 5B, blue), and none with opposite effects. We also hierarchically clustered the pooled 485 mRNAs significant in one or both reprogramming competent mutants: 95% (461/485) of those significantly changed in one reprogramming competent mutant displayed the same directionality of change in the other, even though magnitudes of change differed (Figure 5D). The effects were thus largely similar. We conclude that the reprogramming competent puf-8; lip-1 and fbf-1; lip-1 transcriptomes are more alike than suggested by the overlap of the most stringently defined 84 mRNAs.
mRNAs differentially increased in reprogramming competent mutants include many PUF mRNA targets
To investigate the connection between PUF regulation and mRNA abundance changes, we compared mRNAs differentially expressed in reprogramming competent vs. noncompetent mutants with targets of PUF-8 and FBF-1 (Kershner and Kimble 2010; Mainpal et al. 2011). We first analyzed the 84 mRNAs that met the significance threshold in both reprogramming competent mutants: the 75 differentially increased mRNAs included 15 PUF-8 targets, 30 FBF-1 targets, and 7 targets of both PUF proteins (hypergeometric distribution, FDR < E-16); the differentially reduced cluster included no targets of either PUF-8 or FBF-1 (Figure 5C; File S3). We then looked for targets among the pooled 485 differentially expressed mRNAs from both mutants: only the 122 mRNAs strongly upregulated in both reprogramming competent mutants were significantly enriched for PUF-8 and FBF-1 targets (FDR < 10E-6) (Figure 5D). Those 122 mRNAs included 14 PUF-8 targets, 32 FBF-1 targets, and 6 targets of both. No other cluster identified by hierarchical clustering was enriched for PUF-8 or FBF-1 targets (FDR > 0.97). The differentially increased mRNAs that were enriched for PUF-8 and FBF-1 targets were also enriched for the PUF-8 binding element (UGUANAUA, where N is any nucleotide) and the FBF binding element (UGUDHHAUA, where D is A, G, or U and H is A, U, or C) (FDR < 0.001) (File S3). We conclude that a cohort of PUF-8 and FBF-1 target mRNAs increases in abundance in mutants competent for chemical reprogramming.
Reprogramming competent germlines express both sperm-specific and oocyte-specific proteins
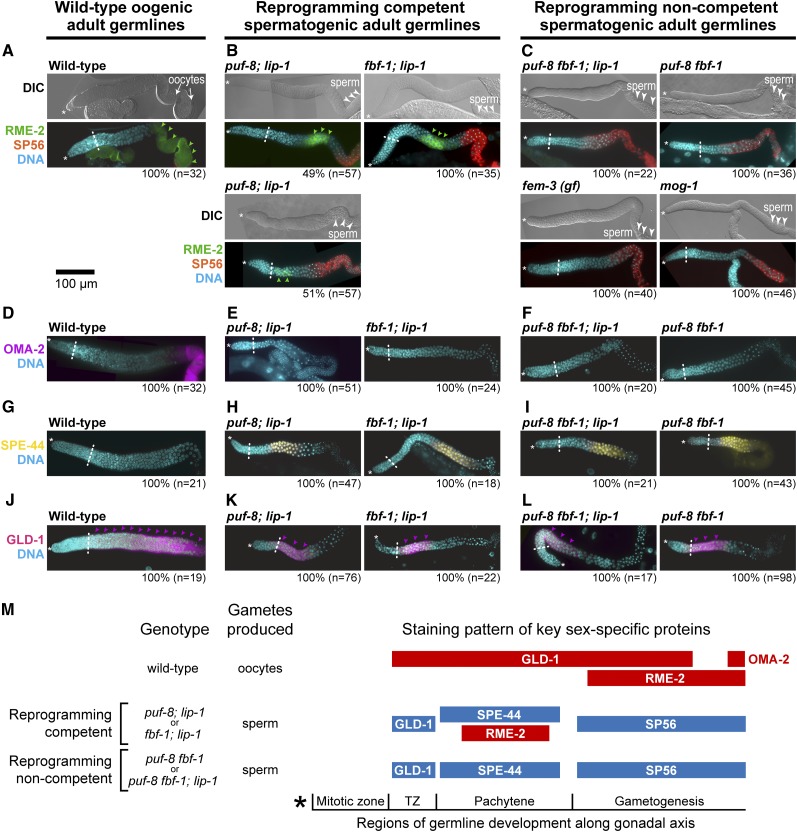
To ask whether reprogramming competent germlines express oocyte proteins in addition to oocyte mRNAs we stained for proteins diagnostic of sex-specific expression in the germline. We first stained for the oogenic RME-2 yolk receptor and spermatogenic SP56 antigen. In wild-type adult oogenic germlines, RME-2 increases in abundance as developing oocytes progress through meiotic prophase (Figure 6A) (Grant and Hirsh 1999), whereas SP56 is expressed in stored sperm (Ward et al. 1986). Both reprogramming competent germlines stained positively for RME-2 and SP56 (100%, puf-8; lip-1, n = 57; 100%, fbf-1; lip-1, n = 35) (Figure 6B). RME-2 abundance was strong in fbf-1; lip-1 germlines, but about equally split between strong and weak staining in puf-8; lip-1 germlines (Figure 6B). Moreover, RME-2 was confined to the pachytene region in both mutants (Figure 6B). We cannot explain the previously reported negative result for RME-2 in puf-8; lip-1 germlines (Morgan et al. 2010), but note that our positive results were found in both puf-8; lip-1 and fbf-1; lip-1 germlines. Germlines not competent for reprogramming, by contrast, expressed SP56 but not RME-2 (Figure 6C). Therefore, RME-2 is uniquely expressed in spermatogenic germlines capable of U0126 reprogramming.
Figure 6.
Expression of sperm-specific and oocyte-specific proteins in wild-type, reprogramming competent, and reprogramming noncompetent sperm-only adult germlines. (A–L) Extruded and immunostained young adult germlines, organized in three columns: left, wild-type adult oogenic germlines; center, reprogramming competent spermatogenic germlines; right, reprogramming noncompetent spermatogenic germlines. Specific genotypes are shown. In each image, * marks the distal end of the germline and a dashed line marks the boundary between the end the mitotic zone and the start of meiotic prophase. Bar, 100 μm for all images. (A–C) Each pair of images shows DIC above and a merged image of three channels [DAPI (DNA), SP56, and RME-2] below. In DIC images, oocytes are marked with arrows and regions of sperm are marked with arrowheads. RME-2 staining (green arrowheads) is strong and extensive in wild-type oogenic germlines (A), can be either strong (above) or weak (below) in puf-8; lip-1 germlines (B, left), is strong in fbf-1; lip-1 germlines (B, right), and is absent from all noncompetent germlines. (D–L) Merged images of DAPI-stained DNA plus one gamete-specific protein. (D–F) OMA-2 is limited to the proximal-most region of wild-type oogenic germlines where mature oocytes form and is not expressed in reprogramming competent or noncompetent spermatogenic germlines. (G–I) SPE-44 is not observed in wild-type oogenic germlines but is seen in both reprogramming competent and noncompetent sperm-only germlines. (J–L) GLD-1 (pink arrowheads) is broadly expressed in wild-type oogenic germlines, but spatially restricted in reprogramming competent and noncompetent spermatogenic germlines. (M) Schematic summary of sex-specific protein expression in XX adult germlines. Blue, expression extents of spermatogenic-specific proteins; red, expression extents of oogenic-specific proteins. (GLD-1 is expressed in either a limited male-specific pattern or an expanded female-specific pattern and is therefore colored according to pattern rather than protein.) Germline development occurs in reproducible regions along the gonadal axis; asterisk marks the distal end. See Figure S1 for positions for each genotype in terms of number of germ cell diameters from distal end.
We next stained for three additional proteins: OMA-2, an oocyte-specific protein normally expressed in the one to two most mature oocytes and therefore a marker of overt oocyte differentiation (Detwiler et al. 2001); SPE-44, a sperm-specific protein normally expressed in nuclei of pachytene germ cells and primary spermatocytes (Kulkarni et al. 2012); and GLD-1, which is normally expressed in either a male or a female-specific pattern, being more abundant and more extensively distributed in oogenic than spermatogenic germlines (Hansen et al. 2004). OMA-2 was observed in mature oocytes of wild-type oogenic germlines (Figure 6D) but not in either reprogramming competent or noncompetent spermatogenic germlines (Figure 6, E and F). By contrast, SPE-44 was not seen in wild-type oogenic germlines (Figure 6G) but was found in spermatogenic germlines of both reprogramming competent and reprogramming noncompetent mutants (Figure 6, H and I). Although both RME-2 and SPE-44 were expressed in the pachytene region in reprogramming competent germlines (Figure 6, B, H, and M; Figure S1, B and C), co-staining of RME-2 and SPE-44 was precluded because both antibodies were generated in rabbit. As previously seen for puf-8; lip-1 (Morgan et al. 2013), GLD-1 was expressed in its female pattern in wild-type oogenic germlines (Figure 6J) but in its male pattern in both reprogramming competent and noncompetent germlines (Figure 6, K and L). Therefore, both reprogramming competent mutants expressed sperm-specific proteins throughout the germline, but they also expressed oocyte-specific RME-2 in the pachytene region. We conclude that competence for U0126 germline sex reprogramming is associated with an intersexual pattern of protein expression.
Finally, we asked whether sperm and oocyte markers behaved as expected upon reprogramming and overt differentiation. After U0126 treatment, we waited 24 hr to ensure oocyte differentiation in the reprogramming competent puf-8; lip-1 and fbf-1; lip-1 mutants and confirmed that their germlines had lost SPE-44 expression (Figure S2, A and B) and gained OMA-2 expression (Figure S2, C and D). We conclude that U0126-induced reprogramming of germline sex switches the expression of sex-specific marker proteins.
Transcriptome analysis during the process of reprogramming
The identification of reprogramming competent and reprogramming noncompetent mutants made it possible to query differential changes to the transcriptome during the process of reprogramming. U0126 reprogramming of gamete sex occurs over an interval of 1–9 hr after addition of the MEK inhibition although overt oocyte differentiation is not seen until 18–24 hr after addition of drug (Morgan et al. 2010, 2013). We therefore generated RNA-Seq data at 1, 2, 4, and 6 hr after drug or vehicle treatment for both puf-8; lip-1 reprogramming competent and puf-8fbf-1 reprogramming noncompetent mutants for a total of 16 samples with two biological replicates each (Table S1; Figure S3, A and B). On average, 24 million reads were uniquely mapped per sample with detection of 16,861 mRNAs across all samples or 80% of all annotated C. elegans mRNAs. We first compared the 8 vehicle-treated to the 8 U0126-treated transcriptomes regardless of genotype and identified 56 mRNAs that were increased or reduced greater than twofold over time in drug-treated animals relative to vehicle controls (FDR < 0.01) (Figure S3C; File S4). This analysis identified a robust transcript-level response to treatment with MEK inhibitor. We next compared drug-treated reprogramming competent animals to either vehicle-treated or reprogramming noncompetent controls, but found no expression changes associated with reprogramming (FDR < 0.01). That negative result may reflect a predominance of post-transcriptional regulation during reprogramming, as suggested previously from negative qPCR results of 8 candidate mRNAs (Morgan et al. 2013), or a lack of experimental resolution.
We also generated RNA-Seq data after 18 hr of drug treatment, when mature oocytes are first formed in reprogrammed germlines. This experiment also compared drug-treated reprogramming competent animals to either vehicle-treated or reprogramming noncompetent controls. We found 1131 significant changes of twofold or more, with 928 increases and 203 reductions in mRNA abundance (FDR < 0.01) (File S5). The increased mRNAs included 32% (298/928) oogenesis-enriched mRNAs (hypergeometric distribution, FDR < E10-170), whereas the reduced mRNAs included only 5.9% (12/203) oogenesis-enriched mRNAs (hypergeometric distribution, FDR > 0.3). The increased mRNAs were enriched for three GO terms: cell cycle, reproduction, and sex differentiation (Table S6), but the reduced mRNAs were not enriched for any GO term. Therefore this final experiment easily detected the shift to an oogenic program.
Discussion
A previous study found that treatment with a MEK inhibitor reprograms a C. elegans adult from sperm to oocyte production (Morgan et al. 2010). This work investigates the underlying genetic and molecular bases of competence for this chemical reprogramming. We find that competence correlates with a sexually intermediate molecular state, despite production of functional sperm, and that either of two PUF proteins, PUF-8 or FBF-1, can confer competence. Below, we discuss these conclusions and place them within the context of a model for how puf-8; lip-1 and fbf-1; lip-1 genotypes confer reprogramming competence.
Competence for chemical reprogramming correlates with a sexually intermediate molecular signature
A major conclusion of this work is that competence for chemical reprogramming correlates with a sexually mixed molecular signature at both the RNA and protein levels. This intersexual signature was found in both of the reprogramming competent mutants (puf-8; lip-1 and fbf-1; lip-1), but not in either mutant lacking reprogramming competence (puf-8fbf-1 or puf-8fbf-1; lip-1). In addition to spermatogenic RNAs and proteins expected for animals making only sperm, the reprogramming competent animals expressed an elevated abundance of oogenic mRNAs and at least one oogenic protein, albeit with a restricted spatial distribution (Figure 6, B and M). Indeed, sperm-specific and oocyte-specific proteins (SPE-44 and RME-2) were coexpressed in a block of cells within the pachytene region (Figure S1, B and C), even though these germlines continuously made sperm. This intersexual molecular signature suggests that the gamete sex determination regulatory network can exist in an intermediate state with its equilibrium shifted from strongly male to a sexually mixed molecular character.
The existence of intermediate molecular signatures associated with reprogramming is best documented for the process of reprogramming, both in vitro and in vivo (for review, see David and Polo 2014). Such states were first discovered using cell surface protein markers to analyze cells during reprogramming of fibroblasts to pluripotency (Brambrink et al. 2008; Stadtfeld et al. 2008) and then characterized more deeply in the same system by analyzing transcriptomes (Sridharan et al. 2009; Polo et al. 2012) and proteomes (Hansson et al. 2012). The emerging view is that reprogramming to pluripotency is a stepwise process with multiple intermediates (David and Polo 2014). A similar phenomenon is observed in cell fate reprogramming by transdifferentiation within an organism, although these in vivo examples are less well defined. For example, murine pancreatic α-cells that are in the process of reprogramming into pancreatic β-cells pass through a state producing both glucagon (an α-cell product) and insulin (a β-cell product) (Thorel et al. 2010). Similarly, Drosophila neural progenitors that are reprogramming into the glial lineage pass through a state producing protein markers of both neural stem cells and glial cells (Flici et al. 2011).
Our discovery of an intermediate molecular signature in mutants competent for gamete sex reprogramming differs from these previous findings. We have discovered an intermediate state that occurs in the absence of reprogramming but signifies competence to change sexual fate in response to ERK/MAPK pathway inhibition, a phenomenon distinct from but perhaps related to the dynamic transitional states that occur during the reprogramming process itself. If the two are indeed related, the apparently stable and homogeneous intersexual state reported here in reprogramming competent germlines could provide an in vivo model for intermediate molecular states that occur transiently during cell fate reprogramming. What is more, the intermediate molecular signature we find associated with competence for reprogramming may represent a specialized case of a broader phenomenon. We speculate that genotypes leading to an intermediate molecular state may exist broadly in a natural population, even though organisms appear “normal.” Those intermediate molecular states might affect sexual or other characteristics and could render an individual sensitive to chemical cues, including exogenous ones.
Either puf-8 or fbf-1 is needed for reprogramming competence
Two double mutants, puf-8; lip-1 and fbf-1; lip-1, are competent for chemical reprogramming of adult gamete sex, but the triple mutant, puf-8fbf-1; lip-1, is not competent (Morgan et al. 2010; this work). Therefore, puf-8 and fbf-1 are functionally redundant for their roles in conferring reprogramming competence. Both puf-8 and fbf-1 genes encode PUF RNA-binding proteins (see Introduction) and are redundant for the hermaphrodite switch from sperm to oocyte fate specification (Bachorik and Kimble 2005). The roles of these two PUF proteins both in the normal sperm/oocyte decision and in competence for U0126-induced reprogramming of that decision provide one line of evidence that chemical reprogramming likely uses the normal sperm/oocyte fate circuitry. Other lines of evidence were described previously (Morgan et al. 2013).
How might removal of one PUF protein render an animal competent for chemical reprogramming? PUF-8 and FBF-1 regulate many mRNAs (PUF-8, >100 mRNAs; FBF-1, >1000 mRNAs), including key components of the ERK/MAPK pathway (Lee et al. 2007a; Kershner and Kimble 2010; Mainpal et al. 2011; Vaid et al. 2013). Intriguingly, mRNAs with elevated abundance in reprogramming competent animals included many PUF-8 and FBF-1 targets (Figure 5, B and C), whereas reduced mRNAs did not. Because PUF-8 and FBF-1 are both mRNA repressors (e.g., Zhang et al. 1997; Mainpal et al. 2011), a simple explanation is that the elevated mRNAs are released from repression. However, these mRNAs were not similarly elevated in puf-8fbf-1 or puf-8fbf-1; lip-1 mutants, which suggests a more complex interpretation. We favor the possibility that PUF-8 and FBF-1 buffer the gamete sex determination network against perturbations. According to this notion, removal of one PUF protein would reduce buffering capacity and render gamete sex determination vulnerable to, for example, an increase in ERK/MAPK activity, whereas removal of both PUF proteins would abolish that buffering capacity. Testing this idea remains a challenge for the future. The current lists of PUF-8 and FBF-1 mRNA targets (Kershner and Kimble 2010; Mainpal et al. 2011) are not comprehensive, and those targets have not been characterized for network-level effects. Regardless, PUF-8 and FBF-1 are key regulators of competence for U0126-induced reprogramming, perhaps as a regulatory hub in the gamete sex determination network (see Figure 7 and below).
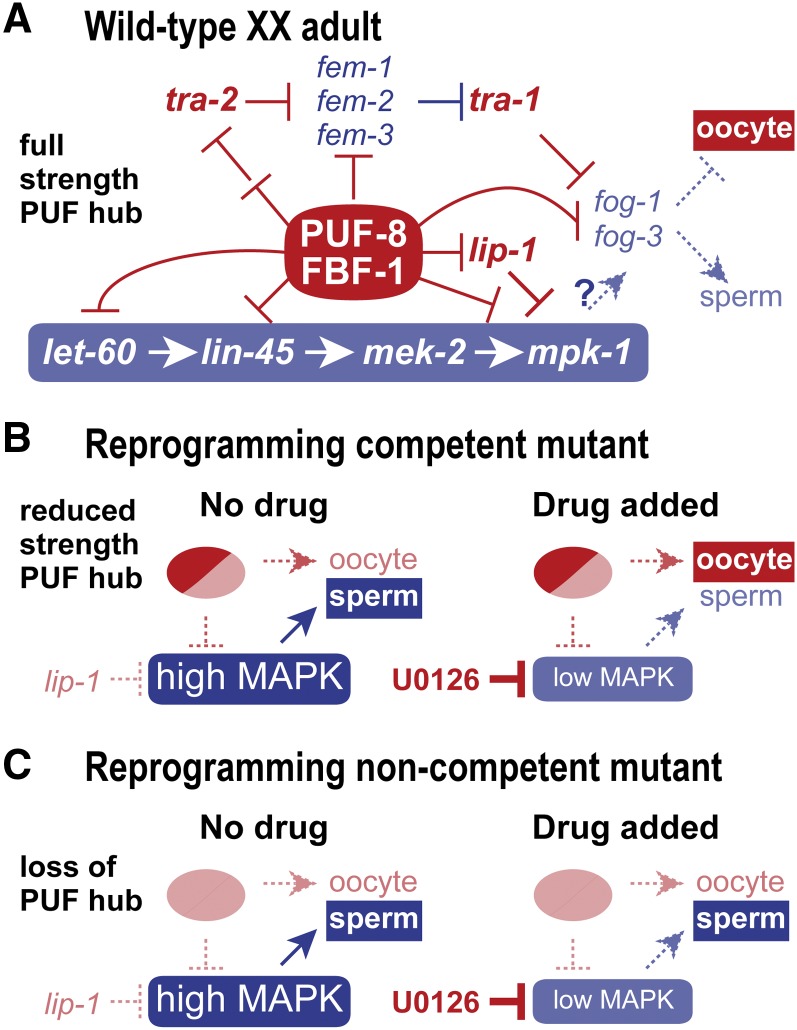
Figure 7.
Model for genotype-specific basis for reprogramming competence. (A) Simplified network for gamete fate specification in wild-type XX adults. The sex determination pathway is set to the female mode and oocyte fate specification; MAP kinase activity is low and therefore does not promote the sperm fate; PUF-8 and FBF-1 promote the oocyte fate via repression of both sex determination and MAP kinase pathways as well as repression of terminal gamete fate regulators fog-1 and fog-3. This model is based largely on established regulatory relationships, but also posits the existence of a PUF hub composed of PUF-8 and FBF-1 RNA-binding proteins. Relevant MPK-1 substrates are not known, but FOG-1 and FOG-3 proteins are candidates. See text for further explanation and references. (B) Model for sperm/oocyte specification in mutants with a reprogramming competent genotype (puf-8; lip-1 and fbf-1; lip-1). Without drug (left), germlines are set to the male mode of sperm fate specification. The PUF hub operates at reduced strength because one PUF protein is removed, and MAP kinase is hyperactivated because lip-1 is removed; the reduced-strength PUF hub is postulated to generate a sexually intermediate molecular signature. With drug (right), the U0126 MEK inhibitor reduces MAP kinase hyperactivation and permits the reduced-strength PUF hub to drive the oocyte fate. (C) Model for sperm/oocyte specification in mutants with a noncompetent genotype (puf-8 fbf-1; lip-1 depicted here). Without drug (left) or with drug (right), elimination of the PUF hub prevents oocyte fate specification regardless of MAP kinase activity. A similar explanation would hold for puf-8 fbf-1, which would not experience the hyperactivated ERK/MAPK depicted here for puf-8 fbf-1; lip-1.
A model for chemical reprogramming and the sperm/oocyte regulatory network
How might the puf-8; lip-1 and fbf-1; lip-1 genotypes modulate the sperm/oocyte cell fate regulatory network to achieve an intersexual molecular state and promote reprogramming competence? A likely answer emerges from a look at how puf-8, fbf-1, and the ERK/MAPK signaling pathway fit into the gamete sex determination network of regulation (Figure 7A). This network comprises three regulatory parts that we propose may all converge on fog-1 and fog-3, the terminal regulators of sperm fate specification (Barton and Kimble 1990; Ellis and Kimble 1995). One part is the core sex determination pathway (Zarkower 2006; Ellis and Schedl 2007), which terminates in TRA-1 to repress fog-1 and fog-3 in XX adults, resulting in oocyte fate specification (Figure 7A, top) (Chen and Ellis 2000); by contrast, XO males downregulate TRA-1, permitting fog-1 and fog-3 expression and sperm specification. A second part is the core ERK/MAPK pathway (Sundaram 2013), which terminates with MPK-1/ERK to promote the sperm fate via unknown substrates (Figure 7A, bottom) (Lee et al. 2007b; Arur et al. 2009, 2011); FOG-1 and FOG-3 both possess motifs typical of ERK/MAPK substrates and therefore are candidate substrates (Lee et al. 2011; E. Sorokin, unpublished results). Indeed, >30 MPK-1 substrates have been identified in the germline, although depleting individual substrates by RNAi resulted in only weak sex determination defects, suggesting redundancy among MPK-1 substrates for germ cell fate control (Arur et al. 2009). The third part relies on PUF RNA-binding proteins to repress expression of key mRNAs in both the sex determination and ERK/MAPK pathways as well as to repress terminal regulators fog-1 and fog-3 (Figure 7A, middle) (Zhang et al. 1997; Crittenden et al. 2002; Thompson et al. 2005; Lee et al. 2007a; Kershner and Kimble 2010; Mainpal et al. 2011). FBF-1 targets include multiple sperm-promoting mRNAs in the sex determination pathway [e.g., fem-3 (Zhang et al. 1997; Kershner and Kimble 2010)] as well as central elements of the MAPK pathway [e.g., mpk-1 (Lee et al. 2007a; Kershner and Kimble 2010)]. PUF-8 targets are less well characterized, but include let-60 mRNA of the ERK/MAPK pathway (Mainpal et al. 2011). Because PUF-8 and FBF-1 are redundant for the hermaphrodite sperm/oocyte switch (Bachorik and Kimble 2005), we propose the existence of a PUF-8/FBF-1 regulatory hub that conflates the roles of FBF-1 and PUF-8 in gamete sex specification. Consistent with that idea is the elevated abundance of mRNAs with both PUF-8 and FBF-1 binding sites in reprogramming competent animals (Figure 5C). However, these two proteins could repress different mRNAs in the same pathway to achieve a similar effect. In sum, gamete fate specification is the binary readout of a complex network composed of at least three regulatory inputs. Most regulatory relationships in Figure 7A are well established (see reviews by Zarkower 2006; Ellis and Schedl 2007; Kimble and Crittenden 2007; Ellis 2010; Sundaram 2013), but the MPK-1/ERK substrates for gamete specification and the full spectrum of PUF-8 and FBF-1 individual and joint target mRNAs remain unknown.
We now consider the reprogramming competent and noncompetent mutants in light of this three-part gamete specification network. These mutants lack either part or all of the proposed PUF regulatory hub and also lack LIP-1, the negative regulator of MPK-1/ERK. The puf-8 and fbf-1 single mutants have a partially active PUF hub, which can maintain the network in its oocyte fate mode for the most part, but with low-penetrance germline masculinization (1–3%) (Lamont et al. 2004; Bachorik and Kimble 2005), suggesting a compromised network of gamete specification. This reduced-strength PUF hub renders the network sensitive to perturbations in the ERK/MAPK pathway, perhaps due to a loss in buffering capacity in the network. Removal of the lip-1 MAP kinase inhibitor on its own hyperactivates MPK-1 activity but does not alter the network sufficiently to affect gamete fate (Berset et al. 2001). However, puf-8; lip-1 and fbf-1; lip-1 double mutants are shifted to the sperm-promoting mode at the cellular level, demonstrating synergy between the weakened PUF hub and hyperactivated MPK-1 (Figure 7B, left). Yet this cellular shift is incomplete at the molecular level, as demonstrated by its intersexual molecular signature (this work), and therefore the network exists in an intermediate state. Addition of the U0126 MEK inhibitor restores ERK/MAPK signaling to its normal basal level and shifts the network to the oocyte fate mode (Figure 7B, right). By contrast, noncompetent mutants lack both puf-8 and fbf-1 and therefore lack the entire PUF hub, a key regulatory module that promotes the oocyte fate by regulation of the sex determination pathway, the ERK/MAPK pathway, and the terminal gamete fate regulators (Figure 7C, left). In contrast to the effect of a partial PUF hub, complete loss of the PUF hub is not associated with an intersexual molecular signature, suggesting that the network in this case has shifted to a more male mode that reduces the ERK/MAPK vulnerability from the network (Figure 7C, right).
In sum, we suggest that the gamete specification network can exist in multiple states despite driving the same gamete fate decision. We also suggest that intermediate molecular signatures are diagnostic of intermediate “network states.” According to this idea, a more oogenic molecular signature or network state can be more readily reprogrammed than a less oogenic network state. This notion begins to probe the gamete specification regulatory network at a global scale not yet possible for most in vivo cell fate reprogramming processes.
Supplementary Material
Acknowledgments
We thank Clint Morgan and Myon-Hee Lee for suggesting fbf-1; lip-1 and puf-8 fbf-1 as possible reprogramming competent and noncompetent genotypes, Mark Meyer and Wes Pike for initial bioinformatics advice and server use, Anne Helsley for help with manuscript preparation, and Laura Vanderploeg for help with figures. We also thank all members of the Kimble laboratory for helpful advice and discussions throughout this work. Antibodies were generous gifts from Barth Grant, Rutgers University, Piscataway, NJ, (RME-2), Susan Strome, UC, Santa Cruz, CA, (SP56), Harold Smith, National Institutes of Health, Bethesda, MD, (SPE-44), Rueyling Lin, UT Southwestern, Dallas, TX, (OMA-2), and Betsy Goodwin (GLD-1). We thank Y. Kohara for maintenance of the NEXTDB database. E.P.S. was supported by the Biotechnology Training Program [National Institutes of Health (NIH) grant T32 GM08349]. This work was supported in part by NIH grants GM083989 (to A.P.G.) and GM069454 (to J.K.). J.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169409/-/DC1.
Data sets have been submitted to the GEO database at NCBI as series GSE54030.
Communicating editor: M. Sundaram
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Reyes A., Huber W., 2012. Detecting differential usage of exons from RNA-seq data. Genome Res. 22: 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., Ohmachi M., Nayak S., Hayes M., Miranda A., et al. , 2009. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl. Acad. Sci. USA 106: 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., Ohmachi M., Berkseth M., Nayak S., Hansen D., et al. , 2011. MPK-1 ERK controls membrane organization in C. elegans oogenesis via a sex-determination module. Dev. Cell 20: 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachorik J. L., Kimble J., 2005. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. USA 102: 10893–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W., 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Kimble J., 1990. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B., Kimble J., 1987. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Hill A. A., Slonim D. K., Brown E. L., Hunter C. P., 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Berkseth M., Ikegami K., Arur S., Lieb J. D., Zarkower D., 2013. TRA-1 ChIP-seq reveals regulators of sexual differentiation and multilevel feedback in nematode sex determination. Proc. Natl. Acad. Sci. USA 110: 16033–16038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset T., Fröhli Hoier E., Battu G., Canevascini S., Hajnal A., 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., et al. , 2008. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun J. S., Gardner K., 2013. Wounds that will not heal: pervasive cellular reprogramming in cancer. Am. J. Pathol. 182: 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M., Nichols A., Arkinstall S., 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14: 6–16 [PubMed] [Google Scholar]
- Chen P.-J., Ellis R. E., 2000. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127: 3119–3129 [DOI] [PubMed] [Google Scholar]
- Chen P.-J., Singal A., Kimble J., Ellis R. E., 2000. A novel member of the Tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev. Biol. 217: 77–90 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663 [DOI] [PubMed] [Google Scholar]
- David L., Polo J. M., 2014. Phases of reprogramming. Stem Cell Res. 12: 754–761 [DOI] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199 [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D., 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R., 2010. The sperm/oocyte decision, a C. elegans perspective, pp. 1–26 in Oogenesis: The Universal Process, edited by Verlhac M.-H., Villeneuve A. John Wiley & Sons, Chichester, UK [Google Scholar]
- Ellis, R., and T. Schedl, 2007 Sex determination in the germ line (March 5, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.82.2, http//www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., Kimble J., 1995. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139: 561–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flici H., Erkosar B., Komonyi O., Karatas O. F., Laneve P., et al. , 2011. Gcm/Glide-dependent conversion into glia depends on neural stem cell age, but not on division, triggering a chromatin signature that is conserved in vertebrate glia. Development 138: 4167–4178 [DOI] [PubMed] [Google Scholar]
- Friedl P., Alexander S., 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147: 992–1009 [DOI] [PubMed] [Google Scholar]
- Friend K., Campbell Z. T., Cooke A., Kroll-Conner P., Wickens M. P., et al. , 2012. A conserved PUF–Ago–eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S. F., 1997. Developmental Biology. Sinauer Associates, Sunderland, MA [Google Scholar]
- Graham P. L., Kimble J., 1993. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D., 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A., Berset T., 2002. The C. elegans MAPK phosphatase LIP-1 is required for the G2/M meiotic arrest of developing oocytes. EMBO J. 21: 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Wilson-Berry L., Dang T., Schedl T., 2004. Control of the proliferation vs. meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131: 93–104 [DOI] [PubMed] [Google Scholar]
- Hansson J., Rafiee M. R., Reiland S., Polo J. M., Gehring J., et al. , 2012. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Reports 2: 1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jungkamp A. C., Stoeckius M., Mecenas D., Grun D., Mastrobuoni G., et al. , 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A. M., Kimble J., 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA 107: 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Wilson T. L., Kimble J., 2010. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc. Natl. Acad. Sci. USA 107: 17445–17450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433 [DOI] [PubMed] [Google Scholar]
- Kudron M., Niu W., Lu Z., Wang G., Gerstein M., et al. , 2013. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol. 14: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M., Shakes D. C., Guevel K., Smith H. E., 2012. SPE-44 implements sperm cell fate. PLoS Genet. 8: e1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Kornfeld K., Miller L. M., Horvitz H. R., Kim S. K., 1994. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 8: 160–173 [DOI] [PubMed] [Google Scholar]
- Lamont L. B., Crittenden S. L., Bernstein D., Wickens M., Kimble J., 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7: 697–707 [DOI] [PubMed] [Google Scholar]
- Lee M.-H., Hook B., Lamont L. B., Wickens M., Kimble J., 2006. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 25: 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Hook B., Pan G., Kershner A. M., Merritt C., et al. , 2007a Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 3: e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007b Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Kim K. W., Morgan C. T., Morgan D. E., Kimble J., 2011. Phosphorylation state of a Tob/BTG protein, FOG-3, regulates initiation and maintenance of the Caenorhabditis elegans sperm fate program. Proc. Natl. Acad. Sci. USA 108: 9125–9130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Zhu S., Russ H. A., Xu S., Xu T., et al. , 2014. Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell 14: 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainpal R., Priti A., Subramaniam K., 2011. PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 360: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C. T., Lee M. H., Kimble J., 2010. Chemical reprogramming of Caenorhabditis elegans germ cell fate. Nat. Chem. Biol. 6: 102–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C. T., Noble D., Kimble J., 2013. Mitosis-meiosis and sperm-oocyte fate decisions are separable regulatory events. Proc. Natl. Acad. Sci. USA 110: 3411–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans G3 (Bethesda) 4: 1765–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otori M., Karashima T., Yamamoto M., 2006. The Caenorhabditis elegans homologue of Deleted in Azoospermia is involved in the sperm/oocyte switch. Mol. Biol. Cell 17: 3147–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J. M., Anderssen E., Walsh R. M., Schwarz B. A., Nefzger C. M., et al. , 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151: 1617–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323 [DOI] [PubMed] [Google Scholar]
- Schedin P., Jonas P., Wood W. B., 1994. Function of the her-1 gene is required for maintenance of male differentiation in adult tissues of C. elegans. Dev. Genet. 15: 231–239 [DOI] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., et al. , 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K., 2008. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram, M. V., 2013 Canonical RTK-Ras-ERK signaling and related alternative pathways (July 1, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.80.2, http//www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Thompson B. E., Bernstein D. S., Bachorik J. L., Petcherski A. G., Wickens M., et al. , 2005. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development 132: 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel F., Népote V., Avril I., Kohno K., Desgraz R., et al. , 2010. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464: 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid S., Ariz M., Chaturbedi A., Kumar G. A., Subramaniam K., 2013. PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development 140: 1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao Y., Wong K., Ehlers P., Kohara Y., et al. , 2009. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Roberts T. M., Strome S., Pavalko F. M., Hogan E., 1986. Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J. Cell Biol. 102: 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D. S., Kimble J., Parker R., 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157 [DOI] [PubMed] [Google Scholar]
- Wong V. W., Gurtner G. C., Longaker M. T., 2013. Wound healing: a paradigm for regeneration. Mayo Clin. Proc. 88: 1022–1031 [DOI] [PubMed] [Google Scholar]
- Xin M., Olson E. N., Bassel-Duby R., 2013. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14: 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower, D., 2006 Somatic sex determination (February 10, 2006), Wormbook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook1.84.1, http//www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., et al. , 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484 [DOI] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., et al. , 2010. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7: 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O., Glaser B., Dor Y., 2013. The plastic pancreas. Dev. Cell 26: 3–7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads and mRNA expression data are available at the Gene Expression Omnibus under accession no. GSE54030.