Abstract
Control of the eukaryotic G2/M transition by CDC2/CYCLINB is tightly regulated by protein–protein interactions, protein phosphorylations, and nuclear localization of CDC2/CYCLINB. We previously reported a screen, in Aspergillus nidulans, for extragenic suppressors of nimX2cdc2 that resulted in the identification of the cold-sensitive snxA1 mutation. We demonstrate here that snxA1 suppresses defects in regulators of the CDK1 mitotic induction pathway, including nimX2cdc2, nimE6cyclinB, and nimT23cdc25, but does not suppress G2-arresting nimA1/nimA5 mutations, the S-arresting nimE10cyclinB mutation, or three other G1/S phase mutations. snxA encodes the A. nidulans homolog of Saccharomyces cerevisiae Hrb1/Gbp2; nonessential shuttling messenger RNA (mRNA)-binding proteins belonging to the serine-arginine-rich (SR) and RNA recognition motif (RRM) protein family; and human heterogeneous ribonucleoprotein-M, a spliceosomal component involved in pre-mRNA processing and alternative splicing. snxAHrb1 is nonessential, its deletion phenocopies the snxA1 mutation, and its overexpression rescues snxA1 and ΔsnxA mutant phenotypes. snxA1 and a second allele isolated in this study, snxA2, are hypomorphic mutations that result from decreased transcript and protein levels, suggesting that snxA acts normally to restrain cell cycle progression. SNXAHRB1 is predominantly nuclear, but is not retained in the nucleus during the partially closed mitosis of A. nidulans. We show that the snxA1 mutation does not suppress nimX2 by altering NIMX2CDC2/NIMECYCLINB kinase activity and that snxA1 or ΔsnxA alter localization patterns of NIMECYCLINB at the restrictive temperatures for snxA1 and nimX2. Together, these findings suggest a novel and previously unreported role of an SR/RRM family protein in cell cycle regulation, specifically in control of the CDK1 mitotic induction pathway.
Keywords: CDC2/CYCLINB, G2/M transition, Hrb1/Gbp2, SR/RRM, cell cycle
CONTROL of the eukaryotic G2/M transition by protein kinases has been widely studied and is highly conserved among all eukaryotes from the budding and fission yeasts and filamentous fungi to metazoans (for review, see Ma and Poon 2011). The CDK1/CYCLINB protein kinase complex is a major regulator of this transition in all eukaryotes and is responsible for the phosphorylations of numerous proteins, leading to massive nuclear and cytoplasmic reorganizations that regulate mitosis (for review, see Lindqvist et al. 2009). The complex itself is tightly regulated, both temporally and spatially, to allow mitotic entry.
Although CDK1/CYCLINB activity is essential for mitotic entry in all eukaryotes, structural differences in the nucleus in various organisms result in “open” mitosis (more complex eukaryotes) or “closed” mitosis (budding yeasts); these differences likely affect the temporo-spatial functioning of CDK1/CYCLINB. The partially closed mitosis of the filamentous fungus Aspergillus nidulans is an evolutionary intermediate between open and closed mitoses and provides a system for studying mitotic entry in organisms intermediate between budding yeasts and more complex eukaryotes. The nuclear pore complexes in A. nidulans partially disassemble at mitotic entry (they are “partially closed”), and proteins not specifically retained in the nucleus diffuse out of the partially closed nuclear pores and may equilibrate across the nuclear envelope (De Souza et al. 2004). In A. nidulans, the activity and proper localization of three protein kinases are required for initiation of mitosis: the CDK1/CYCLINB protein kinase complex (NIMXCDC2/NIMECYCLINB), the NIMA kinase, and a polo-like kinase (PLKA), all three of which must be inactivated to allow mitotic exit (Bachewich et al. 2005; Osmani et al. 2006). The activity of CDK1/CYCLINB is tightly regulated by phosphorylation and is part of an autocatalytic feedback loop (Ye et al. 1995); its activity is inhibited by the ANKAWEE1 kinase and activated by the NIMTCDC25 phosphatase. Furthermore, active NIMA kinase is required for mitotic initiation; in the absence of functional NIMA kinase, cells with fully active CDK1/CYCLINB arrest in late G2 (Osmani et al. 1991). NIMA activity is also regulated by phosphorylation (Ye et al. 1995) and is required for proper localization of CDK1/CYCLINB (Wu et al. 1998) and tubulin (Ovechkina et al. 2003) into the nucleus at the G2/M transition. Specifically, the SONAGLE2 and SONBNUP98 nucleoporins interact with NIMA to regulate the nuclear localization of NIMXCDC2/NIMECYCLINB (Wu et al. 1998; De Souza and Osmani 2009). Wu et al. (1998) demonstrated that NIMXCDC2 colocalizes in the nucleus with NIMECYCLINB during S and G2, that the G2 arrest that occurs in the absence of NIMA activity occurs with predominantly cytoplasmic NIMXCDC2/NIMECYCLINB, and that in nimA1 mutants the sonA1 suppressor of nimA1 re-establishes nuclear localization of NIMXCDC2/NIMECYCLINB and entry into mitosis. These data provide evidence that proper localization of NIMXCDC2/NIMECYCLINB is both regulated and essential for controlling mitotic entry during the partially closed mitosis of A. nidulans.
CDK1 localization and cell cycle progression depend upon the localization of CYCLINB (Nigg 1995). NIMECYCLINB localization has been characterized through the cell cycle in A. nidulans (Wu et al. 1998; De Souza et al. 2009); its nuclear localization is closely mirrored by NIMXCDC2 localization (Nayak et al. 2010). NIMXCDC2 and NIMECYCLINB become visible in the nucleus at or near the G1/S boundary and disappear from the nucleus during mitosis. De Souza et al. (2009) localized NIMECYCLINB in live cells to the nucleoplasm and to the spindle pole bodies (SPBs) during interphase and early mitosis; this work demonstrated that the partial disassembly of the nuclear pore complex (NPC) at mitotic prophase allows most of the NIMECYCLINB to exit the nucleus; however, a nuclear pool remains, concentrated at the SPBs and also in the region of the segregating kinetochores. It is to this pool of NIMECYCLINB that NIMXCDC2 presumably remains bound. The nuclear NIMECYCLINB disappears sequentially during mitotic progression. The SPB pool disappears during anaphase, followed rapidly by the pool at the kinetochores. Surprisingly, NIMXCDC2 exits the nucleus slightly before the complete destruction of nuclear NIMECYCLINB (Nayak et al. 2010).
While phosphorylation/dephosphorylation and cell cycle-regulated localization of mitotic proteins have been shown to play integral roles in controlling the transition from G2 into mitosis in A. nidulans, much remains to be learned. NIMXCDC2 encodes the only known p34CDC2 protein kinase in A. nidulans, and its activity is required at both G1 and G2 (Osmani et al. 1994). NIMECYCLINB also functions at both G1 and G2 in A. nidulans. While nimE6 causes a G2 arrest at restrictive temperature (O’Connell et al. 1992), the nimE10 mutation (originally identified as nimG10) arrests cells in S at restrictive temperature (Supporting Information, Figure S1). The G1-specific functions of NIMXCDC2/NIMECYCLINB are not well understood. To better understand cell cycle regulation in A. nidulans by NIMXCDC2/NIMECYCLINB, an extragenic suppressor screen to identify genes that interact with NIMXCDC2 was undertaken (McGuire et al. 2000). The snxA1 mutation was identified in this screen as an extragenic suppressor of nimX2F223L. snxA1 suppresses the nimX2 heat-sensitive G2 arrest, allowing cells to enter and exit mitosis at the restrictive temperature for nimX2; additionally, and independent of its suppression of nimX2, snxA1 confers cold sensitivity, leading to a G1 arrest at its restrictive temperature (McGuire et al. 2000). Thus, as is the case for nimXcdc2 and nimEcyclinB mutations, the snxA1 mutation has effects on the cell cycle at both G1 and G2. In this article, we report genetic, cytological, and molecular analysis of snxA and demonstrate that it encodes the A. nidulans homolog of Saccharomyces cerevisiae Hrb1/Gbp2 proteins.
Hrb1/Gbp2 and the more divergent but similar protein Npl3 are nonessential shuttling messenger RNA (mRNA)-binding proteins that are similar to members of the mammalian serine-arginine-rich (SR) protein family (Rougemaille et al. 2008). SR proteins are a class of small nuclear ribonucleoprotein splicing factors that harbor two distinct types of domains: RNA recognition motifs (RRMs), responsible for RNA binding, and SR domains, long repeats of serine-arginine or arginine/serine dipeptides that are extensively phosphorylated (Ma and Poon 2011). Some RRM proteins contain RGG/RG tripeptide motifs that serve as sites for arginine methylation (Thandapani et al. 2013). In addition to their functions in spliceosome assembly and catalysis (Sanford et al. 2004), SR proteins are also involved in mRNA transcription and export (TREX) (reviewed in Reed and Cheng 2005; Rougemaille et al. 2008).
In budding yeast, Hrb1/Gbp2 are recruited to and associate cotranscriptionally with mRNA transcripts by physical association with the TREX complex and Ctk1, a member of the cyclin-dependent kinase family that is the catalytic domain of C-terminal domain kinase 1 (Ctdk-1) (Figure S2). Ctdk-1 is a protein kinase required for efficient transcription elongation that phosphorylates the C-terminal domain of RNA polymerase II (RNAPII) (Hurt et al. 2004). The TREX complex consists of the THO complex (composed of Tho2, Hpr1, Mft1, and Thp2), which associates with RNA polymerase II as it transcribes, and Sub2 and Yra1, export factors that bind to THO and function as a platform to recruit Hrb1/Gbp2 (Hurt et al. 2004). Hrb1/Gbp2 associate both with the genes and the growing, unprocessed mRNA transcripts produced from them and remain bound to the mRNA transcripts as part of the messenger RNA–protein complex (mRNP). As shuttling proteins, Hrb1/Gbp2 remain bound to the transcript during export from the nucleus; they also remain associated with the mRNP during translation (Windgassen et al. 2004). Nuclear export of the mRNP requires the THO complex (Hacker and Krebber 2004) and the Mex67-Mtr2 dimer, an export receptor that interacts with nucleoporins (Strasser and Hurt 2000). Yra1 interacts with Mex67 (Strasser and Hurt 2000) and is essential for mRNA export for some, but not all, transcripts (Kim Guisbert et al. 2005). In addition to their functions in mRNA transcription and export, SR-like proteins act as localization signals that aid in delivering the mRNP to the translational machinery (Windgassen et al. 2004). More recently, both Hrb1 and Gbp2 have been shown to associate with the budding yeast spliceosome (Warkocki et al. 2009), with unspliced transcripts close to 5′ intron sequences (Tuck and Tollervey 2013), and with intron sequences and splicing factors (Hackmann et al. 2014).
While Hrb1/Gbp2 remain bound to transcripts during translation, localization studies show that Hrb1 is predominantly nuclear at steady state and that its re-import into the nucleus is mediated by the Mtr10 karyopherin import receptor (Hacker and Krebber 2004). Both Hrb1 and Gbp2 are phosphorylated by the cytoplasmic SR-specific protein kinase Sky1 (Porat et al. 2006; Ma and Poon 2011). This phosphorylation increases the affinity of Hrb1 for the mRNP. However, while Sky1 phosphorylation is required for re-entry of Gbp2 into the nucleus, it is not essential for nuclear import of Hrb1.
Hrb1/Gbp2 appear to have roles in mRNA export; however, deletion of these genes does not disrupt or alter the nucleocytoplasmic distribution of bulk polyadenylated mRNAs (Hacker and Krebber 2004). Interestingly, overexpression of Gbp2 leads to a delay in G1 (Stevenson et al. 2001). As suggested by Rougemaille et al. (2008), Hrb1/Gbp2 may act as dedicated adaptors that recognize specific sets of transcripts. Hackmann et al. (2014) recently demonstrated that Hrb1/Gbp2 function both to recognize and to mediate the elimination of aberrantly spliced transcripts via binding to Mtr4 and to mediate the nuclear export of correctly spliced transcripts via Mex67 (Hackmann et al. 2014).
Given that the A. nidulans Hrb1 homolog, snxA, was identified as a genetic interactor with nimXcdc2 and that the snxA1 mutation arrests cells in G1 at its restrictive temperature, it is possible that SNXAHRB1 interacts with cell cycle-specific transcripts or proteins. The SR/RRM family has not previously been implicated in the regulation of the G2/M transition. These findings may therefore represent a novel regulatory mechanism in cell cycle control of A. nidulans.
Materials and Methods
Strains and general techniques
Strains used in this study are listed in Table S1. PCR primers used for gene deletion, gene tagging, and molecular diagnoses are shown in Table S2. Phusion Polymerase was used for all PCR experiments, and all other DNA-modifying enzymes were from New England BioLabs. Standard methods of Aspergillus culture (Kafer 1977), genetic analysis (Pontecorvo et al. 1953; Kaminskyj 2001), construction and analysis of heterokaryons and diploids (Todd et al. 2007), and Aspergillus transformation (Ballance et al. 1983) were employed. Appropriately supplemented minimal media (1% glucose) were used for all strain construction and genetic mapping. Rich media, composed of minimal medium (Kafer 1977) supplemented with 0.5% yeast extract and containing 2% glucose, were used for kinase assay experiments and to grow cells for DNA-mediated transformation.
Generation of a new snxA allele by a noncomplementation screen
To generate a new snxA allele, we mutagenized a diploid strain, dSWJ 3693, homozygous for nimX2 and heterozygous for snxA1 (riboA1/+ +/yA2; nimX2 snxA1 wA2/nimX2 + +; +/pyroA4; +/methB3; nicB8/+), in which the snxA1 chromosome is marked by the wA2 white-spored mutation. This diploid is green because it carries a wild-type wA allele and it expresses the nimX2 temperature-sensitive (TS) lethality at 39°–44°. Freshly harvested conidia (2 × 107) were treated for 30 min at 30° with 0.8 or 1.2 μg/ml of the UV-mimetic 4-nitroquinoline-1-oxide (4-NQO), inactivated by treatment with 10% Na2S2O3 for 5 min, and washed twice in dH2O, and then 5 × 106 spores were spread on appropriately supplemented minimal medium and grown for 4–6 days at 41° to select for strains that gain the ability to grow at the nimX2 restrictive temperature. Because one copy of snxA is defective, we reasoned that new nimX2-suppressing mutations might occur in the remaining wild-type snxA allele. Alternatively, suppression may occur by back mutation or reversion in a nimX2 allele or by dominant mutations in genes other than nimX or snxA. Thirteen candidate TS+ green colonies were recovered, streaked three times, and grown at 37° to ensure clonal purity.
To recover the new mutations in a haploid background, a parasexual genetic approach was employed (Todd et al. 2007) to break down the diploid strains into their component haploid strains nonmeiotically, i.e., via random assortment of chromosomes without crossing over. Each TS+ strain was streaked in a line on rich media containing the microtubule-destabilizing agent benomyl (Bonide, Trainor and Bonide Products, Inc.) at 60, 67.5, 75, and 82.5 μg/ml and grown at 33° for 8 days until haploid sectors grew out. Haploids were distinguished from diploids by segregation of yellow (yA2 on chromosome I) or white (nimX2 snxA1 wA2 on chromosome II) sectors or by occasional green sectors from the green diploid parent. Thirty-five white, yellow, or green haploid sectors were recovered from each TS+ diploid and streaked three times to clonal purity. The haploids were then screened on minimal media for the auxotrophic markers (pyroA4, nicB8, methB3, riboA1) and for temperature sensitivity. As expected, all wA2 sectors were phenotypically nimX2 snxA1. Among the TS+ candidate diploids, representative yellow and green haploids derived from 7 of these 13 strains were analyzed further, as reported in Results (below).
Fluorescence microscopy
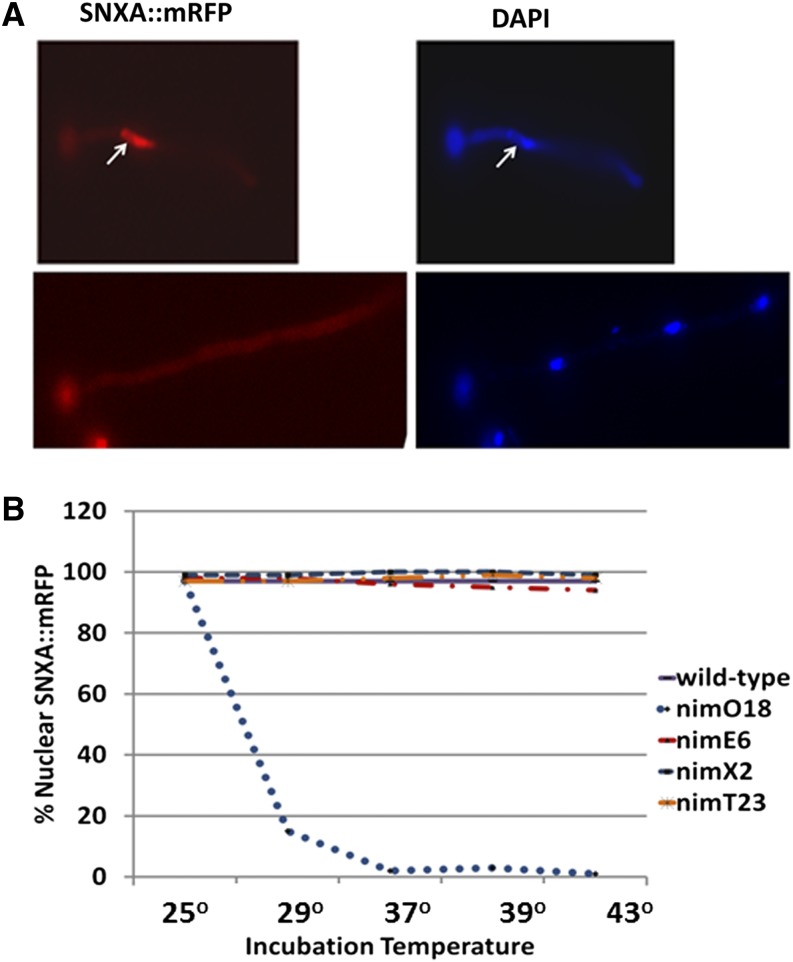
For fluorescence microscopy, cells were grown in minimal media containing 1% glucose or 50 mM glycerol as the carbon source. To visualize fluorescently labeled proteins, conidia were incubated on coverslips as described (Harris et al. 1994) and fixed for 20 min in PEMF (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, 4% formalin). Nuclear staining with 2,4-diamidino-2-phenylindole (DAPI) following fixation was as described (Brody et al. 1991). Experiments were performed in duplicate; 200 germlings (containing fewer than eight nuclei/germling) per replicate were visualized and quantitated using a Zeiss Axioimager.M1 microscope equipped with a Photometrics Coolsnap HQ2 camera. Exit from mitosis was determined by dissolution of the mitotic spindle in strains harboring both GFP-α-tubulin and SNXA::mRFP. For NIMECYCLINB localization experiments, bulk localization of NIMECYCLINB was quantitated; detecting the localization of the small nuclear pool that remains after mitotic entry and is degraded at mitotic exit is beyond the capabilities of our instrumentation.
snxA gene deletion, overexpression, and gene tagging
snxA-predicted gene structure is complex, with as many as 11 coding exons and in which the first 2 exons are very short: the first exon = 34 nt, containing 30 nt of 5′ UTR and 1 1/3 codons, ATGG; and the second exon = 16 nt and encodes 5 1/3 codons [Aspergillus Comparative Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/)]. Furthermore, these short ORFs are interspersed with unusually long and GC-rich introns of 766 and 548 nt, respectively. Moreover, an alternative start codon, 42 nt inside the 3′ end of the second intron and in-frame with the third exon, would eliminate the first 2 short exons and would create a 9-exon protein that differs by 13N-terminal amino acids from the 11-exon protein.
By one-step gene replacement, snxA was deleted by replacing all but the first two short exons and by completely eliminating the alternative nine-exon gene. Three-way fusion PCR was used to create a linear deletion construct (Yu et al. 2004) (Table S2) using the Aspergillus fumigatus pyroA gene as a selectable marker, followed by nested PCR to amplify a sufficient amount of purified product for DNA-mediated transformation. This nested construct was introduced into a snxA+ host strain (tSWJ 2973: pyrG89; pyroA4 Kutr-L::pyrG Af; nicA2; riboB2) in which the nonhomologous end-joining pathway of DNA repair (nkuA gene, encoding Ku70) was transiently disrupted (Nielsen et al. 2008) to facilitate homologous integration (Nayak et al. 2006). Weakly growing pyroA+ transformants were selected at 37° for 6 days, followed by serial streaking to clonal purity. The expected patterns for gene replacement at the snxA locus were observed by Southern blotting (data not shown), and two of these deletions were further verified by trans-locus PCR using primers that lie outside of the region covered by the nested deletion construct (Figure S3).
Overexpression studies were performed with strains carrying one copy of an alcA-driven snxA allele integrated at the argB locus. The alcA::snxA construct was made by amplifying the alternative nine-exon gene from genomic DNA, using PCR primers into which a novel SpeI or BamHI restriction site was incorporated (Table S2). The 2249-nt genomic PCR clone was digested first with SpeI, blunted by treatment with Klenow fragment, and then digested with BamHI. This fragment was cloned into the SmaI and BamHI sites of the plasmid pSDW194 (James et al. 1999) that contains the inducible-repressible A. nidulans alcA alcohol dehydrogenase gene promoter (Waring et al. 1989) and the A. nidulans argB gene as a selectable marker. Southern blotting with probes against argB and snxA was used to diagnose single argB integrations (Figure S4). alcA::snxA expression was strongly induced in minimal medium containing 200 mM ethanol + 0.04% fructose as the carbon source. Basal expression was achieved using 50 mM glycerol, and alcA::snxA expression was repressed on rich media containing 2% glucose.
GFP and mRFP tags were added to the C terminus of the wild-type snxA gene by using universal tagging cassettes (Yang et al. 2004). Linear tagging constructs were generated by three-way fusion PCR (Table S2), followed by nested PCR. These constructs were transformed into a snxA+ host (tSWJ 2353: pyrG89; pyroA4 ΔnkuA::argB; nimO18; riboB2). From each transformation five pyrG+ transformants were recovered and streaked three times to clonal purity. Creation of snxA::GFP and snxA::mRFP was verified in all 10 pyrG+ strains by trans-locus PCR (data not shown). Several of these strains were examined by fluorescence microscopy; all produced strong nuclear signals in interphase cells. These strains were outcrossed to remove the nimO18 mutation and to combine the tagged alleles with other relevant markers and mutations, including GFP-α-tubulin, GFP-nimEcyclinB, nimX2cdc2, nimT23cdc25, and nimE6cyclinB.
A myc9 epitope tag was added to the C terminus of snxA wild-type, snxA1, and snxA2 alleles by using universal tagging cassettes. This tag contains an 11-amino-acid GAGAGAGFDGA N-terminal linker followed by the myc 10-amino-acid repeats; each myc repeat is flanked by a GA dipeptide. The myc9 tag was generated de novo using two self-dimerizing primers as described by Nakajima and Yaoita (1997) (Table S2). The primers were 5′ phosphorylated using T4 Polynucleotide Kinase (New England BioLabs), boiled 3 min to inactivate the kinase and then used as template to amplify myc tags of varying length via 30 cycles of PCR. AmpliTaq DNA Polymerase Stoffel fragment (Applied Biosystems) was used for amplification in 1× Stoffel Buffer with annealing temperature 62° and a 10-min extension at 72°. The largest PCR products were recovered and purified (QIAEX II Gel Extraction Kit, QIAGEN), cloned into the EcoRV site of pBluescript KS(+), and sequenced. One of these clones, containing a myc9 fragment, was amplified further to add, at the 5′ end, a BspDI site followed by an N-terminal GAGAGAGFDGA linker; to the 3′ end, a stop codon followed by a HindIII site were added (Table S2). This amplicon was cloned into the BspDI and HindIII sites of pBluescript KS(+) and then sequenced to verify the (GA)3GFDGA-myc9-STOP tag. Finally, three universal myc9-tagging cassettes were constructed by cloning in the pyrG, pyroA, or riboB selectable marker genes from A. fumigatus. pyrGAf was removed from a plasmid, pCLS366, as an EcoRV–EcoRI fragment and cloned into the corresponding sites in the myc9 plasmid to generate pmyc9-pyrGAf. pyroAAf and riboBAf were removed from pTN01 and pTN02, respectively (Nayak et al. 2006), as HindIII (blunt)–PstI fragments and cloned into the EcoRV and PstI sites of the myc9 plasmid to create pmyc9-pyroAAf and pmyc9-riboBAf (Figure S5).
Using three-way fusion PCR followed by one-step gene replacement, the snxA+, snxA1, and snxA2 genomic loci were C-terminally tagged with myc9::riboBAf (Table S2) in five A. nidulans host strains (snxA+: tSWJ 2888; snxA1: tSWJ 5573/5574; snxA2: tSWJ 5583/5584) and diagnosed by trans-locus PCR (data not shown).
Isolation and characterization of snxA complementary DNAs
A 1.3-kb PCR fragment corresponding to the AN3739 snxA-predicted coding region (spanning exons 6–11) was used to screen an A. nidulans λgt10 complementary DNA (cDNA) library (generously provided by S. Osmani) (Osmani et al. 1988). Of eight independent phage isolates that were amplified and cloned into pBSKS(+), four were judged to be full-length based on their size (1.8–2.1 kb). Sequencing revealed two alternative snxA cDNAs (two clones of each) that were spliced differently at their N termini and composed of either 9 or 11 exons (Figure S6). Both of these isoforms correspond to alternatively spliced AN3739 transcripts identified previously by EST sequencing of A. nidulans cDNA libraries [Aspergillus Comparative Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/)] and as described above. To assess their functions, the cDNAs were fused with the A. nidulans alcA promoter in plasmid pSDW194 (James et al. 1999). Following transformation of snxA1 mutants SWJ 3676 and SWJ 3678, Southern blotting was used to identify strains carrying a single copy of alcA-driven snxA integrated at the argB or snxA loci. Ability to complement snxA1 cold sensitivity was assessed on inducing medium (200 mM ethanol + 0.04% fructose), on medium that allows basal expression (50 mM glycerol), and on repressing medium (2% glucose).
mRNA transcript analysis
RNA isolation and gene expression were determined for wild-type (PCS 439), snxA1 (SWJ 4030), and snxA2 (SWJ 5562) strains using quantitative RT-PCR (qRT-PCR) as described (Alam et al. 2012). Randomly cycling vegetative mycelia grown at 32° were collected by vacuum filtration using Miracloth (Calbiochem), frozen in liquid nitrogen, and lyophilized overnight. Total RNA was extracted using an RNeasy plant kit (Qiagen) per manufacturer’s instructions. RNA concentration and RNA quality were determined using a NanoDrop spectrophotometer to determine 260/280 ratio, followed by native agarose gel electrophoresis. Total RNA was diluted to 400 ng/μl, and genomic DNA (gDNA) elimination and reverse transcription were performed using a QuaniTect reverse transcription kit (Qiagen) according to manufacturer’s instructions, including no reverse transcriptase controls. Samples were diluted 1:5 and Quantitative real-time PCR (qPCR) was performed using iQ SYBR Green Supermix (Qiagen) according to the manufacturer’s instructions. Appropriate negative controls were used in all experiments (no template DNA; no reverse transcriptase) for each gene. Six replicates of each reaction were performed using a 20-μl total volume in 96-well optical plates in a C1000 thermal cycler using a CFX96 real-time detection system (Bio-Rad). actA (actin) was used as the reference gene, using primers actF and actR that amplify across an intron; for snxA, either of two forward primers, one straddling intron 9 (snxA-q1) and one in exon 10 (snxA-q2), was paired with one reverse primer (snxA-q3), located 80 nt into exon 11 (Table S2). qPCR amplification conditions were the following: 95°/3 min for one cycle, 95°/15 sec, 60°/15 sec, 72°/20 sec for 44 cycles, 95°/10 sec. Melt curve analysis was performed starting at 65° and rising by increments of 0.5° every 5 sec to 95°. Expression was normalized to actA and calculated using the ΔΔCt method (Livak and Schmittgen 2001). Five independent replicate experiments were performed with nearly identical results.
Western blot analysis and kinase assays
Protein purification, Western analysis, and kinase assays were performed as previously described (Osmani et al. 1991; Liu et al. 2010). Briefly, total protein extracts were generated from lyophilized mycelia as described (Liu et al. 2010). For NIMX detection, 100 μg of extracts were separated by SDS-PAGE, blotted to nitrocellulose, and probed with anti-NIMX E77 antibody (kind gift of Stephen Osmani). Detection was performed using goat anti-mouse IgG, HRP conjugate (Millipore; #12-349), and GE Healthcare Advance Chemilluminescent Western blotting detection kit. For the histone 1H kinase assay (described in Osmani et al. 1991), anti-NIMX E77 antibody was incubated with 200 μg of extracts, and immune complexes were precipitated with protein A/G sepharose (Pierce). Following precipitation, beads were washed four times and then resuspended in 20 μl kinase assay buffer (Osmani et al. 1991), incubated with substrate (Millipore histone 1H; #14-155) for 5 min, and the reaction was stopped by addition of 20 μl Laemmli sample buffer. After boiling for 5 min, the entire supernatant for each reaction was separated by SDS-PAGE, and the dried gel was exposed to autoradiography film.
For SNXA detection, 2.5 μg of extracts were separated by SDS-PAGE (4–20% TBX gels, Bio-Rad), blotted to PVDF, and probed with either myc.A7 mouse mAb at 1:3000 (Abcam ab18185) or DM1A anti-α-tubulin mouse mAb at 1:1000 (Abcam DM1A) followed by 1:5000 anti-mouse IgG HRP (Promega W4021). Protein complexes were detected by a 1-min exposure in 5 ml of TMB stabilized substrate for HRP (Promega W4121). For GFP-NIMECYCLINB detection, 2.5 μg of extracts was separated by SDS-PAGE (10% Tris-glycine gels, Bio-Rad), blotted to nitrocellulose, and probed with anti-GFP antibody at 1:1000 (Invitrogen A11122) followed by 1:10,000 anti-rabbit IgG HRP (Santa Cruz Biotechnology Sc-2004). The blots were stripped and reprobed with anti-α-tubulin mouse mAb at 1:1000 (Sigma T9026) followed by 1:10,000 anti-mouse HRP (Pierce 32230).
Results
snxA interacts with the CDK1 mitotic regulatory pathway
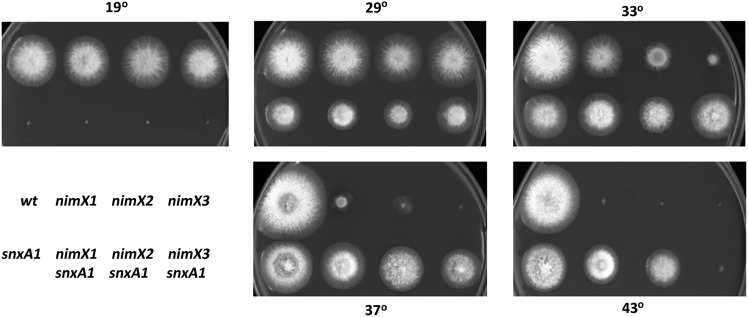
To determine if the snxA1 mutation is allele specific, snxA1/nimX double mutants were generated with three heat-sensitive nimX alleles (nimX1G225S, nimX2F223L, and nimX3Y306H); colony growth at increasing temperatures was compared for single mutants and double mutants (Figure 1). The snxA1 mutation conferred cold sensitivity at 20° and reduced colony size at permissive and semipermissive temperatures (25°–43°) in all strains tested. Suppression of heat sensitivity was evident in all three double-mutant strains at 37° and for snxA1/nimX1 and snxA1/nimX2 double mutants at 43°.
Figure 1.
snxA1 suppresses multiple alleles of nimXcdc2. Growth phenotypes of single and double mutants on minimal medium at the indicated temperatures. Days of growth at each temperature are as follows: 19°, 6 days; 29°/33°/37°/43°, 2.5 days.
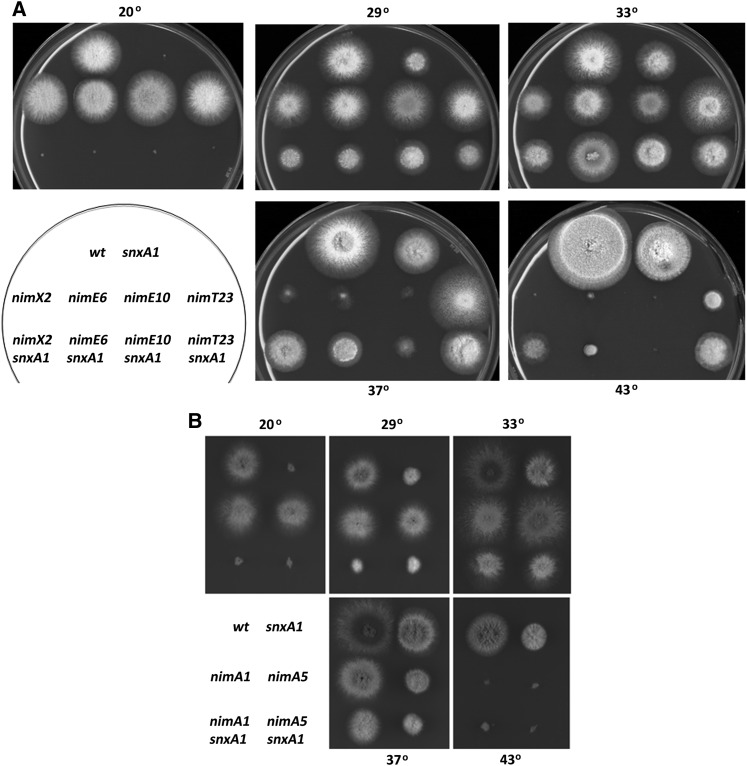
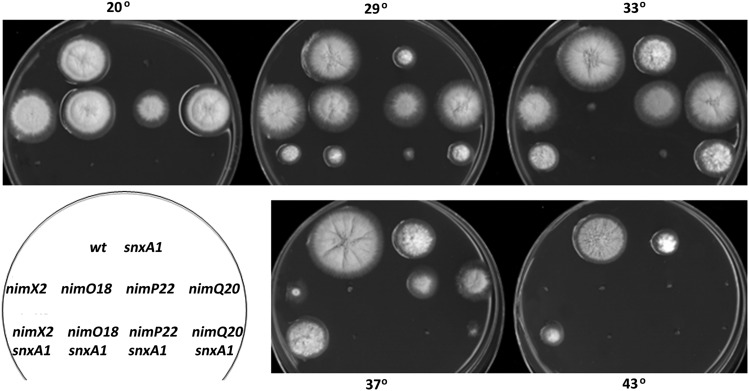
snxA1 double mutants were generated with four additional heat-sensitive G2-arresting mutations [nimE6cyclinB, nimT23cdc25, and nimA1/nimA5 (O’Connell et al. 1992)], two S-phase-arresting nim mutations [nimE10cyclinB (Figure S1) and nimP22DNApolε (S. James, unpublished results)], and two late G1-arresting mutations [nimO18dbf4 (James et al. 1999) and nimQ20mcm2 (James et al. 1995)]. The late-G1-arresting nimO18dbf4 and nimQ20mcm2 mutations fail to initiate DNA synthesis at restrictive temperature. However, they do undergo mitotic catastrophe by attempting to segregate their unreplicated chromatin; i.e., they undergo a pseudomitosis resulting from failure to activate a DNA replication checkpoint at the G1/S transition (James et al. 1999). Thus, these mutants are presumed to activate the CDK1 mitotic induction pathway at restrictive temperature. Growth of double mutants was compared to single mutants at increasing temperatures (Figure 2 and Figure 3). snxA1 suppressed the heat-sensitive G2-arrest phenotypes of nimX2cdc2, nimE6cyclinB, and nimT23cdc25 at 37° and 43° (Figure 2A). In contrast, snxA1 failed to suppress the nimA1 and nimA5 G2-M regulatory defects, and instead the double mutants were more growth-impaired than either single mutant at the permissive temperatures of 29° and 33° (Figure 2B). Similarly, snxA1 exhibited little or no suppression of the nimE10cyclinB and nimP22DNApolε S defects, and snxA1 failed to suppress the heat-sensitive G1 arrest of nimO18dbf4 and nimQ20mcm2. Instead, and contrasting with snxA1 rescue of CDK1 pathway mutations, growth and temperature defects became more severe in these latter four snxA1 double mutants, as expected for mutations that operate in unrelated pathways. Therefore, snxA1 suppression of heat-sensitive cell cycle defects appears restricted to components of the CDK1 mitotic regulatory pathway. Both NIMECYCLINB and NIMTCDC25 are specifically required for the activation of NIMXCDC2 to allow mitotic entry, and NIMA is required for the partial disassembly of nuclear pore complexes, which allows entry of the active NIMXCDC2/CYCLINB complex into the nucleus (De Souza et al. 2003, 2004). The suppression of three different nimXcdc2 alleles and of mutations in multiple activators of the NIMXCDC2/NIMECYCLINB G2-M regulatory pathway, but not of mutations affecting G1- or S-phase control or the NIMA G2-M regulator, strongly suggests a role for SNXA in the regulation of the G2/M transition via the CDC2/CYCLINB regulatory pathway.
Figure 2.
snxA1 suppresses mutations in multiple components of the CDC2/CYCLINB cell cycle regulatory pathway but not mutations in nimA. Growth phenotypes of single and double mutants on minimal medium at the indicated temperatures. Days of growth at each temperature are as follows: 20°, 5 days; 29°/33°/37°, 2.5 days; 43°, 3.5 days. (A) Growth phenotypes of single and double mutants in the CDC2/CYCLINB pathway. (B) Growth phenotypes of single and double mutants in nimA.
Figure 3.
snxA1 is not a general suppressor of cell cycle defects. Growth phenotypes of single and double mutants on rich medium at the indicated temperatures. Three G1- or S-phase cell cycle mutations (nimO18, nimP22, and nimQ20) were tested for suppression by snxA1 and compared with the G2-M cell cycle mutation nimX2. Days of growth at each temperature are as follows: 20°, 7 days; 29°/33°/37°/43°, 2.5 days.
SNXA encodes the A. nidulans homolog of Hrb1Sc/Gbp2Sc and human heterogeneous ribonucleoprotein-M
snxA (suppressor-of-nimXcdc2) was previously assigned to linkage group II (LGII) via parasexual mapping (McGuire et al. 2000). After several unsuccessful attempts to complement snxA1 using a LGII-specific cosmid library (Brody et al. 1991) [obtained from the Fungal Genetics Stock Center (FGSC), Kansas City, MO] or a total genomic library (Osherov and May 2000) (obtained from the FGSC), the location of snxA within LGII was determined by linkage analysis. The initial discovery of linkage to anB8 (17.4 map units, n = 144) was followed by three-factor crosses involving snxA1, cnxE16, and hisG113, in which snxA localized 10.7 map units rightward of cnxE and 1.3 map units rightward of hisG (n = 224).
To isolate the wild-type snxA gene, a group of five overlapping fosmids (Galagan et al. 2005) (obtained from the FGSC) that span the relevant region were cotransformed into a snxA1 argB2 strain (SWJ 3676: riboA1; snxA1; argB2; pyroA4; chaA1) using the A. nidulans autonomously replicating helper plasmid pDHG25 (Brody et al. 1991) (obtained from the FGSC), which carries A. nidulans argB as a selectable marker. Cotransformed cells were grown for 18 hr at 37° to establish thousands of argB+ transformants, then shifted to 20° for 8 days to select for complementation of snxA1 cold sensitivity. Only one fosmid, 8201-D10, complemented snxA1; >100 snxA+ argB+ cotransformants were obtained. 8201-D10 overlapped a noncomplementing fosmid (8199-G2) over two-thirds of its length, thereby eliminating 10 of 16 fosmid-borne genes from further consideration and narrowing the search to a group of 5 protein-coding genes (AN3738 to AN3742) and one leucyl-transfer RNA (tRNA) in the non-overlapping region of 8201-D10. Four fragments covering this region were subcloned into pBluescript KS(+) and transformed into a snxA1 strain. Only one plasmid, containing AN3739 and lacking the tRNA gene, complemented snxA1, thus identifying AN3739 as the putative snxA gene.
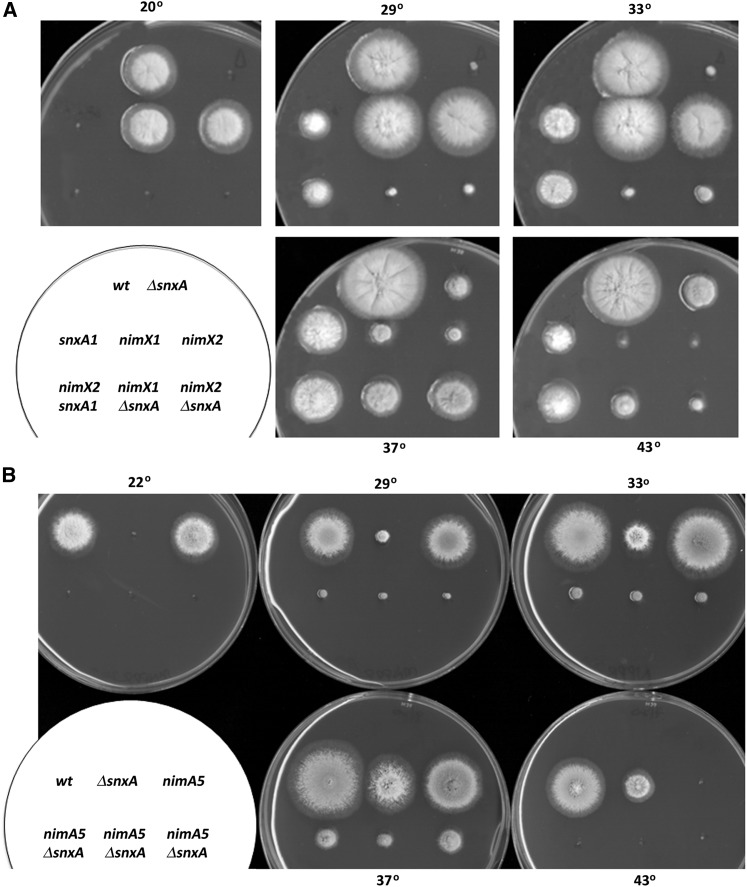
Deletion of AN3739/snxA was accomplished by one-step gene replacement (Figure S3). ΔsnxA strains were viable and slightly more impaired than snxA1 for vegetative growth and conidiation at all temperatures. Deletion of snxA phenocopied the snxA1 mutation, conferring cold-sensitive lethality at 20°, mild heat sensitivity at 43°, and suppressing the heat sensitivity of nimX1 and nimX2 at 37° and 43° (Figure 4A). Conversely, and similar to snxA1 nimA5 and snxA1 nimA1 double mutants (Figure 2B), ΔsnxA did not suppress nimA5 (Figure 4B); these double mutants remained heat sensitive and cold sensitive, with pronounced impairment of growth and conidiation at the permissive temperatures of 29° and 33°, typical of mutations that act through different pathways.
Figure 4.
ΔsnxA suppresses nimXcdc2 mutations but does not suppress the nimA5 G2-M defect. (A) Growth phenotypes of nimX- single and nimX- ΔsnxA double mutants on rich medium at the indicated temperatures. Days of growth at each temperature are as follows: 19°, 6 days; 29°/33°/37°/43°, 3 days. (B) Growth phenotypes of three independent nimA5 ΔsnxA double mutants at the indicated temperatures. Days of growth at each temperature are as follows: 22°, 5 days; 29°, 2.5 days; 33°/37°/43°, 2 days.
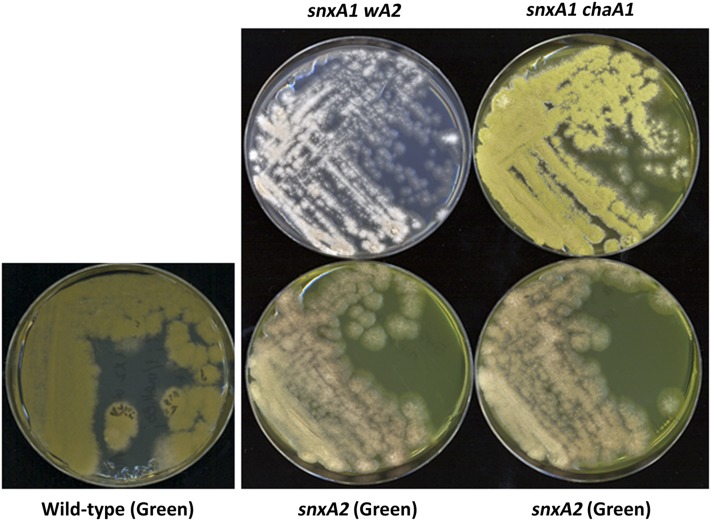
A noncomplementation screen was used to generate an additional snxA mutation in a diploid strain homozygous for nimX2 and heterozygous for snxA1, as described in Materials and Methods. Thirteen diploid revertants that had regained the ability to grow at the nimX2 restrictive temperature of 41° were haploidized by treatment with benomyl. Haploid strains derived from 7 of the 13 candidates were crossed to hisG113, and of these one harbored a cold-sensitive nimX2 suppressor that was tightly linked to hisG (two recombinants in 60 progeny, 3.3 map units). This mutant was named snxA2. To verify the ability of snxA2 to suppress nimX2, snxA2 nimX2 double mutants were resynthesized by crossing snxA2 nimX+ × nimX2 snxA+. In these double mutants, snxA2 suppressed nimX2 temperature sensitivity at 37° and 43° in a manner similar to snxA1 (Figure S7). The snxA2 phenotype differs from snxA1. Whereas snxA1 grows optimally and conidiates strongly at 37°, snxA2 mutants grow and conidiate optimally at 33° and exhibit a defect in conidiation at 37°, suggesting that snxA2 harbors a more severe defect than snxA1 and that snxA1 and snxA2 result from different alterations (Figure 5).
Figure 5.
The snxA2 defect is more severe than that of snxA1. snxA1 strains harboring white (wA2) and chartreuse (chaA1) spore color mutations (top row), and green snxA2 strains (bottom row) were streaked onto minimal medium and grown for 3 days at 37°. The brownish coloration of the snxA2 strains reflects the absence of green conidia, demonstrating weaker growth at 37°.
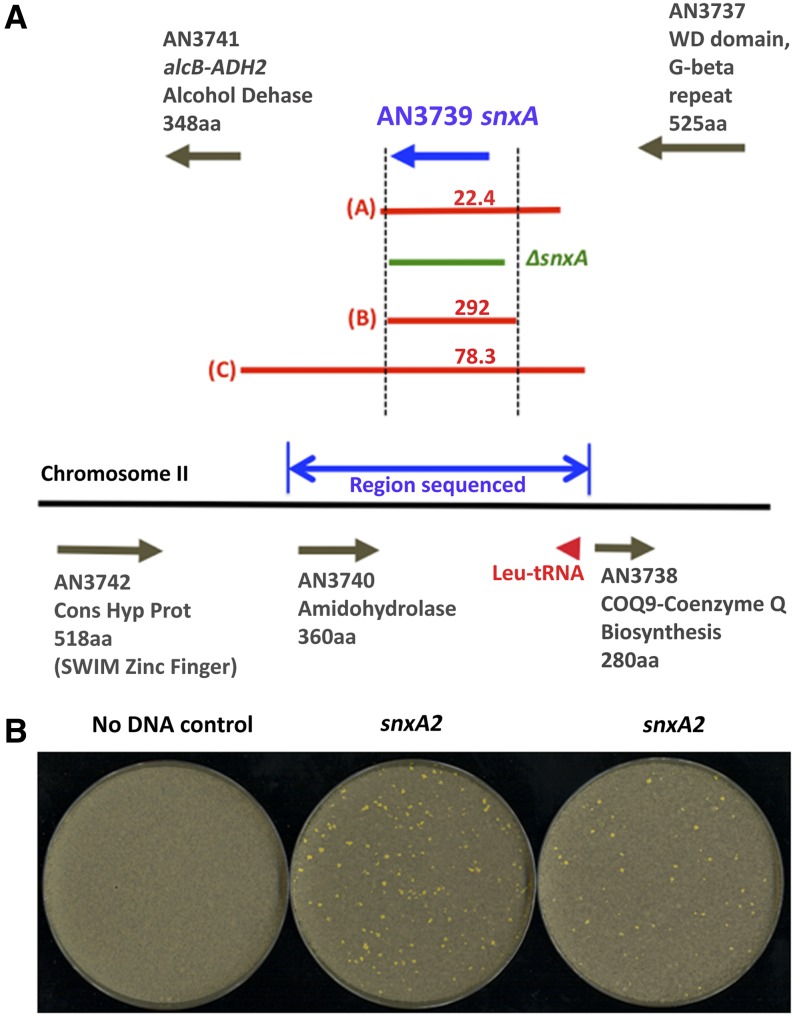
To verify the location of the snxAl and snxA2 mutations, one PCR-derived and two plasmid-derived linear wild-type DNA fragments were used to complement snxA1 in two strains (tSWJ 5573/5574) and snxA2 in two strains (tSWJ 5567/5584). These strains harbored a deletion of the nkuA (Ku70) gene to facilitate targeted integration by homologous recombination (Nayak et al. 2006). Each of these transformations yielded hundreds of transformants that efficiently complemented the cold sensitivity of the mutants (Figure 6). The shortest linear DNA, a 3353-nt EcoRI–SnaBI fragment, begins approximately in the middle of intron 1 and encompasses exons 2–11 and 90% of the AN3739 3′ UTR (263 of 291 nt).
Figure 6.
Complementation of snxA1 snd snxA2 by linear DNA fragments. snxA1 strains tSWJ 5573/5574 and snxA2 strains tSWJ 5567/5584, containing a deletion of nkuA (Ku70), were transformed with a PCR-derived linear DNA fragment of 3897 nt (panel A, red “A”) and two plasmid-derived linear DNA fragments of 3530 nt (panel A, red “B”) and 6686 nt (panel A, red “C”). Transformants were grown at 21° for 11 days to select for complementation of cold sensitivity. (A) Complementation results. Single arrows denote genes in the snxA region. Red lines indicate linear fragments used for transformation. Green line indicates the region deleted in ΔsnxA strains. Numbers in red indicate the number of snxA2-complementing transformants per microgram of DNA. Blue double-arrow indicates the region sequenced in snxA1 and snxA2 mutants. (B) snxA2 transformants complemented by the shortest, 3530-nt linear DNA fragment.
Allelism between snxA1, snxA2, and ΔsnxA was assessed in complementation tests using diploids and heterokaryons (Figure S8). In heterozygous diploids, each snxA mutant was fully recessive (Figure S8A). Only one homozygous diploid, snxA1/snxA1, was isolated (Figure S8A). Although balanced heterokaryons harboring ΔsnxA/snxA1 and ΔsnxA/snxA2 were repeatedly established, diploids could not be derived after three independent attempts. Therefore, complementation was examined in these balanced heterokaryons (Figure S8B). snxA1 and snxA2 failed to complement ΔsnxA, conferring cold-sensitive lethality at 20° and growth impairment at 43° similar to haploid snxA mutants. Interestingly, at 43° the growth defect of ΔsnxA/snxA2 heterokaryons was dramatically enhanced, showing greater impairment than ΔsnxA/snxA1 heterokaryons or haploid ΔsnxA and snxA2 single mutants.
AN3739 shares ∼30% identity with S. cerevisiae Hrb1/Gbp2 (1e-45 and 2e-43, respectively), nonessential shuttling mRNA-binding proteins containing an N-terminal SR domain and three RRMs (Figure S6A). As the only Hrb1/Gbp2-homologous protein in the A. nidulans genome, AN3739 contains a similar overall structure, with three highly conserved RRMs and an N-terminal SR domain that is slightly more similar to Hrb1 than to Gbp2 (34 identities/eight similarities vs. 33 identities/four similarities). The closest human homolog of AN3739, heterogeneous ribonucleoprotein-M (hnRNP-M, 3e−27), is an abundant spliceosomal component involved in pre-mRNA splicing and alternative splicing (Lleres et al. 2010; Xu et al. 2014). hnRNP-M also contains three conserved RRM motifs but lacks an N-terminal SR domain (Figure S6B). SNXA and hnRNP-M share two features that distinguish them from the budding yeast proteins. First, the sequences between RRM1-RRM2 and RRM2-RRM3 are glycine-rich, containing several runs of between two and seven glycines. Second, SNXA and hnRNP-M contain tripeptide motifs that are commonly methylated on arginine. SNXA contains seven RGG tripeptides, of which five are clustered between RRM2 and RRM3; hnRNP-M has undergone an expansion of this same region, which contains 25 RMG/RVG/RIG/RMA/RMV motifs that are known to be methylated (Hung et al. 2009).
snxA function was examined further by overexpressing genomic and cDNA clones. Overexpression of a 9-exon wild-type AN3739/snxA genomic DNA, containing exons 3–11 and 90% of the 3′ UTR, efficiently rescued snxA1 cold sensitivity and abrogated the suppression of nimX2 by snxA1 (Figure S4C; Figure S9). In addition, overexpression of this alcA::snxA+ gDNA complemented ΔsnxA defects in growth and cold sensitivity (Figure S10). Unexpectedly, overexpression of the 9-exon region obtained from snxA1 mutant gDNA also complemented snxA1 efficiently (data not shown), suggesting that (1) the snxA1 mutation lies upstream of exon 3 or (2) overexpression of partially functional snxA1 mutant protein was able to fully rescue snxA1 phenotypes. We also generated alcA-driven cDNAs from the 9- and 11-exon alternatively spliced cDNAs described in Materials and Methods. The 9-exon cDNA contains 27 nt of 5′ UTR. Three different 11-exon cDNA versions were tested, containing 32, 39, or 162 nt of 5′ UTR sequence. No upstream ATGs occur in these four UTR sequences. Overexpression of each cDNA from a single copy at the argB locus complemented snxA1 to varying degrees (Figure S9).
Typically, an exonic mutation can be complemented by integration of promoter-less cDNAs at the endogenous locus to restore a wild-type allele under control of the native promoter. However, in transformations with plasmid-borne alcA::snxA cDNAs, neither the 11- nor the 9-exon cDNAs were able to complement snxA1 on glucose, i.e., in an ethanol-independent manner, in 167 argB+ transformants with the 11-exon cDNA and in a total of 70 argB+ transformants obtained by using the 9-exon cDNA. There is very little difference in length and sequence of the two cDNAs (Figure S6). The two additional N-terminal exons in the 11-exon cDNA encode only nine amino acids; these two exons are interrupted by unusually long introns of 766 and 548 nt. The 9-exon cDNA begins within the 548-nt second intron (42 nt upstream of the intron 2, 3′ splice junction) and encodes 14 N-terminal amino acids by which it differs from the 11-exon cDNA (Figure S6). Given that these two cDNAs are therefore identical except at a small region in the N terminus, the absence among transformants of constitutive restoration of the wild-type phenotype suggests that (1) the snxA1 mutation may lie within an intron or an upstream regulatory region; (2) the mutation may lie so close to the coding region N terminus as to preclude the likelihood of a crossover event between the N terminus of the cDNA and the mutation at the endogenous locus; or (3) the snxA1 alteration may affect chromatin structure and function in the snxA region.
We amplified and sequenced the AN3739 genomic region in snxA1, spanning 5882 nucleotides from nt 3,224,102 to 3,218,220 (Aspergillus Genome Database: http://www.aspgd.org). The sequenced region begins 85 nt before the AN3738 start codon and 671 nt upstream of the start site of the longest 11-exon snxA cDNA (nt 3,213,431). This cDNA start site precedes the exon 1 start codon by 162 nt. The 5′ sequenced region contains a 100-nt-long leucyl-tRNA gene that ends 333 nt upstream of the cDNA start site. The 3′ region includes the AN3739 3′ UTR, 3′ flanking intergenic region, and the adjacent gene AN3740. The snxA1 sequenced region extends >1000 nt beyond both ends of the 3353-nt sequence that fully complemented snxA1 and snxA2 in transformations with a linear wild-type DNA fragment (Figure 6). snxA2 was sequenced across 4842 nt from nt 3,223,063, which lies 82 nt upstream of the 3353-nt snxA1/snxA2 linear complementing DNA region, and ends at 3,218,220, as for snxA1 (above). Three independent PCR products each from one wild-type strain (PCS 439), a minimum of two snxA1 strains (SWJ 3676, SWJ 3678, SWJ 3404, or SWJ 2862), and two snxA2 strains (EAK 5496 and EAK 5497) were sequenced on both strands, for a total of at least six snxA1 and six snxA2 mutant-derived PCR products at each interval, and compared with the published genomic sequence derived from strain FGSC A4 (Aspergillus Comparative Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org/). In addition, snxA1 was sequenced more extensively in the 5′ flank plus exons 1 and 2 and introns 1 and 2 from nt 3,223,572 (141 nt upstream of the startsite of the 11-exon cDNA) to 3,221,861 (1711 nt), over three intervals, with at least 10 templates (20 sequences) and as many as 16 templates (33 sequences) per interval. Without exception, no DNA sequence changes were detected in snxA1 or snxA2.
Multiple lines of genetic and phenotypic evidence indicate that AN3739 is snxA: First, classical genetic mapping localized the mutation to a small interval on chromosome II. Second, plasmid subclones containing only the wild-type AN3739 DNA complemented the snxA1 mutation. Third, both snxA1 and snxA2 were efficiently complemented by a linear 3.353-kb fragment spanning most of the AN3739 locus. Finally, the fully recessive snxA1 and snxA2 mutations failed to complement a deletion of AN3739, indicating that these three mutations are allelic. Phenotypic support for AN3739 being snxA is evident in that deletion of snxA phenocopies snxA1 and snxA2 in both cold sensitivity and suppression of nimX2. Conversely, overexpression of wild-type and snxA1 mutant genomic clones and alternatively spliced AN3739 cDNAs complemented snxA1 and ΔsnxA when integrated as a single copy at the argB locus; and importantly, overexpression of a snxA+ gDNA abrogated the suppression of nimX2 by snxA1. Therefore, despite the absence of DNA sequence changes in the snxA1 and snxA2 ORFs and their upstream and downstream flanking regions, we conclude that snxA (AN3739) encodes the single A. nidulans homolog of S. cerevisiae Hrb1/Gbp2 and human hnRNP-M.
snxA1 and snxA2 mutations decrease snxA expression
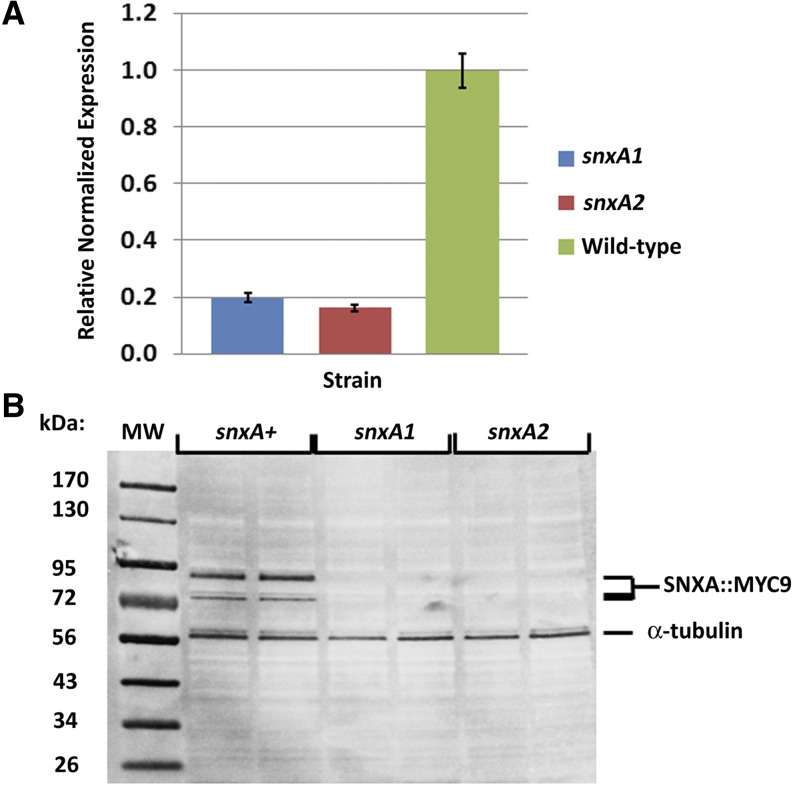
Because deletion of snxA phenocopies both the snxA1 and snxA2 mutations, we hypothesized that the hypomorphic snxA1 and snxA2 phenotypes are due to decreased snxA expression. mRNA transcript levels were quantitated in randomly cycling vegetative mycelia in snxA1, snxA2, and snxA+ strains using qRT-PCR. Both snxA1 and snxA2 exhibited fivefold decreased expression relative to wild type (Figure 7A). To investigate the effects of snxA1 and snxA2 mutations on the levels of protein, the snxA1, snxA2, and snxA+ alleles were tagged with a (GA)3GFDGA-myc9 epitope, and their levels were detected by Western blotting from randomly cycling, vegetative mycelia grown in liquid media. As predicted, SNXA levels were greatly reduced or absent in both mutants (Figure 7B). Thus, the hypomorphic phenotypes caused by both snxA1 and snxA2 are due to decreased expression of snxA.
Figure 7.
snxA mRNA and protein levels are reduced in snxA mutants. (A) To determine relative snxA mRNA expression, qRT-PCR analysis of randomly cycling vegetative mycelia was performed on snxA+, snxA1, and snxA2 strains. Relative expression was normalized to actA and calculated using the ΔΔCt method. Data shown are the average relative normalized expression from six replicates. Bars = SEM. (B) To determine relative SNXA protein levels, colorimetric Western analysis of total protein extracts from randomly cycling vegetative mycelia of myc9-tagged snxA+, snxA1, and snxA2 strains was performed. Blots were probed with mAb mycA7, washed, and reprobed with anti-α-tubulin mAb as a loading control.
SNXA1 does not affect NIMX2CDC2 kinase activity
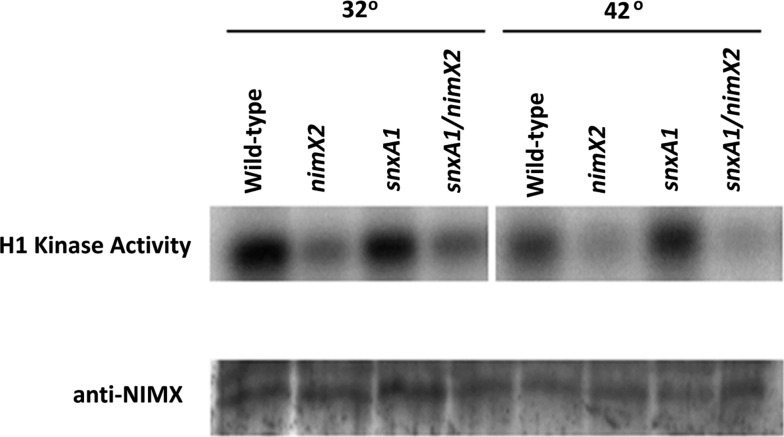
To determine if SNXA affects the levels of NIMXCDC2 protein or NIMXCDC2/NIMECYCLINB kinase activity, histone 1H kinase assays and Western blots were performed using total protein extracts from strains carrying snxA+/nimX+, snxA+/nimX2, snxA1/nimX+, or snxA1/nimX2 grown at 32° or germinated at 32° and then shifted for 3 hr to the nimX2 restrictive temperature of 42° (Figure 8). No differences in NIMXCDC2 protein levels were evident in any of the strains at permissive temperature or after shifting to restrictive temperature. However, strains harboring the nimX2 mutation had lower NIMX2CDC2/NIMECYCLINB kinase activity at permissive temperature. Furthermore, NIMX2CDC2/NIMECYCLINB kinase activity decreased in both the snxA+/nimX2 and snxA1/nimX2 strains after shifting to 42° (Figure 8, lanes 6 and 8). These experiments, which were repeated three times with identical results, strongly suggest that snxA1 suppression of nimX2 does not occur by increasing the activity of NIMX2CDC2/NIMECYCLINB.
Figure 8.
SNXA does not affect NIMXCDC2 histone 1H kinase activity. To determine if snxA1 suppresses nimX2 by affecting NIMX protein levels or activity, snxA+/nimX+ (lanes 1 and 5), nimX2 (lanes 2 and 6), snxA1 (lanes 3 and 7) and snxA1/nimX2 (lanes 4,8) were grown at the nimX2 permissive temperature (32°) or germinated at 32° and shifted to restrictive temperature (42°) for 2 hr. Histone 1H kinase assays were performed on anti-NIMX immunoprecipitates followed by SDS-PAGE. A total of 100 ug total protein was separated by SDS-PAGE, blotted to nitrocellulose, and detected with anti-NIMX antibody E77.
SNXA localizes to the nucleus during interphase
To localize SNXAHRB1 in cells, snxA was C-terminally tagged with green fluorescent protein (GFP) and monomer red fluorescent protein (mRFP) by three-way fusion PCR, followed by one-step gene replacement. A very strong nuclear signal was present in 97% of randomly cycling cells, with diffuse signal in 3% of cells, as demonstrated by colocalization with DAPI-stained nuclei (Figure 9A). SNXA::mRFP was observed in the nucleoplasm but appeared to be excluded from the nucleolus because both SNXA::mRFP and DAPI were excluded from this area. In asynchronous cultures, ∼97% of nuclei were in interphase and 3% were mitotic. The high proportion of cells with nuclear SNXA::mRFP (97%) suggests that SNXA is not concentrated in the nucleus during semiclosed mitosis, during which protein localization has been hypothesized to be regulated by diffusion and localized binding (De Souza et al. 2004). Thus, the observed pattern suggests that SNXAHRB1 nuclear localization is not required at mitosis. To determine if SNXAHRB1 is nuclear during interphase but exits the nucleus at mitosis, strains harboring C-terminally tagged SNXA::GFP or SNXA::mRFP were combined with the following heat-sensitive mutations: nimX2cdc2, nimE6cyclinB, or nimT23cdc25, which are slightly leaky G2-specific mutations; or nimO18dbf4, a highly heat-sensitive G1/S mutation that prevents DNA replication and undergoes mitotic catastrophe, leading to mitotic arrest at restrictive temperature (James et al. 1999). As shown in Figure 9B, localization of SNXAHRB1 was predominantly nuclear in nimX2cdc2, nimE6cyclinB, and nimT23cdc25 cells at all temperatures. However, as nimO18 cells began to enter mitotic arrest at the semipermissive temperature of 29° and were fully arrested at 37°, nuclear localization of SNXAHRB1 decreased to 0%. The entire cell was diffusely stained during mitotic arrest, suggesting that SNXAHRB1 exits the nucleus during mitosis.
Figure 9.
SNXA localizes to the nucleus in 97% of randomly cycling cells. Three-way fusion PCR was used to C-terminally tag SNXA by one-step gene replacement. Following transformation of A. nidulans with PCR products, gene replacement with snxA::mRFP::pyrG or snxA::GFP::pyrG was confirmed by trans-locus PCR and Southern blot. Fluorescence microscopy was performed to visualize SNXA::mRFP and SNXA::GFP. Strains harboring various cell cycle mutations were crossed with transformants to obtain strains with either SNXA::mRFP or SNXA::GFP and specific cell cycle mutations (nimO18, nimE6, or nimX2). (A) Fluorescence micrographs showing nuclear (top) and non-nuclear (bottom) localization of SNXA::mRFP at 32°. Germlings were stained with DAPI after fixation to colocalize SNXA::mRFP to nuclei. Arrows indicate nucleoli. (B) SNXA::mRFP or SNXA::GFP strains with indicated cell cycle mutations were inoculated into liquid minimal media on coverslips, incubated at the indicated temperatures for 16 hr, and visualized by fluorescence microscopy. A total of 200 germlings per strain was counted and scored for nuclear vs. non-nuclear SNXA::mRFP or SNXA::GFP localization.
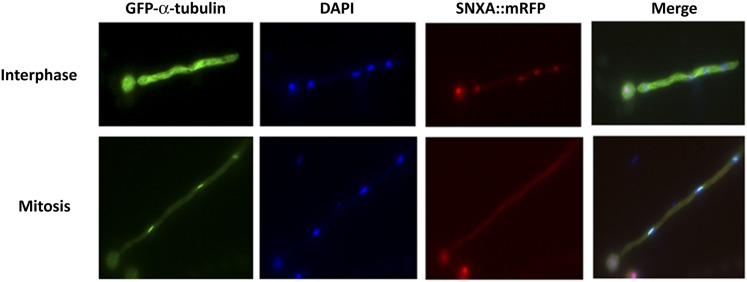
To demonstrate that SNXAHRB1 exits the nucleus during mitosis, a strain with SNXA::mRFP and GFP::α-tubulin was studied. Approximately 97% of randomly cycling cells had nuclear SNXA::mRFP and an interphase array of microtubules (Figure 10). Conversely, 100% of hypha with mitotic spindles (n > 200) were devoid of nuclear SNXA::mRFP and exhibited a diffusely stained cytoplasm. This demonstrates that SNXAHRB1 exits the nucleus during mitosis before formation of the mitotic spindle.
Figure 10.
SNXA::mRFP is absent from the nucleus during mitosis. A strain carrying SNXA::mRFP was crossed with a strain harboring alcA::GFP::α-tubulin. A double-tagged strain was inoculated into minimal media onto coverslips and incubated 14 hr at 29°, followed by fixation, DAPI staining, and visualization by fluorescence microscopy. One hundred percent of hyphae with mitotic spindles (n > 200) were devoid of nuclear SNXA::mRFP and exhibited a diffusely stained cytoplasm.
snxA1 and ΔsnxA affect NIMECYCLINB localization
As discussed earlier, proper localization of mitotic kinases is essential for mitotic progression. Because snxA1 does not suppress nimX2 by affecting either its protein or activity levels, we suspected that it may affect the localization of NIMXCDC2/NIMECYCLINB. The levels of NIMECYCLINB could be monitored using a strain harboring GFP-labeled NIMECYCLINB (Nayak et al. 2010) (kind gift of Berl Oakley). We reasoned that because NIMXCDC2 is active as a protein kinase only when bound to NIMECYCLINB (Osmani et al. 1994), and because its nuclear localization mirrors that of NIMECYCLINB (Nayak et al. 2010), if snxA1 affects the localization of NIMECYCLINB, this could also affect NIMXCDC2/NIMECYCLINB localization. Wu et al. (1998) and Nayak et al. (2010) previously characterized NIMECYCLINB as having predominantly nuclear localization during interphase (∼50–60% of hyphae had nuclear localization at interphase) and localization during mitosis that was characterized by diffuse staining of the entire hypha. To determine if either snxA1 or ΔsnxA affect the localization of NIMECYCLINB, we generated strains harboring the GFP-tagged NIMECYCLINB together with snxA+, snxA1, ΔsnxA, nimX2, or snxA1/nimX2 by crossing strain LO1578 (kind gift of Berl Oakley) with strains having various snxA backgrounds. LO1578 harbors GFP-NIMECYCLINB under control of the endogenous nimE promoter; the fusion protein was reported to be fully functional and to cause no apparent mitotic or cell cycle defects (Nayak et al. 2010). We determined the levels of GFP-NIMECYCLINB in randomly cycling vegetative mycelia from snxA+, snxA1, and ΔsnxA strains grown at 33° and found that mutation or deletion of snxA does not alter the expression of GFP-NIMECYCLINB (Figure S11). The resulting strains were germinated and the percentage of germlings (containing two to eight nuclei) with predominantly nuclear GFP-NIMECYCLINB were quantitated at the snxA1 restrictive temperature (20°), nimX2 restrictive temperature (44°), nimX2 semipermissive temperature (37°), or permissive temperature (32°).
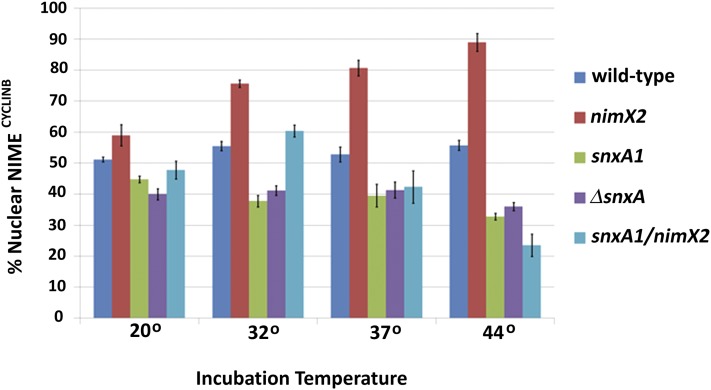
As shown in Figure 11, wild-type germlings (snxA+/nimX+) exhibited ∼50–60% nuclear GFP-NIMECYCLINB at all temperatures tested, in agreement with published data (Wu et al. 1998). Because snxA1 was originally identified as a suppressor of nimX2, we also studied GFP-NIMECYCLINB localization in nimX2 mutants. nimX2/snxA+ germlings exhibited nuclear GFP-NIMECYCLINB similar to that of wild-type germlings at 20°, but the levels increased at permissive and semipermissive temperatures and approached 85% during the slightly leaky G2 arrest (44°). This increase in nuclear GFP-NIMECYCLINB would be expected as the germlings arrest in G2. As in wild-type cells, GFP-NIMECYCLINB localization of nimX+/snxA1 and nimX+/ΔsnxA germlings was consistent over a range of temperatures, but was slightly lower at all temperatures compared to wild-type germlings, ranging from 5% to 20% lower than in wild-type cells. Whereas nimX2/snxA1 germlings generally mirrored wild type at 20°, 32°, and 37°, 50% fewer cells showed nuclear localization at 44°, relative to wild type.
Figure 11.
snxA1 and ΔsnxA strains have decreased nuclear NIMECYCLINB. Strains harboring the GFP-tagged NIMECYCLINB together with snxA+, snxA1, ΔsnxA, nimX2, or snxA1/nimX2 were grown to germling stage (two to eight nuclei) at the indicated temperatures, and the number of germlings with predominantly nuclear GFP-NIMECYCLINB was quantitated. The means of four to six replicates per strain are shown, with 100 germlings scored per replicate. Bars = SEM.
Given that NIMECYCLINB exits the nucleus at mitosis, one possible reason for decreased nuclear localization of GFP-NIMECYCLINB in snxA1/ΔsnxA cells could be an increase in mitotic index. To determine if the localization differences were due to an increased percentage of cells in mitosis, which would lead to a drop in the percentage of cells with nuclear GFP-NIMECYCLINB due to its destruction at the metaphase-to-anaphase transition, the chromosome mitotic index was assessed in these same strains under the same conditions. Due to fluorescence interference, mitotic index could not be assessed in the same slides as nuclear NIMECYCLINB; thus NIMECYCLINB localization and chromosome mitotic index were measured in separate experiments. The percentage of mitotic cells in all strains at permissive temperatures was between 2 and 4%, except at 44°, where nimX2/snxA+ germlings were 0.5% mitotic and nimX2/snxA1 germlings were 6% mitotic. Although we have conservatively reported a small increase in mitotic index (to 6%) in the snxA1/nimX2 double mutant at 44°, repeated observations in these cells (data not shown) gave variable mitotic indices as high as 10–15%. It is not possible to determine the percentage of mitotic cells below 25° in strains containing deleted or mutated snxA due to the accumulation of nuclear abnormalities. These data therefore demonstrate a temperature-dependent perturbation of nuclear NIMECYCLINB localization in snxA-deficient strains.
Discussion
Multiple lines of evidence reveal that in A. nidulans SNXAHRB1 plays an important role in cell cycle progression, functioning both at G1 and G2. At G2, snxA1 and snxA2 affect the A. nidulans CDK1, NIMXCDC2. In addition to isolation of the cold-sensitive, G1-arresting snxA1 mutation as an extragenic suppressor of nimX2 (McGuire et al. 2000), which arrests cells in G2 at elevated temperatures, we demonstrate here that snxA1 suppression is not allele-specific and that snxA1 also suppresses mutations in two heat-sensitive, G2-specific regulators of NIMXCDC2: NIME6CYCLINB and NIMT23CDC25. In contrast, snxA1 exhibits little or no suppression of the G2-arresting nimA1 and nimA5 mutations, of the S-arresting nimE10cyclinB mutation, or of three G1- and S-phase-specific mutations. Functional NIMA is required for the partial disassembly of nuclear pore complexes, which allows mitotic regulators to enter the nucleus (De Souza et al. 2003, 2004); together with the specific suppression of CDK1 pathway mutations, the lack of suppression of nimA mutations by mutation or deletion of snxA suggests that SNXA specifically affects the CDK1 pathway. Deletion of snxA phenocopies the snxA1 and snxA2 mutations in its cold sensitivity and suppression of nimX2, suggesting that snxA1 and snxA2 are hypomorphic alleles and further supporting the finding that snxA interacts genetically with nimX. That snxA1 and snxA2 are hypomorphic alleles was verified by qRT-PCR and immunoblotting.
Our previous results using double reciprocal hydroxyurea block/release assays showed that at reduced temperatures snxA1 leads to a nonreversible cell cycle arrest in G1 that causes gross nuclear abnormalities after prolonged incubation at 19° (McGuire et al. 2000). We repeated these experiments with snxA1 strains used here, with identical results (data not shown). Given that NIMXCDC2 functions both at G1 and G2 (Osmani et al. 1994), it is not surprising that a suppressor of nimX2 has effects at G1 and G2.
The nature of the snxA1 and snxA2 defects is at present mysterious. Given that both alleles failed to complement ΔsnxA, we conclude that they are allelic with the deletion. Moreover, a linear wild-type fragment spanning the 3′ half of intron 1 through exon 11 could efficiently complement snxA1 and snxA2 cold sensitivity. Both alleles were isolated after mutagenesis with the UV-mimetic, 4-NQO, leading to the expectation that they would harbor point mutations or other DNA sequence alterations. Initially, we found that (1) overexpression from a heterologous locus of an alcA::snxA1 9-exon genomic clone complemented snxA1 as efficiently as alcA::snxA+ and (2) both 9- and 11-exon alcA-driven cDNAs, lacking their own promoters, could not rescue snxA1 in an alcA-independent manner, i.e., by crossing over to loop the plasmids into the snxA locus. These somewhat incongruous findings could be explained if the mutations were located in the region spanning the 3′ half of intron 1 to a region slightly downstream of intron 2. However, despite extensive sequencing within and beyond the entire snxA locus, and exhaustive sequencing of the region containing the promoter, 5′ UTR, exons 1–3 and introns 1–2, no DNA sequence changes were found in snxA1 or snxA2.
How, then, to explain the dramatically reduced expression of apparently wild-type SNXA protein in the two mutants, which in turn suppresses defects in the CDK1 mitotic induction pathway? A DNA sequence mutation in a distant snxA/AN3739 regulatory element lying outside the sequenced region could lead to reduced expression and failure to complement among snxA alleles. However, a distant mutation should not be rescued by linear wild-type DNA fragments corresponding to the snxA locus, unless perhaps this mutated element (or gene) alters chromatin structure/function at the snxA locus. Nonetheless, to address this possibility a series of 17 overlapping linear fragments (∼4 kb each) covering 38,180 nt, extending from AN3735 (nt 3,231,933) to AN4748 (nt 3,193,753), were transformed into snxA1 and snxA2 strains. Only AN3739-overlapping fragments complemented the mutations (Figure 6 and data not shown). Although it seems unlikely, we cannot rule out the possibility of a more distant snxA regulatory mutation.
More likely is the possibility that snxA1 and snxA2 defects may result from altered chromatin structure and function in the snxA region, such as by an increase in repressive histone marks or a decrease in activating marks. A. nidulans lacks DNA methylation (Lee et al. 2008) and thus a chromatin immunoprecipitation approach, using antibodies against a variety of modified histones, may be required to reveal the biochemical nature of snxA defects. Of interest in this regard is the recent finding of a functional relationship between the Set1/COMPASS complex (Set1c), an H3K4 methyltransferase, and both the CDK1 and NIMA mitotic kinases, in which Set1c and the two kinases are required for mitotic induction and progression (Govindaraghavan et al. 2014). In general, H3K4 di- and trimethylation by Set1c has been associated with gene activation in euchromatin, and in humans has been suggested to “bookmark” active genes so that they can resume transcription following exit from mitosis (Blobel et al. 2009; Kelly et al. 2010). In A. nidulans, Set1c function has also been shown to mediate repression of certain secondary metabolite gene clusters that occur in subtelomeric regions (reviewed in Gacek and Strauss 2012). Given its newly discovered and poorly understood role in promoting mitosis, and the apparent role of snxA in restraining nuclear division, it will be of interest to determine what kind of functional relationship may exist between Set1c and snxA.
A recent study of Rumplestiltskin (Rump), the Drosophila melanogaster homolog of snxA and hnRNP-M, provides evidence for direct involvement in an epigenetic mechanism affecting higher-order chromatin structure through modulation of insulator function (King et al. 2014). Rump associates physically with Drosophila core insulator proteins, colocalizing to a subset of insulator binding sites. Depletion of Rump improved the enhancer-blocking and barrier function of a gypsy transposable element, suggesting that Rump acts normally to inhibit insulator activity, possibly by competing with insulator–insulator interactions. In addition to this newly discovered role in chromatin dynamics, Rump also autoregulates its own expression (King et al. 2014). These findings suggest a possible mechanisn for snxA in regulating the expression of G2-M regulators in A. nidulans. We show here that levels of NIMXCDC2 and NIMECYCLINB are not affected in snxA mutants. However, depletion of SNXA might, for example, improve insulator function to enhance the expression of a positive regulator such as NIMTCDC25 or to increase the repression of a negative regulator of G2-M, such as SUC1 or ANKAWEE1.
The role of SNXAHRB1 in cell cycle progression is further demonstrated by the observed alterations in NIMECYCLINB distributions in populations of snxA1 and ΔsnxA germlings. The depletion of SNXAHRB1 in these cells diminishes the percentage of cells with nuclear NIMECYCLINB. The lowest levels of nuclear NIMECYCLINB occurred in nimX2/snxA1 germlings at 37° and 44°, the temperatures at which snxA1 suppresses the nimX2 G2 arrest to permit passage through G2/M. The decreased nuclear NIMECYCLINB localization in the nimX2/snxA1 double mutant likely reflects completion of M phase. The consistent and reproducibly lower nuclear NIMECYCLINB localization at 37° and 44° that is less than wild-type levels suggests that these cells progress through mitosis, at which time NIMECYCLINB is degraded and thus no longer visible in the nucleus, and that they may be delayed in G1, giving a further reduction in nuclear NIMECYCLINB. In other words, the observed decreased nuclear NIMECYCLINB at 37° and 44° in snxA1/nimX2 mutants might seem counterintuitive, since snxA depletion rescues the G2 arrest of nimX2CDC2. This rescue could occur by increased movement of NIMXCDC2 and NIMECYCLINB into the nucleus. However, this rescue, by permitting mitotic progression, leads to the destruction of NIMECYCLINB at the metaphase-to-anaphase transition and thus can explain why NIMECYCLINB levels are lowest in this snxA1/nimX2 strain at 37° and 44°.
Mitotic regulation via the CDC2/CYCLINB pathway has been shown to ultimately depend on control of CDC2/CYCLINB activity, which is tightly regulated by phosphorylation/dephosphorylation (Ye et al. 1995), and on proper nuclear localization of the fully active CDC2/CYCLINB complex (Wu et al. 1998). The finding that snxA1 does not increase NIMX2CDC2 protein levels or NIMX2CDC2/NIMECYCLINB activity at the nimX2 restrictive temperature, where snxA1 does suppress the nimX2 G2 arrest, suggests that the effects of SNXA1 on the NIMX2CDC2/NIMECYCLINB mitotic initiation pathway occur by a different mechanism, such as proper localization of the active complex. Given that NIMXCDC2 is active as a protein kinase only when bound to its regulatory subunit, NIMECYCLINB, our findings that NIMECYCLINB has altered localization patterns in both snxA1 and ΔsnxA hyphae support this hypothesis. This is particularly interesting, given that snxA is homologous with S. cerevisiae Hrb1/Gbp2, which encode mRNA shuttling binding proteins similar to the mammalian SR family of proteins. While these proteins have been shown in budding yeast to shuttle mRNAs to the cytoplasm and to remain with the transcript during translation (Windgassen et al. 2004), heretofore no specific cell cycle functions have been suggested for Hrb1/Gbp2, although overexpression of Gbp2 does lead to a delay in G1 (Stevenson et al. 2001). Because deletion of these genes in S. cerevisiae does not disrupt or alter the nucleocytoplasmic distribution of bulk polyadenylated mRNAs (Hacker and Krebber 2004), it is possible that either they function as adaptors for specific subsets of mRNPs, as suggested by Rougemaille et al. (2008), or they may confer additional, previously unidentified functions.
With its partially closed mitosis, A. nidulans represents an evolutionary intermediate between the closed mitosis of budding yeast and the open mitosis of more complex eukaryotes (for review, see De Souza and Osmani 2009). It is therefore not surprising that some evolutionarily conserved proteins involved in cell cycle regulation have additional and/or different functions in different organisms. Given that the G2/M effects of SNXAHRB1 do not occur by modulating NIMXCDC2/NIMECYCLINB activity, but that snxA mutation or deletion alters NIMECYCLINB localization patterns, it is possible that in A. nidulans SNXAHRB1 regulates the localization either of NIMXCDC2/NIMECYCLINB or of mitosis-specific proteins that function upstream or downstream of the activation of NIMXCDC2/NIMECYCLINB.
Using time-lapse imaging of cyclin B-GFP, Nayak et al. (2010) demonstrated that cyclin B becomes visible in the nucleoplasm and at SPBs ∼40% of the way through interphase, during S phase, and confirmed observations by De Souza et al. (2009) that the majority of this cyclin B exits the nucleus when the NPC partially disassembles, leaving a subpool of cyclin B concentrated at SPBs and on the spindle. This subpool disappears gradually as mitosis progresses. Although it is beyond our abilities to detect, it is possible that mutation of SNXAHRB1 allows this subpool to remain in the nucleus longer; if so, this could allow threshold levels of NIMX2CDK1 protein to accumulate in nimX2 mutants, resulting in suppression of the heat-sensitive phenotype. Alternatively, it is possible that nuclear transport is altered in snxA mutants, leading to the accumulation or retention of threshold levels of CDK1/CYCLINB. An additional possibility is that SNXAHRB1 affects proteins involved in mitotic exit; a defect in inactivation of the anaphase-promoting complex/cyclosome, as Nayak et al. (2010) reported in mipAD159 mutants (γ-tubulin), could both allow threshold levels of NIMX2CDC2 activity to be reached and delay G1, resulting in lower nuclear NIMECYCLINB. We are currently investigating these alternatives.
The dramatic decrease in SNXAHRB1 nuclear localization during the highly heat-sensitive pseudomitotic arrest of the nimO18 mutation underscores the nature of the partially closed mitosis of A. nidulans. At mitosis, the nuclear pore complexes partially disassemble, and proteins not specifically retained in the nucleus diffuse out of the partially closed nuclear pores and may equilibrate across the nuclear envelope (De Souza et al. 2004); those proteins destined to return to the nucleus are likely specifically re-imported once the nuclear pores completely reassemble. SNXAHRB1 appears to be one of these proteins.
The identification of snxA as the A. nidulans homolog of Hrb1/Gbp2, the G1 arrest phenotype of snxA1 mutants, the exit of SNXAHRB1 from the nucleus during mitosis and its subsequent re-import after mitotic exit, the NIMXCDC2/NIMECYCLINB activity-independent effects of snxA on the NIMXCDC2/NIMECYCLINB G2/M pathway, and the altered localization of NIMECYCLINB suggest a novel function for the SNXAHRB1 SR family protein in eukaryotes: a function in cell cycle regulation. Moreover, the fact that reducing or eliminating snxA function alleviates heat-sensitive defects in nimXcdc2 mutants and in mutant regulators of nimXcdc2 activity (nimE6cyclinB and nimT23cdc25) suggests that snxA may normally act to restrain mitotic induction by the CDC2/CYCLINB regulatory pathway.
Supplementary Material
Acknowledgments
We thank Jennifer Powell (Gettysburg College), Peter Mirabito (University of Kentucky), and André Walther (Cedar Crest College) for helpful advice and suggestions; the Fungal Genetics Stock Center (Kansas City, MO) for providing genetic mapping strains, libraries, cosmids, and fosmids used in this study; Stephen Osmani (Ohio State University) for providing antibodies and strains; Xin Xiang (Uniformed Services University of the Health Sciences) for providing the GFP-α-tubulin strain; Berl Oakley (University of Kansas) for providing the GFP-NIMECYCLINB strain; Sabrice Guerrier (Millsaps College) for providing antibodies; Roy Duhe and Chetan Patil (University of Mississippi Medical Center) for assistance with kinase assays; Cory Toyota (Millsaps College) for assistance with qRT-PCR analysis; and the reviewers for helpful comments that improved this manuscript. This work was supported by grants from Gettysburg College to S.W.J.; by grants to S.L.A. from Millsaps College; by National Institutes of Health grant R15GM055885 and Mississippi IDeA Networks of Biomedical Research Excellence grant P20RR016476 funded by the National Institute of General Medical Sciences, National Institutes of Health to SLA; and by grants from the Howard Hughes Medical Institute to Gettysburg College and Millsaps College.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167445/-/DC1.
Communicating editor: D. Lew
Literature Cited
- Alam M. K., El-Ganiny A. M., Afroz S., Sanders D. A., Liu J., et al. , 2012. Aspergillus nidulans galactofuranose biosynthesis affects antifungal drug sensitivity. Fungal Genet. Biol. 49: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Bachewich C., Masker K., Osmani S., 2005. The polo-like kinase PLKA is required for initiation and progression through mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 55: 572–587 [DOI] [PubMed] [Google Scholar]
- Ballance D. J., Buxton F. P., Turner G., 1983. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem. Biophys. Res. Commun. 112: 284–289 [DOI] [PubMed] [Google Scholar]
- Blobel G. A., Kadauke S., Wang E., Lau A. W., Zuber J., et al. , 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36: 970–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody H., Griffith J., Cuticchia A. J., Arnold J., Timberlake W. E., 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19: 3105–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani S. A., 2009. Double duty for nuclear proteins: the price of more open forms of mitosis. Trends Genet. 25: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Horn K. P., Masker K., Osmani S. A., 2003. The SONB(NUP98) nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A., 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14: 1973–1984 [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Nayak T., Oakley B., Osmani S. A., 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek A., Strauss J., 2012. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 95: 1389–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Govindaraghavan M., Anglin S. L., Osmani A. H., Osmani S. A., 2014. The Set1/COMPASS histone H3 methyltransferase helps regulate mitosis with the CDK1 and NIMA mitotic kinases in Aspergillus nidulans. Genetics 197: 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker S., Krebber H., 2004. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J. Biol. Chem. 279: 5049–5052 [DOI] [PubMed] [Google Scholar]
- Hackmann A., Wu H., Schneider U. M., Meyer K., Jung K., et al. , 2014. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat. Commun. 5: 3123. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Morrell J. L., Hamer J. E., 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136: 517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. J., Lee Y. J., Chen D. H., Li C., 2009. Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody. Protein J. 28: 139–147 [DOI] [PubMed] [Google Scholar]
- Hurt E., Luo M. J., Rother S., Reed R., Strasser K., 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 101: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S. W., Mirabito P. M., Scacheri P. C., Morris N. R., 1995. The Aspergillus nidulans bimE (blocked-in-mitosis) gene encodes multiple cell cycle functions involved in mitotic checkpoint control and mitosis. J. Cell Sci. 108(Pt 11): 3485–3499 [DOI] [PubMed] [Google Scholar]
- James S. W., Bullock K. A., Gygax S. E., Kraynack B. A., Matura R. A., et al. , 1999. nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J. Cell Sci. 112(Pt 9): 1313–1324 [DOI] [PubMed] [Google Scholar]
- Kafer E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131 [DOI] [PubMed] [Google Scholar]
- Kaminskyj S. G. W., 2001. Fundamentals of growth, storage, genetics and microscopy of Aspergillus nidulans. Fungal Genet. Newsl. 48: 25–31 [Google Scholar]
- Kelly T. K., Miranda T. B., Liang G., Berman B. P., Lin J. C., et al. , 2010. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol. Cell 39: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert K., Duncan K., Li H., Guthrie C., 2005. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. RNA 11: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. R., Matzat L. H., Dale R. K., Lim S. J., Lei E. P., 2014. The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J. Cell Sci. 127: 2956–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Freitag M., Selker E. U., Aramayo R., 2008. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS ONE 3: e2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., Rodriguez-Bravo V., Medema R. H., 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L., Osmani A. H., Ukil L., Son S., Markossian S., et al. , 2010. Single-step affinity purification for fungal proteomics. Eukaryot. Cell 9: 831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lleres D., Denegri M., Biggiogera M., Ajuh P., Lamond A. I., 2010. Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice. EMBO Rep. 11: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. T., Poon R. Y., 2011. How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 435: 17–31 [DOI] [PubMed] [Google Scholar]
- McGuire S. L., Roe D. L., Carter B. W., Carter R. L., Grace S. P., et al. , 2000. Extragenic suppressors of the nimX2(cdc2) mutation of Aspergillus nidulans affect nuclear division, septation and conidiation. Genetics 156: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Yaoita Y., 1997. Construction of multiple-epitope tag sequence by PCR for sensitive Western blot analysis. Nucleic Acids Res. 25: 2231–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., et al. , 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Edgerton-Morgan H., Horio T., Xiong Y., De Souza C. P., et al. , 2010. Gamma-tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J. Cell Biol. 190: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. B., Nielsen M. L., Mortensen U. H., 2008. Transient disruption of non-homologous end-joining facilitates targeted genome manipulations in the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 45: 165–170 [DOI] [PubMed] [Google Scholar]
- Nigg E. A., 1995. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays 17: 471–480 [DOI] [PubMed] [Google Scholar]
- O’Connell M. J., Osmani A. H., Morris N. R., Osmani S. A., 1992. An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO J. 11: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N., May G., 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S. A., Engle D. B., Doonan J. H., Morris N. R., 1988. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52: 241–251 [DOI] [PubMed] [Google Scholar]
- Osmani A. H., McGuire S. L., Osmani S. A., 1991. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell 67: 283–291 [DOI] [PubMed] [Google Scholar]
- Osmani A. H., van Peij N., Mischke M., O’Connell M. J., Osmani S. A., 1994. A single p34cdc2 protein kinase (encoded by nimXcdc2) is required at G1 and G2 in Aspergillus nidulans. J. Cell Sci. 107(Pt 6): 1519–1528 [DOI] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Nile A., Osmani S. A., 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17: 4946–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y., Maddox P., Oakley C. E., Xiang X., Osmani S. A., et al. , 2003. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell 14: 2192–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Bufton A. W., 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238 [DOI] [PubMed] [Google Scholar]
- Porat Z., Erez O., Kahana C., 2006. Cellular localization and phosphorylation of Hrb1p is independent of Sky1p. Biochim. Biophys. Acta 1763: 207–213 [DOI] [PubMed] [Google Scholar]
- Reed R., Cheng H., 2005. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Rougemaille M., Villa T., Gudipati R. K., Libri D., 2008. mRNA journey to the cytoplasm: attire required. Biol. Cell 100: 327–342 [DOI] [PubMed] [Google Scholar]
- Sanford J. R., Gray N. K., Beckmann K., Caceres J. F., 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson L. F., Kennedy B. K., Harlow E., 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 98: 3946–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Hurt E., 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P., O’Connor T. R., Bailey T. L., Richard S., 2013. Defining the RGG/RG motif. Mol. Cell 50: 613–623 [DOI] [PubMed] [Google Scholar]
- Todd R. B., Davis M. A., Hynes M. J., 2007. Genetic manipulation of Aspergillus nidulans: heterokaryons and diploids for dominance, complementation and haploidization analyses. Nat. Protoc. 2: 822–830 [DOI] [PubMed] [Google Scholar]
- Tuck A. C., Tollervey D., 2013. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 154: 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., May G. S., Morris N. R., 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79: 119–130 [DOI] [PubMed] [Google Scholar]
- Warkocki Z., Odenwalder P., Schmitzova J., Platzmann F., Stark H., et al. , 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Windgassen M., Sturm D., Cajigas I. J., Gonzalez C. I., Seedorf M., et al. , 2004. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 24: 10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Osmani S. A., Mirabito P. M., 1998. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 141: 1575–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Gao X. D., Lee J. H., Huang H., Tan H., et al. , 2014. Cell type-restricted activity of hn RNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 28: 1191–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., et al. , 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3: 1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X. S., Xu G., Pu R. T., Fincher R. R., McGuire S. L., et al. , 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14: 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Dominguez Y., et al. , 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.