Abstract
MiRNAs bear an increasing number of functions throughout development and in the aging adult. Here we address their role in establishing sexually dimorphic traits and sexual identity in male and female Drosophila. Our survey of miRNA populations in each sex identifies sets of miRNAs differentially expressed in male and female tissues across various stages of development. The pervasive sex-biased expression of miRNAs generally increases with the complexity and sexual dimorphism of tissues, gonads revealing the most striking biases. We find that the male-specific regulation of the X chromosome is relevant to miRNA expression on two levels. First, in the male gonad, testis-biased miRNAs tend to reside on the X chromosome. Second, in the soma, X-linked miRNAs do not systematically rely on dosage compensation. We set out to address the importance of a sex-biased expression of miRNAs in establishing sexually dimorphic traits. Our study of the conserved let-7-C miRNA cluster controlled by the sex-biased hormone ecdysone places let-7 as a primary modulator of the sex-determination hierarchy. Flies with modified let-7 levels present doublesex-related phenotypes and express sex-determination genes normally restricted to the opposite sex. In testes and ovaries, alterations of the ecdysone-induced let-7 result in aberrant gonadal somatic cell behavior and non-cell-autonomous defects in early germline differentiation. Gonadal defects as well as aberrant expression of sex-determination genes persist in aging adults under hormonal control. Together, our findings place ecdysone and let-7 as modulators of a somatic systemic signal that helps establish and sustain sexual identity in males and females and differentiation in gonads. This work establishes the foundation for a role of miRNAs in sexual dimorphism and demonstrates that similar to vertebrate hormonal control of cellular sexual identity exists in Drosophila.
Keywords: miRNA, sex determination, ecdysteroid, gonad, development, Drosophila, genetics of sex
SEXUAL dimorphism is pervasive throughout the animal kingdom. From insects, fishes, reptiles, and birds to mammals, hormones and genes shape the morphological, behavioral, and reproductive potential of each sex throughout development and adult life. Drosophila is no exception, with males and females differing in many ways: anatomical differences include the number of abdominal segments and their pigmentation, the proboscis, labial parts, dimorphic reproductive organs, the formation of sex combs exclusively in males, and 25% larger size in females. Differences that affect male and female behavior exist also in the nervous system and the brain. Y chromosome aside, male and female cells possess a strictly identical genomic content. Most of the differences between the sexes arise and persist via the regulation of sets of genes in a sex-specific manner.
The question of how hundreds if not thousands of genes are differentially expressed in males and females to produce sexually dimorphic individuals is extensively studied. Refined genomic and genetic studies have converged toward a model of differential expression that requires that both spatial and temporal programs be established throughout development (Arbeitman et al. 2002; Parisi et al. 2004; Lebo et al. 2009; Chatterjee et al. 2011). Probably the most important of these programs in flies is the sex-determination hierarchy (Baker et al. 1989; Christiansen et al. 2002; Camara et al. 2008; Clough and Oliver 2012). The primary determinant of Drosophila sex is the X chromosome to autosome (X:A) ratio (Bridges 1921), which determines the production of alternative splice variants of Sex lethal (Sxl) to generate an active SXL protein in females and a nonfunctional truncation in males (Cline 1978). Sxl activity is sufficient to direct the entire developmental programs of both somatic and germline sex determination (Christiansen et al. 2002; Robinett et al. 2010; Salz 2011; Whitworth et al. 2012). Sxl serves two essential functions: it restricts dosage compensation to males and controls the sex-determination hierarchy in each sex.
Dosage compensation is the process by which males double the transcription of genes on their single X chromosome to match the levels found in diplo-X females. This process requires a ribonucleoprotein complex, the compensasome, composed of two noncoding RNAs (roX1 and roX2) and six proteins (male-specific lethals MSL-1, -2, -3, the helicase/ATPase MLE, histone acetyltransferase MOF, and histone kinase JIL1). In females, SXL represses the production of MSL-2 at both the transcriptional and translational level, therefore preventing dosage compensation. In males, lack of SXL function allows the male-specific expression of MSL-2 and its assembly into compensasomes to initiate dosage compensation (Bashaw and Baker 1997; see Duncan et al. 2006 for review).
At the top of the sex-determination hierarchy, SXL controls which sex-specific isoform is being processed from the doublesex (dsx) transcripts (reviewed in Christiansen et al. 2002). If the X:A ratio is 1, Sxl produces a female-specific splicing factor that causes female-specific splicing of the transformer (tra) transcript. TRA interacts with the transformer-2 (TRA2) splicing factor to produce a female-specific splice variant of dsx (Belote et al. 1989; Sosnowski et al. 1989; Ryner and Baker 1991). The female-specific DSXF protein then activates female and inhibits male development. Because males lack SXL and subsequently TRA, a “default” male-specific splicing of dsx transcript generates the DSXM protein, which inhibits female and promotes male traits. Loss-of-function mutations in Sxl, tra, and tra2 transform XX individuals into males, but have no effect in XY males. In contrast, the dsx gene is important for the sexual differentiation of both sexes—in the absence of dsx, both XX and XY flies are anatomically and behaviorally intersex (Baker and Ridge 1980; Belote et al. 1985).
Only a few transcriptional targets through which DSX ultimately functions are known (Luo et al. 2011). DSX regulates sex-specific pigmentation patterns with abdominal-B (Abd-B) and bric-a-brac1 (bab1), resulting in males’ darker abdomen (Williams et al. 2008). DSXM controls the development of male-specific bristles or sex combs on the forelegs with sex-comb reduced (Scr) (Tanaka et al. 2011). In each sex, DSX orchestrates the differentiation of larval genital discs into mature dimorphic reproductive organs, external genitalia, and analia (Hildreth 1965; Chatterjee et al. 2011). DSXF directly upregulates the expression of yolk proteins (Yp1, Yp2) (Burtis et al. 1991), and DSXM downregulates their transcription.
The thorough dissection of dsx expression reveals that DSX presents two main characteristics (Lee et al. 2002; Hempel and Oliver 2007; Rideout et al. 2010; Robinett et al. 2010). First, the levels of DSX protein vary greatly throughout development within cells and tissues, implying a tight regulation of its steady states. Second, DSX is not present in all cells in a given tissue, so only some cells know their sex while others remain asexual.
MicroRNAs (miRNAs) appear as critical regulators of development and are themselves highly regulated (Ambros and Chen 2007; Bartel 2009; Smibert and Lai 2010; Dai et al. 2012). The interaction of microRNAs with the 3′-UTRs of transcribed mRNAs affects both a transcript’s stability and its translation. Each miRNA can target several different mRNAs and each mRNA can be targeted by multiple miRNAs, generating an intricate network of gene expression regulation. As miRNAs could provide a rapid and tissue-specific means to alter gene expression, they represent ideal candidates for the regulation of spatial and temporal expression patterns of sex-determination genes, their cofactors, and downstream targets. Ultimately, the sex-biased expression of miRNAs could control directly the differential expression of many genes contributing to sexually dimorphic traits at a given time and place during development.
Sexually dimorphic miRNA profiles have been reported in mouse and chicken gonads, and in whole adult Caenorhabditis elegans (Mishima et al. 2008; Kato et al. 2009; Baley and Li 2012). In Drosophila, probing miRNA populations in whole animals during development has revealed widespread developmental regulation (Aravin et al. 2003; Ruby et al. 2007). However, the small RNA libraries generated in these studies came from either mixed-sex samples or single-sex but nonhomogenous tissues, which may mask important sex- and tissue-specific variability in miRNA expression and function. To date, Drosophila lacks a critical examination of miRNA expression in two important contexts: sex-biased expression that may lead to sexually dimorphic function or spatial and temporal heterogeneity in expression that may drive tissue-specific functions. Both are critical to understanding the role of miRNAs across development.
To investigate these issues, we first established the profiles of miRNAs in several male and female adult parts and organs, larval dissected tissues, and embryonic cells. Their comparison reveals, in each tissue, sets of male- and female-biased miRNAs, increasing in number and extent with the complexity and sexual dimorphism of each tissue. We further address two aspects of miRNA functions in the context of sexual identity: first, we test whether X-linked miRNAs are regulated by dosage compensation in males and, second, we explore the role of the steroid-induced miRNA let-7 in regulating sexually dimorphic traits and how its male-biased expression in the gonads affects germline differentiation programs.
Materials and Methods
Fly strains and genetics
Oregon-R flies were used for miRNA profiling. Msl3p, mle1, and pr mle12.17 mutants are described in Fagegaltier and Baker (2004). All chromosomes but the mutant-bearing allele were exchanged to create isogenized lines by back crossing to a w1118; MKRS/TM6B stock for >10 generations. Wandering non-Tb- mutant male and female larvae were identified by their gonads. Overexpression of miRNAs was performed using a dsx–GAL4 driver (Robinett et al. 2010). UAS–NLS–GFP flies are from Bloomington (BL4776); UAS–let-7, UAS-mir-100, UAS–mir-125, and UAS–let-7-C constructs are described in Bejarano et al. (2012). In let-7-C and ecd mutant studies, flies were raised on standard cornmeal–yeast–agar-medium at 25° and fattened on wet yeast paste 1 day before dissection unless otherwise stated. The two knockout alleles let-7-CGK1 and let-7-CKO1 lack the whole let-7-C cluster; let-7-CGK1 contains the transcriptional activator GAL4 under the control of the let-7-C promoter; let-7-CGK1/let-7-CKO1 are referred to as Δlet-7-C (Sokol et al. 2008). Flies with a transgene rescuing the let-7-C cluster (P{W8, let-7-C}; let-7-CGK1/let-7-CKO1) are referred to as let-7-C Rescue. The P{W8, let-7-C Δlet-7} construct restores all let-7-C miRNA members except for let-7. For miRNA loss of function, let-7-CGK1/let-7-CKO1; P{W8, let-7-C Δlet-7}/+ flies referred to as Δlet-7 were used. The following additional fly stocks were used: FRT40A let-7 mir-125/CyO and UAS–let-7-C; Sco/CyO (Caygill and Johnston 2008), UAS–let-7/TM6 (Sokol et al. 2008), Ubi–GFP FRT40A/CyO; bab1–Gal4:UAS–Flp/TM2 (a gift from A. González-Reyes), UAS–CD8GFP:UAS–nuc lacZ (a gift from F. Hirth), Oregon-R, w1118, and ecd1ts (BL4210).
Sample collections for miRNA–Seq and validation
To ensure that miRNA–Seq samples are not contaminated by other tissues, ∼120 Oregon-R heads were individually separated with a scalpel from the rest of the body of ∼24-hr-old males and females, collected on ice and quickly frozen. Salivary glands were dissected from ∼130 wandering late L3 larvae of each sex identified by their gonads. For qPCR validations of miRNA–Seq data sets, at least two additional independent collections were performed as above. We also collected ovaries and testes from 0- to 2- and 2- to 4-day-old Oregon-R individuals, S2 (Invitrogen), and Kc-167 cells (DGRC) washed in 1× PBS. All dissected tissues and cells were quickly snap frozen in liquid nitrogen and RNA preparations enriched for small RNAs using an adapted Trizol protocol.
miRNA–Seq
30-100μg of total RNAs were subject to 2S rRNA depletion and DNAse treated. Size selected 18-29nt sRNAs were cloned according to (Malone et al. 2012). Libraries were clustered and sequenced on the Illumina GAIIx platform.
Cuticle preparations
Three- to 4-day-old flies were placed in ethanol and incubated in 10% NaOH for 1 hr at 70°. Adult abdominal cuticles were mounted and flattened in 30% glycerol. Pictures were taken at the same magnification using a Nikon SMZ150 microscope and Nikon DS-RiI camera.
Perturbation of ecdysone levels
The ecd1ts temperature-sensitive mutation is known to reduce ecdysone levels at the nonpermissive temperature (Garen et al. 1977). Oregon-R and ecd1ts flies were kept at the permissive temperature (18°) and 2- to 4-day-old adults were shifted to the restrictive temperature (29°) for 5–11 days to block ecdysone synthesis. Control Oregon-R and ecd1ts flies were kept at 18° for the same time.
Clonal analysis
Somatic cell clones in CpCs and ECs were induced using mitotic recombination as described previously (König et al. 2011). FRT40A let-7 mir-125/CyO; P{W8, let-7-C Δlet-7} flies were crossed to Ubi–GFP FRT40A/CyO; bab1–Gal4:UAS–Flp/TM2. Third-instar larvae were heat shocked for 2 hr on 2 consecutive days in a 37° water bath and returned to 25°. Mutant clones were identified by the absence of GFP in 5-day-old adult ovaries.
Immunofluorescence and antibodies
Ovaries and testes were fixed in 5% formaldehyde (Polysciences, Inc.) for 10 min and stained as described in König and Shcherbata (2013). We used the following mouse monoclonal antibodies: anti-adducin (1:50), anti-lamin C (1:50), anti-arm (1:50), anti-FasIII (1:50), rat monoclonal antibody anti-DE-cadherin (Developmental Studies Hybridoma Bank), guinea pig anti-Tj (1:3000, D. Godt), rabbit anti-vasa (1:5000, gift from R. Pflanz), anti-β-Gal (1:3000, Cappel), and anti-GFP-directly conjugated with AF488 (1:3000, Invitrogen), Alexa 488, 568, or 633 goat anti-mouse, anti-rabbit (1:500, Molecular Probes), goat anti-rat Cy5 (1:250, Jackson Immunoresearch). Images were obtained with a confocal laser-scanning microscope (Zeiss LSM700) and processed with Adobe Photoshop.
Testis analysis and statistics
To determine the frequency of somatic cell differentiation defects in testis, the percentage of testis with somatic cell clusters (<5, ≥5, and >10) and epithelium appearance at the apex or at the lateral side of the anterior region of testicular tube were quantified. Statistics were calculated using two-way tables and chi-square test.
Determination of let-7 expression
To analyze the expression pattern of let-7-C, let-7-CGK1/CyO flies were crossed to UAS–mCD8–GFP:UAS–nuc–lacZ. To analyze let-7-C levels upon stress, 3- to 5-day-old let-7-CGK1/UAS–mCD8GFP:UAS–nuc–lacZ flies were heat shocked for 1 hr at 37° and their gonads were dissected and assayed for immunohistological analysis.
Quantitative PCR Assays (RT–qPCR)
For qPCR validation of the miRNA–Seq data sets and X-linked miRNA expression studies, 100 ng/μl RNA samples were spiked after DNAse digestion with a synthetic primer at 6.10e9 copies/μl, polyadenylated and reverse transcribed according to the miScript reverse transcriptase kit instructions (Qiagen). Each miRNA was quantified with a specific primer (Supporting Information, Table S1) following the miScript SYBR green PCR kit instructions. All miRNAs were tested in triplicates on two independent biological replicates with the appropriate controls. Ct values were normalized to U6 snRNA levels in miRNA–Seq validation experiments using the ∆∆CT method and 2−ΔΔCT values calculations and to Dspt4 mRNA levels in the dosage compensation mutant studies (see Table S11, Table S12, Table S13, Table S14, Table S15, and Table S16).
For let-7 quantification, reverse transcription and qPCR were performed following the manufacturers’ protocol using TaqMan MicroRNA assay, with 2S rRNA as an endogenous control. Let-7 levels were determined in gonads and carcasses from Oregon-R males and females. To eliminate effects that could possibly arise because ovaries contain different amounts of eggs and late egg chambers, late-stage egg chambers and eggs were removed from the dissected ovaries, leaving only the anterior part of the ovary containing the germaria. To measure let-7 levels upon perturbation of ecdysone, ovaries and testes of Oregon-R and ecd1ts mutants raised at 18° and shifted to 29° for 5 or 11 days (or kept at 18° for control for the same amount of time) were dissected. RNA was extracted in Trizol according to the manufacturer’s instructions before proceeding with RT–qPCR.
Sex-specific mRNA transcript levels were assessed in 5- to 7-day-old Δlet-7 and let-7-C Rescue control whole flies that were raised at 25° and shifted to 33° for 4 days or in Oregon-R and ecd1ts mutants raised at 18° and shifted 1–3 days after hatching to 29° (or kept at 18° for control) for 5 or 11 days. cDNA was generated using the cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s instructions and qPCR performed using the fast SYBR Green Master Mix (Applied Biosystems). A Step One Plus 96 well system (Applied Biosystems) was used for all analyses. All reactions were run in triplicates with appropriate blanks. The reactions were incubated at 95° for 10 min followed by 40 cycles at 95° for 15 sec and 60° for 60 sec (TaqMan MicroRNA). The ∆CT value was determined by subtracting the CT value of the endogenous control from the experimental CT value. ∆∆CT was calculated by subtracting the ∆CT of the control sample from the suspect ∆CT value. The relative RNA levels were calculated using 2−ΔΔCT. Primers are described in Table S1.
Bioinformatics
The libraries produced in this study (NCBI GEO series record GSE57029) were complemented with existing libraries from lymphoid cells (GSM272653, Kc cells; GSM272652, S2 cells) (Chung et al. 2008) and from 2- to 4-day-old Oregon-R adult testis (GSM280085) and ovary (GSM280082) (Czech et al. 2008). Reads were clipped of the adapter sequences, filtered for sequences mapping to viruses and simple repeats, and aligned to the Drosophila melanogaster genome (BDGP R5/dm3) with no mismatches using NexAlign (program available from the OSC: Data & Resource website). Uniquely mapped reads were annotated using a priority pipeline as in (Czech et al. 2008) and FlyBase r5.26, miRbase r.15 and in-house miRNA annotations. Reads corresponding to 184 annotated miRNAs were extracted and counts reported to estimate expression levels (Table S9). For each library, miRNA reads are normalized using the trimmed median ratio used to calculate a correction factor applied to all miRNA counts in a given library. Normalized counts (Table S10) were then input to calculate the relative expression and fold change in expression in pairwise comparisons between male and female samples. The contribution of each miRNA to a library was calculated as the normalized read counts over the total number of miRNA reads in the library (Table S22). The relative abundance of a miRNA across tissues was calculated by dividing the normalized count by the total of normalized counts across all libraries. Heatmaps were generated in R using the hclust() function to perform hierarchical cluster analysis.
Results and Discussion
MiRNA profiling in male and female tissues
We adopted a genomic approach and surveyed the populations of miRNAs in several Drosophila male and female tissues selected at various stages of development. Sexed miRNA–Seq data sets include late-embryo-derived lymphoid cells, larval salivary glands, and ∼1-day-old adult head and body, as well as germline-enriched tissues (ovary and testis) from mature adults (2- to 4-day-old).
Libraries prepared in different ways have been shown to result in variable sequencing efficiencies for individual miRNAs, so that differences in miRNA read counts between libraries may be caused by differential cloning efficiencies rather than reflect differences in their expression. To assess potential biases between sexes as accurately as possible, we prepared libraries and integrated carefully selected existing data sets (see Materials and Methods). All provide sufficient depth and breadth and were prepared with the Rnl2 truncated ligase, identical 5′ and 3′ cloning adaptors, and PCR conditions.
Several methods are available to analyze high-throughput miRNA profiling; however, none has reached a consensus. The choice of a processing and normalization method can affect greatly the estimates of differentially expressed miRNAs between tissues, especially when comparing tissues that differ substantially in their miRNA content (Dillies et al. 2013). We find the trimmed median method most suitable to normalize our data sets, which cover a wide range of tissues (see Materials and Methods). Normalized counts were then input to get empirical estimates of the relative abundance of a given miRNA in a library, in pairwise comparisons between male and female tissues, and across all samples. To assess the accuracy of our analyses, we assayed the expression of miRNAs across these samples using an independent method (qPCR; Figure S1). Of the 22 miRNAs tested for validation, 16 miRNAs show similar trends by qPCR and deep sequencing consistently across all samples. For five additional miRNAs, the enrichment trend and/or sex biases could not be confirmed in one or more tissues, while consistent in others. One miRNA showed systematically different profiles by qPCR and deep sequencing due to the presence of miRNAs with very close sequences that qPCR could not distinguish. Although cloning artifacts that may alter miRNA expression profiles in some libraries cannot be excluded, the highly overlapping trends of our qPCR validation set with those of normalized miRNA-Seq data suggest that our analysis reflects miRNA populations in each tissue and supports our main conclusions.
Samples included in our data set were selected to allow a detailed examination of the biases in miRNA content between male and female in tissues of increasing complexity with regard to the nature and number of different cells they contain and in tissues ranging from poorly to highly sexually dimorphic. We assess sex biases of miRNAs at the level of single cells in culture (lymphoid cells), in a very simple tissue with functions in both sexes (salivary glands), and in several fly parts of variable complexity and sexual dimorphism (head, body, ovary, and testis).
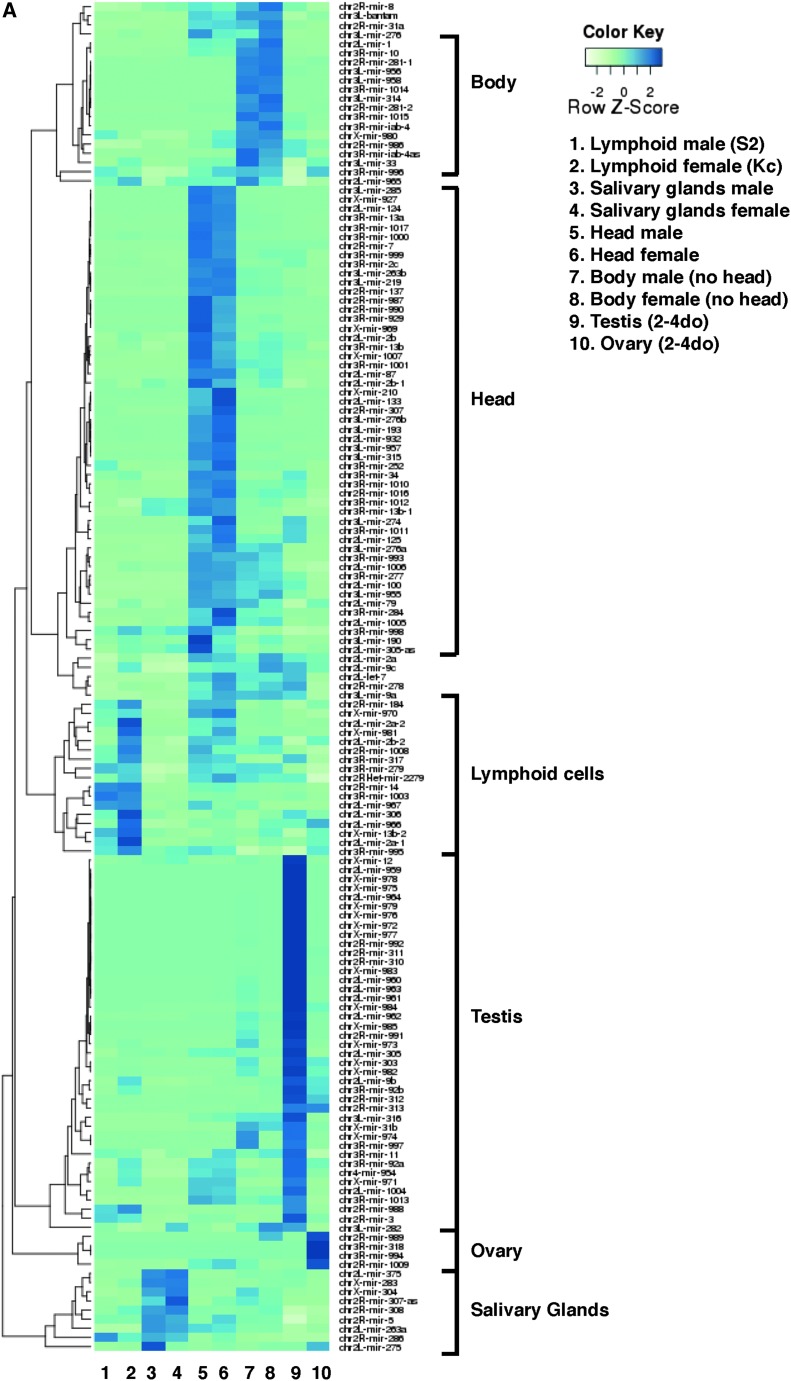
Each tissue is enriched for specific miRNAs, highlighted in clusters in Figure 1A, many of which are enriched in both sexes. Ubiquitous and evolutionary conserved miRNAs (mir-14, bantam, mir-8, mir-1) are highly expressed in most tissues (Table 1). However, even for such prevalent miRNAs, their relative abundance varies greatly depending on the tissue and stage studied (Table 1 and Figure 1A), denoting a high degree of variation in miRNA populations between tissues and the compartmentalization of their functions in a dynamic fashion.
Figure 1.
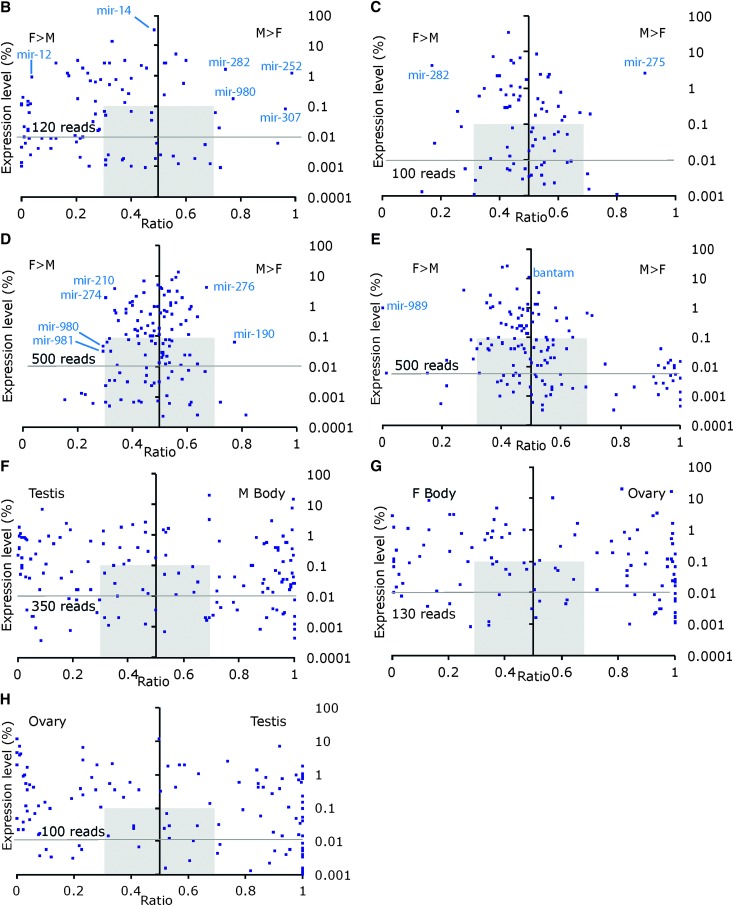
Expression of miRNAs in various male and female tissues. (A) Expression of miRNAs across 10 sexed libraries. Blue color intensity ranging from light to dark indicates an increasing abundance of normalized counts estimates for a given miRNA relative to all miRNAs in that library and to all libraries. Clusters of miRNAs enriched in a given tissue are highlighted. The 147 miRNAs with >30 normalized reads across all libraries were retained. 2–4 do: 2- to 4-day-old flies. The biased expression of miRNAs and their expression levels are shown in pairwise comparisons in each tissue in (B) embryonic hemocyte-derived S2 (male) and Kc-167 (female) cells. (C) Late larvae L3 Salivary Glands. (D) Adult Head. (E) Adult body (head removed). (F) Testis compared to male Body. (G) Ovary compared to female Body. (H) Testis compared to ovary. For each miRNA, the ratio of normalized reads (M/M+F or F/M+F) (x-axis) is plotted against its normalized expression level (%, log scale). Each dot represents a miRNA; miRNA’s present in one sexed tissue have a ratio of 0 or 1. Pearson correlations between male and female samples: 0.95 in lymphoid cells; 0.98 in salivary glands; 0.96 in heads; 0.99 in body; 0.25 in testis/ovary. M, Male; F, Female.
Table 1. Twenty-five most abundant miRNAs in male and female libraries from embryonic lymphoid cells (S2, male; Kc, female), third-instar larval salivary glands, 1-day-old adult head and body, and 2- to 4-day-old adult gonads.
| Embryonic lymphoid cells | Larval salivary glands | Adult heads | Adult body | Adult gonads | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S2 | % | Kc | % | Male | % | Female | % | Male | % | Female | % | Male | % | Female | % | Testis | % | Ovary | % |
| mir-14 | 38.35 | mir-14 | 29.10 | mir-8 | 32.42 | mir-8 | 35.67 | mir-8 | 14.57 | mir-8 | 11.44 | mir-1 | 25.56 | mir-1 | 26.75 | mir-12 | 16.87 | mir-989 | 18.37 |
| mir-184 | 11.35 | mir-184 | 16.38 | mir-263a | 10.45 | mir-12 | 8.49 | mir-1 | 10.00 | mir-1 | 8.18 | mir-8 | 23.01 | mir-8 | 25.64 | mir-8 | 15.35 | bantam | 11.43 |
| bantam | 7.44 | mir-2a | 4.44 | mir-375 | 7.15 | mir-263a | 7.75 | bantam | 8.19 | mir-276a | 7.63 | bantam | 12.56 | bantam | 9.79 | mir-305 | 5.04 | mir-8 | 9.30 |
| mir-996 | 4.85 | mir-8 | 4.15 | mir-12 | 6.25 | mir-375 | 6.72 | mir-276a | 5.81 | bantam | 7.14 | mir-276a | 4.66 | mir-276 | 5.02 | mir-959 | 4.68 | mir-2a | 7.73 |
| mir-279 | 3.47 | bantam | 4.08 | mir-304 | 5.73 | mir-282 | 6.30 | mir-276 | 5.39 | mir-210 | 5.00 | mir-277 | 3.91 | mir-276a | 4.27 | mir-2a | 3.85 | mir-318 | 7.25 |
| mir-11 | 3.29 | mir-34 | 3.40 | mir-275 | 5.19 | mir-304 | 6.01 | mir-277 | 4.66 | mir-277 | 4.90 | mir-276 | 2.47 | mir-2a | 2.93 | let-7 | 3.13 | mir-184 | 6.40 |
| mir-34 | 2.93 | mir-988 | 3.37 | bantam | 4.71 | bantam | 4.82 | mir-184 | 4.06 | mir-184 | 4.17 | mir-2a | 2.20 | mir-277 | 2.68 | mir-34 | 3.11 | mir-996 | 5.92 |
| mir-988 | 2.90 | mir-317 | 3.15 | mir-2a | 3.04 | mir-11 | 2.46 | mir-34 | 3.58 | mir-34 | 3.61 | mir-996 | 1.54 | mir-989 | 1.76 | mir-282 | 3.00 | mir-2b | 3.30 |
| mir-282 | 2.84 | mir-2b | 3.11 | mir-305 | 2.73 | mir-13b | 2.45 | mir-2b | 3.42 | mir-14 | 3.20 | let-7 | 1.49 | mir-282 | 1.68 | mir-964 | 2.74 | mir-994 | 3.01 |
| mir-252 | 2.79 | mir-276 | 2.30 | mir-2b | 2.43 | mir-305 | 1.93 | mir-2a | 3.23 | mir-285 | 2.95 | mir-9a | 1.48 | mir-31a | 1.48 | mir-277 | 2.59 | mir-275 | 2.62 |
| mir-13b | 2.43 | mir-996 | 2.30 | mir-13b | 1.99 | mir-2a | 1.91 | mir-285 | 3.22 | mir-2a | 2.92 | mir-14 | 1.34 | mir-2b | 1.21 | mir-11 | 2.51 | mir-13b | 2.20 |
| mir-2b | 2.09 | mir-279 | 2.26 | mir-11 | 1.91 | mir-2b | 1.55 | mir-14 | 2.46 | mir-2b | 2.73 | mir-31a | 1.28 | mir-9a | 1.18 | mir-978 | 2.29 | mir-9b | 2.20 |
| mir-317 | 1.92 | mir-13b | 2.20 | mir-283 | 1.79 | mir-283 | 1.54 | mir-210 | 2.44 | mir-276 | 2.72 | mir-282 | 1.20 | mir-996 | 1.08 | mir-977 | 2.20 | mir-263a | 1.46 |
| mir-276a | 1.92 | mir-11 | 2.09 | mir-282 | 1.58 | mir-996 | 1.20 | mir-124 | 2.35 | mir-274 | 2.60 | mir-12 | 1.10 | let-7 | 1.08 | mir-274 | 2.15 | mir-79 | 1.40 |
| mir-2a | 1.65 | mir-9b | 1.77 | mir-996 | 1.14 | mir-276 | 1.15 | mir-13b | 2.20 | let-7 | 2.39 | mir-13b | 1.08 | mir-184 | 1.06 | mir-976 | 2.04 | mir-282 | 1.38 |
| mir-276 | 0.95 | mir-276a | 1.62 | mir-308 | 1.06 | mir-308 | 1.00 | let-7 | 1.62 | mir-124 | 2.28 | mir-10 | 0.98 | mir-13b | 0.96 | mir-9a | 1.86 | mir-11 | 1.34 |
| mir-8 | 0.84 | mir-12 | 1.58 | mir-14 | 0.99 | mir-998 | 0.99 | mir-1000 | 1.47 | mir-252 | 1.76 | mir-2b | 0.94 | mir-10 | 0.84 | mir-983 | 1.83 | mir-34 | 1.32 |
| mir-995 | 0.79 | mir-305 | 1.54 | mir-998 | 0.81 | mir-276a | 0.74 | mir-263a | 1.37 | mir-13b | 1.63 | mir-281-1 | 0.91 | mir-11 | 0.75 | mir-9b | 1.82 | mir-305 | 1.08 |
| mir-277 | 0.78 | mir-2a-2 | 1.45 | mir-276 | 0.74 | mir-79 | 0.69 | mir-987 | 1.16 | mir-9a | 1.53 | mir-304 | 0.88 | mir-14 | 0.74 | mir-14 | 1.82 | mir-9c | 0.94 |
| mir-305 | 0.70 | mir-306 | 0.91 | mir-34 | 0.63 | mir-995 | 0.64 | mir-252 | 1.14 | mir-996 | 1.28 | mir-184 | 0.79 | mir-281-1 | 0.72 | mir-988 | 1.79 | mir-12 | 0.92 |
| mir-998 | 0.65 | mir-998 | 0.89 | mir-995 | 0.62 | mir-34 | 0.59 | mir-274 | 1.09 | mir-263a | 1.21 | mir-279 | 0.75 | mir-314 | 0.60 | mir-2b | 1.64 | mir-33 | 0.84 |
| mir-306 | 0.52 | mir-9c | 0.84 | mir-79 | 0.61 | mir-14 | 0.52 | mir-7 | 1.09 | mir-1000 | 1.20 | mir-11 | 0.74 | mir-279 | 0.56 | mir-960 | 1.47 | mir-995 | 0.76 |
| mir-9b | 0.52 | mir-995 | 0.84 | mir-9a | 0.59 | mir-275 | 0.51 | mir-9a | 0.95 | mir-31a | 1.03 | mir-305 | 0.72 | mir-263a | 0.52 | mir-13b | 1.41 | mir-14 | 0.63 |
| mir-79 | 0.43 | mir-79 | 0.81 | mir-9b | 0.48 | mir-33 | 0.45 | mir-929 | 0.95 | mir-11 | 1.00 | mir-79 | 0.62 | mir-305 | 0.50 | mir-317 | 1.22 | mir-1 | 0.62 |
| mir-2a-2 | 0.42 | mir-282 | 0.69 | mir-33 | 0.47 | mir-9a | 0.43 | mir-31a | 0.91 | mir-7 | 0.97 | mir-263a | 0.61 | mir-34 | 0.44 | mir-316 | 1.20 | mir-312 | 0.58 |
miRNAs are ranked in decreasing order and their contribution to the library given as a percentage of all miRNA reads in the library. The full list and percentages are provided in Table S22.
In addition to a temporal and spatial regulation of miRNAs during development (Ruby et al. 2007), a novel layer of complexity arises as we compare miRNA populations between males and females. Regardless of its heterogeneity, each tissue examined reveals miRNAs differentially or even exclusively expressed in one sex (Figure 1, B–H). The example of mir-8 illustrates how a miRNA classically regarded as abundant presents unexpected differences at the cellular level: mir-8 ranks within the top two most abundant miRNAs in heads and salivary glands (10–15% of reads), but fourth in lymphoid Kc cells (4%), and only 17th in S2 cells (<1%; Table 1). Following from this example, we systematically analyzed the miRNAs families present in each tissue and estimate their sex-biased expression.
MiRNAs in lymphoid embryonic-derived cells
We examined miRNAs populations in two embryonic hemocyte-derived cell lines commonly considered as male (S2) and female (Kc-167) based on the status of dosage compensation (Alekseyenko et al. 2012). In S2 cells, the X chromosome is upregulated by compensasomes recruited along the X chromosome at sites identical to those seen in embryos, male salivary glands, and whole larvae (Alekseyenko et al. 2006; Legube et al. 2006; Straub et al. 2008). In Kc cells, dosage compensation does not occur.
In our assays, most miRNAs are common to both Kc and S2 cell lines (Figure 1B). Overall, mir-14, mir-184, and bantam are most abundant and account for about half of all miRNA reads (Table 1) in both cell lines. Our miRNA profiles agree with the medium levels of mir-2 (mir-2a/-2b mainly) and high levels of mir-34 described in S2 cells (Sempere et al. 2003). High expression of mir-14 in these immortalized cell lines is consistent with its role as a cell-death suppressor (Xu et al. 2003). In addition, we do not detect miRNAs from the let-7-C cluster (let-7, mir-100, mir-125) in either cell type. This is consistent with previous reports in which the let-7-C miRNAs are detected only upon addition of the steroid hormone ecdysone, which has the opposite repressive effect on mir-34 (Sempere et al. 2003; Chawla and Sokol 2012).
For sexually dimorphic expression, we identify sex biases for several miRNAs: mir-12, mir-304, mir-92a, and mir-278 are more abundant in female Kc cells, whereas mir-252, mir-307, mir-282, and mir-980 are more prominent in male S2 cells (full lists in Table S2). Simple male and female embryonic cell lines express comparable miRNAs, yet differ in the level at which they express them.
MiRNAs in larval salivary glands
We examined miRNAs in larval salivary glands, organs that secrete the “glue” allowing the larva to attach to a substrate before pupariation. Salivary glands consist of two simple cell types (Kerman et al. 2006): secretory cells and duct cells that connect them to the larval mouth. Each type originates from ∼100 cells in the embryonic ectoderm. The secretory cells initiate multiple rounds of DNA replication and differentiate without subsequent division (endoreplication) to create the giant polytene chromosomes needed to meet the increased metabolic requirements of these cells to produce secretory proteins.
Among all samples, miRNAs with a strong preference for expression in salivary glands are, in descending expression levels, mir-263a, mir-375, mir-304, mir-275, mir-283, mir-308, mir-286, mir-5, and mir-307-as (Figure 1A). Many of these highly expressed miRNAs (e.g., mir-8, mir-263a, mir-275, mir-12, and mir-304) match those whose expression peaks during the larval stages (Ruby et al. 2007). In contrast, others (bantam, mir-13b, mir-11 or mir-14, mir-2b) are expressed more ubiquitously during development.
As expected from a tissue of low complexity with functions essential to both sexes, miRNAs are found in most instances at levels comparable in male and female salivary glands (Figure 1C). This overwhelming similarity is not without exceptions. Salivary glands present a limited number of miRNAs whose expression is biased toward male (mir-275) and female cells (miR-279, mir-282) (Table S3).
For several miRNAs, including mir-304, mir-12, mir-283, mir-314, and mir-981, the relative abundance in male and female salivary glands is matched in whole larvae, suggesting widespread or general functions (D. Fagegaltier, data not shown). Others vary, suggesting more discrete roles—these include miR-210, mir-13b, mir-100, mir-1013, and miR-979 (note that the latter has low reproducibility in salivary glands in our assay). Supporting this view, we detect by qPCR higher amounts of mir-979, mir-210, and mir-314 in whole larvae compared to salivary glands, and mir-979 and mir-314 are expressed in mixed L1-L3 larval imaginal discs (Ruby et al. 2007).
Surprisingly, although let-7 is undetectable by Northern blot on unsexed whole L3 larvae (Sempere et al. 2002), we find let-7 in salivary glands of both males (48 normalized reads) and females (219 reads) (Table S3). The function of let-7 in salivary glands is unknown, but sex-biased expression could reveal interesting differences and asynchrony in tissue homeostasis during male and female transition from larval to pupal stages.
MiRNAs in adult heads
In adult heads from 1-day-old flies, the most prominent miRNAs are mir-8, mir-1, mir-276a, and bantam (Table 1). All were shown to act on neural target genes, genes controlling circadian rhythms, eye development, and growth (see Smibert and Lai 2010 for review; Marrone et al. 2012; Li et al. 2013). Interestingly, whole heads do not differ dramatically in their miRNA profiles between males and females (Figure 1, A–D). Mir-2a, mir-2b, mir-277, mir-278, and, importantly, let-7, mir-34, and mir-124 are expressed at comparable levels in both sexes, in agreement with their general role in specifying neuronal cell fate, plasticity, and neurodegeneration (Kucherenko et al. 2012; Liu et al. 2012; Weng and Cohen 2012; Wu et al. 2012). Conversely, head expression of mir-276 and mir-190 is enriched in males, and mir-210, mir-274, mir-980, and mir-981 in females (Table S4). This finding hints toward sexually dimorphic functions of these miRNAs in neuronal tissues.
Finally, miR-252 and miR-980 were previously detected by qPCR in adult male and female heads 2–4 days after eclosion, while no miR-984 could be detected (Marrone et al. 2012). The present analysis provides evidence that all three miRNAs are expressed in the head and establishes the expression of both mir-252 and miR-984 as slightly more prominent in female heads compared to males. The functional significance of such biases is yet to be determined.
MiRNAs in the adult body, testis, and ovary
Almost all known miRNAs (155 miRNAs) are detected in the body, the most complex sample in our data set that covers all body parts of the adult fly with the exception of the head. We find primarily mir-1, mir-8, bantam, mir-276a, mir-277, mir-276, mir-2a, mir-996, and let-7 in the fly body (Table 1). The adult fly presents striking biases for miRNAs in males and females (Figure 1E and Table S5). Mir-989 represents the strongest female-biased miRNA in the body. Among a larger set of highly male-biased miRNAs, few are detected in females. Of the 22 most strongly male-biased miRNAs in the body (above fivefold), 17 miRNAs appear exclusively in males and the remaining miRNAs (mir-960, mir-961, mir-963, mir-983, and mir-984) yield at best a few tens of reads in females. Valuable insights into the origin of these discrepancies come from the following comparisons.
We compared the populations of miRNAs in testes and ovaries. We have seen so far that from single lymphoid cells to more complex and dimorphic heads and bodies, the most abundant miRNAs remain overall conserved between males and females with a few exceptions such as mir-8. This is strikingly not the case in testes and ovaries in which the most abundant miRNAs differ. In 2- to 4-day-old flies, about half of the miRNAs cloned from ovaries correspond to mir-989, bantam, mir-8, mir-2a, and mir-318 (Table 1). In testes, this set only partially overlaps with that of ovaries and includes mir-12, mir-8, mir-305, mir-959, mir-2a, let-7, and mir-34. Younger males exhibit the same set of abundant miRNAs in testes (D. Fagegaltier, data not shown) indicating that their high expression persists in testes at least over the early adult stages.
Each tissue in our data set presents a set of miRNAs underrepresented in the other tissues examined; although their relative abundance varies between male and female, these tissue-enriched miRNAs are usually overrepresented in both sexes (Figure 1).
Ovaries and testes are enriched for certain miRNAs, but very few are enriched in both tissues. Instead, two distinct sets of miRNAs emerge: a large cluster of testis-enriched miRNAs and a smaller set of ovary-enriched miRNAs (Figure 1A). The miRNA populations of these highly sexually dimorphic tissues are highly skewed and sexually dimorphic (Figure 1H). 47 miRNAs show higher levels of expression in testes compared to ovaries (above fivefold), whereas 29 are preferentially expressed in the ovary (Table S6). Fourteen strongly testis-biased miRNAs are not detected in ovaries and may carry important and specialized functions.
During maturation, oocytes express several classes of miRNAs whose levels vary upon fertilization and later during the initiation of zygotic transcription. Maternal deposition of miRNAs has been reported in Drosophila to serve a variety of functions, including the destabilization of a large number of maternal mRNAs in early embryos via the SMAUG protein (Giraldez et al. 2006; Bushati et al. 2008; Soni et al. 2013).
In whole ovaries from 2- to 4-day-old females, we identify all but one of the reported maternally deposited miRNAs previously described in oocytes, including miRNA populations denoted as unstable (class I, restricted to stage 14 oocytes), and stable (class II, including maternally deposited and zygotically expressed miRNAs). Specifically, we identified mir-318, mir-276a, mir-34, mir-317, mir-284, let-7 (class I); mir-13b, mir-2a, mir-306, mir-184, mir-312, and mir-310 (class II). Zygote-specific miRNAs of class III were not detected, namely the miR-309 cluster zygotically expressed ∼2- to 4-hr-post-fertilization (miR-309, miR-3, miR-286, miR-4, miR-5, miR-6-1, mir-6-2, and mir-6-3). We note, however, that the expression of mir-3 and mir-5 for class III could start earlier than previously described: six to eight reads match mir-3 and mir-5. Supporting this view, low counts for mir-3 and mir-5 in 2- to 4-day-old female ovaries increase in loqs mutants in Czech et al. (2008). A low expression of these two miRNAs in the ovary therefore remains a possibility.
In summary, in addition to expressing highly divergent sets of miRNAs, both testes and ovaries express very different sets of miRNAs compared to other tissues (Figure 1A): their respective miRNA profiles appear as the most divergent of our data set.
To gain insight into the sexually dimorphic functions of miRNAs in somatic tissues, we compared the miRNA populations in testes and male body or ovaries and female body, respectively. We reasoned that a miRNA present in the male body sample (that includes testes) but not enriched in testes is likely to have at least a somatic function outside the testes. Thirty such miRNAs are enriched at least fivefold in the male body (Figure 1F and Table S7), suggesting that they carry distinct functions in the soma.
In a similar comparison between female body and ovaries, we identify 45 miRNAs enriched in female somatic tissues, 28 of which are almost exclusively detected in the female body (Figure 1G and Table S8).
Finally, the tallies of miRNAs enriched in ovaries and testes compared to the body samples establish that most of the differences in miRNA expression between male and female body come primarily from these highly dimorphic internal reproductive tissues.
Male- and female-biased somatic miRNAs
We next looked specifically for miRNAs presenting a consistent bias toward males or toward females in all somatic tissues studied. We considered miRNAs presenting at least a twofold enrichment in one sex in lymphoid cells, salivary glands, head, and/or body. Interestingly, not a single miRNA presented male biases in all four or even three tissues, suggesting that there is no ubiquitous male-biased somatic miRNA. However, a total of 23 miRNAs are male biased in two tissues, generally salivary glands and S2 cells, or head and body (Figure S2A). Of these 23 somatic male miRNAs, 10 are also enriched in testes (compared to the male body) and might therefore repress sex-specific targets in the male soma and gonads. The remaining 13 miRNAs are not enriched in testes and represent somatic male-biased miRNAs.
For females, 32 miRNAs are consistently female biased in the soma (more than twofold; Figure S2B). Interestingly, mir-318 is enriched in females in all four somatic tissues and several other miRNAs in three tissues. Eight of these female-biased miRNAs are also enriched in ovaries. The remaining 24 miRNAs represent somatic female-biased miRNAs.
Ovary and testis miRNAs
We next searched for miRNAs prevalent in ovaries, testes, or both to identify those with possible functions in gametogenesis or the organization of the adult gonads. Compared to same-sex somatic tissue, 32 miRNAs are enriched in the ovary and 51 in testis. Of these, 18 are enriched in both testis and ovary, likely underscoring important processes common to gonads of both sexes. Conversely, 8 miRNAs enriched in ovaries but not in testes and 30 miRNAs enriched in testes but not in ovaries could control sex-specific aspects of gonad development (Figure S2C).
The 30 testis-enriched miRNAs reflect a remarkable genomic homogeneity: 11 of these miRNAs are located on the X chromosome (Figure S2C). This fraction is much higher than expected (P = 0.0018, hypergeometric distribution) and significantly greater than the proportion of X-linked miRNAs found in other tissues (male soma, P = 0.0747; ovaries, P = 0.2115). This enrichment of miRNAs originating from the X chromosome is therefore a phenomenon specific to testes. The prevalence of X-linked miRNAs expression in testes has been reported in mouse and was explained by X-linked gene dosage (Ro et al. 2007; Mishima et al. 2008). It is tempting to speculate that such a mechanism may hold true in Drosophila testes as well. To date, the mechanisms regulating X-linked genes expression in testes such as meiotic sex-chromosome inactivation and dosage compensation remain unclear (Vibranovski et al. 2009; Deng et al. 2011; Meiklejohn et al. 2011; Vibranovski et al. 2012).
Are X-linked miRNAs transcriptionally regulated by dosage compensation?
We next asked whether the transcriptional regulation of X-linked miRNAs depends on dosage compensation in male somatic tissues. At the genomic level, miRNAs are evenly distributed across the Drosophila genome with no obvious pattern on the X chromosomes. The X chromosome represents ∼20% of the Drosophila genome and bears an even share of miRNAs. The 149 genomic miRNA loci queried in this study are distributed evenly across all chromosomes with 34, 33, 22, 29, and 27 miRNAs on chromosomes 2L and 2R, 3L and 3R, and the X chromosome. Together, the fourth chromosome and heterochromatin of chromosome 3 (3h) account for 4 additional miRNAs. To date, no miRNA has been reported on the Y chromosome.
Two classes of miRNAs are present as typical genic miRNAs and mirtrons. A total of 37 mirtrons (24.8%) reside within introns in the genome. Compared to miRNAs regulated by their own promoter, mirtrons are slightly enriched on chromosome 2L (32.4% of chromosome 2L miRNAs) and underrepresented on chromosome 3L (13.6%); on the X chromosome, 22.2% are encoded as mirtrons.
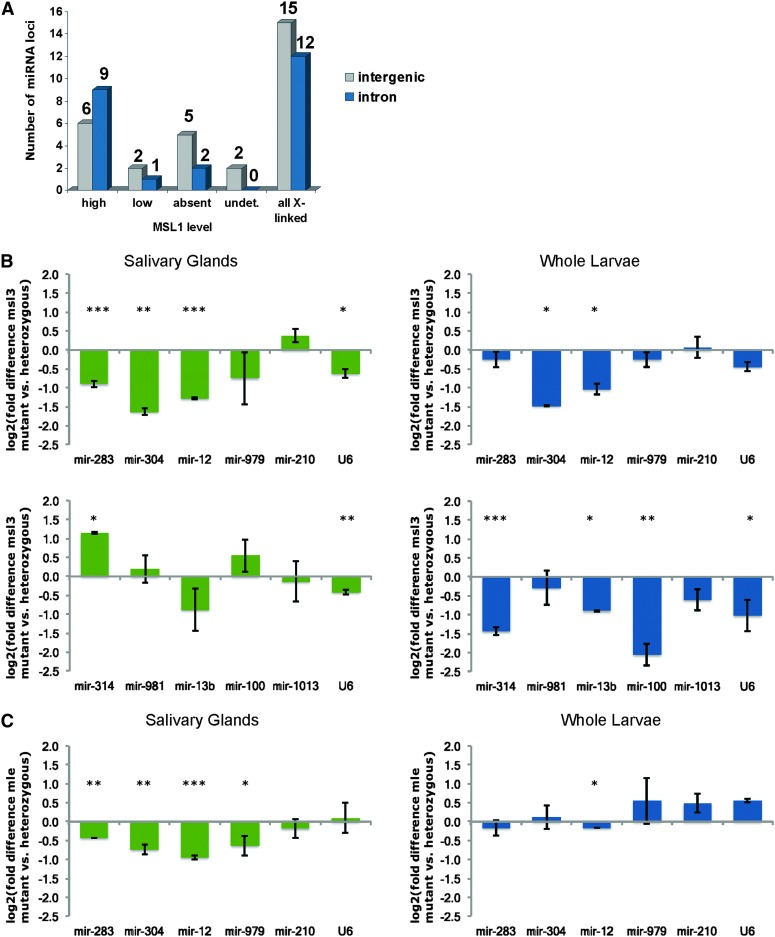
To address whether dosage compensation controls miRNA expression directly on the X chromosome, we first examined whether X-linked miRNAs reside in regions bound by MSL-1 in ChIP–Chip experiments reported by Straub et al. (2008) in S2 cells and embryos. We note that X-linked miRNAs reside outside the primary “high affinity sites” defined in these studies and are therefore not primary strong binding sites for compensasomes. MSL-1 levels were further divided into high, medium, low, or MSL-1 absent subtypes (Figure 2A). With the exception of two miRNA loci residing in regions not addressed in this study, 67% of X-linked miRNA loci reside in regions bound by compensasomes, indicating that some miRNAs could be dosage compensated.
Figure 2.
Effects of dosage compensation on X-linked miRNAs expression. (A) Compensasome association at the 27 X-linked miRNA loci vary, with 15 miRNAs located in regions highly populated by compensasomes (high), three miRNAs with low MSL coverage and seven miRNAs residing in regions deprived of compensasomes. Two miRNAs loci in regions not covered by the arrays (undet.) could not be assessed. We observe no difference in MSL occupancy for miRNAs residing in intronic or intergenic regions. The level of expression of miRNAs in the absence of a functional dosage-compensation complex was examined in male salivary glands (green charts) and whole larvae (blue charts) mutant for msl3 (B) or mle (C). Of the six X-linked miRNAs expressed in these tissues, mir-304, mir-12, and mir-13b show the expected twofold decrease in all mutant samples compared to controls. MiRNA levels remain unchanged, however, for X-linked mir-979, mir-210, or mir-283. Autosomal mir-981, mir-100, or mir-1013 levels did not change. Mir-314 levels increase in msl3 mutant salivary glands and decrease in whole larvae. All miRNAs were tested in triplicates on two independent biological replicates. Values were normalized to the autosomal gene standard, Dspt4, whose levels remain unchanged between males, females, or compensasome mutants (Chiang and Kurnit 2003). We observe similar trends between male and female salivary glands miRNAs in wild-type Oregon-R and msl3 heterozygous mutant controls for all 10 miRNAs tested (mir-100, mir-979, mir-12, mir-314, mir-981, mir-210, mir-1013, mir-283, mir-13b, mir-304). Error bars represent standard deviations. P-values: (*) P < 0.05; (**) P < 0.005; (***) P < 0.0005. Calculations are provided in Table S11, Table S12, Table S13, Table S14, Table S15, and Table S16.
For those X-linked miRNAs residing in regions bound by MSL1, dosage compensation could provide a direct means to equalize their expression in both sexes. To test this possibility, we assayed by quantitative PCR the levels of X-linked miRNAs expressed in salivary glands, a tissue in which dosage compensation has been extensively studied, and compared miRNA levels in wild-type animals and mutants for compensasomes function (msl-3 and mle; Figure 2, B and C). In mutant males, X-linked mir-304, mir-13b, and mir-12 levels are reduced approximately twofold, as expected for genes whose expression depends on compensasomes; autosomal controls mir-100 (2L), mir-981, and mir-1013 (3R) were unaffected. However, levels of two miRNAs mir-979 and mir-210 did not change significantly in mutants, implying that some X-linked miRNAs escape dosage compensation—these join the short list of X-linked protein coding genes (Lsp1-α, dpr8, CG9650) that similarly avoid compensation. Interestingly, mir-12 and mir-304 belong to the same cluster as mir-283 but behave differently in mutants. The milder reduction of mir-283 levels in mutant animals supports previous observations that mir-12 and mir-304 expression patterns show little correlation with that of mir-283. Although they share a common Pol II promoter, post-transcriptional processing could account for these differences (Ryazansky et al. 2011).
In summary, only a few X-linked miRNAs are influenced to some extent by dosage compensation. Two X-chromosome miRNAs clearly escape dosage compensation (mir-979, mir-210). One major difference between miRNA and protein-coding genes escaping dosage compensation is that the latter tend to reside outside MSL-bound domains. Overall, it is tempting to speculate that dosage compensation presents only limited advantages for short, rapidly regulated miRNA genes.
A role for the let-7-C locus in sexual dimorphism
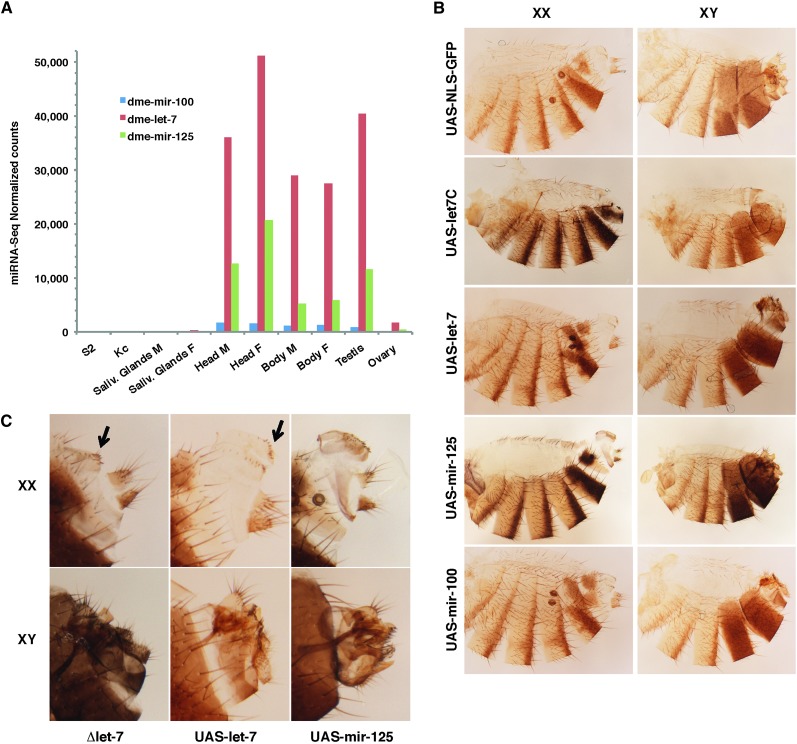
We investigated the functional relevance of sex-biased expression of miRNAs using the example of let-7. Two miRNAs in the let-7-C cluster, mir-125 and let-7, are highly expressed in the fly head, body, and testis and poorly in the ovary (Figure 4A). Mir-100 levels are consistently lower in all tissues expressing let-7 and mir-125. From worms to flies and humans, the let-7-C locus has an important role throughout development (Pasquinelli et al. 2000). In flies, let-7-C is under the control of ecdysone and mimics the hormone peaks required for each of the developmental transitions that turn an embryo into an adult (reviewed in Ambros 2011). We set out to investigate the functional implications of the male-biased expression of the let-7-C miRNAs in the gonads and more generally its function in adult males and females.
Figure 4.
Let-7-C miRNAs abundance across tissues and overexpression. (A) The three miRNAs of the let-7-C locus are almost undetectable in salivary glands and lymphoid cells and highly expressed in male and female heads and body. All let-7-C miRNAs are highly expressed in testes compared to ovaries. Let-7 is consistently more abundant than mir-125 in all tissues expressing let-7-C, while mir-100 remain comparably low. (B) Cuticle preparations of dsx–Gal4 > UAS–let-7-C males (XY) and females (XX): both fail to transform A8–A10 segments into a proper genitalia and analia. Increased abdominal pigmentation in let-7-C and mir-125 overexpressing flies (compare UAS–let-7-C or UAS–mir-125 to UAS–NLS–GFP). (B and C) Terminalia phenotypes of flies overexpressing single miRNAs of the locus: additional vaginal teeth toward the posterior section of the vulva in dsx–Gal4 > UAS–let-7 females, similar to Δlet7 flies (arrows in closeups in C); additional row of vaginal teeth and overgrowth of the anterior vulva in dsx–Gal4 > UAS–mir-125 females; overgrowth of the genital ring/arch dsx–Gal4 > UAS–let-7 male genitalia; dsx–Gal4 > UAS–mir-125 male genital ring collapses into a more compact bulged ring (lateral view in C). Note the reduced size of male external genital and anal structures in UAS–let-7, UAS–mir-125, and Δlet7 males. Dsx–Gal4> UAS–NLS–GFP, and dsx–Gal4 > UAS–mir-100 males and females appear normal.
Sex-biased let-7 miRNA expression in gonadal somatic cells sustains germline differentiation
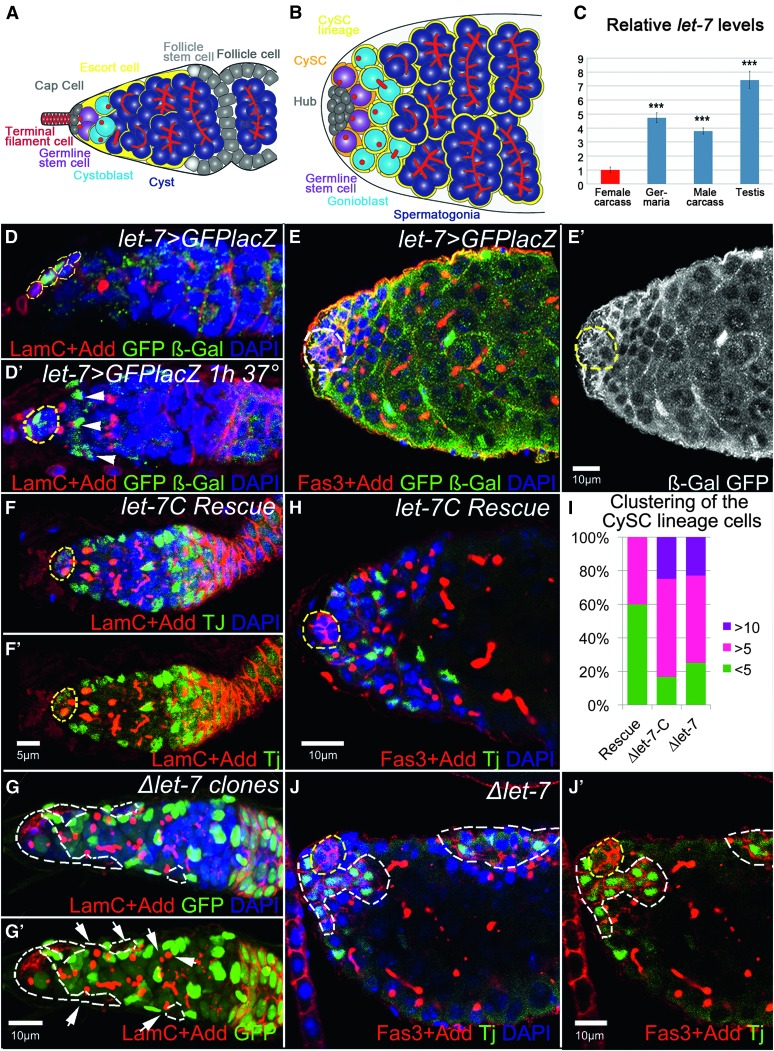
We analyzed let-7-C expression pattern in the adult Drosophila gonads using Gal4 under the control of the intrinsic let-7-C promoter. In the germarium, let-7-C is expressed in some of the somatic escort (ECs) and cap cells (CpCs; Figure 3, A, B, and D). The number of let-7-C-expressing cells and the activity of the let-7-C promoter per se fluctuate in CpCs and ECs in different germaria and within the same germarium (compare Figure 3, D and D′), demonstrating that the expression pattern of let-7-C is highly dynamic.
Figure 3.
let-7 deficiency affects somatic cells behavior in ovaries and testes, which cell-non-autonomously influences early germline differentiation. Drosophila ovaries and testes present commonalities in their general organization and the type of cells they comprise (Fuller and Spradling 2007). The ovary is a paired organ consisting of 16–20 ovarioles, each representing a string of progressively developing egg chambers. (A) At the apex of an ovariole, the germarium comprises somatic cells (terminal filament, TF; cap cells, CpC’s; escort cells, ECs; follicle stem cells, FSCs; follicle cells, FCs) and germline cells (germline stem cells, GSCs; cystoblast, CB; differentiating cyst, Cyst). GSCs are physically attached to the somatic cluster of cap cells that, with the terminal filament cells, represent the GSC niche. GSCs divide into a differentiating cystoblast, which then divides four times with incomplete cytokinesis, producing a 16-cell cyst in which 1 cell becomes the oocyte while the others develop as nurse cells. These different germline cell populations can be easily identified by their location and specific markers in the germarium. For example, Drosophila adducin homolog antibody marks an actin-rich cellular organelle represented as a dot-like structure in single cells (GSCs and CBs) and as a branched fusome in the developing cysts. There is another class of somatic cells at the anterior of the germarium, called the escort cells. These squamous cells are mitotically quiescent and envelop differentiating cysts to protect them from niche signaling, an important role for germline differentiation (Chen et al. 2011). ECs guide differentiating cysts to the posterior end of the germarium, where the germline becomes encapsulated by the follicular epithelium and pinched off from the germarium. (B) The Drosophila testes are a paired tubular organ that consists of somatic and germline cells. Scheme depicting the testis apex somatic cells (hub cells, Hub; cyst stem cells, CySC’s; cyst stem cells lineage, CySC lineage) and germline cells (germline stem cells, GSCs; gonioblast, GB; differentiating spermatocysts and spermatogonia). Attached to the stem cell niche, termed the hub, reside two types of stem cells: GSCs and CySC’s. While the hub is made by a cluster of postmitotic somatic cells, both stem cell types divide in synchrony to produce differentiating germline cysts, each of which is encapsulated by two somatic CySC’s lineage cells. This encapsulation is critical for proper germline differentiation (Leatherman and Dinardo 2010). Similar to the ovarian GSC progeny, the germline gonioblast undergoes four rounds of incomplete cytokinesis to produce 16 primary spermatocytes in a cyst, eventually generating 64 sperm cells. (C) The relative expression levels of miRNA let-7 in female and male carcasses, ovaries, and testes show that let-7 miRNA is sex biased and expressed at the higher levels in testes (see also Table S17). (D, D′, E, E′) Localization of let-7-C miRNAs in the ovaries and testes, detected via membrane GFP and nuclear lacZ expressed under the control of the let-7-C promoter (let-7-CGK1–Gal4/UAS–CD8-GFP::nuc–lacZ). In the ovary, let-7-C is expressed in the somatic cells of the germarium, CpCs and ECs (D). (D′) More ECs express let-7-C, shown by the presence of lacZ (β-Gal, green) when adult flies were subject to a heat shock for 1 hr prior to dissection. (E and E′) In the testis, let-7-C is broadly expressed in all somatic cells, CySC’s and their lineage and the hub cells (let-7-CGK1-Gal4/UAS-CD8-GFP::nuc-lacZ). (F and F′) Control let-7-C rescue germaria (P{W8, let-7-C}/+; let-7-CGK1/KO1) show typical numbers of germline SSCs and developing cysts (marked by the spectrosome and fusome marker Adducin, Add, red) as well as somatic ECs (marked by Traffic jam, Tj, green). The GSC niche (marked by Lamin C, LamC, red) is outlined by yellow dashed lines. (G and G′) Somatic cell ∆let-7 clones (Ubi–GFP, FRT40A/ FRT40A, let-7 miR-125; bab1–Gal4, UAS–Flp/P{W8, let-7-CΔlet-7}) are marked by the absence of GFP and outlined by white dashed lines. Upon let-7 depletion in the soma of the germarium, the number of somatic ECs and germline SSCs increases, while the number of fusome-containing cysts decreases. The magnification is the same in F and G. (H) The testicular apex of a control let-7-C rescue (P{W8, let-7-C}; let-7-CGK1/KO1) displays typical numbers of germline CBs and cysts (marked by spectrosome and fusome marker Add, red) and the CySC lineage cells (marked by Tj, green); yellow dashed lines outline the hub (marked by Fasciclin III, Fas3, red). (J) In Δlet-7 mutant testis (let-7-CGK1/KO1; P{W8, let-7-CΔlet-7}/+) the CySC lineage cells cluster in larger groups and express the ovarian follicular epithelium marker, Fas3. These clusters (outlined by white dashed lines) can be found at the apex or the side of the testicular tube. (I) Percentages of mutant testes containing large ≥5 or >10 somatic cell clusters in comparison to control let-7-C Rescue. Ten to 20 testes were analyzed for each genotype. Red: LamC + Add in D, D′, F, F′, G, G′ ; Fas3 + Add in E, H, J, J′. Green: GFP + β-Gal in D and E; GFP in G, G′; Tj in F, F′, H, J, J′. White: GFP + β-Gal in E′. Blue: DAPI in D–H, G–J′. D, E + E′, H, J + J′ are single confocal sections while D′, F + F′, and G + G′ are projections of several z-stacks.
In testes, somatic cells, namely cyst stem cells (CySC’s), and their lineage express let-7-C (Figure 3, E–E′). We find consistently higher levels of let-7-C per cell and higher numbers of let-7-C-positive cells per testicular apex compared to positive cells in the germarium (compare Figure 3, D–D′ and E, green).
We confirmed by RT–qPCR the male-biased expression of let-7 in the gonads. Let-7 levels are about eight times higher in testes compared to those in ovaries (Figure 3C). In addition, let-7 is enriched in the gonads of both sexes compared to carcasses. Together, these findings support the biased expression of let-7-C and let-7 reported here by miRNA profiling. Further, they establish a stronger transcriptional activity of the let-7-C promoter in the testicular soma compared to the activity in the ovarian soma.
In addition, let-7 has been detected in the hub cells of old male testes, demonstrating its responsiveness to aging in males (Toledano et al. 2012). The dynamic expression of let-7 and its responsiveness to external and internal conditions (temperature—see below, aging) in both sexes prompted us to ask whether let-7 regulates gonadal somatic cell behavior.
We analyzed the tissue architecture of the germaria and testes upon let-7 depletion. In the ovary, ∆let-7 clones induced specifically in somatic ECs result in a peculiar germarium architecture where both the germline and soma are abnormal (Figure 3G). Mutant germaria contain supernumerary single spectrosome germline cells (arrows) and oddly shaped somatic ECs expressing higher amounts of cell adhesion molecules (Figure 3G). Let-7 deficiency results in excessive single spectrosome germline cells (GSCs, CBs) and reduced numbers of differentiating cysts (compare Figure 3, F and G), reflective of a delayed early germline differentiation. Altered ECs lacking let-7 likely fail to protect the differentiating germline cysts from the niche signaling. The phenotypes observed imply that let-7 controls somatic ECs behavior, which in turn non-cell-autonomously modulates the efficiency of early ovarian germline differentiation.
In the testes too, let-7 deficiency affects somatic cell behavior. Normally, two squamous cells encapsulate one gonioblast, such that only small clusters of somatic cells (fewer than five cells) marked by a nuclear marker can be detected (Figure 3, H and I). Instead, CySC lineage cells lacking let-7 form large aggregates of >10 cells (Figure 3, I, J, and J′). Interestingly, these clustered cells accumulate the cell adhesion molecule FasIII, indicative of a columnar epithelium that resembles the ovarian follicular epithelium. Such aggregates do not develop in let-7-C rescue flies (Figure 3, H and I).
Let-7 levels are important to specify male and female sexual identify
Let-7 levels fluctuate at each developmental transition of the fly, including a major pulse in late pupae. During development, let-7 miRNA first appears in L3 larvae and has been detected in genital discs of L3 male and female larvae (Sokol et al. 2008). Dsx levels vary as well during development and are especially important from the L3 larval to the late pupal stages to transform genital discs into adult genital and anal structures. To ask whether let-7 influences the transformation of these structures, we expressed UAS–let-7-C in patterns that mimic pulses of the major controller of the sex-determination hierarchy DSX using the dsx–Gal4 driver. Animals overexpressing let-7-C in dsx-expressing cells develop severe phenotypes: males and females fail to develop segments A8–A10 and lack a terminalia (analia and genitalia) (Figure 4B). Overexpressing individual miRNAs from the cluster alters the same segments (Figure 4, B and C): females overexpressing mir-125 form additional vaginal teeth, up to a full second row of teeth, and a vulva overgrowth. Additional vaginal teeth appear toward the posterior part of the vulva in females overexpressing let-7. Males overexpressing let-7 show an overgrowth of the genital ring/arch. In males overexpressing mir-125, this structure proliferates yet collapses into a more compact bulged ring (Figure 4, B and C). In addition, overexpressing let-7 or mir-125 in dsx-expressing cells results in the overall reduction of the male external genital structures. Importantly, the external terminalia of males and females lacking let-7 present similar defects (Figure 4C). Finally, in animals overexpressing mir-125, the typical female abdominal pigmentation darkens in the posterior stripes of segments A3–A5 and in the terminal A6 and A7 segments where it widens (Figure 4B). Females overexpressing the whole let-7-C cluster show a similar increase in pigment, but not those overexpressing let-7 alone, pointing at mir-125 as the miRNA responsible for the pigment defects. Controls expressing UAS–NLS–GFP or UAS–mir-100 constructs do not present such phenotypes. Together, these results suggest that the ectopic expression of let-7-C, let-7, or mir-125 in dsx-expressing cells represses gene(s) necessary for pigmentation in most abdominal segments and importantly, gene(s) critical for the formation of the highly dimorphic segments A8–A10 in both sexes. The transformation of the genital disc into a male or female adult terminalia and the sexually dimorphic pigmentation pathway are both orchestrated by DSX during the pupal stages. It is likely that the respective timing and levels of expression of let-7-C miRNAs and dsx at this stage are important. None of the miRNAs in the let-7-C cluster, however, are predicted to target the dsx transcripts, suggesting that the phenotypes reflecting alterations of the levels of DSX or of the genes it acts with are generated upstream or in parallel to DSX action rather than from a direct interaction.
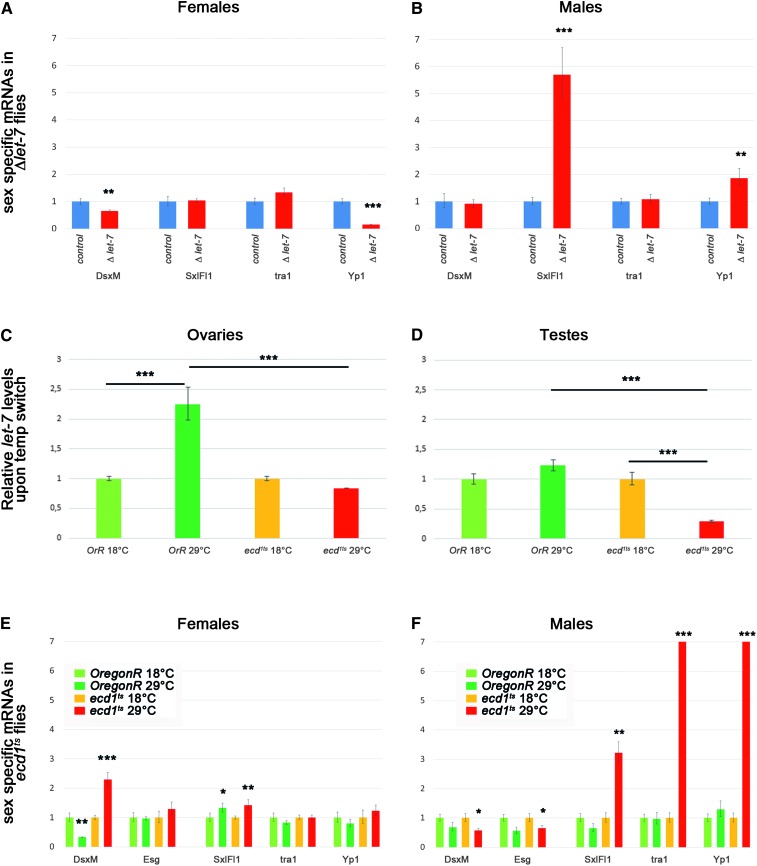
To test whether sex determination is affected by let-7, we quantified in animals lacking let-7 the male- (dsxM) and female-specific transcripts (Sxl, tra) of the sex-determination hierarchy and DSX downstream target Yp1 (Figure 5, A and B). In let-7 mutant females, the levels of Sxl and tra remain unchanged but Yp1 mRNA levels are significantly lower. The background levels of dsxM remain stable in females lacking let-7, implying that the levels of the direct activator of Yp1, DSXF, but not its repressor DSXM, are compromised. A general depletion of let-7 during development generates more dramatic effects in males. Specifically, males deficient for let-7 present a spurious expression of two genes normally restricted to females, Sxl and Yp1. Altogether these results point at a role of let-7 in modulating the sex-determination cascade during development, at least during the late-larval to late-pupal stages.
Figure 5.
Levels of sex-specific mRNAs are altered due to loss of function of steroid-dependent miRNA let-7 and ecdysone deficiency. (A and B) Relative levels of sex-specific mRNAs in Δlet-7 mutant (let-7-CGK1/KO1; P{W8, let-7-CΔlet-7}/+) and control rescue (P{W8, let-7-C}/+; let-7-CGK1/KO1) females (A) and males (B). (C and D) Relative levels of let-7 miRNA expression in wild-type (Oregon-R) and ecd1ts mutants kept at permissive (18°) and restrictive (29°) temperatures measured in the anterior parts of ovaries (C) and testes (D). (E and F) Relative levels of sex-specific mRNAs in wild-type (Oregon-R) and ecd1ts females (E) and males (F) kept at permissive (18°) and restrictive (29°) temperatures. Samples at restrictive temperature were compared to the respective genotype and sex at permissive temperature. Error bars represent the range of mRNA levels; P-values are calculated by Student t-test: (*) P < 0.05, (**) P < 0.005, (***) P < 0.0005. See also Table S18, Table S19, and Table S20.
Ecdysone signaling via let-7 maintains sexual identity during adulthood
We attempted to determine what signaling pathway acts upstream of let-7 in the process of sexual identity establishment but also its maintenance. Hormonal signaling is a strong candidate for this type of regulation, since it may coordinate the sex-specific differentiation of different tissues in the whole organism. The ecdysteroid signaling cascade governs various biological responses during Drosophila lifetime. Its specificity depends on the differential spatiotemporal expression of downstream components specific to various cell types and developmental stages. Steroid-coupled regulation of let-7 expression takes place during the developmental transition from larval-to-reproductive animals (Sempere et al. 2002, 2003; Garbuzov and Tatar 2010; Chawla and Sokol 2012; Kucherenko et al. 2012). During this period, DSX most actively controls the transformation of the A8–A10 genital primordia into dimorphic male and female terminal structures. In addition, we and others have reported that a deficit in ecdysone signaling generates non-cell-autonomous defects in early female germline differentiation (König et al. 2011; Morris and Spradling 2012). These defects resemble the let-7 loss-of-function defects described here in ovaries. Moreover, we report similar germline differentiation defects in let-7 mutant testes. Together, the steroid-dependent onset of let-7 expression during development and the similarity of the germline phenotypes observed in adult males and females lacking let-7 converge toward a model in which ecdysone signaling acts upstream of the let-7 miRNA to modulate (i) sex determination in tissues and (ii) germ-cell differentiation via the gonadal soma.
Whether let-7 also depends on ecdysone signaling in adults has not been addressed previously. To address this, we took advantage of a temperature-sensitive ecdysoneless mutation, ecd1ts, that blocks the production of ecdysone at restrictive temperature (29°) (Garen et al. 1977). After impairing ecdysone signaling specifically in adults, we analyzed let-7 levels in germaria and testes of Oregon-R and ecd1ts flies. Shifting Oregon-R male and female adults to 29° results in increased let-7 levels, showing that let-7 expression is dependent on temperature (Figure 5, C and D). Enhanced activity of the let-7-C promoter is also visible at the cellular level in the ovarian CpC’s and ECs upon heat shock (Figure 3D′). Contrary to wild-type flies, let-7 levels drop at the restrictive temperature in ecd1ts mutant female and more significantly in male gonads, demonstrating that let-7 expression depends on ecdysone signaling in the adult germaria and testes (Figure 5, C and D).
While the signaling cascade that establishes sexual identity has been studied extensively, the question of whether certain cues are needed to actively maintain sexual identity throughout the adult life has not been addressed. We quantified the expression of female- and male-specific components of the sex-determination hierarchy 3 days after inducing an ecdysone deficit in mature adults. Females lacking ecdysone begin to express the male-specific isoform of dsx, dsxM (Figure 5E). In males lacking ecdysone, conversely, male-specific mRNA levels of dsxM and escargot (esg) fall significantly while the female Sxl, tra, and Yp1 transcripts undergo a dramatic burst in expression (Figure 5F). Because Sxl and tra become aberrantly produced in males, and both are required to produce the female-specific isoform of dsx, it is likely that Yp1 hyperactivation in males is a consequence of the production of DSXF when ecdysone signaling is disrupted.
Together, our data support the hypothesis that ecdysone is required to maintain the sexual fate of adult cells. Interestingly, ecdysone effect on sex-specific mRNAs expression is significantly stronger than that of let-7 alone (compare Figure 5, A, B, E, and F), indicating that this systemic hormonal signaling regulates in addition to let-7 other key players in the maintenance of sexual identity during adulthood. Critically, these data suggest that ecdysone signaling plays an essential role in the maintenance of sexual identity in the adult Drosophila, primarily in males, and that this function is mediated at least in part by let-7 miRNA.
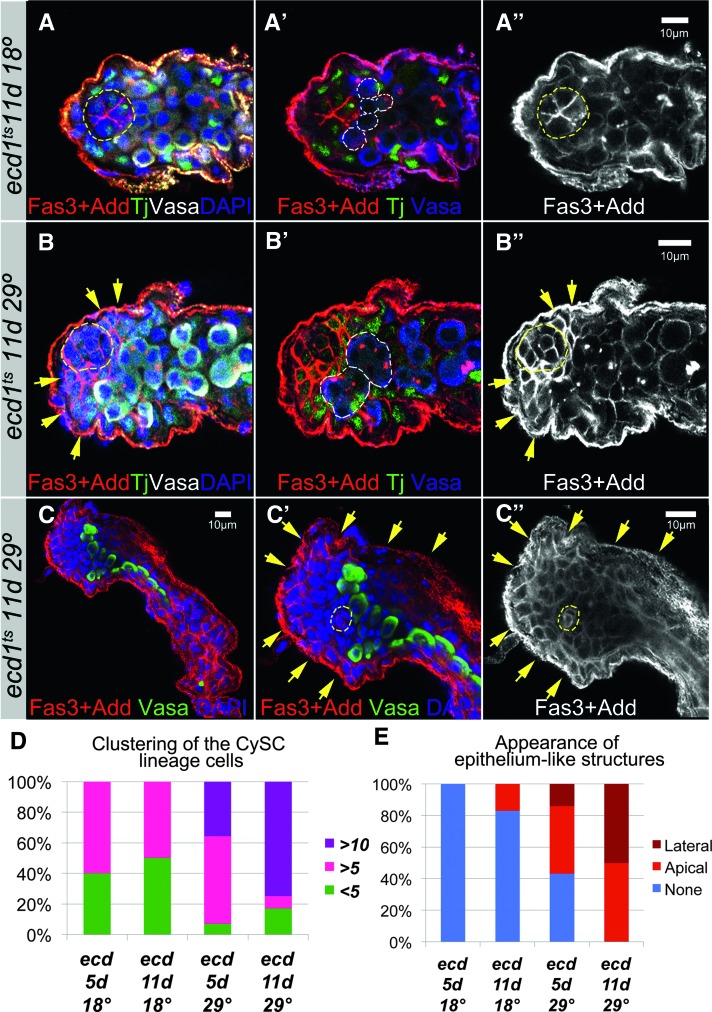
Ecdysone signaling via let-7 maintains male cell fate of the testicular soma during adulthood
Ecdysone-deficient (ecd1ts) males have reduced fertility and most become completely sterile after 3 days at restrictive temperature (Garen et al. 1977). At the tissue level, ecdysone signaling appears important to maintaining the proper behavior and function of the somatic CySC’s lineage in the adult testis (Figure 6, A–C). In adult males subject to ecdysone deficiency, the CySC’s lineage overproliferates, expresses epithelial markers, and non-cell-autonomously affects germline differentiation. Massive clusters of somatic cells appear at the testis apex, forming occasionally epithelial-like sheaths surrounding the testicular tube (Figure 6, B and C). The appearance of aggregates coincides with defects in germline differentiation at two levels. First, somatic cells clustering at the apex displace germline stem cells from the hub, forcing their premature differentiation (Figure 6B′). Second, somatic cells accumulating in lateral sheaths further disrupt their progression through germline differentiation programs (Figure 6C′). Flies deprived of ecdysone for a longer period show more severe phenotypes (Figure 6, D and E). Still, these alterations remain partial sex transformations. We never observed a full transformation resulting in the production of sperm or egg in mutants of the opposite sex. These findings suggest that sustained steroid activity is required to maintain let-7 levels within testicular somatic cells.
Figure 6.
Ecdysone signaling controls testicular soma behavior and function. (A) In control (ecd1ts, 11 days at 18°), the hub (outlined in yellow, A, A′′) is surrounded by proportional numbers of CySC’s and GSCs (outlined in white, A′). (B and C) In mutants (ecd1ts, 11 days at 29°), somatic cells aggregate in large epithelium-like clusters next to the hub (yellow arrows, B, B′′). In contrast, GSCs that would be seen as germline cells with a single spectrosome are no longer attached to the hub, indicating that they were pushed away from the niche and their differentiation was induced. Differentiating cysts are outlined in white, B′. Sometimes these abnormal epithelial-like sheets are found around the whole testicular tube (yellow arrows, C′, C′′), affecting germline differentiation: differentiating cysts that would be marked by branched fusomes are not seen. Note that the aberrant somatic cells express the Fas3 marker of hub cells and ovarian follicular epithelium. (D and E) The frequency of testes displaying clustering of the CySC lineage cells (D) and the appearance of epithelium sheets (E) increase with the time flies were deprived of ecdysone (5 or 11 days). See also Table S21. Red: Fas3 + Add in A, A′, B, B′, C, C′. White: Vasa in A, B; Fas3 + Add in A′′, B′′, C′′. Green: Tj in A, A′, B, B′; Vasa in C, C′. Blue: DAPI in A, B, C; Vasa in A′, B′, C′. All pictures are single confocal sections.
Taken together, our analyses show that steroid signaling is involved in the maintenance of sexual identity in adult flies and in the maintenance of germline differentiation programs in the gonads. This hormonal signal engages miRNAs to execute this regulation in a gender-specific manner. Therefore the maintenance of sexual identity in the adult life requires a systemic signaling that strongly depends on the general state of the organism and external conditions. This type of regulation is common in higher vertebrates including humans, implying that the analysis of sex-biased miRNAs and their targets will be of great importance to better understanding of sexual identity safeguarding throughout life at the cellular level in different organisms.
Evidence supporting the existence of a long-range gonadal axis in flies has emerged recently. Ecdysone is metabolized in the fat body in both sexes and is also present exclusively in the female germline late follicles. As a result, ecdysone titers are higher in females (Bownes et al. 1983; Parisi et al. 2010). Germline ablation largely decreases ecdysone titers and affects sex-biased somatic genes, including ecdysone biosynthesis genes exclusively in females (Parisi et al. 2004, 2010). In fact, half of the sex-biased genes in the female soma are estimated to be germline-dependent genes that respond to ecdysone, the other half comprising germline-independent sex-biased genes regulated by the sex-determination hierarchy. The absence of germline, however, does not affect somatic expression of the canonical sex-determination genes in either sex. Our study shows that sex-determination gene expression depends largely on ecdysone-mediated let-7 signaling in the soma, primarily in males. The pronounced effects on dsx and Sxl in males suggest that ecdysone and let-7 constitute the system that supports the expression of somatic sex-biased genes in males.
The effects of let-7 depletion on Yp1 and, importantly, the influence of ecdysone signaling on the sex-determination hierarchy in males raise the possibility that, although more weakly, all sex-biased genes eventually depend on hormones in females as well. If true, their sex-biased expression would result from two pathways regulated by ecdysone: a germline-dependent influence of ecdysone known as the gonadal sex hormone axis and a second pathway regulated by the sex-determination hierarchy that is germline independent but, as shown here, influenced mildly by hormones and let-7. Supporting this view, females require ecdysone receptor (EcR) expression in FRU expressing neurons to modulate precopulatory behavior just like males (see below), and ecdysone deficient females present male behaviors (Dalton et al. 2009; Ganter et al. 2012).
Intrinsic sex-specific specific factors and the proper sexual identity of the fat body are important for courtship behavior orchestrated by DSX and FRU in the brain, suggesting a fine interplay between signaling from the fat body and the sex-specific regulation of the nervous system (Lazareva et al. 2007; Camara et al. 2008). In particular, the EcR-A isoform that may interact with let-7 signaling is required in the FRUM neural circuit for male courtship behavior (Sanders and Arbeitman 2008; Dalton et al. 2009).
FRUM and DSX establish neural circuits differentially in each sex at mid-metamorphosis, a time at which they show strongest expression. Let-7 pulses could affect the FRU and DSX branches of the sex-determination hierarchy in the neural circuitry during this period. Let-7 may well contribute to a feedback loop in the nervous system since FRU targets include a preponderance of genes regulated by the steroid hormone ecdysone (Dalton et al. 2009, 2013; Neville et al. 2014). However, the reported FRUM and DSX binding sites in Luo et al. (2011) do not include the let-7-C locus, suggesting an indirect interaction.
In addition to ecdysone titers, let-7 levels and its mode of action are particularly important to understanding the impact of hormones in each sex and how they may regulate sex determination. Let-7 expression is dependent on ecdysteroids in both male and female adult gonads; however, ecdysone signaling facilitates cell gender maintenance in the organism via let-7 primarily in males, suggesting that let-7 response to ecdysone signaling and/or its effectors differ between the sexes. During metamorphosis, let-7 is induced by ecdysone signaling to control the timing of neuronal differentiation by way of BTB transcription factors. Two of them, Abrupt and Chinmo, act as negative regulators of ecdysone signaling, therefore creating a feedback loop (Zhu et al. 2006; Wu et al. 2012). By targetting repressors, let-7 finely modulates and reinforces steroid hormone signaling pulses in the brain. Interestingly, Abrupt modulates ecdysone signaling in the ovary as well (Jang et al. 2009). Increasing ecdysone titers established during oocyte development coincide with the gradual decrease of Abrupt concentration in the ovary. When ecdysone signaling is very high, Abrupt fails to bind the EcR coactivator Taiman, resulting in ecdysone signaling block. Whether let-7 controls Abrupt in the gonads and contributes to reinforcing ecdysone signaling via a feedback loop has not been studied. Nonetheless, naturally low levels of let-7 in the ovary where ecdsyone levels are plenty, contrary to testes, allows let-7 to generate in the germarium soma a sharper threshold response to the systemic signaling only when ecdysone titers are high (A. Konig and H. Shcherbata, unpublished results). In turn, because ecdysone and let-7-deficient phenotypes are just as strong in male and female gonads, higher levels of let-7 seem required to respond to lower ecdysone titers in males.
The two bona fide targets of let-7 in the brain present intriguing parallels in the gonads. Under the control of ecdysone, Chinmo and Abrupt control the differentiation of the gonadal soma specifically in one sex: Abrupt acts in the germarium and Chinmo in testis (Jang et al. 2009; Flaherty et al. 2010). Both are effectors of the JAK/STAT pathway, known to act differently in male and female gonads (Decotto and Spradling 2005). As examplified by the JAK/STAT pathway in the gonads, sex-specific transcription factors controlled by differential levels of let-7 may respond differently to ecdysone signaling and lead to sex-specific functions. This may hold true in the brain where Chinmo is a direct target of FruM in males (Neville et al. 2014).
Another important aspect of our findings is the temporal character of ecdysone-let-7 involvement in the gonadal soma. It has been shown previously that there are differential requirements for the miRNA pathway in preadult and adult stages for the maintenance of germline stem cells (Shcherbata et al. 2007). In addition, augmentation of JAK/STAT signaling via let-7 miRNA in the testicular stem cell niche is essential for male germline stem cell preservation in aging flies (Toledano et al. 2012).
The temporal ecdysteroid–let-7 signaling cascade also cooperates with the JAK/STAT cytokine pathway in neuronal cell fate determination (Kucherenko and Shcherbata 2013). One possibility is that the ecdysteroid–let-7 effect on somatic gender identity also has a temporal character and incorporates JAK/STAT cytokine signaling to control sex determination in the somatic cells.
Interestingly, one of the JAK/STAT pathway ligands, Unpaired (Upd), is an X-linked signal element gene that affects Sxl expression (Sefton et al. 2000). Upd is a genuine target of miR-279 that regulates circadian rhythms (Luo and Sehgal 2012). Knockdown of Upd rescues the behavioral phenotype of miR-279 mutants. Further studies of the ecdysone–miRNA-JAK/STAT signaling cascade will determine upstream and downstream components sustaining cellular sexual identity during the postembryonic stages, adulthood, and aging.
Finally, the significant upregulation of Sxl in both wild-type and ecd1ts females upon temperature shift (Figure 5E) indicates that Sxl expression is temperature dependent in flies and also modulated by ecdysteroid hormones. This provides a direct role for hormones in regulating sex determination in females. Clearly our understanding of this unprecedented complexity of sex determination requires both temporal and cell-specific examinations of the interplay between miRNAs, steroid hormones, and sex determination.
These results comprehensively demonstrate an important role for miRNAs as regulators of sexual dimorphism and identify several sets of candidate miRNAs to achieve differential functions in each sex. The function of let-7 provides a proof of concept of the importance of sex-biased miRNA expression in the developing and aging adult gonad and reproductive apparatus. In mammals, sex hormones produced in the gonads coordinate gene expression in distant tissues and organs. The existence of a similar hormonal axis in Drosophila has remained elusive. We show here for the first time that ecdysone signaling controls sex-determination genes in flies, via the miRNA let-7. Misregulation of let-7 in mammals has been shown to impair differentiation and leads to the development of diseases including cancer. Let-7 appears as a master regulator of the cell-proliferation pathways altered in lung, breast, colon, and prostate cancer (Takamizawa 2004; Johnson et al. 2007; Trang et al. 2009; reviewed in Lo et al. 2013). Notably, such cancer cells present unusually low levels of let-7. In addition, connections between let-7 and hormone signaling have been established in mammals. Let-7c plays an important role in the regulation of androgen signaling in castration-resistant prostate cancers, acting by downregulation of the androgen receptor. In turn, androgen signaling further increases cellular levels of miR-125b (Shi et al. 2007), suggesting that both mir-125 and let-7 respond to hormonal imbalances. Whether sex determination is affected in these contexts will require future studies.
We provide an extensive repertoire of sex-biased and gender neutral miRNAs that will help address the varied functions of miRNA activity across development and aging, as well as in the context of human diseases.
Supplementary Material
Acknowledgments
We thank Laura Johnston, Nicholas Sokol, Frank Hirth, Ralf Pflanz, Bruce Baker, Carmen Robinett, Acaimo Gonzalez-Reyes, the Bloomington Stock Center for providing flies and reagents, and Mark Siegal for advice on cuticle preparations. We thank all members of the Hannon lab, Ilaria Falciatori, the Department of Herbert Jäckle and the Shcherbata Lab for helpful discussions, the two reviewers and Alexander Vaughan for providing constructive comments that helped improve this manuscript. This work was supported by grants from the National Institutes of Health, a kind gift from Kathryn W. Davis to G.J.H., and by the Max Planck Society to A.K. and H.R.S.
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169268/-/DC1.
miRNA libraries from male and female tissues have been submitted to the GEO database at NCBI as series GSE57029.
Communicating editor: B. Andrews
Literature Cited
- Alekseyenko A. A., Larschan E., Lai W. R., Park P. J., Kuroda M. I., 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20: 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko A. A., Ho J. W. K., Peng S., Gelbart M., Tolstorukov M. Y., et al. , 2012. Sequence-specific targeting of dosage compensation in Drosophila favors an active chromatin context. PLoS Genet. 8: e1002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., 2011. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Chen X., 2007. The regulation of genes and genomes by small RNAs. Development 134: 1635–1641. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., et al. , 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Arbeitman M. N., Furlong E. E. M., Imam F., Johnson E., Null B. H., et al. , 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. [DOI] [PubMed] [Google Scholar]
- Baker B. S., Ridge K. A., 1980. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94: 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Burtis K., Goralski T., Mattox W., Nagoshi R., 1989. Molecular genetic aspects of sex determination in Drosophila melanogaster. Genome 31: 638–645. [DOI] [PubMed] [Google Scholar]
- Baley J., Li J., 2012. MicroRNAs and ovarian function. J. Ovarian Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw G. J., Baker B. S., 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798. [DOI] [PubMed] [Google Scholar]
- Bejarano F., Bortolamiol-Becet D., Dai Q., Sun K., Saj A., et al. , 2012. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139: 2821–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote J. M., McKeown M. B., Andrew D. J., Scott T. N., Wolfner M. F., et al. , 1985. Control of sexual differentiation in Drosophila melanogaster. Cold Spring Harb. Symp. Quant. Biol. 50: 605–614. [DOI] [PubMed] [Google Scholar]
- Belote J. M., McKeown M., Boggs R. T., Ohkawa R., Sosnowski B. A., 1989. Molecular genetics of transformer, a genetic switch controlling sexual differentiation in Drosophila. Dev. Genet. 10: 143–154. [DOI] [PubMed] [Google Scholar]
- Bownes M., Blair M., Kozma R., Dempster M., 1983. 20-Hydroxyecdysone stimulates tissue-specific yolk-protein gene transcription in both male and female Drosophila. J. Embryol. Exp. Morphol. 78: 249–268. [PubMed] [Google Scholar]
- Bridges C. B., 1921. Triploid intersexes in Drosophila melanogaster. Science 54: 252–254. [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Coschigano K. T., Baker B. S., Wensink P. C., 1991. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10: 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M., 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18: 501–506. [DOI] [PubMed] [Google Scholar]
- Camara N., Whitworth C., Van Doren M., 2008. The creation of sexual dimorphism in the Drosophila soma. Curr. Top. Dev. Biol. 83: 65–107. [DOI] [PubMed] [Google Scholar]
- Caygill E. E., Johnston L. A., 2008. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 18: 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. S., Uppendahl L. D., Chowdhury M. A., Ip P.-L., Siegal M. L., 2011. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development 138: 1099–1109. [DOI] [PubMed] [Google Scholar]
- Chawla G., Sokol N. S., 2012. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development 139: 1788–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wang S., Xie T., 2011. Restricting self-renewal signals within the stem cell niche: multiple levels of control. Curr. Opin. Genet. Dev. 21: 684–689. [DOI] [PubMed] [Google Scholar]
- Chiang P.-W., Kurnit D. M., 2003. Study of dosage compensation in Drosophila. Genetics 165: 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen A. E., Keisman E. L., Ahmad S. M., Baker B. S., 2002. Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 18: 510–516. [DOI] [PubMed] [Google Scholar]
- Chung W.-J., Okamura K., Martin R., Lai E. C., 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18: 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., 1978. Two closely linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics 90: 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Oliver B., 2012. Genomics of sex determination in Drosophila. Brief Funct Genomics 11: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., et al. , 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Smibert P., Lai E. C., 2012. Exploiting Drosophila genetics to understand microRNA function and regulation. Curr. Top. Dev. Biol. 99: 201–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J. E., Lebo M. S., Sanders L. E., Sun F., Arbeitman M. N., 2009. Ecdysone receptor acts in fruitless: expressing neurons to mediate Drosophila courtship behaviors. Curr. Biol. 19: 1447–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J. E., Fear J. M., Knott S., Baker B. S., McIntyre L. M., et al. , 2013. Male-specific Fruitless isoforms have different regulatory roles conferred by distinct zinc finger DNA binding domains. BMC Genomics 14: 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decotto E., Spradling A. C., 2005. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev. Cell 9: 501–510. [DOI] [PubMed] [Google Scholar]
- Deng X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D., et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M.-A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., et al. , 2013. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 14: 671–683. [DOI] [PubMed] [Google Scholar]
- Duncan K., Grskovic M., Strein C., Beckmann K., Niggeweg R., et al. , 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 20: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Baker B. S., 2004. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2: e341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty M. S., Salis P., Evans C. J., Ekas L. A., Marouf A., et al. , 2010. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev. Cell 18: 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M. T., Spradling A. C., 2007. Male and female Drosophila germline stem cells: two versions of immortality. Science 316: 402–404. [DOI] [PubMed] [Google Scholar]
- Ganter G. K., Desilets J. B., Davis-Knowlton J. A., Panaitiu A. E., Sweezy M., et al. , 2012. Drosophila female precopulatory behavior is modulated by ecdysteroids. J. Insect Physiol. 58: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzov A., Tatar M., 2010. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly 4: 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen A., Kauvar L., Lepesant J. A., 1977. Roles of ecdysone in Drosophila development. Proc. Natl. Acad. Sci. USA 74: 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., et al. , 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. [DOI] [PubMed] [Google Scholar]
- Hempel L. U., Oliver B., 2007. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev. Biol. 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth P. E., 1965. Doublesex, recessive gene that transforms both males and females of Drosophila into intersexes. Genetics 51: 659–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang A. C.-C., Chang Y.-C., Bai J., Montell D., 2009. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat. Cell Biol. 11: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., et al. , 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67: 7713–7722. [DOI] [PubMed] [Google Scholar]
- Kato M., de Lencastre A., Pincus Z., Slack F. J., 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman B. E., Cheshire A. M., Andrew D. J., 2006. From fate to function: the Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation 74: 326–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König A., Shcherbata H. R., 2013. Visualization of adult stem cells within their niches using the Drosophila germline as a model system. Methods Mol. Biol. 1035: 25–33. [DOI] [PubMed] [Google Scholar]
- König A., Yatsenko A. S., Weiss M., Shcherbata H. R., 2011. Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J. 30: 1549–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherenko M. M., Shcherbata H. R., 2013. Steroids as external temporal codes act via microRNAs and cooperate with cytokines in differential neurogenesis. Fly 7: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherenko M. M., Barth J., Fiala A., Shcherbata H. R., 2012. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31: 4511–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareva A. A., Roman G., Mattox W., Hardin P. E., Dauwalder B., 2007. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 3: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman J. L., Dinardo S., 2010. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell Biol. 12: 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo M. S., Sanders L. E., Sun F., Arbeitman M. N., 2009. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics 10: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Hall J. C., Park J. H., 2002. Doublesex gene expression in the central nervous system of Drosophila melanogaster. J. Neurogenet. 16: 229–248. [DOI] [PubMed] [Google Scholar]
- Legube G., McWeeney S. K., Lercher M. J., Akhtar A., 2006. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 20: 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cressy M., Qin H., Fulga T., Van Vactor D., et al. , 2013. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J. Neurosci. 33: 5821–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Landreh M., Cao K., Abe M., Hendriks G.-J., et al. , 2012. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo U., Yang D., Hsieh J. T., 2013. The role of microRNAs in prostate cancer progression. Transl. Androl. Urol. 2(3): 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S. D., Shi G. W., Baker B. S., 2011. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138: 2761–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Sehgal A., 2012. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148: 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C., Brennecke J., Czech B., Aravin A., Hannon G. J., 2012. Preparation of small RNA libraries for high-throughput sequencing. Cold Spring Harb Protoc 2012: 1067–1077. [DOI] [PubMed] [Google Scholar]
- Marrone A. K., Edeleva E. V., Kucherenko M. M., Hsiao N.-H., Shcherbata H. R., 2012. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol. 13: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Landeen E. L., Cook J. M., Kingan S. B., Presgraves D. C., 2011. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 9: e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T., Takizawa T., Luo S.-S., Ishibashi O., Kawahigashi Y., et al. , 2008. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction 136: 811–822. [DOI] [PubMed] [Google Scholar]
- Morris L. X., Spradling A. C., 2012. Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PLoS ONE 7: e46109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. C., Nojima T., Ashley E., Parker D. J., Walker J., et al. , 2014. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr. Biol. 24: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Edwards P., Minor J., Naiman D., et al. , 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M. J., Gupta V., Sturgill D., Warren J. T., Jallon J.-M., et al. , 2010. Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC Genomics 11: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., et al. , 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. [DOI] [PubMed] [Google Scholar]
- Rideout E. J., Dornan A. J., Neville M. C., Eadie S., Goodwin S. F., 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13: 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S., Park C., Sanders K. M., McCarrey J. R., Yan W., 2007. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 311: 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Vaughan A. G., Knapp J.-M., Baker B. S., 2010. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8: e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., et al. , 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazansky S. S., Gvozdev V. A., Berezikov E., 2011. Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 12: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S., 1991. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 5: 2071–2085. [DOI] [PubMed] [Google Scholar]
- Salz H. K., 2011. Sex determination in insects: a binary decision based on alternative splicing. Curr. Opin. Genet. Dev. 21: 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. E., Arbeitman M. N., 2008. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320: 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefton L., Timmer J. R., Zhang Y., Béranger F., Cline T. W., 2000. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature 405: 970–973. [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Dubrovsky E. B., Dubrovskaya V. A., Berger E. M., Ambros V., 2002. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244: 170–179. [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., Ambros V., 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259: 9–18. [DOI] [PubMed] [Google Scholar]
- Shcherbata H. R., Ward E. J., Fischer K. A., Yu J.-Y., Reynolds S. H., et al. , 2007. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell 1: 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.-B., Xue L., Yang J., Ma A.-H., Zhao J., et al. , 2007. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. USA 104: 19983–19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Lai E. C., 2010. A view from Drosophila: multiple biological functions for individual microRNAs. Semin. Cell Dev. Biol. 21: 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N. S., Xu P., Jan Y.-N., Ambros V., 2008. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22: 1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K., Choudhary A., Patowary A., Singh A. R., Bhatia S., et al. , 2013. miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 41: 4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski B. A., Belote J. M., McKeown M., 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58: 449–459. [DOI] [PubMed] [Google Scholar]
- Straub T., Grimaud C., Gilfillan G. D., Mitterweger A., Becker P. B., 2008. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 4: e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J., 2004. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64: 3753–3756. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Barmina O., Sanders L. E., Arbeitman M. N., Kopp A., 2011. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9: e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano H., D’Alterio C., Czech B., Levine E., Jones D. L., 2012. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 485: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P., Medina P. P., Wiggins J. F., Ruffino L., Kelnar K., et al. , 2009. Regression of murine lung tumors by the let-7 microRNA. Oncogene 29: 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Lopes H. F., Karr T. L., Long M., 2009. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5: e1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Zhang Y. E., Kemkemer C., Lopes H. F., Karr T. L., Long M., 2012 Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the Drosophila X chromosome. BMC Biol. 10: 49–50. [DOI] [PMC free article] [PubMed]
- Weng R., Cohen S. M., 2012. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development 139: 1427–1434. [DOI] [PubMed] [Google Scholar]
- Whitworth C., Jimenez E., Van Doren M., 2012. Development of sexual dimorphism in the Drosophila testis. Spermatogenesis 2: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. M., Selegue J. E., Werner T., Gompel N., Kopp A., et al. , 2008. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134: 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-C., Chen C.-H., Mercer A., Sokol N. S., 2012. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev. Cell 23: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A., 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C.-F., Awasaki T., Chiang A.-S., et al. , 2006. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127: 409–422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.