Abstract
The relationship between quantitative genetics and population genetics has been studied for nearly a century, almost since the existence of these two disciplines. Here we ask to what extent quantitative genetic models in which selection is assumed to operate on a polygenic trait predict adaptive fixations that may lead to footprints in the genome (selective sweeps). We study two-locus models of stabilizing selection (with and without genetic drift) by simulations and analytically. For symmetric viability selection we find that ∼16% of the trajectories may lead to fixation if the initial allele frequencies are sampled from the neutral site-frequency spectrum and the effect sizes are uniformly distributed. However, if the population is preadapted when it undergoes an environmental change (i.e., sits in one of the equilibria of the model), the fixation probability decreases dramatically. In other two-locus models with general viabilities or an optimum shift, the proportion of adaptive fixations may increase to >24%. Similarly, genetic drift leads to a higher probability of fixation. The predictions of alternative quantitative genetics models, initial conditions, and effect-size distributions are also discussed.
Keywords: two-locus model, stabilizing selection, polygenic adaptation, pleiotropy
QUANTITATIVE genetics assumes that selection on an adaptive character involves simultaneous selection at multiple loci controlling the trait. This may cause small to moderate allele-frequency shifts at these loci, in particular when traits are highly polygenic (Barton and Keightley 2002). Therefore, adaptation does not require new mutations in the short term. Instead, selection uses alleles found in the standing variation of a population. Genome-wide polymorphism data and association studies suggest that this quantitative genetic view is relevant (Pritchard and Di Rienzo 2010; Pritchard et al. 2010; Mackay et al. 2012).
Different types of selection on a trait, such as directional, stabilizing, or disruptive selection, modify the genetic composition of a population and favor either extreme or intermediate genotypic values of the trait. In this study we focus on stabilizing selection, which drives a trait toward a phenotypic optimum. Many models of this common form of selection have been analyzed. The central question in all these studies has been whether stabilizing selection can explain the maintenance of genetic variation. This is considered an important question, as it has been known for long that stabilizing selection exhausts genetic variation (Fisher 1930; Robertson 1956), yet quantitative traits exhibit relatively high levels of genetic variation in nature (Endler 1986; Falconer and Mackay 1996; Lynch and Walsh 1998).
Here we ask a different question, namely how much adaptive evolution is predicted by quantitative genetic models of stabilizing selection? In contrast to quantitative genetics, in population genetics and genomics our understanding of the genetics of adaptation has revolved in recent years around the role of selective sweeps, i.e., signatures of positive directional selection in the genome. The model underlying this population genetic view of adaptation is that of the hitchhiking effect developed for sexual species by Maynard Smith and Haigh (1974). The hitchhiking process describes how the fixation of a beneficial allele (starting from very low frequency) affects neutral or weakly selected variation linked to the selected site. For single and recurrent advantageous alleles appearing in a population, the model has been further developed by Kaplan et al. (1989), Stephan et al. (1992), Stephan (1995), and Barton (1998). Later this model was generalized to sweeps from standing variation or soft sweeps taking into account that sweeps may also occur if the initial frequency of the beneficial allele is not very low (Innan and Kim 2004; Hermisson and Pennings 2005).
Yet, despite its simplicity, the use of the hitchhiking model was quite successful in recent years in detecting evidence for positive directional selection in the genomes of plant and animal species. Typically, many studies of populations with large effective sizes have revealed numerous genes or gene regions showing selective footprints (reviewed in Stephan 2010). The detection of such footprints is based on statistical tests that utilize several features of the hitchhiking model such as reduced variation around the selected site and shifts in the neutral site-frequency spectrum (Kim and Stephan 2002; Nielsen et al. 2005; Pavlidis et al. 2010). However, it is remarkable that none of these tests incorporates phenotypic information. If functional studies were performed to reach an understanding of the effects of selection, they were done in most cases only post hoc, after a gene or gene region has been identified by a sweep.
Recent theoretical work has addressed the question whether and to what extent the quantitative and population genetics views of adaptation are compatible with each other. For instance, the following question was asked: Can the quantitative genetics models of stabilizing selection also be used to predict observed levels of selective fixations? Chevin and Hospital (2008) presented a model for the footprint of positive directional selection at a quantitative trait locus (QTL) in the presence of a fixed amount of background genetic variation due to other loci. This approach is based on Lande’s (1983) model that consists of a locus of major effect on the trait and treats the remaining loci of minor effect as genetic background. This analysis predicts that QTL of adaptive traits under stabilizing selection exhibit patterns of selective sweeps only very rarely. de Vladar and Barton (2011, 2014) studied a polygenic character under stabilizing selection, mutation, and genetic drift. They found sweeps after abrupt shifts of the phenotypic optimum, without quantifying how often such signatures occurred. Furthermore, Pavlidis et al. (2012) analyzed a classical multilocus model with two to eight loci controlling an additive quantitative trait under stabilizing selection (with and without genetic drift). Using simulations, they showed that multilocus response to selection often prevents trajectories from going to fixation, particularly for the symmetric viability model. They also found that the probability of fixation strongly depends on the genetic architecture of the trait.
To understand these results in greater depth, we present here an analysis of the two-locus model of an adaptive trait under stabilizing selection and drift. This model is sufficiently simple that analytical approximations can be used to back up the simulations. We concentrate on the case of weak selection. Given that we are interested in a comparison between quantitative and population genetics and selection coefficients estimated in molecular population genetics are generally small (<10−1), this parameter range appears to be justified (Orr 2005; Turchin et al. 2012). We show that in this realm quasi-linkage equilibrium (QLE) approximations are possible. Furthermore, we address the question of initial conditions, a subject that was neglected in all of the above-mentioned studies. These, however, are important as the two- and multilocus models of stabilizing selection have generally multiple stable equilibrium states and hence various basins of attraction, which may make the ensuing dynamics sensitive to the initial conditions.
Models
Symmetric fitness model
We consider two loci with two alleles each, A1 and A2 at locus A and B1 and B2 at locus B. The effects of the alleles A1, A2, B1, and B2 on a quantitative trait are , and , respectively. Assuming additivity, the effects of the gametes A1B1, A1B2, A2B1, and A2B2 are , and , respectively. The resulting genotypic values are given by
| (1) |
Without loss of generality, we assume 0 ≤ γB ≤ γA; i.e., locus A is the major and locus B the minor locus.
Under sufficiently weak selection (and/or for sufficiently small genotypic effects), it may be assumed that the trait is under quadratic stabilizing selection; i.e., the relative fitness w(G) of individuals with genotypic value G is
| (2) |
where s is the selection coefficient. The relative fitnesses of all genotypes are then given by
| (3) |
where , and (Bürger 2000, p. 204). Neglecting mutation, the ordinary differential equations (ODEs) determining the dynamics of the system can then be written as
| (4) |
where x1, x2, x3, and x4 denote the frequencies of the gametes A1B1, A1B2, A2B1, and A2B2, respectively, and D = x1x4 – x2x3 is the linkage disequilibrium (LD). The parameter r is the product of the recombination fraction and the birth rate of the double heterozygotes (Crow and Kimura 1970, pp. 196–197). By setting the latter to 1, r is allowed to vary between 0 and 0.5. The marginal fitnesses wi (i = 1,..., 4) of the gametes (Bürger 2000, p. 51) are
| (5) |
and the mean fitness of the population is
| (6) |
The equilibria of the system of ODEs (4) and their stability properties are identical to those of the corresponding discrete-time model, which has been studied by Gavrilets and Hastings (1993) and Bürger and Gimelfarb (1999). As reviewed in Bürger (2000, pp. 204-208), there are four possible types of equilibria: (i) four monomorphic equilibria (henceforth also called corner equilibria) corresponding to the fixation of the gametes A1B1, A1B2, A2B1, and A2B2; (ii) two edge equilibria with the major locus polymorphic and the minor locus fixed; (iii) a symmetric (internal) equilibrium for which both loci are polymorphic, and (iv) two unsymmetric equilibria. The conditions for the existence of these equilibria, their explicit expressions, and their local stability properties are summarized in Bürger (2000, pp. 205–208).
QLE approximation:
To gain insight into the qualitative behavior of the ODEs (4), it is convenient to reduce the system from three to two dimensions. This is possible if, in addition to weak selection, linkage is sufficiently loose (relative to epistasis). This is the so-called QLE assumption, which was introduced by Kimura (1965a). In other words, we transform the gamete frequencies xi (i = 1,...,4) into the frequencies p = x1 + x2 and q = x1 + x3 of the alleles A1 and B1, respectively, and introduce linkage disequilibrium D as the third new variable. Then we treat the latter as a fast variable on the time scale of changes in p and q. The xi can be expressed by the new variables as
| (7) |
Next, following Kimura we use Z = x1x4/x2x3 as a measure of LD. Then with the help of the ODEs (4), the time derivative of Z can be written as (Crow and Kimura 1970, p. 198)
| (8) |
where E = w1 − w2 − w3 + w4 measures the strength of epistasis. Note that in our case
| (9) |
In the QLE, Equation 8 can be evaluated to give the leading-order term
| (10) |
By inserting D into the variables xi in (7), the reduced system of ODEs can be written in the form
| (11) |
The local stability properties of the equilibria of Equations 11 are quite similar to those of the original system (4). The two corner equilibria and are always unstable, as the respective eigenvalues are always positive. The biological explanation would be that the extreme phenotypes are selected against when the optimum is the double heterozygote. The corner equilibria and are stable if the condition γA/2 < γB < γA holds.
Edge equilibria exist for , and for (Bürger 2000, p. 205). The stability condition is identical to that of the full system, as for both edge equilibria are stable.
The main difference from the full system (4) is that the internal symmetric equilibrium is always unstable. This can be seen by linearizing the reduced system about . Neglecting higher-order terms in s, the Jacobian of (11) becomes
| (12) |
with its eigenvalues
| (13) |
Thus, for the eigenvalues of this matrix have opposite signs, showing that this equilibrium point is unstable. In fact, since the eigenvalues are real, is a saddle point. The two separatrices of this saddle divide the pq phase plane into four regions with different dynamics. For each of these regions the locally stable equilibrium points are globally attractive. Unfortunately, the ODEs for the separatrices cannot be solved explicitly. However, numerical analysis of the vector field of ODE (11) in the phase plane shows that for each of these regions the locally stable equilibrium points are globally attractive.
The parameter ranges of stability for the different systems depend on γB/γA and r/s. Inner stable equilibria exist only in the full system (4). For r/s sufficiently large, the stability properties of the edge equilibria are equivalent in the full and the QLE system. Monomorphic equilibria are also qualitatively equal in both systems.
Accuracy of the QLE approximation:
We estimated the range of parameter values for which we obtain a reasonably good QLE approximation of the two-locus model. We generated a set of random parameter values as described in Table 1 and initial conditions for which we estimated the trajectory from the full system and the QLE system. Then we compared both trajectories by calculating the mean relative error using the distance
summed over all times t. Here and denote the trajectories from the full model and p(t) and q(t) are the trajectories of the QLE approximation.
Table 1. Parameter values used for simulations.
| Parameter | Range | Condition |
|---|---|---|
| Uniform(0, 2) | ||
| Uniform(0, 2) | ||
| r | Uniform(0, 0.5) | |
| s | Uniform(0, 0.1) |
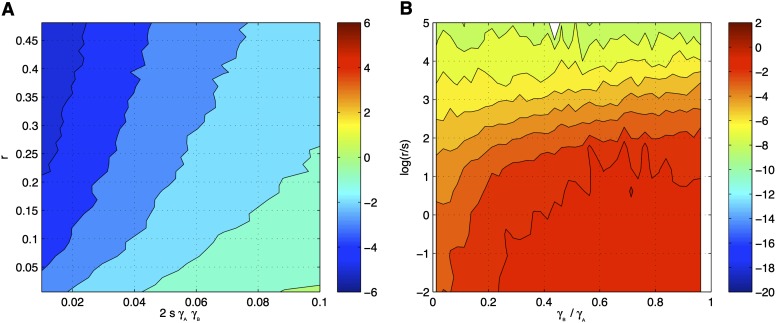
In Figure 1 it can be seen that for increasing recombination rates the mean error is <5%, which corresponds to a threshold of −3 (blue to dark blue areas in Figure 1A and green to yellow in Figure 1B). For example, assuming 2sγAγB = 0.04 would require a recombination rate above 0.07 to achieve an approximation of the two-locus model with a relative error <5%. Thus, when assuming a relatively low selection coefficient of 0.01, the effects could be moderately high, such as γA = γB = 1.5. For many parameter ranges of interest this is a sufficiently good approximation. Figure 1B demonstrates that the ratio of the effects has only a marginal impact on the quality of the approximation.
Figure 1.
Quality of the QLE approximation. (A) Logarithm of the mean relative error for recombination rate r vs. –E. (B) Logarithm of the mean relative error for log(r/s) vs. γB/γA. Broadly, log(r/s) must be >2 to achieve an error <5%.
Generalizing the symmetric fitness model
We have studied two generalizations of the symmetric fitness model. First, in the general fitness model we relaxed the restrictions on the relations of allelic contributions by allowing for arbitrary values of the effects γA1, γA2, γB1, and γB2 for the alleles A1, A2, B1, and B2, respectively. However, by assuming that the absolute values of γA1 and γA2 are equal or larger than those of γB1 and γB2, we further consider locus A as the major and locus B as minor locus, and the phenotypic optimum is again 0. In this case the genotypic values are
| (14) |
Second, in the shifted optimum model we consider the quadratic optimum model with an arbitrary position θ of the optimum, yet assign the effects as in the symmetric fitness model. Note that in both cases the genotypic fitnesses may no longer conform to the symmetric viability model.
General fitness model:
We established the ODEs in accordance to those of system (4) and investigated this model numerically. Furthermore, we derived the ODEs using the QLE approximation as
| (15) |
Thus, if the effects γij are chosen as in the symmetric fitness model, Equations 15 reduce to Equations 11. Furthermore, if the parameter values follow constraints similar to those in the symmetric fitness model, the equilibrium structure of (15) is similar to that of (11). In particular, if the absolute values of the effects of both loci are comparable, but their signs are opposite for each locus, the corner equilibria (1, 0) and (0, 1) are locally stable and the other two are unstable. If the absolute values of the effects of the major locus are sufficiently large relative to those of the minor locus, we find again two edge equilibria. For instance, the eigenvalues of with
| (16) |
are approximately
| (17) |
Hence, is locally stable if the parameters γA1 + γA2 and γB1 – γB2 have the same sign. Finally, the interior equilibrium exists and remains a saddle point if and only if γA1 + γA2 + γB1 + γB2 = 0. Under more general conditions, however, this equilibrium does no longer exist, as we show in our simulations (Table 2).
Table 2. Proportions (%) of trajectories that lead to one of the equilibria in the deterministic case.
| Equilibrium | Descriptiona | Constant environment, weak selection | Constant environment, strong selection | Changing environmentb | Optimum changec | General fitnessd |
|---|---|---|---|---|---|---|
| E1 | Polymorphic/symmetric | 1.97 | 21.21 | 2.63 | 0 | 0 |
| E2 + E3 | Polymorphic/unsymmetric | 4.6 | 4.46 | 2.73 | 1.91 | 2.19 |
| E4 | A polymorphic, loss at B | 34.89 | 25.81 | 39.09 | 22.07 | 17.32 |
| E5 | A polymorphic, fixation at B | 0.92 | 0.68 | 2.9 | 3.63 | 4.28 |
| E6 | Fixation at A, loss at B | 31.44 | 31.16 | 41.58 | 7.76 | 14.53 |
| E7 | Loss at A, fixation at B | 10.75 | 10.8 | 9.15 | 25.54 | 12.13 |
| E8 | Loss at A, loss at B | 0 | 0 | 0 | 0 | 14.99 |
| E9 | Fixation at A, fixation at B | 0 | 0 | 0 | 16.22 | 23.99 |
| Sweep at A (fixation in E6)e | 48.03 | 47.79 | 0.48 | 44.85 | 54.44 | |
| Sweep at B (fixation in E5)e | 1.09 | 0 | 0 | 44.08 | 49.07 | |
| Sweep at B (fixation in E7)e | 9.67 | 11.85 | 0.22 | 40.6 | 51.69 | |
| Sweep at A and B (fix. in E9)e | 0 | 0 | 0 | 31.81 | 32.56 | |
| Transient | 13.56 | 4.53 | 0 | 9.79 | 0 | |
| Unclassified | 1.87 | 1.35 | 1.92 | 13.08 | 10.57 | |
| Sweeps total | 16.15 | 16.17 | 0.22 | 20.61 | 24.09 |
The equilibria E1–E9 are defined following Bürger (2000, p. 205).
Environmental changes were defined by random change of the original effects and selection coefficient.
In the shifted optimum model (see Models) the phenotypic optimum takes an arbitrary position 0 < θ < 1.
In the general fitness model the effects have arbitrary values sampled from (0, 2) such that A is major locus.
Selective sweeps are defined here as trajectories that lead to fixation from very low initial frequencies (<0.01). Numbers denote the proportions of fixations with initial frequencies <0.01.
Shifted optimum model:
Here we use the fitness function
| (18) |
for individuals with genotypic value G (Bürger 2000, p. 213). Then assuming that the effects follow those used in the symmetric fitness model, the ODEs under the QLE approximation are
| (19) |
The equilibrium structure of this model is discussed in Bürger (2000, pp. 213–214) as a function of θ > 0. For θ sufficiently small, either two stable corner equilibria or two stable edge equilibria exist, depending on the ratio γB/γA. For the corner equilibrium (0, 1) becomes unstable and the stable edge equilibrium
| (20) |
arises. For both loci are under directional selection driving the most extreme gamete to fixation.
Simulations
Simulation settings were kept identical throughout this section. The model parameters were drawn according to Table 1. Initial gamete frequencies were produced under the assumption of no initial LD with the constraint . Furthermore, initial conditions of the gamete frequencies were drawn from allele frequencies that were distributed as the standard neutral site-frequency spectrum (i.e., for constant population size). To do so, we sampled the locus frequencies from the neutral site-frequency spectrum (Griffiths and Tavaré 1998, Equation 1.3) for a large sample size (n = 50). This choice of initial conditions is biologically more plausible than the random initial frequency values used in Pavlidis et al. (2012). The initial gamete frequencies are in our case clustered near the corner equilibrium (E8) such that the frequencies of both alleles A1 and B1 are low.
For each parameter set the simulation was run for 10,000 generations forward in time. In total, 10,000 simulations were produced. Various environmental settings were analyzed: (i) a constant environment under which weak stabilizing selection is acting on the trait, (ii) a constant environment with strong stabilizing selection, (iii) a change in the environmental parameter values describing stabilizing selection (i.e., effects and selection coefficient), (iv) a change in the optimum of the trait, and (v) a general fitness model as described above. For each environmental setting we studied the equilibrium structure of the dynamical system under uniformly distributed parameter values. In addition, to be biologically more realistic, we used specific distributions for some parameters (e.g., effect sizes).
Deterministic simulations
For a given parameter set we solved the ODEs (4) using the “ode23s” command in Matlab (v. 6). We refer to the parameter setting as constant environment when the parameters of the symmetric fitness model are fixed during evolution. In the following, we report the percentage of trajectories that converge to a certain equilibrium point of the symmetric model defined by Bürger (2000, p. 205).
First, in the case of a trait under stabilizing selection according to the symmetric model (constant environment) and weak selection (s < 0.1), we observed that only 2% of all simulations end up in a symmetric, polymorphic equilibrium (E1, see Table 2). About 5% of all trajectories reached one of the unsymmetric equilibria (E2 + E3). Edge equilibria with locus A polymorphic and a loss at locus B were reached in 35% of the cases (E4), whereas only 1% of the runs ended at B with A polymorphic (E5). We also observed that more trajectories (31%) approach the corner where fixation occurs at A (and loss at B, E6) compared to fixation at B (and loss at A, E7) (11%). The higher proportion of fixations at A (rather than locus B) is due to the fact that A has been chosen as major locus. Thus, in total 42% of the trajectories were fixed at locus A (E6) or B (E7), whereas 43% converged to the polymorphic equilibria (E1–E5). Note that the proportions of trajectories approaching the equilibria E1–E9 do not add up to 100%. A substantial percentage (∼14%) did not converge within the simulated 10,000 generations, particularly when the effects and selection coefficients are close to zero, and a small percentage of ∼2% could not be assigned exactly to one of the possible equilibrium points due to numerical errors in the approximation of the trajectory or unreached equilibrium states. Furthermore, some corner equilibria (E8 and E9) are not approached at all as they are unstable (see Bürger 2000, pp. 205–206).
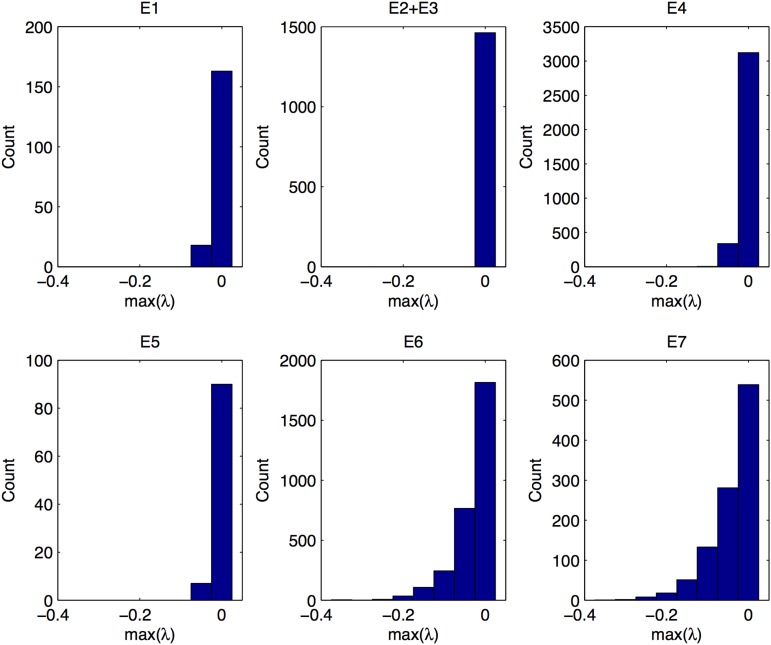
To obtain a better insight into the stability of the equilibrium states (Table 2), which we need to interpret some of the simulation results, we calculated the distribution of eigenvalues of the stable equilibrium points (Figure 2). For simplicity we report only the maximum eigenvalue of each equilibrium point that decides the stability and calculate the mean of the maximum eigenvalues across all simulations that reached the equilibrium point. The corner equilibria where either fixation or loss at a locus occurred (E6 and E7) have on average the lowest eigenvalues (E6, −0.033; E7, −0.043) and are therefore most stable. The eigenvalues in the edge equilibria where locus A remains polymorphic (E4 and E5) are much higher (E4, −0.009; E5, −0.0085) than those of E6 and E7, and the same is true for the symmetric equilibria (E1, −0.01). The unsymmetric equilibria have the highest mean of the maximum eigenvalues (E2 + E3, −0.00034). The unsymmetric equilibria are therefore the weakest of all stable equilibrium states. However, we observe that the degree of stability is not related to the percentage of trajectories ending up in a state. For instance, we found about 2.5 times more trajectories converging to unsymmetric equilibria compared to symmetric ones, which may be attributed to the fact that they are randomly distributed in the gamete space, whereas the symmetric ones are located on a line.
Figure 2.
Distribution of the maximum eigenvalues in the equilibrium states (see Table 2) obtained from 10,000 simulations under stabilizing selection in a constant environment.
Selective sweeps occur at loci when a beneficial allele rises rapidly to fixation from a very low frequency. Fixation of alleles can be observed in corner equilibria at one of the loci (E6 and E7) or at both (E9). In Table 2 we report the proportion of trajectories that reached these equilibria from initially low frequencies (0.01). We observed that for 48% of the trajectories that were going to fixation at A (and loss at B, E6) the initial allele frequency was <0.01 at locus A. Hence, 48% of the fixations in E6 were classical sweeps (according to our definition). In contrast, only 11% of the 12% observed fixations at B (from E5 and E7) were selective sweeps. In total, ∼16% of all trajectories that ended in one of the equilibria E1–E9 were selective sweeps. This proportion of sweeps is much higher than that observed by Pavlidis et al. (2012) who found that 4% of the trajectories were sweeps when the initial conditions are randomly drawn from [0, 0.2] × [0, 1.0] and selection is strong (0.1 ≤ s ≤ 1). Joint sweeps at both A and B do not occur in the deterministic case under stabilizing selection, as this state is unstable. Selective sweeps occur exclusively for γA ≤ 2γB (as predicted in the Models section).
Second, to better understand why the number of sweeps we observed is increased in comparison to Pavlidis et al. (2012), we simulated the distribution of trajectories for strong selection (with s sampled from [0.1,1]) and the same initial conditions as used above. We found that 10 times more trajectories converge to a polymorphic symmetric equilibrium (E1) because the double heterozygote is most fit. The proportions of sweeps at the edge equilibrium with A polymorphic and fixation at B (E5, 0%), the corner with fixation at A and loss at B (E6: 48%), and the other corner with loss at A and fixation at B (E7: 12%) remain about the same (16%). Hence, the difference from our result is largely attributable to the fact that in the standard neutral frequency spectrum that we used here to determine the initial conditions, most initial frequencies are small.
We also estimated the mean time to equilibrium under weak selection. For the trajectories that lead to adaptive fixations the mean time to equilibrium was ∼476 generations, whereas for trajectories that lead to other equilibria (including the ones that remained transient) it was ∼1105 generations (P_ranksum = 2.8 × 10−82). As depicted in Figure 3, for a large range of parameter values, we observe a relatively short mean time to equilibrium of ∼200 generations if recombination rate is sufficiently large (corresponding roughly to the range in which QLE holds; see Figure 1). Figure 3 also shows that the mean time to reach an equilibrium (including fixation) gets longer for low values of the selection coefficient or the effect sizes. This means that for low values of these parameters the speed of fixation may become too small to cause sweeps. For the constant-environment model with strong selection we observe a mean time to equilibrium for trajectories that lead to adaptive fixations of ∼50 generations, whereas all other trajectories need ∼60 generations (P_ranksum = 2.9 × 10−12). Hence, larger effects decrease on average the time to reach an equilibrium.
Figure 3.
Mean speed of adaptation for the constant-environment model. Mean speed is measured by the mean time to equilibrium as a function of the recombination rate and the absolute value of the strength of epistasis. The scale on the right-hand side provides the mean time in generations.
Third, we simulated a change in the environment for the symmetric fitness model as follows. We assume that a population has evolved first in a constant environment and reached one of the possible equilibria E1 to E7 in the proportions reported in Table 2 for the constant-environment model. From these proportions we then sample the initial allele and gamete frequencies. Furthermore, we change randomly the effects and selection coefficient, thus reflecting an extreme case of an environmental change (see below for biologically more realistic cases). Then we let the system converge to its new equilibrium points. More than 98% of the trajectories ended in one of the equilibria listed in Table 2. We observe that ∼42% of the trajectories are driven to fixation at A and loss at B (E6). Compared to the constant environment (with weak selection), we observe a slightly higher amount of trajectories that approached the edge equilibrium E4 (39%). The chance of detecting selective sweeps as fixation at locus A (E6, 42%) is very low (0.5%). At locus B the proportion of selective sweeps is ∼0.2%. Thus, the total number of sweeps is much lower than in the constant-environment model (0.2%). The reason is that in contrast to an initial condition under standard neutrality, the population is in this scenario preadapted before the environmental change occurs; i.e., it sits in a state in which the allele frequencies are not clustered near the corner (E8). However, soft sweeps may occur if the initial state is an edge equilibrium (e.g., E4).
Fourth, we simulated a change in the optimum according to Equation 18. In addition to the parameter values sampled according to Table 1, we sampled the new optimum θ from a uniform distribution of the interval (0, 1). The initial condition for the allele frequencies was chosen as for the constant-environment model (standard neutral allele frequency spectrum). Compared to the symmetric fitness model, >13% of the trajectories did not end up in one of the states E1–E9, suggesting that the equilibrium structure of this model is rather different from the symmetric fitness model (discussed further below). We found an increase of the proportion of trajectories (26%) that led to fixation at the minor locus and loss of the allele at the major locus (E7). Furthermore, in accordance with theory, fixations occur at both loci in 16% (E9) of the cases, in particular when θ is large. Indeed, in all fixations the condition was met (see remark below Equation 20). Hence, selective sweeps are expected to be relatively frequent at both the major and the minor locus, as observed in 44% of the trajectories reaching the edge with fixation at locus B (E5, 4%), 45% of the trajectories that reach the corner E6 (8%), and 41% of the trajectories that reach the corner E7 (26%). From the simulations that go to the corner with fixation at both loci (E9, 16%) 32% generate a selective sweep. Note that a sweep occurring at both loci has been counted as one sweep, as it is produced by merely one trajectory. In total, ∼21% of all trajectories ending in E1–E9 led to selective sweeps at A or B, which is a slight increase relative to the symmetric fitness model (16%) and can be attributed to the change of the optimum. The amount of trajectories leading to an edge equilibrium with A polymorphic and loss at B is much lower (22%) compared to the previously discussed models.
Fifth, we simulated the general fitness model and observed that ∼89% of the trajectories ended in one of the states E1–E9. We found an increase of simulations that led to corner equilibria compared to the symmetric fitness model. Convergence to extreme phenotypes is quite likely compared to the symmetric fitness model (loss at A and B, 15%; fixations at both A and B, 24%). Furthermore, the internal symmetric equilibrium is generally unstable (as predicted in the Models section). In total, ∼24% of all trajectories ending in E1–E9 led to selective sweeps at A or B. These results agree roughly with those of Pavlidis et al. (2012) and are expected because in the general fitness model the double heterozygote is not necessarily associated with the highest fitness as in the symmetric fitness model.
For all four models we also estimated the amount of LD between locus A and B using the squared correlation coefficient r2 (Table 3). This coefficient that is defined only for the internal polymorphic equilibria was measured at the end of each run. We note that the values in column 2 of Table 3 appear to be primarily determined by the symmetric equilibrium rather than the unsymmetric ones. Thus, for strong selection we found 47% LD relative to 9% for weak selection, and LD is low if the polymorphic/symmetric equilibrium (E1) does not exist as in the shifted optimum model. The reason is that LD is generally much stronger in the symmetric equilibrium. In our stochastic simulations (discussed below) we observed generally higher LD, due to the action of drift.
Table 3. Average amount of linkage disequilibrium (r2) between the major and minor locus at the end of each run.
| Modela | Deterministic (%) | Stochastic (N = 1000, %) |
|---|---|---|
| Constant environment, weak selection | 9.38 | 64.41 |
| Constant environment, strong selection | 46.84 | 61.17 |
| Changing environment | 24.37 | 62.93 |
| Optimum change | 4.06 | 16.97 |
| General fitness | 9.24 | Not applicableb |
Random sets of parameter values were chosen according to Table 1.
Internal polymorphic equilibria were not found.
Finally, we studied the probability of observing adaptive fixations under biologically more realistic assumptions (see Table 4). The initial conditions were derived from a mutation–selection equilibrium (Orr and Betancourt 2001), with 0 < s < 0.1 and mutation rates of 8.4 × 10−9 per site per generation for Drosophila (Haag-Liautard et al. 2007) and of 2.5 × 10−8 for humans (Nachman and Crowell 2000). The effects were sampled from a gamma distribution with the shape parameter k = 0.2 and k = 0.35, which have been previously measured for Drosophila and humans, respectively (Keightley and Eyre-Walker 2007). Furthermore, k = 1 was chosen as an extreme case. We report the proportion of adaptive fixations observed from 10,000 simulations after 10,000 generations for the various combinations of models, initial conditions, and distributions of effect sizes (Table 4). Overall we observe that using the biologically more reasonable gamma-distributed effects, the proportion of selective sweeps is reduced in the constant-environment model (e.g., from 16 to 1% comparing the uniformly distributed effects with gamma-distributed effects and k = 0.2). Similarly, in the general fitness model the percentage of fixations is lower for gamma-distributed effects and realistic values of the shape parameter than for the effects sampled form the uniform distribution. In contrast, using the mutation–selection balance as initial condition strongly increases the chance of observing selective sweeps for the constant-environment model, as much more low-frequency variants are initially available that might go to fixation. For the changing-environment model, we also analyzed the impact of moderate environmental changes by altering the effects by 10 or 50%. In both cases the observed number of sweeps remains quite low, independent of the chosen effect size distribution. A more moderate optimum shift than in the original model reduced the proportion of sweeps to some extent compared to the results shown in Table 2.
Table 4. Percentage of selective sweeps for various models, initial conditions, and distributions of effects.
| Effects |
|||||
|---|---|---|---|---|---|
| Model | Initial condition | Uniform | k = 0.2 | k = 0.35 | k = 1 |
| Constant environment | Neutral | 16.15 | 1.16 | 3.05 | 12.28 |
| Constant environment | Mut.-sel. balance (μ = 2.5 × 10−8) | 43.48 | 29.45 | 18.22 | 30.31 |
| Constant environment | Mut.-sel. balance (μ = 8.4 × 10−9) | 43.67 | 32.16 | 21.54 | 29.44 |
| Changing environment (<0.1)a | Equilibriumb | 0.26 | 0.36 | 0.4 | 0 |
| Changing environment (<0.5)a | Equilibriumb | 0.28 | 0.43 | 0.35 | 0.04 |
| Optimum change (θ < 0.1) | Neutral | 14.67 | 0.71 | 2.09 | 9.46 |
| Optimum change (θ < 0.5) | Neutral | 17.2 | 5.21 | 9.15 | 16.05 |
| Optimum change (θ < 0.1) | Equilibriumb | 0.05 | 0.03 | 0.09 | 0.16 |
| Optimum change (θ < 0.5) | Equilibriumb | 2.27 | 1.27 | 2.8 | 3.97 |
| General fitness | Neutral | 24.09 | 6.22 | 12.98 | 26.77 |
Effects were changed randomly relative to the initial effects by 10 or 50%.
The equilibrium states of the constant-environment model were used as initial condition.
We also tested the constant-environment model under the assumptions that the effects are derived from the initial frequencies using an exponential distribution a exp(−bp). We have chosen the parameter a = 0.5 and b = 1 to match the distribution measured in Mackay et al. (2012). We observe a high proportion of trajectories going to fixation from initially low frequencies (30%). Hence, alleles with high effect sizes and initially low frequencies reach fixation quickly.
Stochastic simulations
We ran stochastic simulations in a similar way as described above to study the impact of genetic drift on the resulting proportions of equilibria approached by the trajectories. Instead of ODEs, we used the system of corresponding difference equations (Willensdorfer and Bürger 2003). Gamete frequencies are sampled after each generation using the “mnrnd” function in Matlab (v. 6.1). As in the deterministic case we let the system evolve for 10,000 generations. Due to the random nature of genetic drift, equilibrium points can be approached, yet the trajectories may not stay at the deterministic equilibria. Therefore, we measured the proportion of trajectories that reside in an ε interval (with ε =10−3) of one of the respective equilibria after 10,000 generations (see Table 2 and Table 5).
Table 5. Proportions (%) of trajectories that lead to one of the equilibria in stochastic simulations (N = 1000) after 10,000 generations.
| Equilibrium | Descriptiona | Constant environment, weak selection | Constant environment, strong selection | Changing environmentb | Optimum changec | General fitnessd |
|---|---|---|---|---|---|---|
| E1 | Polymorphic/symmetric | 0 | 0 | 0 | 0 | 0 |
| E2 + E3 | Polymorphic/unsymmetric | 0.59 | 3.75 | 0.07 | 0.79 | 0.00 |
| E4 | A polymorphic, loss at B | 38.07 | 32.33 | 27.41 | 26.83 | 1.28 |
| E5 | A polymorphic, fixation at B | 1.56 | 1.25 | 1.62 | 2.09 | 0.05 |
| E6 | Fixation in A, loss at B | 26.48 | 15.77 | 37.39 | 18.15 | 20.40 |
| E7 | Loss at A, fixation at B | 25.97 | 12.08 | 32.14 | 19.23 | 18.86 |
| E8 | Loss at A, loss at B | 0 | 0 | 0 | 0.01 | 15.66 |
| E9 | Fixation at A, fixation at B | 0.46 | 32.94 | 0.40 | 20.66 | 42.19 |
| Sweep at A (fixation in E6)e | 45.39 | 49.21 | 0.4 | 44.3 | 48.48 | |
| Sweep at B (fixation in E5)e | 12.82 | 14.4 | 0 | 22.97 | 40.0 | |
| Sweep at B (fixation in E7)e | 40.35 | 36.67 | 1.56 | 39.16 | 48.57 | |
| Sweep at A and B (fix. in E9)e | 65.22 | 31.36 | 100 | 33.4 | 31.62 | |
| Transient | 0 | 4.09 | 0 | 0 | 0 | |
| Unclassified | 6.87 | 2.21 | 0.97 | 12.24 | 1.56 | |
| Sweeps total | 23.0 | 22.7 | 1.05 | 22.95 | 32.41 |
The equilibria E1 to E9 are defined following Bürger (2000, p. 205).
Environmental changes were defined by random change of the original effects and selection coefficient.
In the shifted optimum model (see Models) the phenotypic optimum takes an arbitrary position 0 < θ < 1.
In the general fitness model the effects have arbitrary values sampled from (0, 2) such that A is major locus.
Selective sweeps are defined here as trajectories that lead to fixation from very low initial frequencies (<0.01). Numbers denote the proportions of fixations with initial frequencies <0.01.
For the constant-environment model and a population size of N = 1000, the trajectories reside in the neighborhood of the unsymmetric polymorphic equilibria at low proportion (E2 + E3, 1%) compared to the edge equilibria (E4, 38%). This is not surprising given that their eigenvalues are closest to zero compared to the eigenvalues of the other equilibria. However, in the constant-environment model due to drift, losses at A and fixations at B (E7, 26%; see Table 5) occur more frequently than in the deterministic case (E7, 11%; see Table 2), and the same is true under the changing-environment model. Consequently, sweeps at the minor locus are observed almost as often as at the major locus. This involves frequent crossings of the separatrices described above (for the QLE approximation). For example, assuming a constant environment and equal effects, the separatrices are defined as the diagonals in the pq plane. From 10,000 stochastic simulations with equal effects and a neutral allele frequency distribution as initial condition we observe 11% crossings of the separatrices. Selective sweeps at the B locus are possible. Increasing the population size to >10,000 makes the relative proportions of time spent in the equilibria similar to the percentages found in the deterministic case (data not shown).
Discussion
We have analyzed four versions of the two-locus model of stabilizing selection on a phenotypic trait to understand the signatures of selection at the molecular level. For symmetric fitnesses we found for realistic parameter ranges (i.e., weak selection and loose linkage) essentially two evolutionary outcomes: fixation or polymorphism at the major locus (while the minor locus was monomorphic). At the genomic level, fixations may be observed as classical selective sweeps if the initial frequency of the beneficial allele was very low and selection sufficiently strong or as sweeps from standing variation (soft sweeps). Polymorphic equilibria may be detected as allele frequency shifts. However, this is possible only if these events occurred relatively recently (Kim and Stephan 2000). In the following we discuss how the different versions of the model, the initial conditions of the ensuing evolutionary trajectories, and population size influence the relative abundance of these signatures.
Models
The symmetric viability model produces sweeps for ∼16% of the trajectories if the initial frequencies were chosen according to the standard neutral site-frequency spectrum (Tajima 1989). This number is considerably higher than in the case of randomly drawn initial frequencies (Pavlidis et al. 2012). The main reason is that most polymorphisms in the neutral frequency spectrum are singletons (i.e., occur once in a sample) and are therefore centered around the state in which both loci are monomorphic (E8). The locus with the larger effects exhibits more selective sweeps than the locus with the minor effects. The amount of sweeps remains about the same for high-selection coefficients. However, more trajectories are found in the polymorphic equilibrium than for weak selection.
In the other two generalized models in which the trajectories started from initial frequencies sampled from the neutral frequency spectrum, the proportion of sweeps is even higher than that for the symmetric fitness model. For the model with an optimum θ > 0, we found that ∼20% of the trajectories reach fixation starting from an initial frequency < 0.01, although the parameter values (other than θ) were drawn from the same distributions as for the symmetric fitness model. This increase in the number of sweeps is due to the fact that a shift of the optimum to a positive value reflects directional selection on the phenotype. Under standard neutral initial conditions, the general fitness model predicts that ∼24% of the trajectories lead to sweeps. The reason is that, as in the shifted optimum model, the double heterozygote is not necessarily associated with the highest fitness.
Initial conditions
The initial conditions of the trajectories studied in this article are usually drawn from the standard neutral site-frequency spectrum. This appears to be biologically more plausible than random initial frequencies. However, this difference of initial conditions may have a large influence on the ensuing trajectories. In other words, the evolutionary dynamics of the models are relatively sensitive to the basins of attraction of the underlying system. As we have emphasized above, in the case of initial conditions following the standard neutral frequency spectrum, the number of sweeps is much higher than for random initial frequencies, as the initial frequencies are more centered around state in which both loci are monomorphic (E8). As expected, we found that the proportion of sweeps is even higher if the initial frequencies are drawn from a mutation–selection balance (Orr and Betancourt 2001), which show an excess of low-frequency alleles (Ohta 1973). The choice of the initial conditions appears to be more important for the symmetric fitness model, whereas the general fitness model appears to be less sensitive to whether the initial frequencies are < 0.0001 or 0.2 (see Pavlidis et al. 2012, Figure 4).
On the other hand, assuming that a sudden environmental shift occurs from an equilibrium state to which a population has already been adapted, the number of sweeps observed may be drastically reduced. We observed this in our model of an environmental change in which we sampled the model parameters from the same distributions as for the other models. Many trajectories are driven from edges into a corner equilibrium. Thus, we found a very strong reduction of classical sweeps and instead a large proportion of allele frequency shifts or fixations from standing variation.
Population size
All results discussed above were generated using the deterministic version of the models. In our stochastic simulations, the effect of genetic drift due to a finite population size is twofold. First, the equilibrium points of the deterministic system, in particular, the less stable internal ones, are no longer attractive. As a consequence, more trajectories tend to approach the corner equilibria in which drift can no longer operate. Second, drift facilitates crossing of separatrices, which is not possible in the deterministic system. For this reason, we occasionally observed for small population sizes (of N = 1000) relatively large differences between the outcomes of the deterministic and stochastic simulations. For instance, in the changing-environment model, many more trajectories have approached the equilibrium E7 (loss at A, fixation at B) in the stochastic case than in the deterministic one. Similarly, we observe in all models a slightly higher proportion of sweeps in the stochastic case compared to the deterministic case. For larger population sizes (N > 10,000) the system approaches the deterministic version (data not shown).
Predictions of alternative models
We have quantified the frequency of selective fixations (leading to selective sweeps) in one of the most widely studied models of quantitative genetics, the two-locus model of stabilizing selection. In a similar analysis, this model has been extended from two to eight loci by Pavlidis et al. (2012). Using the general fitness scheme, these authors found that the frequency of sweeps declines with the number of loci. However, several classes of alternative models of stabilizing selection have been analyzed with respect to the maintenance of polygenic variation, for which we do not know to what extent they predict selective fixations.
One of the simplest ways to maintain genetic variation in natural populations is to include mutation (Kimura 1965b; Turelli 1984; Barton 1986). In Barton’s model, selective fixations have been detected after an optimum shift was introduced. Although the frequency of sweeps was not estimated (de Vladar and Barton 2011, 2014), their simulations indicate that larger shifts of the optimum increase the fixation probability, as discussed above for the shifted optimum model. However, in none of the other mutation-stabilizing selection models was this issue addressed.
Polygenic variation could also be caused by pleiotropic effects of balanced polymorphisms (Turelli 1985; Barton 1990; Keightley and Hill 1990). An extension of Barton’s (1986) model including pleiotropy was proposed by Gimelfarb (1996). In this model, n diallelic loci contribute additively to n quantitative traits under stabilizing selection, such that each locus has a major effect on one trait and minor effects on all other traits. It can be shown analytically that adaptive fixations are facilitated if the shift of the optimum of a trait after an environmental change is relatively large and the effects of the locus on the other traits small (unpublished results). Recently, pleiotropy was combined with mutation and stabilizing selection, and it was demonstrated that this is a sufficient mechanism to explain observed levels of genetic variation (Zhang and Hill 2002; 2005). In this class of models, real stabilizing selection works directly on the trait under study and apparent stabilizing selection is caused by the deleterious pleiotropic side effects of mutations on fitness. Therefore, these models are expected to predict extremely low fixation probabilities of new mutations, unless the degree of pleiotropy is much lower than generally thought (Wagner and Zhang 2011).
Acknowledgments
A.W. was supported by Volkswagen Foundation (ref. 86042). W.S. was supported from the German Research Foundation (Priority Program 1590, grant Ste 325/14-1). Part of this article was written at the Simons Institute for the Theory of Computing (University of California—Berkeley).
Footnotes
Communicating editor: M. K. Uyenoyama
Literature Cited
- Barton N. H., 1986. The maintenance of polygenic variation through a balance between mutation and stabilizing selection. Genet. Res. 47: 209–216 [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1990. Pleiotropic models of quantitative variation. Genetics 124: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133 [Google Scholar]
- Barton N. H., Keightley P. D., 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3: 11–21 [DOI] [PubMed] [Google Scholar]
- Bürger R., 2000. The Mathematical Theory of Selection, Recombination, and Mutation. Wiley, New York [Google Scholar]
- Bürger R., Gimelfarb A., 1999. Genetic variation maintained in multilocus models of additive quantitative traits under stabilizing selection. Genetics 152: 807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L. M., Hospital F., 2008. Selective sweep at a quantitative trait locus in the presence of background genetic variation. Genetics 180: 1645–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J., Kimura M., 1970. An Introduction to Population Genetics Theory. Burgess, Minneapolis [Google Scholar]
- de Vladar H. P., Barton N. H., 2011. The statistical mechanics of a polygenic character under stabilizing selection, mutation and drift. J. R. Soc. Interface 8: 720–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vladar H. P., Barton N., 2014. Stability and response of polygenic traits to stabilizing selection and mutation. Genetics 197: 749–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler J., 1986. Natural Selection in the Wild. Princeton University Press, Princeton [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Prentice Hall, London: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford [Google Scholar]
- Gavrilets S., Hastings A., 1993. Maintenance of genetic variability under strong stabilizing selection: a two-locus model. Genetics 134: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelfarb A., 1996. Some additional results about polymorphisms in models of an additive quantitative trait under stabilizing selection. J. Math. Biol. 35: 88–96 [DOI] [PubMed] [Google Scholar]
- Griffiths R., Tavaré S., 1998. The age of a mutation in a general coalescent tree. Commun. Statist. Stoch. Models 14: 273–295 [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 [DOI] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kim Y., 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl. Acad. Sci. USA 101: 10667–10672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Hill W. G., 1990. Variation maintained in quantitative traits with mutation-selection balance: pleiotropic side-effects on fitness traits. Proc. Biol. Sci. 242: 95–100 [Google Scholar]
- Kim Y., Stephan W., 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1965a Attainment of quasi linkage equilibrium when gene frequencies are changing by natural selection. Genetics 52: 875–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1965b A stochastic model concerning the maintenance of genetic variability in quantitative characters. Proc. Natl. Acad. Sci. USA 54: 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1983. The maintainance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 26: 221–235 [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetic Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Nachman M. W., Crowell S. L., 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98 [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2005. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6: 119–127 [DOI] [PubMed] [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Jensen J. D., Stephan W., 2010. Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations. Genetics 185: 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Metzler D., Stephan W., 2012. Selective sweeps in multilocus models of quantitative traits. Genetics 192: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Di Rienzo A., 2010. Adaptation: not by sweeps alone. Nat. Rev. Genet. 11: 665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A., 1956. The effect of selection against extreme deviants based on deviation or on homozygosis. J. Genet. 54: 236–248 [Google Scholar]
- Stephan W., 1995. An improved method for estimating the rate of fixation of favorable mutations based on DNA polymorphism data. Mol. Biol. Evol. 12: 959–962 [DOI] [PubMed] [Google Scholar]
- Stephan W., 2010. Genetic hitchhiking vs. background selection: the controversy and its implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H. E., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254 [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin M. C., Chiang C. W., Palmer C. D., Sankararaman S., Reich D., et al. , 2012. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 44: 1015–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., 1984. Heritable genetic variation via mutation-selection balance: Lerch’s zeta meets the abdominal bristle. Theor. Popul. Biol. 25: 138–193 [DOI] [PubMed] [Google Scholar]
- Turelli M., 1985. Effects of pleiotropy on predictions concerning mutation-selection balance for polygenic traits. Genetics 111: 165–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213 [DOI] [PubMed] [Google Scholar]
- Willensdorfer M., Bürger R., 2003. The two-locus model of Gaussian stabilizing selection. Theor. Popul. Biol. 64: 101–117 [DOI] [PubMed] [Google Scholar]
- Zhang X. S., Hill W. G., 2002. Joint effects of pleiotropic selection and stabilizing selection on the maintenance of quantitative genetic variation at mutation-selection balance. Genetics 162: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S., Hill W. G., 2005. Evolution of the environmental component of the phenotypic variance: stabilizing selection in changing environments and the cost of homogeneity. Evolution 59: 1237–1244 [PubMed] [Google Scholar]