Abstract
Small ubiquitin-like modifier (SUMO1–3) conjugation plays a critical role in embryogenesis. Embryos deficient in the SUMO-conjugating enzyme Ubc9 die at the early postimplantation stage. Sumo1−/− mice are viable, as SUMO2/3 can compensate for most SUMO1 functions. To uncover the role of SUMO2/3 in embryogenesis, we generated Sumo2- and Sumo3-null mutant mice. Here, we report that Sumo3−/− mice were viable, while Sumo2−/− embryos exhibited severe developmental delay and died at approximately embryonic day 10.5 (E10.5). We also provide evidence that SUMO2 is the predominantly expressed SUMO isoform. Furthermore, although Sumo2+/− and Sumo2+/−;Sumo3+/− mice lacked any overt phenotype, only 2 Sumo2+/−;Sumo3−/− mice were found at birth in 35 litters after crossing Sumo2+/−;Sumo3+/− with Sumo3−/− mice, and these rare mice were considerably smaller than littermates of the other genotypes. Thus, our findings suggest that expression levels and not functional differences between SUMO2 and SUMO3 are critical for normal embryogenesis.
Keywords: Embryonic development, Knockout, SUMO conjugation, SUMO2, SUMO3
Introduction
SUMO conjugation (SUMOylation) is a protein modification that profoundly influences the stability, activity, and subcellular localization of target proteins, and modulates their interactions with DNA or partner proteins through SUMO-interacting motifs 1. Three SUMO isoforms—SUMO1, SUMO2, and SUMO3—are ubiquitously expressed. SUMO2 and SUMO3 are almost identical in amino acid sequence and are therefore usually referred to as SUMO2/3. SUMO2/3 share about 50% identity with SUMO1. SUMOylation requires the sequential function of activating (E1), conjugating (E2), and ligating (E3) enzymes, a process mechanistically similar to conjugation pathways of other ubiquitin-like proteins. SUMOylation is a dynamic and reversible process, as SUMO-conjugated proteins are deconjugated by sentrin/SUMO-specific proteases (SENPs).
The role of SUMO conjugation in embryonic development has been investigated in many species 2. SUMO conjugation is indispensible for embryonic viability, as embryos deficient in the SUMO-conjugating enzyme Ubc9 die during early development 3,4. Indeed, Ubc9-deficient mouse embryos show severe defects in chromosome segregation and die at the early postimplantation stage 3. On the other hand, SUMO1 is not a critical determinant of embryonic development, as SUMO1-deficient mice are viable and lack any overt phenotype 5,6. Notably, in SUMO1-deficient mice, SUMO2/3 compensates for most SUMO1 functions. For example, RanGAP1, the most prominent SUMO1-conjugated protein in wild-type embryos, is conjugated to SUMO2/3 in Sumo1-null mutants 5. Whether SUMO1 can compensate for SUMO2 or SUMO3, and whether SUMO2 and SUMO3 are functionally redundant for embryonic development have yet to be determined. To better understand the SUMOylation system during embryonic development, and verify the role of SUMO2 and SUMO3 in this process, we have generated Sumo2- and Sumo3-null mutant mice.
Results and Discussion
Generation of Sumo2- and Sumo3-null mutant mice
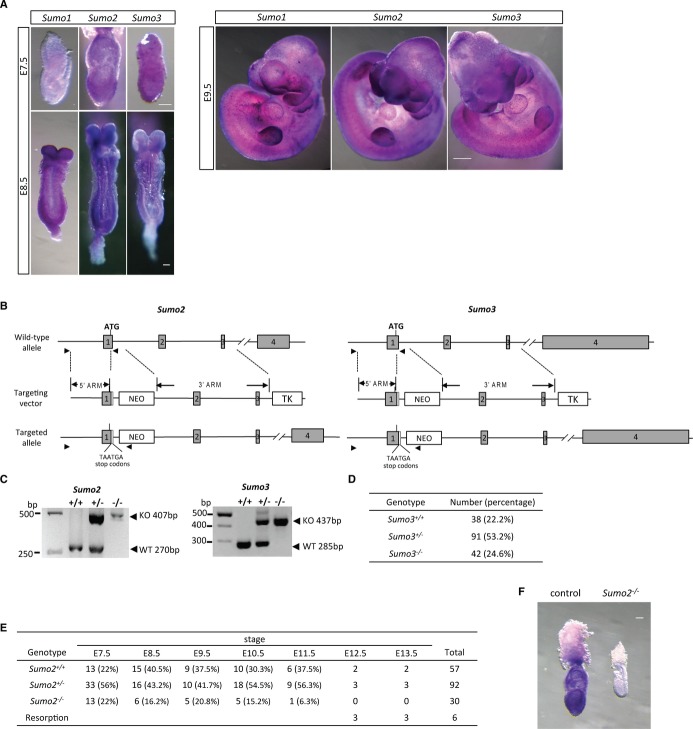
Several studies have demonstrated that SUMO conjugation plays a critical role in embryonic development 2, but the embryonic expression patterns of SUMO isoforms have not yet been investigated. We first examined by whole-mount in situ hybridization the expression of Sumo1, Sumo2, and Sumo3 in embryos using validated probes (Supplementary Fig S1). This confirmed that all 3 SUMO isoforms were ubiquitously expressed in E7.5, E8.5, and E9.5 embryos (Fig. 1A). As SUMO1 is indispensable in normal mouse development due to functional compensation by SUMO2/3 6, the contribution of SUMO2 and SUMO3 remains intriguing in that the proteins differ by only three amino acids. In order to determine the roles of SUMO2 and SUMO3 in embryogenesis, we generated Sumo2- and Sumo3-null mutant mice. Sumo2 and Sumo3 genes are located on mouse chromosome 11 and 10, respectively, and have a similar genomic structure, both containing four exons (Fig. 1B). Therefore, we used the same targeting strategy to generate Sumo2- and Sumo3-null mice (Fig. 1B). In the targeted allele, 2 premature stop codons and a neomycin cassette (NEO) were placed in exon 1 to disrupt the Sumo2 or Sumo3 gene, thereby creating a functional null allele. The null alleles were confirmed by whole-mount in situ hybridization analysis of Sumo2−/− and Sumo3−/− embryos (Supplementary Fig S1).
Figure 1. Targeted disruption of Sumo2 but not Sumo3 is lethal to embryos.
A Ubiquitous expression of Sumo1, Sumo2, and Sumo3 in E7.5, E8.5, and E9.5 wild-type embryos. Whole-mount in situ hybridization analysis of Sumo1, Sumo2, and Sumo3 expression was performed in C57BL/6 wild-type mouse embryos. The specificity of all probes was validated by comparing whole-mount in situ hybridization in wild-type to Sumo1-, Sumo2-, and Sumo3-null embryos, as demonstrated in Supplementary Fig S1. Scale bars: 100 μm (left panel), 500 μm (right panel).
B Targeting strategy for Sumo2 and Sumo3. The targeting vectors were designed to disrupt exon 1 by in-frame insertion of 2 stop codons and a neomycin (NEO) cassette downstream of the ATG codon. Arrowheads mark the location of primers for genotyping. Gray boxes show exons. TK, thymidine kinase cassette.
C Representative genotyping results of embryos obtained from timed Sumo2+/− or Sumo3+/− intercrosses. Genomic DNA prepared from extraembryonic tissue was used for PCR amplification with primers as described in (B) and Supplementary Table S1.
D Normal Mendelian distribution of newborn pups from Sumo3+/− intercrosses.
E Summary of genotyping analysis of staged embryos from Sumo2+/− intercrosses.
F Representative image of Sumo2 expression by whole-mount in situ hybridization in a wild-type embryo (control, left) and a homozygous null embryo (Sumo2−/−, right) at E7.5. Scale bar: 100 μm.
Genotypes were determined by two PCRs with a 5′ primer located outside the 5′ homologous arm, and two 3′ primers, one located immediately outside the 3′ homologous arm and one within the NEO cassette (Fig. 1B,C). Both Sumo2+/− and Sumo3+/− mice were viable and fertile and showed no gross abnormalities in size or weight compared to the wild-type littermate controls. To generate the homozygous mutants, heterozygous mice were intercrossed. Genotypes of the progeny derived from Sumo3+/− intercrosses showed a normal Mendelian distribution (Fig. 1D). The Sumo3 homozygous mutants (Sumo3−/−) were fertile and did not display phenotypic abnormalities. However, no viable Sumo2−/− homozygous pups from Sumo2+/− intercrosses were found. This indicated that SUMO2 deficiency in embryos is lethal.
Sumo2−/− embryos exhibited severe growth retardation and died at about E10.5
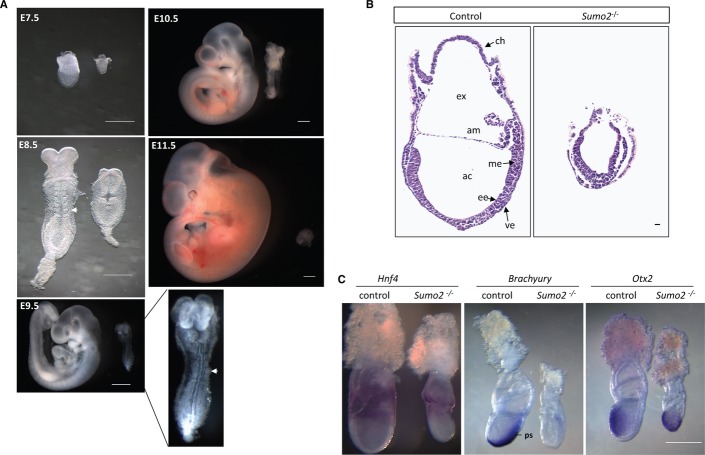
To determine the temporal profile of the lethality caused by SUMO2 deficiency, Sumo2+/− mice were mated, and embryos were isolated and genotyped at embryonic days 7.5 to 13.5 (E7.5–E13.5). At E7.5, Sumo2−/− embryos were detected at the expected Mendelian frequency (Fig. 1E). However, their size was smaller than that of their Sumo2+/+ littermates (Figs. 1F and 2A). At E8.5, the Sumo2−/− embryos had developed a head fold and neural groove, and the allantoic bud was also formed. However, the allantois was found loosely attached to the chorion at E9.5 and later stages as compared to the control littermates. Furthermore, the somite structures were not visible in Sumo2−/− embryos before E9.5, and a heart tube-like structure was not found until E10.5 (Fig. 2A), indicating severe growth retardation. A vascular system was detected in Sumo2−/− embryos at E9.5 when somite structures were visible, using the endothelial cell-specific marker CD31 (Supplementary Fig S2). By E11.5, only resorbed debris and necrotic tissue were observed and no Sumo2−/− embryos were recovered, suggesting that Sumo2-null mutants die at approximately E10.5.
Figure 2. Developmental arrest of Sumo2−/− embryos.
A Severe growth retardation of Sumo2−/− embryos. Representative gross morphology of wild-type (left) and Sumo2−/− littermate (right) embryos at different developmental stages (E7.5, E8.5, E9.5, E10.5, and E11.5). Sumo2−/− embryo at E9.5 was enlarged to show somites (arrow). Scale bar: 500 μm.
B Representative images of hematoxylin and eosin (H&E) staining of control and Sumo2−/− embryos at E7.5. ee, embryonic ectoderm; me, mesoderm; ve, visceral endoderm; ch, chorion; ac, amniotic cavity; ex, exocoelomic cavity; am, amnion. Scale bar: 20 μm.
C Representative images of whole-mount in situ hybridization analysis of Hnf4 (endoderm marker), Brachyury (mesoderm marker), and Otx2 (ectoderm marker) of wild-type and Sumo2−/− littermate embryos at E7.5. ps, primitive streak.
Histological analysis at E7.5 showed that control embryos had a well-defined anterior–posterior axis with the proper formation of the amniotic cavity and a primitive streak, indicating that the embryo had initiated gastrulation (Fig. 2B). However, in Sumo2−/− embryos, the amnion was not seen, although an early primitive streak with mesodermal cells was detectable between the epiblast and extraembryonic endoderm (Fig. 2B). Three germ layers (endoderm, mesoderm, and ectoderm) were confirmed in Sumo2−/− embryos at E7.5 by whole-mount in situ hybridization using the germ layer-specific markers Hnf4 (endoderm), Brachyury (mesoderm), and Otx2 (ectoderm) (Fig. 2C). This suggested that gastrulation was initiated in Sumo2−/− embryos, but the mesoderm was delayed in migrating into the extraembryonic region to form the beginning of the amnion.
To determine whether the distinct small size and growth retardation of Sumo2−/− embryos resulted from a decrease in cellular proliferation or an increase in cell death, we used EdU incorporation and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) to analyze cell proliferation and cell death, respectively. The number of EdU-positive cells was reduced in Sumo2−/− embryos, while the number of TUNEL-positive cells was significantly increased (Fig. 3). Therefore, both reduced cell proliferation and increased cell death likely contributed to the small size and delayed growth of Sumo2−/− embryos.
Figure 3. Compromised cell proliferation and enhanced apoptotic cell death in Sumo2−/− embryos.
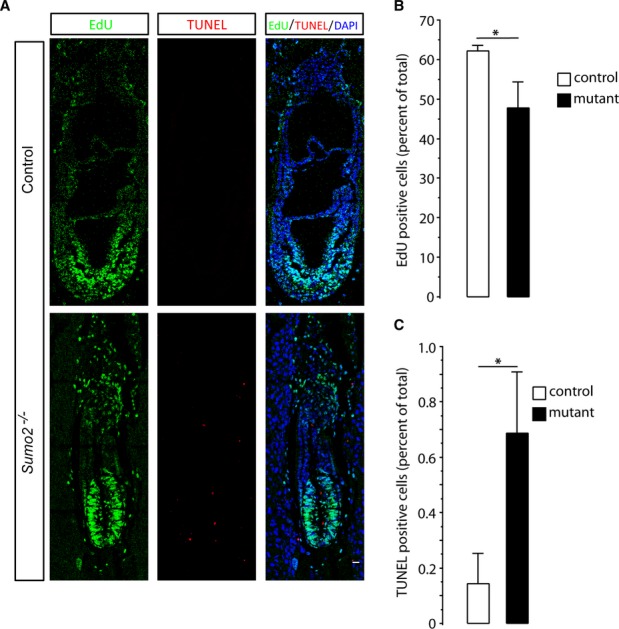
A Representative images of EdU (green) and TUNEL (red) staining of control and Sumo2−/− embryos at E7.5. Nuclei were stained with DAPI. Scale bar: 20 μm.
B, C Quantitative analysis of EdU and TUNEL staining. EdU-positive, TUNEL-positive, and total cells (DAPI staining) were counted, and percentages were calculated (n = 8–10 sections from 3 embryos/group). Data were presented as means ± SD. Student’s t-test was used to calculate statistical significance; * P < 0.05.
SUMO2 is the predominant SUMO isoform
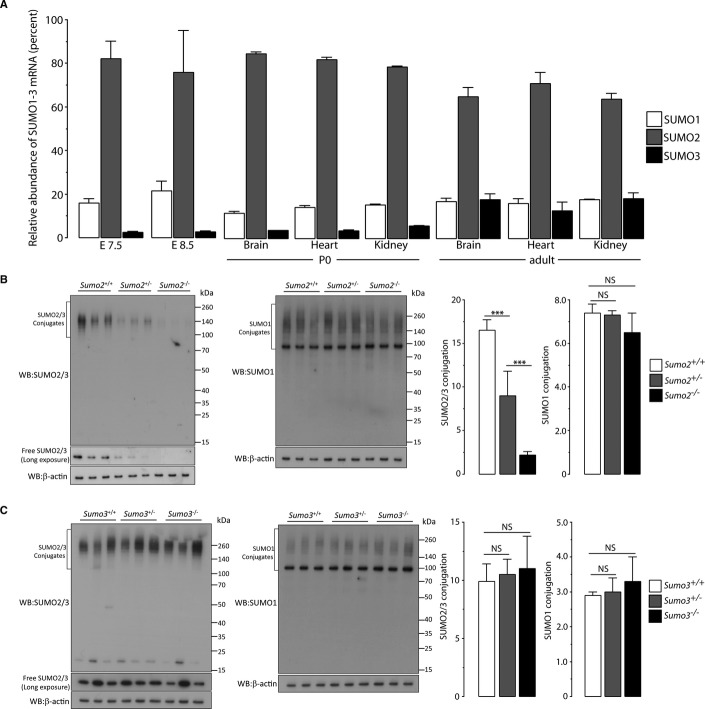
SUMO2 and SUMO3 show a high degree of similarity, as they differ by only three amino acids and cannot be distinguished by available antibodies. Therefore, it was surprising that the phenotypes of Sumo2- and Sumo3-null mutant mice were dramatically distinct. We hypothesized that expression levels of Sumo2 and Sumo3 differ considerably during embryogenesis and that the sum of SUMO2/3, rather than the level of individual proteins, plays the pivotal role in determining the fate of the embryo at early stages. To test this hypothesis, we used quantitative RT–PCR (qPCR) to quantify relative expression levels of Sumo2 and Sumo3 as well as Sumo1 in wild-type C57BL/6 embryos at E7.5 and E8.5. Based on an expression level of 100% for all three Sumo isoforms combined, Sumo2 accounted for about 80 and 75% at E7.5 and E8.5, respectively (Fig. 4A). In contrast, Sumo3 accounted for only 2 and 3%, while Sumo1 accounted for 16 and 21% at E7.5 and E8.5, respectively (Fig. 4A). Thus, SUMO2 was indeed the dominant isoform at early embryonic stages. We also evaluated expression levels of SUMO1-3 in brains, hearts, and kidneys of postnatal mice. An expression pattern similar to embryos was found at postnatal day 0 (Fig. 4A, P0). However, in adult animals, Sumo3 levels were markedly higher and accounted for almost 20% of total Sumos (Fig. 4A, adult).
Figure 4. SUMO2 is the dominant SUMO isoform.
A Relative expression levels of Sumo1, Sumo2, and Sumo3 in C57BL/6 wild-type embryos at E7.5 and E8.5, and in brains, hearts, and kidneys at postnatal day 0 (P0) and the adult state. Data are presented as means ± SD (n = 3).
B Quantitative Western blot analysis results showed the dramatic decrease in levels of SUMO2/3-conjugated proteins in Sumo2−/− E8.5 embryos compared to Sumo2+/− and Sumo2+/+ E8.5 embryos (***P < 0.001). Notably, SUMO1 conjugation did not increase in the absence of SUMO2. The high molecular weight regions marked by brackets were used to quantify SUMO1 or SUMO2/3 conjugation. Data are presented as means ± SD (n = 3). NS, not significant.
C Quantitative Western blot analysis results showed no major difference in levels of SUMO2/3- or SUMO1-conjugated proteins in Sumo3+/+, Sumo3+/−, and Sumo3−/− E8.5 embryos. The high molecular weight regions marked by brackets were used to quantify SUMO1 or SUMO2/3 conjugation. Data are presented as means ± SD (n = 3). NS, not significant.
Source data are available online for this figure.
Next, we used Western blot to analyze the relative protein levels of SUMO2 and SUMO3 in E8.5 embryos. The SUMO2/3 antibody used in this study was generated by immunizing rabbits with purified SUMO2 protein. We confirmed that this antibody had a comparable affinity to both SUMO2 and SUMO3 (Supplementary Fig S3). Levels of SUMO2/3-conjugated proteins were markedly reduced in Sumo2−/− compared to Sumo2+/− and Sumo2+/+ embryos (Fig. 4B). In contrast, no significant differences in levels of SUMO2/3-conjugated proteins were found among Sumo3−/−, Sumo3+/−, and Sumo3+/+ embryos (Fig. 4C), thus confirming our qPCR results that SUMO2 was the predominantly expressed SUMO isoform during mouse embryogenesis. To investigate whether SUMO1 can compensate for SUMO2 or SUMO3 loss, SUMO1 Western blot analysis was performed. Notably, SUMO1 conjugation did not increase significantly in the absence of SUMO2 or SUMO3 (Fig. 4B,C), suggesting that SUMO1 cannot compensate for SUMO2 or SUMO3 loss.
Whether SUMO2 or SUMO3 is the more prominent SUMO isoform in adult mice has not been determined because available SUMO2/3 antibodies cannot distinguish between SUMO2 and SUMO3. To address this question, we collected brains, hearts, and kidneys from adult Sumo2 or Sumo3 mutant and wild-type mice. In contrast to embryos, there are very few SUMO2/3 conjugates in these organs in adult animals (Supplementary Fig S4). We therefore compared levels of free SUMO2/3 in those organs (Sumo2+/− vs Sumo2+/+ and Sumo3−/− vs Sumo3+/+). The data revealed a more pronounced reduction of free SUMO2/3 in Sumo2+/− mice compared with Sumo3−/− mice (Supplementary Fig S4). Together, these results demonstrate that SUMO2 makes up the vast majority of SUMO2/3 in both embryo and adult tissues.
SUMO2/3 levels but not isoforms define embryogenesis
The most prominent findings of this study were the observations that embryos deficient in SUMO3 were viable and lacked any overt phenotype, while Sumo2-null mutants showed severe developmental delay and died at about E10.5. Based on earlier observations that Ubc9-null mutants die at the early postimplantation stage while Sumo1-null mutants are viable, we expected that conjugation by SUMO2/3 plays the pivotal role in embryogenesis. Therefore, the viability of Sumo3 but not of Sumo2-null mutants was an unexpected finding since SUMO2 and SUMO3 are highly homologous. This raises the question of whether SUMO2 and SUMO3 are functionally different despite the high homology of more than 95%, or whether it is the total level of SUMO2/3 that drives embryogenesis. Indeed, results from this study support the latter assumption: Sumo3 mRNA, which accounted for only about 2% of total Sumo mRNAs in E7.5 and E8.5 embryos (Fig. 4A), was dispensable for embryogenesis, whereas Sumo2, which accounted for about 80% of total Sumo mRNAs, was essential.
To address the question of functional redundancy of SUMO2 and SUMO3 conjugation during embryogenesis in more detail, we first mated Sumo2+/− with Sumo3−/− mice. Sumo2+/−;Sumo3+/− mice were obtained from this mating with no overt phenotype. Then, we asked whether SUMO2 is the only critical SUMO isoform required for normal embryonic development. If the specific lethal phenotype of Sumo2-null mutants resulted from the sole role of SUMO2 conjugation during embryonic development, we expected that reducing SUMO3 levels in heterozygous Sumo2+/− embryos would not result in a lethal phenotype, because SUMO3 was found to be dispensable. To test this possibility, we mated Sumo2+/−;Sumo3+/− with Sumo3−/− mice. To our surprise, analysis of 145 progeny from a total of 35 litters revealed that 41 (28.3%) were Sumo2+/−;Sumo3+/−, and 102 (70.2%) were Sumo2+/+;Sumo3+/− or Sumo2+/+;Sumo3−/−, whereas only 2 were Sumo2+/−;Sumo3−/−, and these 2 mice were considerably smaller than littermates of other genotypes (Supplementary Fig S5). These data suggested that Sumo2+/−;Sumo3−/− genotype is potentially lethal at late stage of embryogenesis. Indeed, Sumo2+/−;Sumo3−−/− embryos did not show a obvious phenotype at E15.5 and E17.5, but were considerably smaller at E19.5 (Supplementary Fig S5). Collectively, these findings support our hypothesis that the total levels of SUMO2/3 are critical for embryogenesis. An alternative interpretation of results would be that all 3 SUMO isoforms are dispensable and that the total level of SUMO1-3 is critical for embryogenesis. It has indeed been reported in an earlier study that SUMO isoforms are indispensible but functionally redundant in zebrafish during early development 4. The severe defects in zebrafish embryonic development induced by SUMO1-3 deficiency could be rescued by any human or zebrafish SUMO1, SUMO2, or SUMO3 4. Our data can also not exclude that SUMO2 and SUMO3 have some distinct functions, and SUMO2 can compensate for SUMO3 loss only when SUMO2 levels are high. Since, however, SUMO2 and SUMO3 are highly homologous, it is unlikely that they are functionally distinct.
The results of this and earlier studies highlight the pivotal role of global SUMOylation during embryogenesis. However, embryos deficient in individual components of the SUMOylation machinery die at different embryonic stages. Embryos deficient in Ubc9 die during an early postimplantation stage 3 when embryos deficient in SUMO1, SUMO2, or SUMO3 are still viable. Sumo2-null embryos failed to develop beyond the head-fold stage (E8.5), while embryos deficient in the SUMO proteases SENP1 and SENP2 die at about E12.5–E14.5 and E10, respectively 7,8. SENP1 is specific for SUMO1-conjugated proteins in vivo 9, while SENP2 de-SUMOylates both SUMO1 and SUMO2/3 conjugates 10. Taken together, these observations suggest that SUMO homeostasis must be tightly controlled during embryogenesis.
Materials and Methods
Generation of mutant mice and genotyping
This study was approved by the Duke University Animal Care and Use Committee. Sumo1-null mutant mice were obtained from Dr. Kuehn 5, and C57BL/6 mice were purchased from The Jackson Laboratory (Maine). The same strategy was used to disrupt Sumo2 and Sumo3 genes, as illustrated in Fig. 1B. Note that the nomenclature of SUMO2 and SUMO3 used here is in accordance with the NCBI database—protein entry number P61957 for SUMO2 and Q9Z172 for SUMO3. Two BAC clones, bMQ258k14 and bMQ457C17, containing Sumo2 gene and Sumo3 gene respectively, were isolated from a 129Sv mouse genomic library (Source BioScience, Nottingham, UK) and were used to retrieve the homologous arms. The targeting vectors were constructed by inserting a 5′ homologous arm (Sumo2, 1.2 kb; Sumo3, 1.4 kb), 2 in-frame stop codons in exon 1 (mutated sequences are listed in Supplementary Table S1), a neomycin cassette (NEO), and a 3′ homologous arm (Sumo2, 8 kb; Sumo3, 8 kb). A thymidine kinase cassette (TK) was used for negative selection. After verification by restriction analysis and partial sequencing, the targeting vectors were linearized and electroporated into mouse embryonic stem (ES) cells derived from the 129Sv strain at the Duke Neurotransgenic Laboratory. Positive ES clones were identified by PCR screening and confirmed by Southern blot analysis. Two ES clones were microinjected into blastocysts derived from C57BL/6 mice to produce chimeric mice. After the germline transmission was confirmed, heterozygous mice (Sumo2+/− and Sumo3+/−) were generated and crossed to produce homozygous offspring (Sumo2−/− or Sumo3−/−). For genotypic analysis of embryos and mice, genomic DNA was prepared from yolk sacs, whole embryos, or mouse tails. All primers for genotyping are listed in Supplementary Table S1.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was conducted as described previously 11. Briefly, antisense riboprobes were generated by in vitro transcription using the respective cDNA templates and were then labeled with digoxigenin (DIG)-UTP (Roche). The primers used are listed in Supplementary Table S1. Embryos were washed in PBS with 1% Tween 20 (PBT) and dehydrated by steps (25, 50, 75, and 100%) in methanol before freezing. After rehydration, embryos were treated with proteinase K (20 μg/ml) at room temperature for 15 min followed by several rinses in PBT. Embryos were then transferred to prehybridization buffer (50% formamide, 5× SSC, 2% blocking powder, 0.1% Tween 20, 0.1% CHAPS, 50 μg/ml yeast RNA, 5 mM EDTA, and 50 μg/ml heparin) at 70°C for 1 h before hybridization with probes diluted in fresh prehybridization buffer at 70°C overnight. The next day, embryos were washed 3 times with 2× SSC buffer plus 0.1% CHAPS, and 3 times with buffer (0.2× SSC and 0.1% CHAPS) at 70°C. After blocking in 10% sheep serum in KTBT buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10 mM KCl, and 1% Tween 20) for 2 h at room temperature, embryos were incubated with anti-DIG antibody (1:3,000) overnight at 4°C. After intensive washes with KTBT buffer, embryos were equilibrated in NTMT buffer (100 mM NaCl, 100 mM Tris pH 9.5, 50 mM MgCl2, and 0.1% Tween 20), and color reactions were initiated by adding the NBT/BCIP solution (1:50, Roche). Embryos were photographed on a Leica MZLFIII microscope.
Histology and immunohistochemistry analysis
Paraffin-embedded embryos were used for analysis. They were serially sectioned at 7 μm and stained with hematoxylin and eosin (H&E) by standard procedures. To analyze for cell proliferation and apoptosis, timed pregnant heterozygous females were injected intraperitoneally with EdU (5-ethynyl-2′-deoxyuridine, 20 mg/kg, Life Technologies). Two hours later, embryos were collected and fixed in 4% paraformaldehyde. EdU staining was performed using the Click-iT EdU kit (Life Technologies). Apoptotic cells were detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), using the In Situ Cell Death Detection kit (Roche, IN) according to manufacturer’s instructions. Confocal images were taken using a Zeiss 710 inverted confocal microscope. The percentage of proliferating and apoptotic cells was determined by counting EdU- and TUNEL-positive cells as a percentage of the total number of cells, as determined by DAPI staining, respectively. Eight to ten sections from 3 embryos in each group (control and mutant) were used for quantification.
Quantitative RT–PCR
Total RNA was extracted from C57BL/6 embryos at E7.5 and E8.5 stages and brains, hearts, and kidneys of postnatal mice (n = 3/group) using TRIzol reagent (Life Technologies). Quantitative RT–PCR was performed as previously described 12. In order to directly compare copy numbers of Sumo1, Sumo2, and Sumo3, an absolute quantification method based on the standard curve was used. To achieve high accuracy of quantification, a plasmid harboring the coding sequences of Sumo1, Sumo2, and Sumo3 was constructed and used to generate all 3 standard curves from a single template. The copy number of each gene was calculated based on the corresponding standard curve. Relative levels of each gene are expressed as percentage of total levels of the three genes.
Western blotting
Embryos and mouse tissues were snap-frozen in liquid nitrogen. Proteins were extracted by sonication of frozen samples in 1× Laemmli sample buffer, followed by boiling for 10 min. Western blotting was performed as previously described 13. Polyclonal antibody against SUMO2/3 was raised in rabbits through Custom Immunology Services provided by Covance, using purified SUMO2 proteins (BostonBiochem). Other antibodies include rabbit anti-GADPH (Cell Signaling) and mouse anti-β-actin (Sigma, MO, USA).
Statistical analysis
Data are presented as means ± SD. Statistical significance was assessed by two-tailed Student’s t-test (EdU and TUNEL) or ANOVA followed by Fisher’s PLSD test (SUMO mRNA and protein levels). The level of significance was set at P < 0.05.
Acknowledgments
The authors thank Dr. Brigid Hogan, Department of Cell Biology, Duke University Medical Center for helpful discussions and suggestions, and Kathy Gage for her excellent editorial contribution. This study was funded by NIH RO1 grants HL095552 and NS081299 to W.P. and an American Heart Association Scientist Development Grant 12SDG11950003 to W.Y.
Author contributions
LW, CW, SZ, PM, and WY performed the experiments and data analysis; WP and WY conceived the study; LW, WP, and WY wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Vazquez M. Emerging roles of the SUMO pathway in development. Cell Mol Life Sci. 2011;68:4045–4064. doi: 10.1007/s00018-011-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Yuan H, Zhou J, Deng M, Liu X, Le Bras M, et The H, Chen SJ, Chen Z, Liu TX, Zhu J. Small ubiquitin-related modifier paralogs are indispensable but functionally redundant during early development of zebrafish. Cell Res. 2010;20:185–196. doi: 10.1038/cr.2009.101. [DOI] [PubMed] [Google Scholar]
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Yamada S, Lualdi M, Dasso M, Kuehn MR. Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep. 2013;3:1640–1650. doi: 10.1016/j.celrep.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J. 2010;430:335–344. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansleeben C, Feitsma H, Tertoolen L, Kroon C, Guryev V, Cuppen E, Meijlink F. A novel mutant allele of Ncx1: a single amino acid substitution leads to cardiac dysfunction. Int J Dev Biol. 2010;54:1465–1471. doi: 10.1387/ijdb.093051cw. [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- Wang L, Ma Q, Yang W, Mackensen GB, Paschen W. Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J Neurochem. 2012;123:349–359. doi: 10.1111/j.1471-4159.2012.07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.