Abstract
Human surfactant protein A (SP-A) plays an important role in surfactant metabolism and lung innate immunity. SP-A is synthesized and secreted by alveolar type II cells (ATII), one of the two cell types of the distal lung epithelium (ATII and ATI). We have shown that miRNA interactions with sequence polymorphisms on the SP-A mRNA 3′UTRs mediate differential expression of SP-A1 and SP-A2 gene variants in vitro. In the present study, we describe a physiologically relevant model to study miRNA regulation of SP-A in human ATII. For these studies, we purified and cultured human ATII on an air-liquid interface matrix (A/L) or plastic wells without matrix (P). Gene expression analyses confirmed that cells cultured in A/L maintained the ATII phenotype for over 5 days, whereas P-cultured cells differentiated to ATI. When we transfected ATII with siRNAs to inhibit the expression of Drosha, a critical effector of miRNA maturation, the levels of SP-A mRNA and protein increased in a time dependent manner. We next characterized cultured ATII and ATI by studying expression of 1,066 human miRNAs using miRNA PCR arrays. We detected expression of >300 miRNAs with 24 miRNAs differentially expressed in ATII vs. ATI, 12 of which predicted to bind SP-A 3′UTRs, indicating that these may be implicated in SP-A downregulation in ATI. Thus, miRNAs not only affect SPA expression, but also may contribute to the maintenance of the ATII cell phenotype and/or the trans-differentiation of ATII to ATI cells, and may represent new molecular markers that distinguish ATII and ATI.
Keywords: alveolar type II cells, lung epithelium, surfactant protein A, miRNAs
Introduction
Surfactant protein A (SP-A) is synthesized and secreted in the lung by alveolar type II cells (ATII), one of the two cell types of the distal alveolar epithelium (ATII and ATI) (1, 2). Two homologous genes encode human SP-A: SFTPA1 (SP-A1) and SFTPA2 (SP-A2). Based on sequence differences at the coding region, several coding variants have been identified and characterized for SP-A1 and SP-A2, some of which have been associated with lung disease susceptibility and/or pathogenesis in neonates, children, and adults (3). We have previously reported biochemical and functional differences among SP-A1 and SP-A2 variants in several experimental models (4-13). We have also shown that the ratio of SP-A1 to total SP-A protein is altered in the bronchoalveolar lavage fluid (BALF) of subjects with lung disease vs. healthy individuals, indicating that the overall SP-A functional activity may depend on the relative amounts of SP-A1 and SP-A2 gene products (14, 15). Together, these findings indicate that a controlled regulation of SP-A1 and SP-A2 expression is critical to maintain lung homeostasis and prevent disease.
The specific mechanisms that regulate expression of human SP-A1 and SP-A2 are still not well understood. Previous work from our laboratory has demonstrated that differential regulation of SP-A variants occurs at both transcriptional and translational levels (16-19). Recently, using the NCI-H441 cell line, we have reported that differential miRNA regulation of SP-A variants depends on sequence polymorphisms at the 3′UTR of SP-A variants (20). However, differences may exist in the physiology, regulatory pathways, and overall gene expression between NCI H441 and ATII cells. Thus, a more physiologically relevant model is needed to elucidate and/or confirm the molecular mechanisms that control the expression of SP-A. We aim to accomplish this by using primary cultures of human ATII cells from lung donors.
In previous studies, investigators have successfully purified and cultured ATII from various animal models, and have demonstrated that the optimal culture conditions are different for ATII cells obtained from mouse, rat, and human (21, 22). Moreover, human ATII cells have been shown to trans-differentiate to alveolar type I cells (ATI) in cultures from various species (23, 24). Differentiation of ATII to ATI can be detected by changes in the overall cell morphology, the loss of lamellar bodies (detected by modified Papanicolaou staining, ultrastructure analysis, etc.), or by detection of molecular markers that allow the identification of these two distinct cell types (25). The most commonly used ATII cell specific markers are: Surfactant Protein C (STPC), ATP-binding cassette, sub-family A, member 3 (ABCA3), and thyroid transcription factor-1 (TTF-1); whereas the molecular markers commonly used for identification of ATI are Caveolin-1 (CAV1), T1α (PDPN), Receptor for Advanced Glycation End-products (RAGE), and Aquaporin-5 (AQP-5) (2, 26, 27).
The goal of this study was to define the role of miRNAs in the regulation of SP-A expression in human ATII cells. MicroRNAs (miRNAs) are recently discovered small non-coding RNAs that control gene expression via translation suppression by binding sequence motifs on mRNA 3′UTRs. MiRNA biogenesis initiates in the nucleus, where precursors (pri-miRNAs) are transcribed by the RNA polymerase II, and processed onto pre-miRNAs via cleavage by a complex of enzymes including Drosha (28). Pre-miRNAs are transported by exportin-5 to the cytosol, where further processing by the ribonuclease Dicer results in double-stranded 19-25nt molecules containing the mature miRNA (miR), and the complimentary miRNA (miR*). The mature miR interacts with the mRNA target by sequence complementarity of the seed region (7 8nt) to the 3′UTR. The miR-mRNA complex interacts with proteins of the Argonaut family leading to the assembly of the RNA-induced silencing complex (RISC) (28). The degree of sequence complementarity between miRNA and mRNA targets is known to determine whether repression of protein expression is mediated by mRNA degradation (perfect match), or inhibition of translation (imperfect match) (29).
Due to their multiple target potential, miRNAs have been identified as powerful regulators of a wide spectrum of biological functions, including cell cycle, apoptosis, cell differentiation, and development (30). Furthermore, extracellular secretion of miRNAs has been demonstrated and studied extensively as a novel mechanism of cell regulation and communication (31). Numerous studies have shown that expression of intracellular and secreted miRNAs is altered during the progression of a variety of human diseases, indicating that miRNAs are involved in the control of important physiological processes, and may play a role in the pathogenesis and resolution of disease (32).
In the present study, we have isolated and cultured ATII cells from a human lung donor, and have characterized the alveolar epithelial cell phenotype under different culture conditions to investigate mechanisms of miRNA regulation of human SP-A1 and SP-A2 gene variants. By using a knockdown approach to inhibit miRNA biogenesis, we provide evidence that miRNAs regulate expression of SP-A. Moreover, we show for the first time that differences exist in the miRNA cellular content of human ATII and ATI cultured cells. Our findings support the notion that differential expression of miRNAs may regulate respiratory functions not only through regulation of SP-A gene expression, but also differentiation of alveolar epithelial cells, as well as point to a possible role of miRNAs in SP-A downregulation in ATI cells.
Materials and Methods
Cell purification
Human ATII cells were isolated from a donor lung (The Gift of Life Donor Program, Philadelphia, PA) by modification of previously published protocols (22, 33, 34). All procedures were approved by the Penn State College of Medicine Institutional Review Board. The lung was lavaged, instilled with 12.9U/ml elastase (Roche, Indianapolis, IN), and incubated at 37°C for 50min. The tissue was homogeneized, filtered, and treated with DNAase (Sigma, St. Louis, MO). Cell depletion was performed with anti-CD-14 magnetic beads (Life technologies, Carlsbad, CA) and by incubation with IgG (Sigma) on petri dishes, followed by treatment with RBC lysis buffer (BD Biosciences, San Jose, CA). ATII cells were suspended in DMEM containing 10% FBS, 2mM glutamine, 2.5μg/ml amphotericin B, 100μg/ml streptomycin, penicillin G and gentamicin (D10). Cell purity was confirmed by Papanicolaou stain and electron microscopy (35). The lung donor was a 20 year old male with the SP-A1/SP-A2 haplotype 6A26A3/1A01A1.
Cell culture
Cells were cultured as described elsewhere (34, 36). ATII were plated at a density of 1x106 cells/well in 12-well plates with Minicell inserts pre-coated with a mixture of matrigel (65%) (BD Biosciences) and rat tail collagen (35%) (BD Biosciences) (air-liquid interface, A/L). A second set of cells was plated directly in wells (plastic, P). Cells were incubated for 48h at 37°C with 5% CO2, and the medium was switched to KIA: DMEM containing 1% charcoal stripped FBS, 10ng/ml keratinocyte growth factor (R&D systems, Minneapolis, MN), 0.1mM isubutylmethylxanthine (Sigma), and 0.1mM 8-Br-cAMP (Sigma). One day later, the medium was switched to KIA supplemented with 10nM dexamethasone (KIAD) to enhance expression of surfactant proteins (37, 38), and the cells were maintained in culture for two more days. Cells were harvested daily, and expression of cell markers was determined.
siRNA knockdown
Cells were plated and incubated in A/L for 48h. Medium was switched to KIA, and cell transfection was performed with the GenomONE™-Neo EX HVJ Envelope transfection kit (Cosmo Bio, Carlsbard, CA) and stealth siRNA RNAi: Drosha (50nM), negative control med-GC (50nM), and Block-it Fluorescent oligo (10nM) (Life technologies). Cells were harvested 48h and 72h post-transfection.
Western Blot
Cells were harvested by addition of RIPA buffer. Protein concentration was determined by BCA assay (Thermo, Rockford, IL). Western blots were performed in 30μg (A/L) or 2.5μg (P) of total protein with the following: an SP-A antibody that recognizes SP-A1 and SP-A2 (14), pro-SP-B and pro-SP-C antibodies (Seven Hills bioreagents, Cincinnati, OH), Drosha (Abcam, Cambridge, MA), and GAPDH (Millipore, Billerica, MA).
Gene expression
Cells were harvested with Trizol (Life Technologies). Total RNA was purified with the Direct-zol™ RNA-MiniPrep-kit (Zymo Research, Irvine, CA). RNA concentration and quality were confirmed by nanodrop and Bioanalyzer. Expression of SP-A1, SP-A2, SP-B, SP-C, SP-D, ABCA3, CAV1, PDPN, RAGE and Drosha was detected by Real Time PCR with TaqMan assays (Life Technologies). Results were analyzed by relative quantification and normalized to 18s (39).
miRNA PCR arrays
miRNAs were purified from A/L and P cultured cells with the miRVana miRNA isolation kit (Life Technologies). Expression of 1,066 miRNAs was determined with the Human miRNome miRNA PCR Array (QIAGEN, Valencia, CA) in ATI and ATII cells (n=3). Data analysis was performed with the manufacturer's software and the TAM software (40). Prediction analysis for miRNA binding to SP-A1 and SP-A2 3′UTRs was performed by TargetScan.
Results
The phenotype of ATII cells is maintained in air-liquid interface (A/L) cultures, but not in cells cultured in plastic (P)
Human ATII cells were obtained from one lung transplant donor, as described previously (22, 33). The cell purity was confirmed by Papanicolaou staining, and by ultrastructural visualization of lamellar bodies (Supplementary Figure 1). Cells were plated in cell inserts covered with a mix of matrigel/rat tail collagen (air-liquid interface, A/L), or directly in wells without matrix (P), and maintained in culture for five days, as depicted on Figure 1A. Cells were harvested daily, and the expression of surfactant protein A (SP-A), and precursors of surfactant proteins B, and C (proSP-B, pro-SPC) was monitored by Western blotting (Figure 1B). Cells cultured in A/L showed increased levels of all three proteins over time in cells cultured in A/L, reaching maximum levels on day 4 and 5, consistent with ATII phenotype. On the other hand, cells cultured in P showed a decrease in SP-A expression over time. ProSP-B levels decreased on D2 and D3 vs. D1, and slightly increased following addition of dexamethasone (Dex) to the culture media. We did not observe significant changes in proSP-C during D1-D3, however a slight increase was observed on D4, followed by a decrease on D5 (Figure 1B).
Figure 1. Expression of surfactant proteins in primary ATII cell cultures.
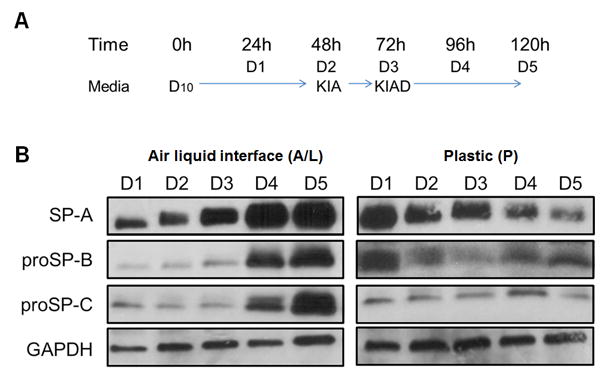
A. Human ATII cells were cultured in air-liquid interface (A/L) or plastic (P) in DMEM-based media for five days, as described in materials and methods, and harvested every 24h. B. Expression of surfactant proteins by Western Blot. Cells cultured in A/L expressed higher levels of SP-A, proSP-B, and proSP-C at 96h, and 120h. Cells cultured in P showed a reduction of SP-A and pro-SP-B expression, but no changes in proSP-C. GAPDH was used for normalization. D10: DMEM + 10% FBS; KIA: D10 + KGF + IBMX + 8-Br-cAMP; KIAD: KIA+Dex.
At the mRNA level, cells cultured in A/L had significantly higher expression of SP-A1 on D2 compared to D1, and these levels were maintained over time (Supplementary Figure 2). SP-A2 expression, however, only showed a significant increase on D4 and D5, i.e. after addition of Dex. These results are in support of previous findings on differential regulation of the two SP-A genes (41-43). Expression of SFTPB (SP-B), SFTPC (SP-C) and SFTPD (SP-D) mRNAs was also significantly higher on D4 and D5 in A/L cultures. On the other hand, ATII cells cultured in P showed a significant decrease in the mRNA expression of all surfactant protein genes over time, and these were not affected by the addition of Dex (Supplementary Figure 2). Together, these results indicate that ATII cells are able to maintain the phenotype over time when cultured in A/L, as shown previously (34, 36), and represent a suitable experimental model for the study of surfactant protein expression.
ATII cells cultured in P trans-differentiate to ATI cells
We next analyzed the expression of known ATII and ATI molecular markers in order to confirm the cell phenotype in both culture conditions. For this, we measured the mRNA levels of one ATII cell marker, the ATP-binding cassette 3 sub-family A, member 3 (ABCA3), and three ATI specific markers: caveolin-1 (CAV1), T1-α/podoplanin (PDPN), and the Receptor for Advanced Glycation End-products (RAGE) in cells cultured in A/L or P. Expression of ABCA3 was significantly higher on D2 and maintained over time in A/L cultures (Supplementary Figure 3). The levels of the ATI specific markers were decreased over time, indicating a selection of ATII cells occurs under these conditions. However, in cells cultured in P, ABCA3 mRNA significantly decreased over time, as previously observed for the expression of surfactant proteins (Supplementary Figures 2 and 3).
Previous studies have reported trans-differentiation of cultured ATII to ATI cells (23, 44). As expected, in cells cultured in P, a significant increase in the mRNA levels of CAV1 and PDPN was observed on D4 and D5, and RAGE expression was significantly higher on D3, D4, and D5. Together, these results indicate that, in our model, cells maintained on A/L for 5 days may represent primarily an ATII population, whereas P cultures may represent predominantly ATI cells, and/or cell intermediates (i.e. transitioning from ATII to ATI cells).
Knockdown of Drosha in ATII cells affects SP-A mRNA and protein levels
Following characterization of the culture, we inhibited the expression of Drosha, a nuclear ribonuclease critical for miRNA maturation, by transfection of ATII cells cultured in A/L, with stealth siRNAs. Efficient delivery of siRNAs was confirmed by microscopic visualization of a fluorescent oligonucleotide (Figure 2A), and knockdown of Drosha was confirmed by Real Time PCR analysis and Western Blot at 48h and 72h post-transfection (Figure 2B). We found that Drosha mRNA levels were reduced significantly (50% and 70%, respectively) when compared with levels of cells transfected with a control siRNA. Similarly, Drosha protein expression was reduced at 72h following siRNA transfection (Figure 2C).
Figure 2. Drosha knockdown.
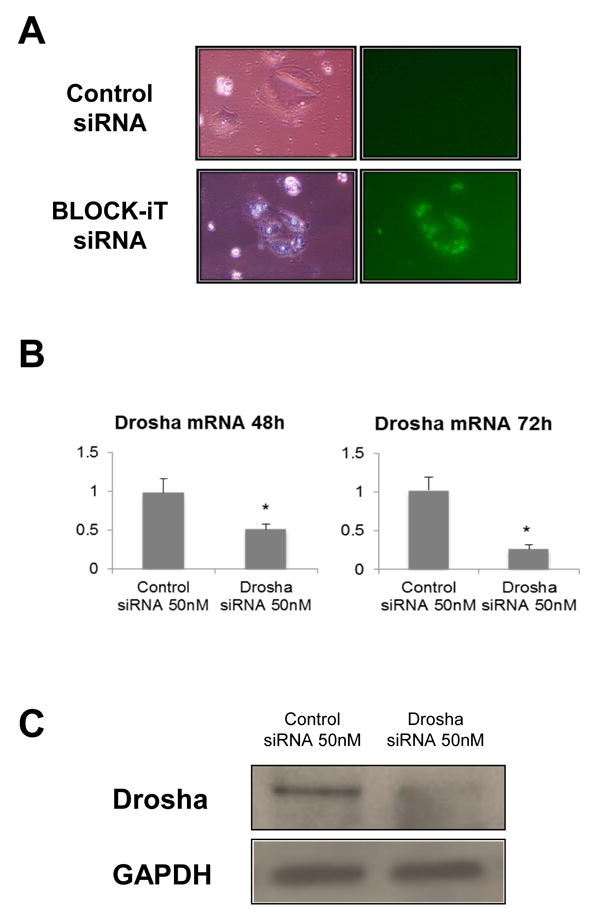
A. ATII cells cultured in A/L were transfected with a control siRNA (upper panels) or a fluorescent oligonucleotide (lower panels). Fluorescence microscopy confirmed absence of background fluorescence (upper right panel), and intracellular localization of siRNA (lower right panel). B. Knockdown of Drosha was confirmed by Real Time PCR. The results shown are normalized to 18s. A 50% and 70% reduction in Drosha mRNA was observed at 48h and 72h post-transfection, respectively (*p<0.05 vs. control siRNA, n=8). C. Protein levels of Drosha in ATII cells 72h post-transfection with Drosha and control siRNAs.
In order to analyze the effects of Drosha knockdown on SP-A expression, we transfected ATII cells on D2 in A/L, and cells were harvested at 48h and 72h post-transfection (D4 and D5) (Figure 3A). We measured the expression of SP-A1 and SP-A2 mRNA levels (Figure 3B) and total SP-A protein (Figure 3C) by Real Time PCR and Western blot, respectively. We observed that 48h and 72h after transfection with Drosha siRNA, cells expressed significantly higher levels of SP-A1 and SP-A2 mRNAs, as well as total SP-A protein than cells transfected with the control siRNA. Similar results were observed for SP-A mRNA expression when cells were treated with siRNAs that target enzymes downstream of Drosha in the miRNA maturation pathway (Dicer and Argonaut-2) (Figure 4). Furthermore, expression of surfactant proteins B, C (mRNA and protein precursor levels) and D (mRNA) was also increased following knockdown of Drosha (Supplementary Figure 4).
Figure 3. Effect of Drosha knockdown on SP-A mRNA and protein levels.
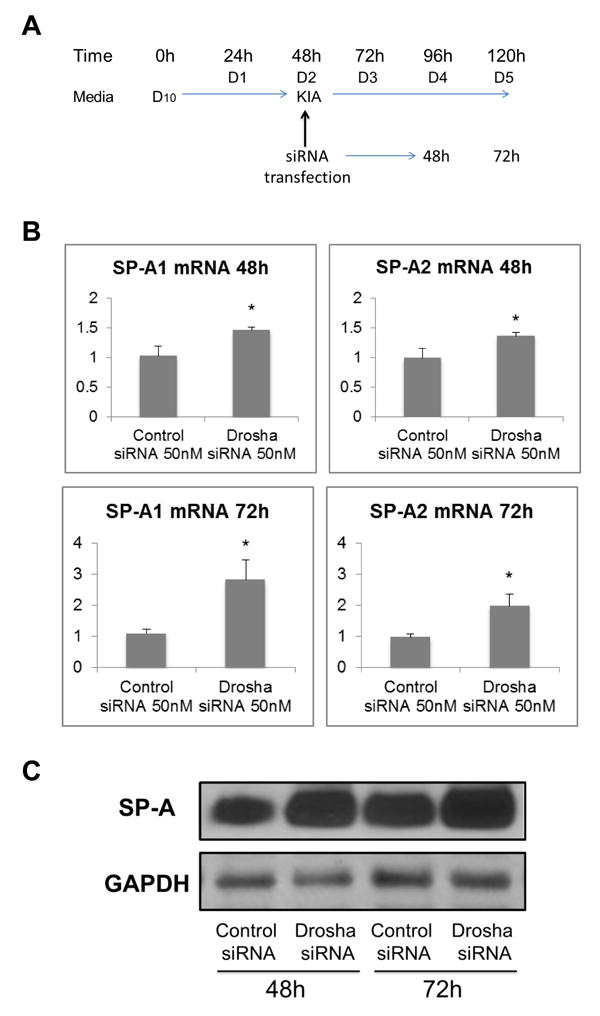
A. ATII cell transfection protocol and culture conditions. B. Relative mRNA levels of SP-A1 (left) and SP-A2 (right) measured by Real Time PCR at 48h and 72h after transfection with Drosha siRNA or control siRNA. The results shown are normalized to 18s. C. Total SP-A protein levels measured by Western Blot post-transfection. Both SP-A mRNA and protein levels were significantly higher in cells transfected with Drosha siRNA vs. control siRNA (*p<0.05, n=8). D10: DMEM + 10% FBS; KIA: D10 + KGF + IBMX + 8-Br-cAMP.
Figure 4. SP-A1 and SP-A2 mRNA expression following Dicer and Argonaut-2 knockdown in ATII cells.
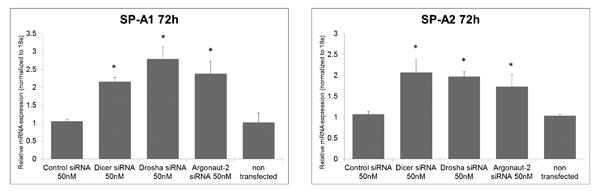
Relative mRNA levels of SP-A1 (left) and SP-A2 (right) measured by Real Time PCR at 72h post transfection with Dicer, Drosha, Argonaut-2 siRNA and control siRNA, and untreated cells. The results shown are normalized to 18s.
MicroRNAs are differentially expressed in human ATII and ATI cells
Next, in order to characterize the miRNA content of ATII and ATI cells in our model, we harvested cells cultured on A/L and P on D5, and extracted the small RNA fraction. By using the Human miRNome miRNA PCR Array (QIAGEN), we characterized the expression of 1,066 human miRNAs and compared differences in expression between the two cell models. Expression of 451 miRNAs was detected in ATII cells, and of 615 miRNAs in ATI cells (Supplementary Tables 1 and 2). We have also identified distinct miRNA expression profiles for ATII and ATI cells, with higher expression of miRNAs in ATI cells (Figure 5), as well as miRNA signatures for the two cell types: 37 miRNAs only expressed in ATII cells, and 180 miRNAs exclusively expressed in ATI cells (Table 1). Prediction target analysis using the online tool TargetScan showed that eleven miRNAs exclusively expressed in ATII cells were predicted to bind SP-A transcripts (3 for SP-A1, 3 for SP-A2, and 5 for both genes), and 52 miRNAs expressed only in ATI cells had predicted binding to SP-A 3′UTRs (14 for SP-A1, 13 for SP-A2, and 15 for both genes) (Table 2). Finally, quantitative analysis revealed that 24 of the miRNAs expressed in ATI and ATII cells had a significantly higher expression in ATI vs. ATII cells (p<0.05), and half of them were predicted to bind SP-A 3′UTR sequences (5 miRNAs for SP-A1, 4 miRNAs for SP-A2, and 2 miRNAs for both genes) (Table 3).
Figure 5. MicroRNA expression in ATII vs. ATI cells.
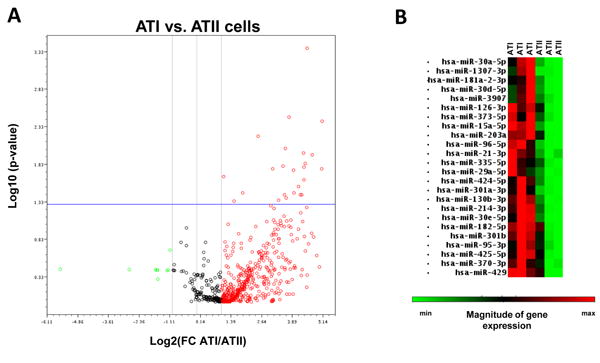
A. Volcano plot of 1,066 miRNAs expressed in ATI and ATII cells (n=3). Red dots indicate miRNAs up-regulated in ATI vs. ATII cells. Green dots represent down-regulated miRNAs in ATI vs. ATII cells. Twenty-four miRNAs were significantly up-regulated in ATI cells vs. ATII (upper right) (p<0.05). B. Heatmap of the 24 differentially expressed miRNAs. Each column represents an independent sample. Data were obtained with the miScript miRNA PCR array data analysis online tool (http://pcrdataanalysis.sabiosciences.com).
Table 1. MiRNA expression in human ATII and ATI cells.
| miRNAs expressed in ATII only | miRNAs expressed in ATI only | |||
|---|---|---|---|---|
| hsa-miR-1182 | hsa-let-7f-1* | hsa-miR-196a | hsa-miR-374c | hsa-miR-542-5p |
| hsa-miR-1184 | hsa-let-7f-2* | hsa-miR-1976 | hsa-miR-376b | hsa-miR-543 |
| hsa-miR-1256 | hsa-miR-103a-2* | hsa-miR-199a-5p | hsa-miR-377 | hsa-miR-545* |
| hsa-miR-1288 | hsa-miR-1183 | hsa-miR-199b-5p | hsa-miR-378 | hsa-miR-548aa |
| hsa-miR-1297 | hsa-miR-1193 | hsa-miR-204 | hsa-miR-378* | hsa-miR-548d-5p |
| hsa-miR-1304 | hsa-miR-1204 | hsa-miR-2113 | hsa-miR-378b | hsa-miR-548e |
| hsa-miR-147 | hsa-miR-1227 | hsa-miR-2116* | hsa-miR-379 | hsa-miR-548h |
| hsa-miR-19b-1* | hsa-miR-1228* | hsa-miR-212 | hsa-miR-380 | hsa-miR-548s |
| hsa-miR-19b-2* | hsa-miR-1236 | hsa-miR-2355-3p | hsa-miR-3914 | hsa-miR-548v |
| hsa-miR-2115* | hsa-miR-124 | hsa-miR-296-3p | hsa-miR-3922-3p | hsa-miR-548w |
| hsa-miR-2117 | hsa-miR-1255b | hsa-miR-299-5p | hsa-miR-3934 | hsa-miR-548y |
| hsa-miR-216a | hsa-miR-125b-1* | hsa-miR-302c | hsa-miR-3938 | hsa-miR-550a |
| hsa-miR-219-1-3p | hsa-miR-125b-2* | hsa-miR-302e | hsa-miR-411 | hsa-miR-550a* |
| hsa-miR-300 | hsa-miR-1261 | hsa-miR-3065-3p | hsa-miR-4252 | hsa-miR-551a |
| hsa-miR-302a* | hsa-miR-1262 | hsa-miR-3120-3p | hsa-miR-4253 | hsa-miR-551b |
| hsa-miR-302b* | hsa-miR-1265 | hsa-miR-3126-5p | hsa-miR-4258 | hsa-miR-556-5p |
| hsa-miR-3125 | hsa-miR-1267 | hsa-miR-3127-5p | hsa-miR-4267 | hsa-miR-566 |
| hsa-miR-3130-5p | hsa-miR-1269 | hsa-miR-3130-3p | hsa-miR-4272 | hsa-miR-577 |
| hsa-miR-3167 | hsa-miR-1271 | hsa-miR-3135 | hsa-miR-4273 | hsa-miR-586 |
| hsa-miR-3680 | hsa-miR-1273 | hsa-miR-3147 | hsa-miR-4276 | hsa-miR-590-5p |
| hsa-miR-3918 | hsa-miR-1277 | hsa-miR-3149 | hsa-miR-4280 | hsa-miR-592 |
| hsa-miR-3945 | hsa-miR-1283 | hsa-miR-3150a-3p | hsa-miR-4294 | hsa-miR-601 |
| hsa-miR-4251 | hsa-miR-1289 | hsa-miR-3150b-3p | hsa-miR-4300 | hsa-miR-613 |
| hsa-miR-4255 | hsa-miR-129-3p | hsa-miR-3157-5p | hsa-miR-4304 | hsa-miR-617 |
| hsa-miR-4279 | hsa-miR-129-5p | hsa-miR-3174 | hsa-miR-4314 | hsa-miR-622 |
| hsa-miR-4283 | hsa-miR-1296 | hsa-miR-3186-5p | hsa-miR-4315 | hsa-miR-624* |
| hsa-miR-4297 | hsa-miR-1301 | hsa-miR-3191 | hsa-miR-4318 | hsa-miR-628-5p |
| hsa-miR-4317 | hsa-miR-1323 | hsa-miR-329 | hsa-miR-449a | hsa-miR-639 |
| hsa-miR-451 | hsa-miR-134 | hsa-miR-330-5p | hsa-miR-450a | hsa-miR-642b |
| hsa-miR-573 | hsa-miR-136 | hsa-miR-331-5p | hsa-miR-452 | hsa-miR-643 |
| hsa-miR-610 | hsa-miR-136* | hsa-miR-337-5p | hsa-miR-455-5p | hsa-miR-645 |
| hsa-miR-616 | hsa-miR-137 | hsa-miR-339-3p | hsa-miR-483-3p | hsa-miR-653 |
| hsa-miR-621 | hsa-miR-138 | hsa-miR-3605-3p | hsa-miR-485-3p | hsa-miR-662 |
| hsa-miR-675 | hsa-miR-138-1* | hsa-miR-363 | hsa-miR-486-5p | hsa-miR-668 |
| hsa-miR-92a-2* | hsa-miR-142-5p | hsa-miR-363* | hsa-miR-487a | hsa-miR-7-1* |
| hsa-miR-937 | hsa-miR-1468 | hsa-miR-3652 | hsa-miR-488* | hsa-miR-764 |
| hsa-miR-943 | hsa-miR-147b | hsa-miR-3654 | hsa-miR-492 | hsa-miR-765 |
| hsa-miR-149 | hsa-miR-3655 | hsa-miR-495 | hsa-miR-767-3p | |
| hsa-miR-153 | hsa-miR-3661 | hsa-miR-497 | hsa-miR-875-3p | |
| hsa-miR-16-1* | hsa-miR-3666 | hsa-miR-499-5p | hsa-miR-9 | |
| hsa-miR-187 | hsa-miR-3667-3p | hsa-miR-506 | hsa-miR-920 | |
| hsa-miR-187* | hsa-miR-3671 | hsa-miR-510 | hsa-miR-924 | |
| hsa-miR-190b | hsa-miR-3678-5p | hsa-miR-511 | hsa-miR-934 | |
| hsa-miR-1914 | hsa-miR-369-3p | hsa-miR-512-3p | hsa-miR-935 | |
| hsa-miR-193a-3p | hsa-miR-3691-5p | hsa-miR-514 | hsa-miR-941 | |
Table 2. MiRNAs predicted to bind SP-A1 and SP-A2.
| miRNAs expressed in ATII | miRNAs expressed in ATI |
|---|---|
|
| |
| SP-A1: | SP-A1: |
| hsa-miR-302a* | hsa-miR-212 |
| hsa-miR-302b* | hsa-miR-302c |
| hsa-miR-3125 | hsa-miR-302e |
| hsa-miR-3130-3p | |
| hsa-miR-3135 | |
| hsa-miR-330-5p | |
| hsa-miR-3922-3p | |
| hsa-miR-3938 | |
| hsa-miR-4252 | |
| hsa-miR-4314 | |
| hsa-miR-548s | |
| hsa-miR-645 | |
| hsa-miR-668 | |
| hsa-miR-875-3p | |
| SP-A2: | SP-A2: |
| hsa-miR-1184 | hsa-miR-1236 |
| hsa-miR-4283 | hsa-miR-296-3p |
| hsa-miR-621 | hsa-miR-337-5p |
| hsa-miR-4267 | |
| hsa-miR-4300 | |
| hsa-miR-4318 | |
| hsa-miR-449a | |
| hsa-miR-455-5p | |
| hsa-miR-483-3p | |
| hsa-miR-642b | |
| hsa-miR-765 | |
| hsa-miR-767-3p | |
| hsa-miR-920 | |
| SP-A1 and SP-A2: | SP-A1 and SP-A2: |
| hsa-miR-147 | hsa-miR-1193 |
| hsa-miR-219-1-3p | hsa-miR-1262 |
| hsa-miR-3918 | hsa-miR-1267 |
| hsa-miR-573 | hsa-miR-1323 |
| hsa-miR-616 | hsa-miR-134 |
| hsa-miR-3120-3p | |
| hsa-miR-3127-5p | |
| hsa-miR-3652 | |
| hsa-miR-3654 | |
| hsa-miR-3661 | |
| hsa-miR-369-3p | |
| hsa-miR-4276 | |
| hsa-miR-488* | |
| hsa-miR-512-3p | |
| hsa-miR-653 | |
Table 3. Differences in miRNA expression in ATI vs. ATII cells.
| miRNA | Fold change | p-value |
|---|---|---|
| hsa-miR-126-3p c | 15.3 | 0.018 |
| hsa-miR-1307-3p | 8.54 | 0.017 |
| hsa-miR-130b-3p | 13.48 | 0.003 |
| hsa-miR-15a-5p | 22.59 | 0.0004 |
| hsa-miR-181a-2-3p a | 20.77 | 0.024 |
| hsa-miR-182-5p a | 18.55 | 0.037 |
| hsa-miR-203a | 26.16 | 0.011 |
| hsa-miR-21-3p | 5.62 | 0.006 |
| hsa-miR-214-3p b | 12.22 | 0.009 |
| hsa-miR-29a-5p | 15.47 | 0.036 |
| hsa-miR-301a-3p | 20.67 | 0.016 |
| hsa-miR-301b | 11.39 | 0.016 |
| hsa-miR-30a-5p b | 20.36 | 0.01 |
| hsa-miR-30d-5p b | 19.42 | 0.038 |
| hsa-miR-30e-5p b | 21.41 | 0.014 |
| hsa-miR-335-5p | 11.85 | 0.038 |
| hsa-miR-370-3p a | 3.66 | 0.035 |
| hsa-miR-373-5p a | 2.13 | 0.022 |
| hsa-miR-3907 a | 2.87 | 0.046 |
| hsa-miR-424-5p | 34.03 | 0.017 |
| hsa-miR-425-5p | 12.53 | 0.041 |
| hsa-miR-429 c | 17.78 | 0.029 |
| hsa-miR-95-3p | 10 | 0.044 |
| hsa-miR-96-5p | 34.54 | 0.004 |
targets SP-A1,
targets SP-A2,
targets both SP-A1 and SP-A2
Discussion
Alveolar type II cells synthesize and secrete surfactant, a lipoprotein complex essential for life. Surfactant protein A, the most abundant protein of this complex, plays an important role in innate immunity, and its function appears to be dependent on the relative content of two gene products (SP-A1 and SP-A2) (14, 15, 45). Using in vitro models, we have previously identified several regulatory mechanisms that control SP-A1 and SP-A2 expression. Here, we have characterized a physiologically relevant model to study and validate important findings on SP-A regulation, with emphasis on miRNA regulatory pathways. In addition, we characterized ATII cells cultured in two different conditions, by measuring expression of surfactant proteins, ATII and ATI cell markers, and miRNAs. We identified differentially expressed miRNAs in ATI vs. ATII cells that could potentially serve as cell markers of ATI and ATII cells.
Primary culture of human ATII cells represents a powerful tool that can be used for the study of SP-A expression, and/or to confirm key findings obtained from the current available models that include animal fetal lung explants, lung adenocarcinoma cell lines, and stably transfected cell lines (16, 17, 20, 41, 46-48). Our goal here was to develop a model that will allow the study of the regulation of human SP-A variants in a physiologically relevant system (i.e. in a normal, non cancerous cell model where SP-A is naturally expressed). We used a combination of published protocols and techniques to obtain ATII cells from a donor lung, and tested two cell culture conditions that resulted in two distinct phenotypes after 5 days. In A/L cultures, addition of keratinocyte growth factor, isubutylmethylxanthine, and 8-Br-cAMP resulted in increased levels of total SP-A. Media supplementation with Dex, on the other hand, significantly increased mRNA and protein levels of all surfactant proteins. These changes were not observed in cells cultured in the absence of matrix (P). These results were not unexpected, as both matrigel (primarily composed of Engelbreth-Holm-Swarm tumor matrix), and rat tail collagen have been shown to stimulate synthesis and secretion of surfactant phospholipids and maintainance of SPA expression in cultured ATII cells (22, 49, 50).
Trans-differentiation of ATII to ATI was previously reported in murine cell models, as a spontaneous process that occurs in culture (44, 51). Currently, the mechanisms involved in this process are unknown, although recent studies have identified a role of TGF-β, and bone morphogenic protein (BPM) signaling pathways in the control of the trans-differentiation rate (44). In the present study, we have shown that ATII cells cultured for 5 days in plastic wells are able to trans-differentiate to ATI, as indicated by surfactant protein expression and three ATI specific markers, and cell morphology consistent with the ATI phenotype. In addition, we have shown differences in the expression of miRNAs in ATII and ATI cells (Table 1), indicating that miRNAs could play a role in the trans-differentiation process, by affecting the regulation of multiple genes, as it has been previously shown for bronchial epithelial cell differentiation (52). It is also possible that the differential miRNA composition of ATI and ATII cells, as well as the differential expression rates for the 24 miRNAs identified (Table 3) may represent a novel molecular marker for identification of these two distinct cell phenotypes. Moreover, given the fact that a number of miRNAs that were highly expressed in ATI vs. ATII cells were predicted to bind SP-A 3′UTRs, it is possible that these pay a role in the downregulation of SP-A1 and SP-A2 in the ATI phenotype.
MicroRNA biosynthesis is a well-regulated event that involves multiple processing steps facilitated by a number of enzymes. The nuclear protein Drosha is a key regulator of this process, as its cleavage of miRNA precursors allows them to enter the cytosol and continue the miRNA biogenesis process. Therefore, by depleting Drosha from ATII cells, one can decrease the miRNA biogenesis rate, and thus minimize the effects of mature miRNAs in the cell. In the current study, we successfully inhibited the expression of Drosha by using a siRNA-mediated approach in ATII cells maintained in A/L, and were able to show for the first time that a) ATII cells can be efficiently transfected in cell monolayers; b) knock-down of Drosha results in significantly higher mRNA and protein levels of surfactant proteins, indicating that miRNAs are involved (directly or indirectly) in the regulation of surfactant protein expression, and c) alveolar epithelial type I and II cells differentially express miRNAs predicted to regulate the expression of SP-A genes. Future research is needed to confirm the mechanisms by which miRNAs affect SP-A translation and mRNA stability.
MicroRNAs are powerful regulators of gene expression, as they have the ability of controlling multiple targets simultaneously, and affect various cellular functions and biological processes, including cell differentiation in various tissues (53-56). In the present study, the miRNA profile of ATII cells was significantly different from that of ATI cells after 5 days in culture (Figure 5). Qualitative analysis revealed 37 ATII-specific miRNAs and 180 specific for ATI cells (Table 1), and differential expression of 24 miRNAs in ATI vs. ATII (Table 3). Together, these indicate a role of miRNAs in ATII and ATI phenotypes. In order to determine some of the regulatory pathways that may be under the control of these miRNAs, we used the online Tool for Annotations of miRNAs (TAM) (40) to evaluate associations of differentially expressed miRNAs with various biological functions. Among the miRNAs with exclusive expression in ATII cells, two (hsa-mir-92a, hsa-mir-19b) belong to the hsa-mir-17-92 family, previously shown to inhibit lung epithelial cell differentiation (57). Furthermore, fourteen miRNAs with exclusive expression in ATI cells are members of the hsa-mir-1185 cluster (hsa-mir-134, hsa-mir-299-5p, hsa-mir-329, hsa-mir-369-3p, hsa-mir-376b, hsa-mir-377, hsa-mir-379, hsa-mir-380, hsa-mir-411, hsa-mir-485-3p, hsa-mir-487a, hsa-mir-495, hsa-mir-543, hsa-mir-668), indicating that they are likely to share common expression patterns, regulation of transcription and processing of pri-miRNA precursors, and regulatory functions (58, 59). However, the underlying mechanisms for these putative functions are currently unknown. Finally, of the 24 commonly expressed miRNAs with differential expression in ATI and ATII, five were predicted to bind SP-A1 3′UTRs (hsa-mir-181a-2-3p, hsa-mir-182-5p, hsa-mir-370-5p, hsa-mir-373-5p, hsa-mir-3907), four were predicted to bind SP-A2 3′UTRs (hsa-mir-214-3p, hsa-mir-30a-5p, hsa-mir-30d-5p, hsa-mir-30e-5p), and two (hsa-mir-126-3p, hsa-mir-429) were predicted to bind regulatory sequences of both SP-A1 and SP-A2 genes. These observations indicate that an increase in the expression of these miRNAs in ATI cells may contribute to the inhibition of expression of SP-A1 and SP-A2 in cells cultured in plastic. Furthermore, TAM analysis of the 24 differentially expressed miRNAs in Table 3 revealed that two (hsa-mir-15a-5p, hsa-mir-424-5p), and three (hsa-mir-15a-5p, hsa-mir-21-3p, hsa-mir-373-5p) were associated with cell differentiation and cell proliferation, respectively. Seven miRNAs (hsa-mir-21-3p hsa-mir-29a-5p, hsa-mir-30a-5p, hsa-mir-30d-5p, hsa-mir-30e-5p, hsa-mir-370-3p, and hsa-mir-429) were associated with epithelial-mesenchymal transition, and two (hsa-mir-96-5p, hsa-mir-182-5p) with cell fate determination. This analysis also identified miRNAs related to apoptosis (hsa-mir-15a-5p, hsa-mir-21-3p, hsa-mir-29a-5p, hsa-mir-96-5p, hsa-mir-181a-2-3p, hsa-mir-182-5p), and cell death (hsa-mir-21-3p, hsa-mir-30d-5p, hsa-mir-130b-3p, hsa-mir-181a-2-3p, hsa-mir-182-5p). Together, these predictions indicate that multiple biological pathways may be contributing to the ATII to ATI differentiation process observed in our model, and miRNAs may play an important role in the control of these functions, by regulation of specific gene expression. Further analyses on miRNA-mRNA pathways, as well as functional analysis of miRNA-gene interactions may reveal specific regulatory mechanisms involved in the control of alveolar cell differentiation, and determine whether these miRNAs could be used as biomarkers for alveolar cell differentiation in vivo. A limitation of the present study is that the alveolar epithelial cells studied were derived from a single lung of a young donor, with a specific SP-A1 and SP-A2 genotype. It is possible that lungs from older donors and different SP-A1 and SP-A2 genotypes may behave slightly different in terms of the parameters measured here.
In summary, we characterized isolated human alveolar cells in culture under two different conditions, and we studied the role of miRNAs in SP-A gene expression, and trans differentiation of ATII to ATI cells cultured in vitro. Our results provide novel data on the miRNA profile of ATII and ATI cells, and the potential role of miRNAs as master regulators of mediating both differentiation of alveolar epithelial cells, as well as expression of SP-A1 and SP-A2 genes during trans-differentiation.
Supplementary Material
Supplementary Figure 1. ATII cells cultured in air liquid interface (A/L). Visualization of Lamellar Bodies on cultured Type II cells by Electron Microscopy.
Supplementary Figure 2. mRNA levels of surfactant proteins in ATII cells cultured in air liquid interface (A/L) or plastic (P). Expression of SP-A1, SP-A2, SP-B, SP-C, and SP-D mRNAs was measured by Real Time PCR and normalized to 18s. Results are expressed as fold changes versus D1 expression. Cells cultured in A/L (left panel) showed higher mRNA levels of surfactant proteins on D4 and D5 (*p<0.05 vs. D1). Surfactant protein mRNA levels in cells cultured in P (right panel) were significantly lower after 48h in culture (*p<0.05 vs. D1, n=8).
Supplementary Figure 3. Expression of ATII and ATI cell markers in cells cultured in air-liquid interface (A/L) or plastic (P). Daily mRNA levels of ABCA3 (ATII cell marker), and CAV1, PDPN, and RAGE (ATI markers) in cells cultured in A/L or P, measured by Real Time PCR and normalized to 18s. Results are shown as fold changes in expression vs. D1. Expression of the ATII marker increased over time in cells cultures in A/L, and decreased in P cultures (*p<0.05 vs.D1). Expression of ATI markers was significantly higher in cells cultured in P, and significantly lower in A/L cultures on D4 and D5 (*p<0.05 vs.D1). ABCA3: ATP-binding cassette, sub-family A, member 3; CAV1: caveolin-1; PDPN: T1-α /podoplanin; RAGE: Receptor for Advanced Glycation End-products (n=8).
Supplementary Figure 4. Expression of SP-B, SP-C, and SP-D following knockdown of Drosha in ATII cells. Left: Relative mRNA levels of SP-B, SP-C, and SP-D measured by Real Time PCR at 72h after transfection with Drosha siRNA or control siRNA. Right: Expression of surfactant proteins and GAPDH levels post-transfection, measured by Western Blot. Cells transfected with Drosha siRNA expressed higher levels of SP-B, SP-C and SP-D mRNA, and proSP-B and proSP-C proteins 72h post transfection (n=6, p<0.05).
Acknowledgments
Funding: This work was supported by grants from NIH (HL-34788, JF), Children's Miracle Network (PS), and Sigma Delta Epsilon-Graduate Women in Science (Adele Lewis Grant Fellowship, PS). Dr. Silveyra's research is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under BIRCWH award number K12HD055882, “Career Development Program in Women's Health Research at Penn State.” The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors thank the Pennsylvania State University College of Medicine core facility for Real Time PCR services and Dr. Robert Mason for technical advice on ATII cell purification and culture.
Footnotes
Declaration of Interests: The authors report no conflicts of interest.
References
- 1.Khubchandani KR, Snyder JM. Surfactant protein A (SP-A): the alveolus and beyond. FASEB J. 2001 Jan;15(1):59–69. doi: 10.1096/fj.00-0318rev. [DOI] [PubMed] [Google Scholar]
- 2.McElroy MC, Kasper M. The use of alveolar epithelial type I cell-selective markers to investigate lung injury and repair. Eur Respir J. 2004 Oct;24(4):664–73. doi: 10.1183/09031936.04.00096003. [DOI] [PubMed] [Google Scholar]
- 3.Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci. 2012;17:407–29. doi: 10.2741/3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function. Crit Rev Eukaryot Gene Expr. 2009;19(2):125–37. doi: 10.1615/critreveukargeneexpr.v19.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, et al. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008 Jan;294(1):L121–30. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, et al. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. 2007 Mar;75(3):1403–12. doi: 10.1128/IAI.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikerov AN, Umstead TM, Huang W, Liu W, Phelps D, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2005;288:L150–L8. doi: 10.1152/ajplung.00135.2004. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Umstead T, Phelps D, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. 2002 Jan;110(1):79–84. doi: 10.1289/ehp.0211079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Bates-Kenney S, Tao J, Phelps D, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. 2004 Apr;43(14):4227–39. doi: 10.1021/bi036023i. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Taneva S, Keough K, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. 2007 Sep;1768(9):2060–9. doi: 10.1016/j.bbamem.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. 2004 Mar;286(3):L546–53. doi: 10.1152/ajplung.00267.2003. Epub 2003/11/18. eng. [DOI] [PubMed] [Google Scholar]
- 12.García-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry. 2002 Nov;41(47):14041–53. doi: 10.1021/bi026540l. [DOI] [PubMed] [Google Scholar]
- 13.Wang G, Guo X, Diangelo S, Thomas N, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: Formation of tubular myelin in vivo requires both gene products. J Biol Chem. 2010 Jan; doi: 10.1074/jbc.M109.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, et al. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. 2007 May;292(5):L1052–63. doi: 10.1152/ajplung.00249.2006. Epub 2006/12/26. eng. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, et al. Surfactant Protein-A is Defective in Abrogating Inflammation in Asthma. Am J Physiol Lung Cell Mol Physiol. 2011 Jul; doi: 10.1152/ajplung.00381.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silveyra P, Wang G, Floros J. Human SP-A1 (SFTPA1) variant-specific 3' UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene. Am J Physiol Lung Cell Mol Physiol. 2010 Oct;299(4):L523–34. doi: 10.1152/ajplung.00113.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silveyra P, Raval M, Simmons BP, Diangelo S, Wang G, Floros J. The Untranslated Exon B of Human Surfactant Protein A2 (SFTPA2) mRNAs is an Enhancer for Transcription and Translation. Am J Physiol Lung Cell Mol Physiol. 2011 Aug; doi: 10.1152/ajplung.00439.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5'-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005 Sep;289(3):L497–508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 19.Noutsios GT, Silveyra P, Bhatti F, Floros J. Exon B of human surfactant protein A2 mRNA, alone or within its surrounding sequences, interacts with 14-3-3; role of cis-elements and secondary structure. Am J Physiol Lung Cell Mol Physiol. 2013 Jun;304(11):L722–35. doi: 10.1152/ajplung.00324.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silveyra P, DiAngelo S, Floros J. Lung Cellular and Molecular Physiology. American Journal of Physiology; An 11-nt sequence polymorphism at the 3′UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in vitro. Accepted on 04/29/2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res. 2012 Sep;38(7):363–73. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 22.Ito Y, Mason RJ. The effect of interleukin-13 (IL-13) and interferon-γ (IFN-γ) on expression of surfactant proteins in adult human alveolar type II cells in vitro. Respir Res. 2010;11:157. doi: 10.1186/1465-9921-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Edeen K, Manzer R, Chang Y, Wang S, Chen X, et al. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am J Respir Cell Mol Biol. 2007 Jun;36(6):661–8. doi: 10.1165/rcmb.2006-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell L, Hollins AJ, Al-Eid A, Newman GR, von Ruhland C, Gumbleton M. Caveolin-1 expression and caveolae biogenesis during cell transdifferentiation in lung alveolar epithelial primary cultures. Biochem Biophys Res Commun. 1999 Sep;262(3):744–51. doi: 10.1006/bbrc.1999.1280. [DOI] [PubMed] [Google Scholar]
- 25.Hermans C, Bernard A. Lung epithelium-specific proteins: characteristics and potential applications as markers. Am J Respir Crit Care Med. 1999 Feb;159(2):646–78. doi: 10.1164/ajrccm.159.2.9806064. [DOI] [PubMed] [Google Scholar]
- 26.Shirasawa M, Fujiwara N, Hirabayashi S, Ohno H, Iida J, Makita K, et al. Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells. 2004 Feb;9(2):165–74. doi: 10.1111/j.1356-9597.2004.00712.x. [DOI] [PubMed] [Google Scholar]
- 27.Yee M, Buczynski BW, O'Reilly MA. Neonatal Hyperoxia Stimulates the Expansion of Alveolar Epithelial Type II Cells. Am J Respir Cell Mol Biol. 2013 Nov; doi: 10.1165/rcmb.2013-0207OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009 Jun;21(3):452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011 Mar;67(1):129–39. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 31.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007 Jun;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 32.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013 Apr;3(2):315–28. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, et al. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol. 2013 Jun;48(6):742–8. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Oberley-Deegan R, Wang S, Nikrad M, Funk CJ, Hartshorn KL, et al. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J Immunol. 2009 Feb;182(3):1296–304. doi: 10.4049/jimmunol.182.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996 Apr;14(4):309–15. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez SR, Travanty EA, Qian Z, Mason RJ. Human coronavirus HKU1 infection of primary human type II alveolar epithelial cells: cytopathic effects and innate immune response. PLoS One. 2013;8(7):e70129. doi: 10.1371/journal.pone.0070129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramin SM, Vidaeff AC, Gilstrap LC, Bishop KD, Jenkins GN, Alcorn JL. The effects of dexamethasone and betamethasone on surfactant protein-B messenger RNA expression in human type II pneumocytes and human lung adenocarcinoma cells. Am J Obstet Gynecol. 2004 Apr;190(4):952–9. doi: 10.1016/j.ajog.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Liley HG, White RT, Benson BJ, Ballard PL. Glucocorticoids both stimulate and inhibit production of pulmonary surfactant protein A in fetal human lung. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9096–100. doi: 10.1073/pnas.85.23.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karinch A, Deiter G, Ballard P, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta. 1998 Jun;1398(2):192–202. doi: 10.1016/s0167-4781(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 42.Kumar AR, Snyder JM. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am J Physiol. 1998;274(2 Pt 1):L177–85. doi: 10.1152/ajplung.1998.274.2.L177. [DOI] [PubMed] [Google Scholar]
- 43.McCormick SM, Mendelson CR. Human SP-A1 and SP-A2 genes are differentially regulated during development and by cAMP and glucocorticoids. Am J Physiol. 1994 Apr;266(4 Pt 1):L367–74. doi: 10.1152/ajplung.1994.266.4.L367. [DOI] [PubMed] [Google Scholar]
- 44.Zhao L, Yee M, O'Reilly MA. Transdifferentiation of alveolar epithelial type II to type I cells is controlled by opposing TGF-β and BMP signaling. Am J Physiol Lung Cell Mol Physiol. 2013 Sep;305(6):L409–18. doi: 10.1152/ajplung.00032.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Ovidio F, Kaneda H, Chaparro C, Mura M, Lederer D, Di Angelo S, et al. Pilot Study Exploring Lung Allograft Surfactant Protein A (SP-A) Expression in Association With Lung Transplant Outcome. Am J Transplant. 2013 Sep; doi: 10.1111/ajt.12407. [DOI] [PubMed] [Google Scholar]
- 46.Hoover RR, Floros J. SP-A 3'-UTR is involved in the glucocorticoid inhibition of human SP-A gene expression. Am J Physiol. 1999 Jun;276(6 Pt 1):L917–24. doi: 10.1152/ajplung.1999.276.6.L917. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Guo X, Floros J. Human SP-A 3'-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol. 2003 May;284(5):L738–48. doi: 10.1152/ajplung.00375.2002. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, Zhang JP, Huang H, Wang ZH, Cheng R, Cai WB. Leptin promotes fetal lung maturity and upregulates SP-A expression in pulmonary alveoli type-II epithelial cells involving TTF-1 activation. PLoS One. 2013;8(7):e69297. doi: 10.1371/journal.pone.0069297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice WR, Conkright JJ, Na CL, Ikegami M, Shannon JM, Weaver TE. Maintenance of the mouse type II cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol. 2002 Aug;283(2):L256–64. doi: 10.1152/ajplung.00302.2001. [DOI] [PubMed] [Google Scholar]
- 50.Demaio L, Tseng W, Balverde Z, Alvarez JR, Kim KJ, Kelley DG, et al. Characterization of mouse alveolar epithelial cell monolayers. Am J Physiol Lung Cell Mol Physiol. 2009 Jun;296(6):L1051–8. doi: 10.1152/ajplung.00021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhaskaran M, Kolliputi N, Wang Y, Gou D, Chintagari NR, Liu L. Trans-differentiation of alveolar epithelial type II cells to type I cells involves autocrine signaling by transforming growth factor beta 1 through the Smad pathway. J Biol Chem. 2007 Feb;282(6):3968–76. doi: 10.1074/jbc.M609060200. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Anton A, Sokolowska M, Kern S, Davis AS, Alsaaty S, Taubenberger JK, et al. Changes in microRNA and mRNA expression with differentiation of human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2013 Sep;49(3):384–95. doi: 10.1165/rcmb.2012-0368OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang G, Malmuthuge N, McFadden TB, Bao H, Griebel PJ, Stothard P, et al. Potential Regulatory Role of MicroRNAs in the Development of Bovine Gastrointestinal Tract during Early Life. PLoS One. 2014;9(3):e92592. doi: 10.1371/journal.pone.0092592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montagncer S, Deho L, Monticelli S. MicroRNAs in hematopoietic development. BMC Immunol. 2014 Mar;15(1):14. doi: 10.1186/1471-2172-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevanato L, Sinden JD. The effects of microRNAs on human neural stem cell differentiation in two- and three-dimensional cultures. Stem Cell Res Ther. 2014 Apr;5(2):49. doi: 10.1186/scrt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersson T, Rahman S, Sansom SN, Alsiö JM, Kaneda M, Smith J, et al. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One. 2010;5(10):e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007 Oct;310(2):442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, Zhao Y, Yang S, Zhang H, Chen F. Integrative Analysis of miRNA-mRNA and miRNA-miRNA Interactions. Biomed Res Int. 2014;2014:907420. doi: 10.1155/2014/907420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molina-Pinelo S, Pastor MD, Suarez R, Romero-Romero B, González De la Peña M, Salinas A, et al. MicroRNA clusters: dysregulation in lung adenocarcinoma and COPD. Eur Respir J. 2014 Apr; doi: 10.1183/09031936.00091513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. ATII cells cultured in air liquid interface (A/L). Visualization of Lamellar Bodies on cultured Type II cells by Electron Microscopy.
Supplementary Figure 2. mRNA levels of surfactant proteins in ATII cells cultured in air liquid interface (A/L) or plastic (P). Expression of SP-A1, SP-A2, SP-B, SP-C, and SP-D mRNAs was measured by Real Time PCR and normalized to 18s. Results are expressed as fold changes versus D1 expression. Cells cultured in A/L (left panel) showed higher mRNA levels of surfactant proteins on D4 and D5 (*p<0.05 vs. D1). Surfactant protein mRNA levels in cells cultured in P (right panel) were significantly lower after 48h in culture (*p<0.05 vs. D1, n=8).
Supplementary Figure 3. Expression of ATII and ATI cell markers in cells cultured in air-liquid interface (A/L) or plastic (P). Daily mRNA levels of ABCA3 (ATII cell marker), and CAV1, PDPN, and RAGE (ATI markers) in cells cultured in A/L or P, measured by Real Time PCR and normalized to 18s. Results are shown as fold changes in expression vs. D1. Expression of the ATII marker increased over time in cells cultures in A/L, and decreased in P cultures (*p<0.05 vs.D1). Expression of ATI markers was significantly higher in cells cultured in P, and significantly lower in A/L cultures on D4 and D5 (*p<0.05 vs.D1). ABCA3: ATP-binding cassette, sub-family A, member 3; CAV1: caveolin-1; PDPN: T1-α /podoplanin; RAGE: Receptor for Advanced Glycation End-products (n=8).
Supplementary Figure 4. Expression of SP-B, SP-C, and SP-D following knockdown of Drosha in ATII cells. Left: Relative mRNA levels of SP-B, SP-C, and SP-D measured by Real Time PCR at 72h after transfection with Drosha siRNA or control siRNA. Right: Expression of surfactant proteins and GAPDH levels post-transfection, measured by Western Blot. Cells transfected with Drosha siRNA expressed higher levels of SP-B, SP-C and SP-D mRNA, and proSP-B and proSP-C proteins 72h post transfection (n=6, p<0.05).


