Abstract
Embryonic heart formation requires the production of an appropriate number of cardiomyocytes; likewise, cardiac regeneration following injury relies upon the recovery of lost cardiomyocytes. The basic helix-loop-helix (bHLH) transcription factor Hand2 has been implicated in promoting cardiomyocyte formation. It is unclear, however, whether Hand2 plays an instructive or permissive role during this process. Here, we find that overexpression of hand2 in the early zebrafish embryo is able to enhance cardiomyocyte production, resulting in an enlarged heart with a striking increase in the size of the outflow tract. Our evidence indicates that these increases are dependent on the interactions of Hand2 in multimeric complexes and are independent of direct DNA binding by Hand2. Proliferation assays reveal that hand2 can impact cardiomyocyte production by promoting division of late-differentiating cardiac progenitors within the second heart field. Additionally, our data suggest that hand2 can influence cardiomyocyte production by altering the patterning of the anterior lateral plate mesoderm, potentially favoring formation of the first heart field at the expense of hematopoietic and vascular lineages. The potency of hand2 during embryonic cardiogenesis suggested that hand2 could also impact cardiac regeneration in adult zebrafish; indeed, we find that overexpression of hand2 can augment the regenerative proliferation of cardiomyocytes in response to injury. Together, our studies demonstrate that hand2 can drive cardiomyocyte production in multiple contexts and through multiple mechanisms. These results contribute to our understanding of the potential origins of congenital heart disease and inform future strategies in regenerative medicine.
Keywords: Hand2, Zebrafish, First heart field, Second heart field, Cardiac regeneration
INTRODUCTION
The assembly of the embryonic heart is a complex procedure involving the differentiation, migration and organization of the proper number of cardiomyocytes in order to form a functional contractile organ. Regulation of the genesis of cardiomyocytes requires the specification of heart fields with appropriate boundaries, as well as the controlled proliferation of the progenitor cells that emerge from these fields. An understanding of the genetic pathways that influence cardiomyocyte production may illuminate mutations that underlie congenital heart disease (CHD) (Fahed et al., 2013). Moreover, cardiomyocyte production, in vitro or in vivo, is a primary goal of cardiovascular regenerative medicine, and insight into genes that drive cardiomyocyte formation can enhance strategies for repairing hearts damaged by myocardial infarction (Laflamme and Murry, 2011; Choi and Poss, 2012; Xin et al., 2013). However, our understanding of the precise functions of factors that facilitate cardiomyocyte production remains incomplete.
The bHLH transcription factor Hand2 has been implicated as an important regulator of cardiomyocyte production. In mice, loss of Hand2 function results in hypoplasia of the right ventricle and outflow tract (Srivastava et al., 1997), suggesting that Hand2 promotes the development of cardiomyocytes derived from the second heart field (SHF). Furthermore, removal of Hand2 function from the SHF via tissue-specific deletion of a conditional allele interferes with survival of this progenitor population (Tsuchihashi et al., 2011). The effects of loss of Hand2 function are exacerbated when combined with either a conditional knockout or a hypomorphic allele of the related gene Hand1, suggesting functional redundancy between Hand2 and Hand1 that could mask earlier or broader roles of these factors in regulating cardiomyocyte production in mice (McFadden et al., 2005; Firulli et al., 2010).
In zebrafish, an early requirement for hand2 during cardiomyocyte production has been clearly demonstrated. Zebrafish hand2 mutants display a striking cardiac phenotype that features a dramatic deficit of cardiomyocytes (Yelon et al., 2000). This defect is evident from an early stage, as indicated by a substantial reduction in the number of cells that initiate expression of myocardial differentiation markers. Fate mapping has shown that hand2 expression demarcates the heart-forming region within the anterior lateral plate mesoderm (ALPM), and, in hand2 mutants, the progenitors residing in this region are ineffective at generating differentiated cardiomyocytes (Schoenebeck et al., 2007). Thus, hand2 is important for facilitating cardiomyocyte production. However, it is not clear whether hand2 regulates cardiomyocyte production by influencing cell fate decisions or by affecting proliferative capacity, nor is it known whether hand2 plays an instructive or a permissive role in this setting.
The possibility that Hand2 instructively directs cardiomyocyte production has been highlighted by recent studies in which forced expression of Hand2 was shown to increase the efficiency of reprogramming mammalian fibroblasts toward a myocardial fate (Song et al., 2012; Nam et al., 2013). Introduction of Hand2, Gata4, Mef2c and Tbx5 into neonatal mouse fibroblasts increased the frequency of reprogramming approximately three- to fourfold over that observed with introduction of only Gata4, Mef2c and Tbx5 (Song et al., 2012). Additionally, in human fibroblasts, Hand2 has been shown to be essential for initiation of cardiac contractile gene expression programs during reprogramming, whereas other factors, such as Gata4, Mef2c, Tbx5 and Myocd, play redundant roles in this regard (Nam et al., 2013). Thus, Hand2 can play a pivotal part in enhancing protocols for cardiac regeneration and repair; however, little is known about how forced expression of Hand2 mediates this role.
To gain new insight into the ability of Hand2 to drive cardiomyocyte production, we have examined the effects of hand2 overexpression on the embryonic and adult zebrafish heart. We find that overexpression of hand2 in the early embryo can increase cardiomyocyte numbers, with a particularly striking impact on the size of the cardiac outflow tract. Our data suggest that this cardiac enlargement results from increased progenitor proliferation within the SHF, as well as from increased cardiomyocyte specification within the first heart field (FHF). In addition, we find that overexpression of hand2 in the adult heart enhances myocardial proliferation following injury. Together, our data suggest that hand2 can play important and instructive roles in elevating cardiomyocyte production in multiple contexts: by promoting specification of FHF cardiomyocytes, by enhancing proliferation of SHF cardiac progenitors and by augmenting proliferation of regenerating cardiomyocytes. These results refine our understanding of the origins of CHD, particularly in the context of partial trisomy distal 4q, which involves duplication of HAND2 (Tamura et al., 2013). Moreover, our findings highlight ways in which hand2 overexpression could facilitate future approaches for cardiac regeneration and repair.
RESULTS
Overexpression of hand2 increases cardiomyocyte production
To determine whether increased hand2 function can enhance cardiomyocyte production, we injected zebrafish embryos with hand2 mRNA. Although we have previously injected hand2 mRNA into wild-type embryos (Yelon et al., 2000; Garavito-Aguilar et al., 2010), our prior studies did not closely examine its impact on heart size. Here, we found that injected embryos exhibited expanded expression of cmlc2, a marker of differentiated cardiomyocytes, within the ALPM at 18 somites (Fig. 1A,B). Measurements of the region of cmlc2 expression revealed a significant increase in area within the hand2-overexpressing embryos (Fig. 1E). To examine whether increased hand2 expression leads to enhanced heart size at later stages, we injected hand2 mRNA into embryos carrying Tg(-5.1myl7:nDsRed2) (Mably et al., 2003), which facilitates quantification of fluorescent cardiomyocyte nuclei, and counted the number of cardiomyocytes present in the heart at 36 h post-fertilization (hpf). We found a significant increase in cardiomyocyte number in embryos overexpressing hand2 (Fig. 1F). Together, these data suggest that increased expression of hand2 can expand the heart-forming region within the ALPM, leading to a larger heart. However, hand2 overexpression did not induce cmlc2 expression outside the ALPM, indicating that cardiomyocyte production depends on the interaction of Hand2 with other ALPM factors.
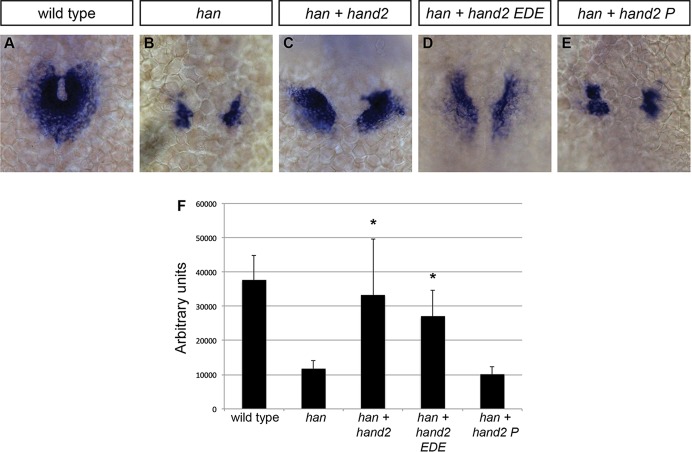
Fig. 1.
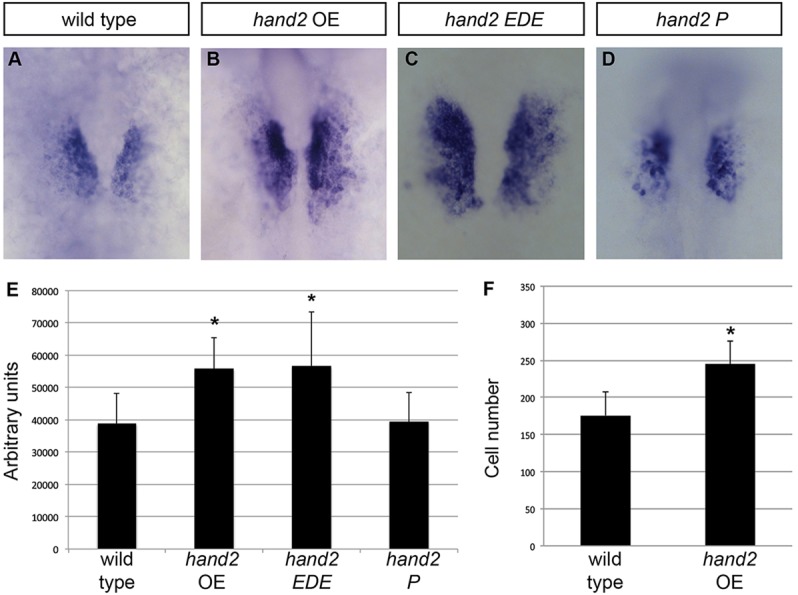
hand2 overexpression increases cardiomyocyte production. (A-D) In situ hybridization depicts cmlc2 expression at 18 somites in (A) wild-type embryos, (B) hand2-overexpressing embryos (hand2 OE), (C) embryos expressing a DNA binding-deficient form of hand2 (hand2 EDE) and (D) embryos expressing a dimerization-deficient form of hand2 (hand2 P); dorsal views, anterior upwards. Broader expression of cmlc2 is found within the ALPM of embryos overexpressing hand2 or hand2 EDE. (E) Average area of cmlc2 expression, in arbitrary units, in wild-type embryos and in embryos overexpressing hand2, hand2 EDE or hand2 P. Error bars indicate s.d.; asterisks indicate significant differences from wild type. Overexpression of hand2 or hand2 EDE increases area of cmlc2 expression (n=13-25; *P<0.001), whereas overexpression of hand2 P does not (n=11; P=0.87). (F) Bar graph compares average number of cardiomyocytes at 36 hpf in wild-type embryos and in embryos overexpressing hand2. Error bars indicate s.d.; asterisk indicates a significant increase compared with wild type (n=17-19; *P<0.001).
The influence of Hand2 on cardiomyocyte production is dependent on protein-protein interactions and not on direct DNA binding
Previous studies have shown that Hand2 can function independently of direct DNA binding in some contexts (McFadden et al., 2002; Liu et al., 2009). Mice in which the Hand2 gene was replaced with a DNA binding-deficient form of Hand2 exhibited relatively normal hearts at E11.5 (Liu et al., 2009), in contrast to the severe ventricular hypoplasia seen in Hand2 mutants by E10.5 (Srivastava et al., 1997). It is presumed that the DNA binding-deficient form of Hand2 can influence transcription through dimerization with other bHLH factors, as well as through interactions with larger protein complexes (Rychlik et al., 2003; Xu et al., 2003). To test whether the early role of hand2 during cardiomyocyte production is dependent on DNA binding or dimerization, we evaluated whether previously characterized DNA binding-deficient and dimerization-deficient versions of Hand2 can expand cmlc2 expression.
Replacement of three arginines (residues 109-111) in the basic domain of mouse Hand2 with acidic residues [glutamic acid, aspartic acid and glutamic acid (EDE)] has been shown to abolish DNA binding (McFadden et al., 2002; Liu et al., 2009). This region is highly conserved between mouse and zebrafish (supplementary material Fig. S1): 98% of the amino acids in the basic helix-loop-helix domain are identical, including these three arginine residues (100-102 in zebrafish). In addition, replacement of a phenylalanine (residue 119) with a proline in the first helix of mouse Hand2 has been shown to disrupt its dimerization (McFadden et al., 2002). This highly conserved amino acid is present in zebrafish Hand2 (F110; supplementary material Fig. S1). Extrapolating from prior work in mouse, we constructed corresponding variants of zebrafish hand2 and injected mRNA encoding each variant into wild-type embryos. Overexpression of the DNA binding-deficient form of hand2 (hand2 EDE) expanded the area of cmlc2 expression in a manner similar to the overexpression of wild-type hand2 (Fig. 1A-C,E). However, overexpression of the dimerization-deficient version of hand2 (hand2 P) did not expand the cmlc2 expression domain (Fig. 1A,D,E). Furthermore, injection of hand2 EDE mRNA substantially rescued cmlc2 expression in embryos homozygous for a null allele of hand2 (hans6) (Yelon et al., 2000), reminiscent of the effects of injecting wild-type hand2 into hans6 mutants (Fig. 2A-D,F). By contrast, injection of hand2 P mRNA did not alter cmlc2 expression in hans6 mutants (Fig. 2B,E,F). These results suggest that Hand2 dimerization, but not its direct binding to DNA, is necessary for its influence on cardiomyocyte production.
Fig. 2.
Influence of Hand2 on cardiomyocyte production is dependent on dimerization and not on direct DNA binding. (A-E) In situ hybridization depicts cmlc2 expression at 21 somites in (A) wild-type embryos, (B) hans6 mutant embryos, (C) hans6 mutant embryos injected with hand2 mRNA, (D) hans6 mutant embryos injected with hand2 EDE mRNA and (E) hans6 mutant embryos injected with hand2 P mRNA; dorsal views, anterior upwards. The hans6 mutation is a null allele of hand2 resulting from a deletion that removes the entire hand2 gene (Yelon et al., 2000). At this stage, wild-type cardiomyocytes have formed a ring (A), whereas few cardiomyocytes have formed in the hans6 mutants (B). Cardiomyocyte formation is substantially rescued with the injection of hand2 (C) or hand2 EDE (D) mRNA, but not of hand2 P mRNA (E). (F) Average area of cmlc2 expression, as in Fig. 1E. Asterisks indicate significant differences between the phenotype of hans6 mutant embryos and the phenotypes of hans6 mutant embryos overexpressing versions of hand2. Overexpression of hand2 or hand2 EDE partially rescues the area of cmlc2 expression in hans6 mutants (n=14; *P<0.001), whereas hand2 P does not (n=14; P=0.085).
Induction of hand2 overexpression after gastrulation expands cardiomyocyte production
In some embryos injected with hand2 mRNA, we observed morphological defects that could result from abnormal gastrulation, such as shortened body axis or abnormal body curvature. To bypass effects on gastrulation, we established an inducible system for hand2 overexpression. We constructed a series of transgenes in which hand2 is transcribed under the control of the hsp70 heat shock promoter (Halloran et al., 2000). In these transgenes, we connected Hand2 to mCherry using the viral 2A peptide sequence that allows separate stoichiometric translation of both proteins in order to facilitate monitoring of transgene expression after heat shock (Provost et al., 2007; Covassin et al., 2009).
In embryos carrying Tg(hsp70:FLAG-hand2-2A-mCherry) [hereafter referred to as Tg(hsp70:hand2)], induction of hand2 overexpression after gastrulation was sufficient to cause a cardiac phenotype similar to that caused by mRNA injection at the one-cell stage. After heat shock at 10 hpf (tail bud stage), transgenic embryos overexpressing hand2 exhibited a significant expansion in cmlc2 expression at 18 somites (Fig. 3A,B,E), as well as a significant increase in cardiomyocyte number at 36 hpf (Fig. 3F), compared with their heat-shocked nontransgenic siblings. Heat-shocked transgenic embryos also displayed a pronounced pericardial edema (supplementary material Fig. S2A,B) and an enlarged heart with a noticeably extended outflow tract (Fig. 3G,I). In contrast to the consequences of hand2 induction at 10 hpf, induction of hand2 overexpression at 24 hpf resulted in an embryo without evident pericardial edema (supplementary material Fig. S2C) or outflow tract enlargement (Fig. 3H). These data suggest that hand2 has a potent influence on cardiomyocyte production between 10 and 24 hpf, after gastrulation and before the initial assembly of the heart tube.
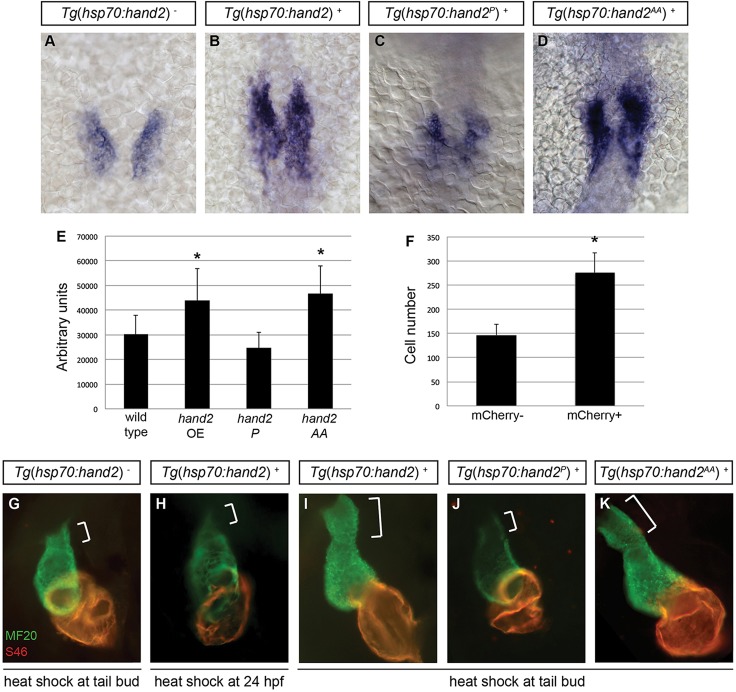
Fig. 3.
Cardiac expansion induced by hand2 overexpression requires phosphorylation-independent dimerization of Hand2. (A-D) In situ hybridization depicts cmlc2 expression at 18 somites in (A) nontransgenic embryos, (B) Tg(hsp70:hand2) embryos, (C) Tg(hsp70:hand2P) embryos and (D) Tg(hsp70:hand2AA) embryos, heat-shocked at 10 hpf; dorsal views, anterior upwards. (E) Average area of cmlc2 expression, as in Fig. 1E, in nontransgenic and transgenic embryos, following heat shock at 10 hpf. Asterisks indicate significant differences from nontransgenic embryos. Overexpression of hand2 or hand2 AA increases the area of cmlc2 expression (n=15-32; *P<0.001), whereas hand2 P does not (n=13; P=0.05). (F) Average number of cardiomyocytes at 36 hpf, as in Fig. 1F, in nontransgenic embryos and in Tg(hsp70:hand2) embryos, following heat shock at 10 hpf. Asterisk indicates a significant difference from nontransgenic embryos (n=16-20; *P<0.001). (G-K) Immunofluorescence at 36 hpf for MF20 (green, visible in the ventricle) and S46 (red, visible in the atrium) shows cardiac morphology. Frontal views; brackets mark the outflow tract. Hearts of Tg(hsp70:hand2) embryos heat-shocked at 24 hpf (H) resemble nontransgenic hearts heat-shocked at 10 hpf (G). Tg(hsp70:hand2) embryos heat-shocked at 10 hpf (I) show an overall increase in cardiac size and an enlarged outflow tract. Similar morphology is seen after heat shock at 10 hpf in Tg(hsp70:hand2AA) embryos (K), but not in Tg(hsp70:hand2P) embryos (J).
The cardiac expansion induced by hand2 overexpression requires phosphorylation-independent dimerization of Hand2
The consequences of injection of the hand2 EDE and hand2 P mRNAs suggest that the role of Hand2 during cardiomyocyte production requires its dimerization but not its direct binding to DNA (Fig. 2). Phosphorylation of conserved threonine and serine residues within the first helix of Hand factors has previously been shown to be important for their choice of dimerization partners (Firulli et al., 2003, 2005). For example, changes in phosphorylation of T107 and S109 of mouse Hand1 influence the affinity for formation of Hand1-Hand1 homodimers relative to the formation of Hand1 heterodimers with E proteins (Firulli et al., 2003). To determine whether the dimerization interactions that influence cardiomyocyte production require Hand2 phosphorylation, we constructed the transgene Tg(hsp70:FLAG-hand2AA-2A-mCherry) [referred to as Tg(hsp70:hand2AA)]. The zebrafish Hand2 variant expressed by this transgene features substitution of the residues T103 and S105 with alanines (supplementary material Fig. S1), thereby preventing their phosphorylation; extrapolating from prior studies of Hand1 and Twist1, these changes may promote affinity for homodimerization rather than heterodimerization with E proteins (Firulli et al., 2003, 2005).
We compared the effects of inducing expression of hand2, hand2AA and hand2P, using inducible transgenes to overexpress each variant (supplementary material Fig. S3). Induction of expression of the phosphorylation-deficient version of hand2 at 10 hpf resulted in expansion of cmlc2 expression at 18 somites, similar to the effects of inducing wild-type hand2 expression (Fig. 3A,B,D,E). Likewise, overexpression of the phosphorylation-deficient version of hand2 caused formation of an enlarged heart and an elongated outflow tract by 36 hpf, as is the case for overexpression of wild-type hand2 (Fig. 3G,I,K). By contrast, induction of expression of the dimerization-deficient version of hand2 did not expand the area of cmlc2 expression (Fig. 3C,E) and did not yield a larger heart or outflow tract (Fig. 3J). These data suggest that an unphosphorylated form of Hand2 mediates its dimerization and role in the context of cardiac expansion.
Increased proliferation in late-differentiating cells contributes to cardiac expansion in embryos overexpressing hand2
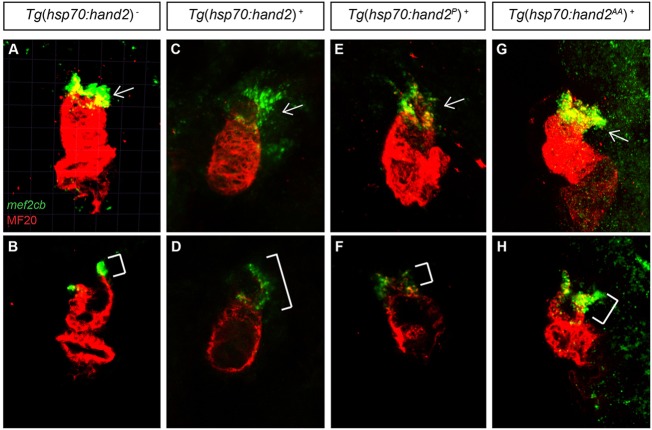
The enlarged hearts in embryos overexpressing hand2 led us to investigate the cellular mechanism through which hand2 influences cardiomyocyte expansion. As overexpression of Hand1 has been shown to enhance proliferation within the outflow tract in mice (Risebro et al., 2006), we were especially interested in determining the origins of the elongated outflow tract in hand2-overexpressing embryos. First, we examined the expression of mef2cb at 36 hpf; at this stage, mef2cb marks a population of SHF-derived cells that contributes to the outflow tract (Lazic and Scott, 2011; Hinits et al., 2012). We found excess mef2cb expression at the arterial pole of the heart in hand2-overexpressing embryos (Fig. 4A-D), suggesting that the observed outflow tract expansion is the product of excess SHF-derived cells, rather than simply a morphological anomaly. In addition, mef2cb expression in embryos overexpressing the phosphorylation-deficient and dimerization-deficient versions of hand2 correlated with the observed outflow tract morphologies: embryos overexpressing hand2 AA exhibited an excess of mef2cb-expressing cells (Fig. 4G,H), whereas embryos overexpressing hand2 P did not (Fig. 4E,F). In contrast to their expanded mef2cb expression at the arterial pole, hand2-overexpressing embryos exhibited a normal pattern of islet1 expression at the venous pole (supplementary material Fig. S4), suggesting that hand2 has a more potent influence on outflow tract formation than on inflow tract formation.
Fig. 4.
Overexpression of hand2 causes outflow tract expansion. (A-H) MF20 antibody staining (red) and mef2cb in situ hybridization (green) at 36 hpf in (A,B) nontransgenic, (C,D) Tg(hsp70:hand2), (E,F) Tg(hsp70:hand2P) and (G,H) Tg(hsp70:hand2AA) embryos, all of which were heat-shocked at 10 hpf. Frontal views; (A,C,E,G) partial reconstructions of confocal z-stacks with arrows indicating outflow tract; (B,D,F,H) single slices with brackets marking outflow tract. Embryos overexpressing hand2 (C,D) or hand2 AA (G,H) exhibit expanded populations of mef2cb-expressing cells, whereas embryos overexpressing hand2 P (E,F) do not.
Next, we employed an EdU incorporation assay to investigate whether the expansion of the outflow tract or the general increase in cardiomyocyte number could be the result of increased proliferation in hand2-overexpressing embryos. Following heat shock at 10 hpf, we pulsed embryos with EdU for 30 min when they reached 17 hpf, thereby labeling proliferating cells during that interval (Fig. 5G). We then compared the numbers of EdU-positive and EdU-negative cardiomyocytes at 36 hpf in Tg(hsp70:hand2) embryos and their nontransgenic siblings (Fig. 5A-F). Strikingly, we found a significantly higher proliferation index in the hand2-overexpressing cardiomyocytes (Fig. 5H), indicating that increased proliferation contributes to the enhanced cardiomyocyte production in these embryos. By contrast, in a parallel set of experiments in which we pulsed embryos with EdU at 23 hpf (supplementary material Fig. S5A-G), we did not detect an increased proliferation index in hand2-overexpressing cardiomyocytes (supplementary material Fig. S5H), suggesting that hand2 overexpression influences proliferation prior to 23 hpf.
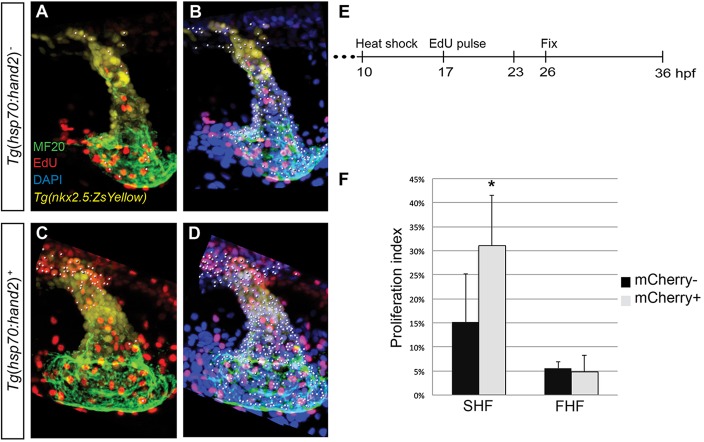
Fig. 5.
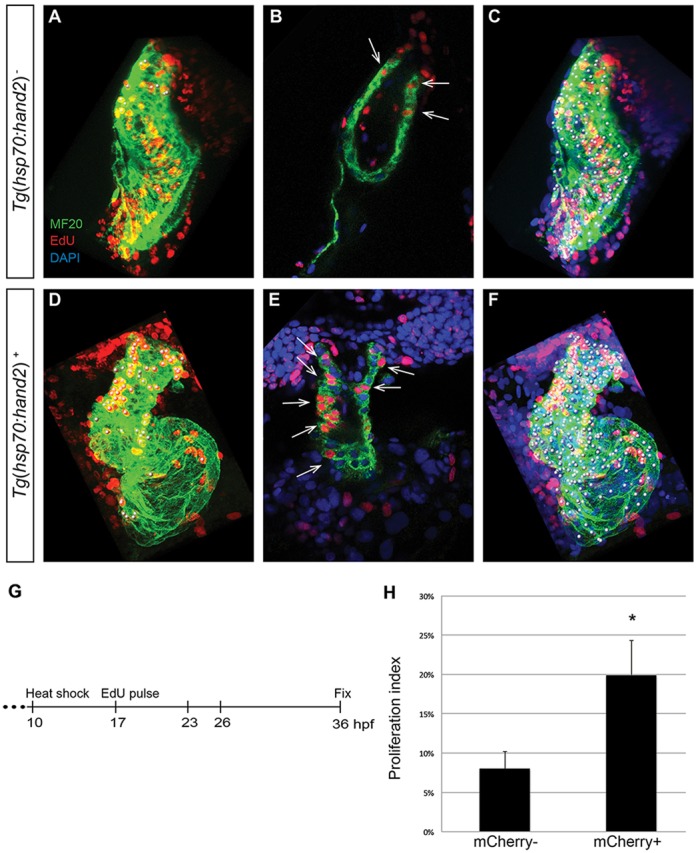
Increased proliferation contributes to cardiac expansion in hand2-overexpressing embryos. (A-F) EdU incorporation in hearts of (A-C) nontransgenic and (D-F) Tg(hsp70:hand2) embryos at 36 hpf, following heat shock at 10 hpf and EdU pulse at 17 hpf. Partial reconstructions of confocal z-stacks with ventricle upwards (A,C,D,F) and representative single slices (B,E). (A,D) White dots indicate EdU-positive (red) cells that are also MF20-positive (green) differentiated cardiomyocytes. (B,E) Arrows indicate EdU-positive cells that are also MF20 positive; DAPI (blue) marks all nuclei. (C,F) White dots indicate all myocardial nuclei. Qualitative assessment suggests increased numbers of EdU-positive cardiomyocytes in hand2-overexpressing hearts (D,E), particularly in the distal region of the ventricle and the outflow tract. (G) Timeline of experimental design. (H) Proliferation indices in nontransgenic (mCherry-negative) and Tg(hsp70:hand2) (mCherry-positive) embryos; error bars indicate s.d. Proliferation index was calculated by dividing the number of EdU-positive cardiomyocytes by the total number of cardiomyocytes. A significant increase in proliferation index was evident in hand2-overexpressing embryos (n=8 or 9; *P<0.001).
Based on this series of results, we hypothesized that increased proliferation in hand2-overexpressing embryos occurs between 17 hpf and the time of initial heart tube assembly. To test this, we pulsed embryos with EdU at 17 hpf and analyzed the distribution of EdU-positive cells at 26 hpf in hand2-overexpressing and nontransgenic embryos (Fig. 6A-E). In addition to scoring EdU-positive cardiomyocytes, we used Tg(nkx2.5:ZsYellow) to facilitate scoring of EdU labeling in SHF-derived progenitor cells that reside near the arterial pole of the early heart tube and ultimately contribute to the outflow tract (Zhou et al., 2011); these progenitor cells express Tg(nkx2.5:ZsYellow) but do not yet express high levels of other myocardial differentiation markers. In these experiments, we found that hand2 overexpression caused a significant increase in the proliferation index of the SHF-derived cardiac progenitor cells near the arterial pole (Fig. 6F). However, hand2 overexpression did not enhance the proliferation index of the first heart field-derived (FHF-derived) cardiomyocytes that form the initial heart tube (Fig. 6F); we obtained similar results when assessing the cardiomyocyte proliferation index in embryos pulsed with EdU at 17 hpf and fixed at 23 hpf, and in embryos pulsed with EdU at 14 hpf and fixed at 26 hpf (supplementary material Fig. S6). Altogether, our data demonstrate that overexpression of hand2 has a potent and early impact on the proliferation of the SHF-derived, late-differentiating progenitor cells that will create the outflow tract.
Fig. 6.
Overexpression of hand2 increases proliferation of SHF-derived progenitor cells. (A-D) EdU incorporation in (A,B) nontransgenic and (C,D) Tg(hsp70:hand2) embryos at 26 hpf, following heat shock at 10 hpf and EdU pulse at 17 hpf; partial reconstructions of confocal z-stacks with ventricle upwards. (A,C) White dots indicate EdU-positive (red) cells that are MF20 positive (green) and/or expressing Tg(nkx2.5:ZsYellow) (yellow). (B,D) White dots indicate all nuclei (DAPI, blue) of cells that are MF20 positive and/or expressing Tg(nkx2.5:ZsYellow). (E) Timeline of experimental design. (F) Proliferation indices, as in Fig. 5H, for two populations of cells: SHF-derived cells, defined as Tg(nkx2.5:ZsYellow)-expressing cells with very low or no MF20 staining; and FHF-derived cells, defined as cells with clearly detectable MF20 staining. Proliferation index was calculated for each population independently by dividing the number of EdU-positive cells by the total number of cells in the population. A significant increase in proliferation index was evident in the SHF-derived cells in hand2-overexpressing embryos (n=10 or 11; *P=0.003), but not in the FHF-derived cells (n=10 or 11; P=0.528).
Overexpression of hand2 alters ALPM patterning
Although our results suggest that enhanced proliferation of SHF-derived progenitors contributes to the enlarged outflow tract seen when hand2 is overexpressed, increased proliferation within the SHF may not account for all of the enhanced cardiomyocyte production in hand2-overexpressing embryos. Notably, the increased area of cmlc2 expression observed at 18 somites (Fig. 3A,B,E) seems to reflect increased production of cardiomyocytes by the FHF, as only early-differentiating, FHF-derived cells are thought to express cmlc2 at this stage (de Pater et al., 2009). Additionally, we have found increased numbers of differentiated cardiomyocytes in hand2-overexpressing embryos at 23 hpf (nontransgenic embryos, 127±25 cardiomyocytes; hand2-overexpressing embryos, 164±39 cardiomyocytes; n=9 or 10, P<0.05) and at 26 hpf (nontransgenic embryos, 154±25 cardiomyocytes; hand2-overexpressing embryos, 224±47 cardiomyocytes; n=10 or 11, P<0.001). As is the case at 18 somites, it is thought that cardiomyocytes have not yet been added from the SHF at 23 or 26 hpf (de Pater et al., 2009; Lazic and Scott, 2011). Furthermore, we did not detect an increased proliferation index in the cardiomyocyte population at 23 or 26 hpf (Fig. 6F; supplementary material Fig. S6F). Together, these data suggest that the early expansion of cardiomyocytes in hand2-overexpressing embryos reflects an influence of hand2 on the early-differentiating FHF, and that this effect is more likely to be the result of altered specification than of altered proliferation.
To evaluate whether overexpression of hand2 alters the patterning of the ALPM, we examined markers of lineages that are neighbors of the heart fields at early stages. scl and etsrp are expressed in progenitors of the blood and vessel lineages that are rostral to the heart-forming region that expresses hand2 (Schoenebeck et al., 2007). When blood and vessel lineage specification are inhibited by loss of scl and etsrp function, hand2 expression expands into the rostral ALPM and an increased number of cardiomyocytes form (Schoenebeck et al., 2007; Palencia-Desai et al., 2011). These findings suggest that scl and etsrp act to limit the extent of myocardial specification in the ALPM. Conversely, we found that overexpression of hand2 limited the expression of anterior hematopoietic and endothelial markers at 12 somites, including scl, etsrp and pu.1 (Fig. 7A-F); even so, these gene expression defects did not seem to cause major anomalies in vascular patterning or in endocardial development (supplementary material Fig. S7). At the same time, hand2-overexpressing embryos exhibited expanded expression of mef2cb (Fig. 7G,H), which is found in myocardial progenitor cells at this stage (Hinits et al., 2012). However, not all myocardial progenitor markers are similarly expanded, as we found that hand2-overexpressing embryos exhibit normal expression of nkx2.5 at 12 somites (Fig. 7I,J). This result suggests that hand2 functions downstream of nkx2.5, which is consistent with the normal nkx2.5 expression pattern observed in hand2 mutant embryos (Yelon et al., 2000; Schoenebeck et al., 2007). Finally, gata4 expression also appeared similar in nontransgenic and hand2-overexpressing embryos (Fig. 7K,L), indicating that increased hand2 expression does not alter the overall dimensions of the ALPM. Thus, in addition to a later capacity to promote proliferation within the SHF, hand2 overexpression can influence early patterning processes within the ALPM, possibly by promoting myocardial specification while limiting hematopoietic and endothelial specification.
Fig. 7.
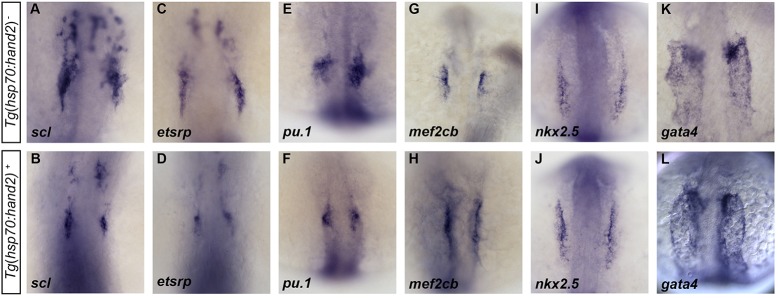
hand2 overexpression limits expression of blood and vessel genes within the ALPM. (A-L) In situ hybridization depicts expression of (A,B) scl, (C,D) etsrp, (E,F) pu.1, (G,H) mef2cb, (I,J) nkx2.5 and (K,L) gata4 in (A,C,E,G,I,K) nontransgenic embryos and (B,D,F,H,J,L) Tg(hsp70:hand2) embryos following heat shock at 10 hpf. Dorsal views, anterior upwards, at (A-J) 12 somites and (K,L) 10 somites. Overexpression of hand2 results in (A-F) decreased distribution of scl, etsrp and pu.1 expression, as well as (G,H) increased expression of mef2cb. By contrast, expression of (I,J) nkx2.5 and (K,L) gata4 appear relatively normal in hand2-overexpressing embryos at these stages.
Overexpression of hand2 augments cardiomyocyte proliferation during cardiac regeneration
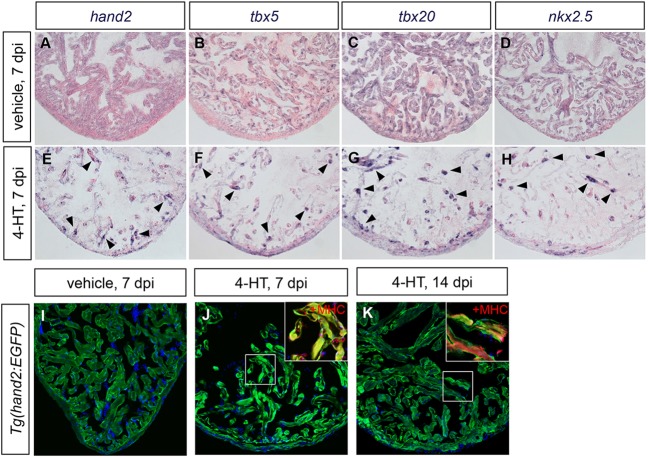
Mechanisms of embryonic heart development have served as necessary guides for understanding cardiac regeneration. The potent effects of hand2 in the early embryo raise the possibility that its expression could influence the efficacy of regeneration in the injured adult heart. In the zebrafish, cardiac injuries are repaired through a process in which spared mature cardiomyocytes reduce indicators of differentiation and proliferate to regenerate lost muscle, involving induction and function of gata4 in source cardiomyocytes (Jopling et al., 2010; Kikuchi et al., 2010; Gupta et al., 2013). In previous studies, we also observed increased expression of nkx2.5, tbx5 and tbx20 in the injury site following resection of the adult zebrafish ventricle (Lepilina et al., 2006), although the induction of these genes was not detectable by others (Raya et al., 2003; Jopling et al., 2010). We additionally found activation of hand2 and gata5 regulatory sequences in endocardial cells after resection injury (Kikuchi et al., 2011a). To assess gene expression after a more severe injury, we employed transgene-driven cardiomyocyte ablation in Z-CAT transgenic fish, in which the combination of Tg(cmlc2:CreER) and Tg(β-actin2:loxp-mCherry-STOP-loxp-DTA) transgenes permits 4-HT-inducible expression of diphtheria toxin in cardiomyocytes (Wang et al., 2011). Enhanced expression of hand2, as well as nkx2.5, tbx5 and tbx20, was evident in spared cardiomyocytes throughout the injured Z-CAT ventricle (Fig. 8A-H). Additionally, fluorescence in Tg(hand2:EGFP) myocardium and endocardium was boosted during regeneration (Fig. 8I-K), indicating that cardiac injury can activate hand2 regulatory sequences in multiple cell types.
Fig. 8.
Cardiac injury induces hand2 expression. (A-H) In situ hybridization depicts expression of (A,E) hand2, (B,F) tbx5, (C,G) tbx20 and (D,H) nkx2.5 in ventricles of adult Z-CAT fish 7 days post-injection (dpi) of vehicle (A-D) or 4-HT (E-H). Expression of each gene is enhanced in spared myocardium following injury (E-H, arrowheads). (I-K) Expression of Tg(hand2:EGFP) (green) highlights the activation of hand2 regulatory sequences following injury. Images are single confocal slices of ventricular tissue at 7 dpi (I,J) and 14 dpi (K); insets demonstrate myosin heavy chain (MHC; red) localization in hand2-expressing cells.
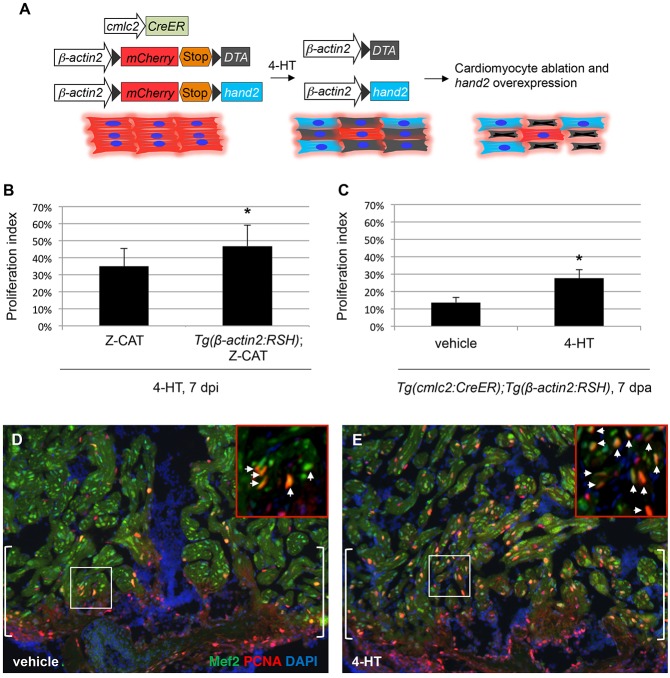
To test whether induced hand2 expression in adult cardiomyocytes affects their regenerative capacity, we first examined cardiomyocyte proliferation in injured Z-CAT animals heterozygous for the hans6 mutation (Yelon et al., 2000). Heterozygosity for hans6 did not alter levels of ablation-induced cardiomyocyte proliferation, although we also did not detect reduced levels of hand2 mRNA in injured heterozygous hearts (data not shown). To artificially increase hand2 expression in cardiomyocytes after injury, we generated a new transgene that permits 4-HT-inducible hand2 expression, Tg(β-actin2:loxp-mCherry-STOP-loxp-hand2) [hereafter referred to as Tg(β-actin2:RSH)]. In fish carrying Tg(β-actin2:RSH) in combination with Tg(cmlc2:CreER), a single 4-HT injection sharply increased myocardial hand2 expression but had no effect on cardiomyocyte proliferation in the absence of injury (data not shown). In Z-CAT animals carrying Tg(β-actin2:RSH), we experimentally elevated hand2 levels in spared cardiomyocytes in concert with inducing ablation injury (Fig. 9A). Under these conditions, we observed a 34% increase in cardiomyocyte proliferation in Z-CAT; Tg(β-actin2:RSH) animals at 7 days post-injection (dpi), compared with that seen in Z-CAT animals at the same timepoint (Fig. 9B). To confirm effects of hand2 on regenerative proliferation, we induced hand2 overexpression in Tg(cmlc2:CreER); Tg(β-actin2:RSH) animals by injecting 4-HT 5 days before performing ventricular resection. In these experiments, we observed a 102% increase in cardiomyocyte proliferation at the resection plane, compared with the proliferation observed in vehicle-injected Tg(cmlc2:CreER); Tg(β-actin2:RSH) animals (Fig. 9C-E). In total, our results indicate that elevation of hand2 expression helps boost cardiomyocyte production during regeneration, mirroring its effect in the embryo.
Fig. 9.
Overexpression of hand2 boosts cardiomyocyte proliferation in response to injury. (A) Schematic representation of transgenes used for cardiomyocyte ablation and myocardium-specific hand2 overexpression in Z-CAT fish. (B) Proliferation indices for ventricular cardiomyocytes; error bars indicate s.e.m. Proliferation was assessed 7 days after 4-HT injection in Z-CAT fish with or without induced cardiomyocyte-specific hand2 overexpression. hand2-overexpressing fish exhibited significantly increased proliferation following ablation injury (n=9 or 10; *P<0.05). (C) Proliferation indices in injured cmlc2:CreER; β-act2:RSH fish; error bars indicate s.e.m. Proliferation was assessed at the resection plane 7 days post-amputation (dpa) in animals that were injected with either vehicle or 4-HT 5 days before injury. hand2-overexpressing fish exhibited significantly increased proliferation following resection injury (n=4 or 5; *P<0.005). (D,E) Representative examples of cardiomyocyte proliferation at 7 dpa at the resection plane (brackets) in injured cmlc2:CreER; β-act2:RSH fish that were treated with vehicle (D) or 4-HT (E). Mef2 (green) marks cardiomyocytes and PCNA (red) marks proliferating cells; arrows in insets indicate double-positive cells.
DISCUSSION
Taken together, our studies demonstrate three distinct mechanisms through which hand2 overexpression can promote enhanced cardiomyocyte production. First, overexpression of hand2 results in expansion of cardiomyocyte production within the early differentiating FHF, potentially by promoting myocardial specification at the expense of neighboring blood and vessel lineages. Second, hand2 overexpression causes excess proliferation of cardiac progenitor cells within the late-differentiating SHF, leading to the formation of an abnormally elongated outflow tract. Third, induced myocardial expression of hand2 enhances regenerative proliferation of cardiomyocytes in response to injury. Our previous studies have shown that loss of hand2 function dramatically reduces the cardiomyocyte population, emphasizing the necessity of hand2 for efficient cardiomyocyte production (Yelon et al., 2000; Schoenebeck et al., 2007). Integrating our current and past work, we conclude that hand2 plays important and instructive roles in promoting cardiomyocyte production via influences on both specification and proliferation.
Our data indicate that overexpression of hand2 is sufficient to induce ectopic cardiomyocyte formation in a region proximal to the typical FHF, but we do not find ectopic cardiomyocytes elsewhere within hand2-overexpressing embryos. Additionally, while hand2 overexpression is capable of hindering the expression of blood and vessel markers, hand2-null embryos do not exhibit expanded expression of blood and vessel genes (Schoenebeck et al., 2007). These findings suggest that other factors partner with Hand2 to promote myocardial specification in the FHF and that expression of these factors is restricted to a particular region of the ALPM. The significance of the partnerships between Hand2 and other factors is reinforced by our data demonstrating that the effects of hand2 overexpression on cardiomyocyte production appear to be independent of direct binding of DNA by Hand2 and dependent on Hand2 dimerization. It will be valuable for future studies to investigate which Hand2 dimerization partners are relevant to its influences on ALPM patterning.
In addition to an instructive role for hand2 during specification of the FHF, our data suggest a previously unappreciated role for hand2 in promoting progenitor proliferation within the SHF. Hand factors have been shown to have roles in both promoting and inhibiting proliferation (Risebro et al., 2006; Li et al., 2011), but this particular role of hand2 in the SHF has not been demonstrated by prior studies. The impact of hand2 on SHF progenitor proliferation contrasts with previous work that found a role for Hand2 in promoting survival of SHF progenitor cells (Tsuchihashi et al., 2011); although proliferation defects were not documented in these studies of conditional Hand2 knockout mice, it is possible that deficient SHF proliferation preceded the observed death of SHF cells in these embryos. Additionally, it is interesting to compare the effects of hand2 overexpression on the zebrafish SHF with the phenotype of mouse mutants in which Hand1-expressing cells were engineered to overexpress Hand1 (Risebro et al., 2006). In these mice, increased proliferation was seen in the distal outflow tract, but no expansion of SHF progenitor markers was observed, suggesting that Hand1 overexpression did not increase proliferation within the SHF itself but instead within the cardiomyocytes present or arriving at the outflow tract (Risebro et al., 2006). These effects of Hand1 overexpression on outflow tract proliferation seem distinct from our observed effects of hand2 overexpression on the SHF progenitor population outside the heart tube; this difference may relate to the conditional regulation of Hand1 expression in these mice, which do not overexpress Hand1 within the SHF.
Finally, our data indicate that hand2 overexpression can impact regenerative cardiomyocyte proliferation in the adult heart. These findings implicate the induction of hand2 expression during regeneration as a key component of the regenerative mechanism. We speculate that the ability of Hand2 to increase reprogramming efficiency of fibroblasts toward a cardiac fate (Song et al., 2012; Nam et al., 2013) could be related to our observation that Hand2 can enhance cardiomyocyte proliferation; alternatively, the function of Hand2 during reprogramming could be a reflection of our observed influences of Hand2 on myocardial progenitor specification or proliferation. To distinguish between these possibilities, it will be important to elucidate and compare the pathways regulated by Hand2 in each of these contexts, as well as in other settings, such as the Hand2-driven induction of hypertrophy in the postnatal mouse myocardium (Dirkx et al., 2013). In the long term, these investigations may allow further improvements in strategies for cardiac reprogramming, regeneration and repair.
In addition to their implications regarding future applications for Hand2 in regenerative medicine, our results provide intriguing insight into possible causes of CHD. Duplications and deletions of the 4q33 chromosomal region, which contains HAND2, have been associated with CHD (Borochowitz et al., 1997; Byatt et al., 1997). Moreover, individuals with partial trisomy distal 4q (4q+ syndrome), a translocation that involves duplication of HAND2 and neighboring genes, exhibit striking cardiac and limb defects (Tamura et al., 2013). In a mouse model for 4q+ syndrome, rebalancing levels of Hand2 by breeding to Hand2-deficient animals ameliorated the heart and limb phenotypes, implicating overexpression of Hand2 as a cause of these defects (Tamura et al., 2013). Our data suggest possible ways in which HAND2 overexpression could cause CHD through alteration of cardiomyocyte production; future studies that elucidate the specific Hand2 partners and targets involved in FHF specification, SHF proliferation and cardiomyocyte proliferation will provide a deeper understanding of the mechanisms through which Hand2 influences the origins of CHD.
MATERIALS AND METHODS
Zebrafish and genotyping
We bred wild-type zebrafish, zebrafish heterozygous for the hand2 mutant allele hans6 (Yelon et al., 2000) or zebrafish carrying Tg(-5.1myl7:nDsRed2)f2 (Mably et al., 2003). In addition, we bred zebrafish carrying Tg(nkx2.5:ZsYellow) (Zhou et al., 2011) or Tg(kdrl:GRCFP) (Cross et al., 2003) to zebrafish carrying the newly generated transgenes described below, and we employed adult zebrafish carrying Tg(hand2:EGFP)pd24 (Kikuchi et al., 2011a). PCR genotyping of hans6 mutant embryos was conducted as previously described (Yelon et al., 2000).
Injection of mRNA
We synthesized capped mRNA from a pCS2-hand2 plasmid using the Ambion mMESSAGE mMACHINE kit, and we injected 160 pg hand2 mRNA into embryos at the one- to two-cell stage. The hand2 dimerization-deficient (hand2 P) and DNA binding-deficient (hand2 EDE) constructs were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies).
Creation of stable transgenic lines
To generate transgenes suitable for heat-activated overexpression of hand2, we first amplified the hand2-coding sequence from the plasmids pCS2-hand2, pCS2-hand2 P and pCS2-hand2 AA, and cloned these amplicons into the p3xFLAG-CMV vector (Sigma) at the HindIII and XbaI restriction sites. Each version of FLAG-hand2 was then introduced into the pENTR/D-TOPO vector (Invitrogen). Each final transgene was achieved through a Gateway LR reaction containing four plasmids: p5E-hsp70l (Kwan et al., 2007), pDestTol2pA2 (Kwan et al., 2007), p3E-2A-mcherry-pA (Covassin et al., 2009) and the appropriate pENTR/D-TOPO FLAG-hand2 plasmid.
We employed standard protocols to create transgenic founders (Fisher et al., 2006). The F1 progeny of prospective founder fish were screened for mCherry fluorescence following heat shock for 1 h at 37°C, and phenotypic analysis was performed on the F1 and F2 progeny of promising founders. We compared the phenotypes generated by heat shock of four lines carrying Tg(hsp70:hand2), three lines carrying Tg(hsp70:hand2AA) and one line carrying Tg(hsp70:hand2P), all of which exhibited qualitatively similar levels of mCherry fluorescence. Following heat shock at 10 hpf, embryos from all of the Tg(hsp70:hand2) and Tg(hsp70:hand2AA) lines exhibited similarly abnormal cardiac morphology at 36 hpf. In addition, expression of Tg(hsp70:hand2) and expression of Tg(hsp70:hand2AA) were both able to rescue heart tube formation in hans6 mutant embryos, whereas expression of Tg(hsp70:hand2P) did not rescue. Results shown here use three specific transgenic lines: Tg(hsp70:FLAG-hand2-2A-mCherry)sd28, Tg(hsp70:FLAG-hand2AA-2A-mCherry)sd29 and Tg(hsp70:FLAG-hand2P-2A-mCherry)sd30.
To generate the transgenic line Tg(β-actin2:loxp-mCherry-STOP-loxp-hand2)pd38, 5.3 kb of genomic DNA immediately upstream of the β-actin2 transcriptional start site was subcloned into a modified pBSK vector with a multiple cloning site flanked by I-SceI restriction sites. A loxP-mCherry-STOP-loxP-hand2 cassette was then subcloned downstream of the β-actin2 promoter. The mCherry-STOP cassette serves as a marker for the transgene and also prevents read-through translation of Hand2 protein.
In situ hybridization
Whole-mount in situ hybridization was performed as previously described (Thomas et al., 2008) using probes for cmlc2 (myl7; ZDB-GENE-991019-3), mef2cb (ZDB-GENE-040901-7), islet1 (isl1; ZDB-GENE-980526-112), scl (tal1; ZDB-GENE-980526-501), etsrp (etv2; ZDB-GENE-050622-14), pu.1 (spi1b; ZDB-GENE-980526-164), nkx2.5 (ZDB-GENE-980526-321) and gata4 (ZDB-GENE-980526-476). Embryonic stages were determined by counting somites prior to fixation. Area measurements were made by using the selection tool in ImageJ64 to score stained pixel area. Statistical analysis of data sets was performed using Microsoft Excel to conduct unpaired t-tests. We have found that cmlc2 area measurements at 18-20 somites correlate well with counts of cell outlines in the same images (data not shown), making area measurement a reasonable strategy for comparing estimated sizes of cardiomyocyte populations.
Immunofluorescence
Whole-mount immunofluorescence was performed as previously described (Thomas et al., 2008), using the monoclonal antibodies MF20 and S46 (Developmental Studies Hybridoma Bank), a polyclonal antibody against Islet1 (GeneTex; GTX128201L) and the secondary antibodies goat anti-mouse IgG2b FITC (Southern Biotech), goat anti-mouse IgG1 TRITC (Southern Biotech) and goat anti-rabbit Alexa Fluor 488 (Molecular Probes). The numbers of Islet1-positive nuclei within MF20-positive cells were counted both in three-dimensional reconstructions and in optical sections.
Fluorescent in situ hybridization with immunofluorescence
Embryos were fixed in 2% formaldehyde for 20 min and then gently agitated in 0.5% Triton and 0.2% saponin to dissolve the yolk. Embryos were then fixed in 4% paraformaldehyde overnight at 4°C. An antisense mef2cb probe was detected by deposition of TSA Plus fluorescein solution (PerkinElmer), using an established protocol (Brend and Holley, 2009), followed by MF20 antibody staining.
Cell counting and EdU incorporation
Cardiomyocyte counting in Tg(-5.1myl7:nDsRed2) embryos was performed using an established protocol (Schoenebeck et al., 2007). Cell counting in Tg(hsp70:FLAG-hand2-2A-mCherry) embryos was performed subsequent to EdU incorporation, using a modification of a described protocol (Zeng and Yelon, 2014). Dechorionated embryos were exposed to 10 mM EdU in 0.3× Danieau buffer with 15% DMSO for 30 min on ice. After a series of washes, embryos were incubated at 28.5°C until fixation. Embryos were fixed in 2% formaldehyde for 20 min and then gently agitated in 0.5% Triton and 0.2% saponin to dissolve the yolk, followed by fixation in 2% formaldehyde overnight at 4°C. Embryos were then stained with the primary antibody MF20 followed by an anti-mouse IgG secondary antibody conjugated with either Alexa Fluor 488 or 647 (Invitrogen). EdU incorporation was visualized using a Click-iT imaging kit (Invitrogen) with either Alexa Fluor 594 or 647. Samples were then placed in SlowFade Gold anti-fade reagent with DAPI (Molecular Probes). Cardiomyocyte number was determined by examining DAPI-stained nuclei within MF20-positive cells both in three-dimensional reconstructions and in optical sections, and proliferation index was calculated as the percentage of these cells that exhibited EdU localization. A similar method was used to determine the number and proliferation index of Tg(nkx2.5:ZsYellow)-expressing MF20-negative cells clustered near the arterial pole of the heart tube. Statistical analysis of data sets was performed using Microsoft Excel to conduct unpaired t-tests.
Imaging
Bright-field and fluorescent images were captured with a Zeiss Axiocam on a Zeiss Axiozoom or a Zeiss Axioplan microscope, and processed using Zeiss AxioVision and Adobe Creative Suite. Confocal z-stacks were collected by a Leica SP5 confocal microscope and analyzed using Imaris software (Bitplane).
Cardiac injury and histology
For cardiomyocyte ablation injury, we induced myocyte expression of diphtheria toxin A in zebrafish carrying both the Tg(cmlc2:CreER)pd10 (Kikuchi et al., 2010) and Tg(β-actin2:loxp-mCherry-STOP-loxp-DTA)pd36 (Wang et al., 2011) transgenes; this combination is referred to as Z-CAT (zebrafish cardiomyocyte ablation transgenes) (Wang et al., 2011). Anesthetized Z-CAT fish were injected with 0.5 mg/ml 4-hydroxytamoxifen (4-HT) in 10% ethanol or with vehicle alone, as previously described (Kikuchi et al., 2010). For ventricular resection injury, we performed surgeries as described previously (Poss et al., 2002).
For analysis of paraformaldehyde-fixed hearts, in situ hybridization on 10 μm cryosections was performed as previously described (Poss et al., 2002), using probes for hand2 (ZDB-GENE-000511-1), tbx5 (ZDB-GENE-991124-7), nkx2.5 (ZDB-GENE-980526-321) and tbx20 (ZDB-GENE-000427-7). Immunofluorescence with primary antibodies that recognize myosin heavy chain (F59; Developmental Studies Hybridoma Bank) and GFP (Invitrogen) was performed as previously described (Kikuchi et al., 2011b). To determine cardiomyocyte proliferation indexes, we employed established techniques for staining sections with antibodies against Mef2 (Santa Cruz Biotechnology) and PCNA (Sigma), and assessing the proportion of Mef2-positive cells that are also PCNA positive (Wang et al., 2011).
Supplementary Material
Acknowledgements
We thank K. Birnbaum, N. Chi, S. Evans, T. Evans, H. Knaut, R. Lehmann, N. Tanese and members of the Yelon laboratory for thoughtful input.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
Y.L.S., J.W., K.D.P. and D.Y. designed these studies; Y.L.S., K.M.G. and J.W. performed experiments; B.A.F. and A.B.F. generated reagents; Y.L.S., K.M.G., J.W., K.D.P. and D.Y. analyzed the data; and Y.L.S. and D.Y. wrote the manuscript with input from all authors.
Funding
This work was supported by grants to D.Y. from the National Institutes of Health (NIH) [R01HL069594 and R01HL108599], the American Heart Association and the March of Dimes; by a grant to K.D.P. from the NIH [R01HL081674]; and by grants to A.B.F. from the NIH [R01HL122123, R01HL120920 and R01AR061392]. Y.L.S. received support from an American Heart Association predoctoral fellowship [10PRE3510044]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.106336/-/DC1
References
- Borochowitz Z., Shalev S. A., Yehudai I., Bar-El H., Dar H., Tirosh E. (1997). Deletion (4)(q33 --> qter): a case report and review of the literature. J. Child Neurol. 12, 335-337 10.1177/088307389701200510 [DOI] [PubMed] [Google Scholar]
- Brend T., Holley S. A. (2009). Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J. Vis. Exp. 25, 1229 10.3791/1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byatt S. A., Baker E., Richards R. I., Roberts C., Smith A. (1997). Unbalanced t(4;11)(q32;q23) in a 34-year-old man with manifestations of distal monosomy 11q and trisomy 4q syndromes. Am. J. Med. Genet. 70, 357-360 [DOI] [PubMed] [Google Scholar]
- Choi W.-Y., Poss K. D. (2012). Cardiac regeneration. Curr. Top. Dev. Biol. 100, 319-344 10.1016/B978-0-12-387786-4.00010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covassin L. D., Siekmann A. F., Kacergis M. C., Laver E., Moore J. C., Villefranc J. A., Weinstein B. M., Lawson N. D. (2009). A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev. Biol. 329, 212-226 10.1016/j.ydbio.2009.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross L. M., Cook M. A., Lin S., Chen J.-N., Rubinstein A. L. (2003). Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler. Thromb. Vasc. Biol. 23, 911-912 10.1161/01.ATV.0000068685.72914.7E [DOI] [PubMed] [Google Scholar]
- de Pater E., Clijsters L., Marques S. R., Lin Y.-F., Garavito-Aguilar Z. V., Yelon D., Bakkers J. (2009). Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633-1641 10.1242/dev.030924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkx E., Gladka M. M., Philippen L. E., Armand A.-S., Kinet V., Leptidis S., el Azzouzi H., Salic K., Bourajjaj M., da Silva G. J. J., et al. (2013). Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat. Cell Biol. 15, 1282-1293 10.1038/ncb2866 [DOI] [PubMed] [Google Scholar]
- Fahed A. C., Gelb B. D., Seidman J. G., Seidman C. E. (2013). Genetics of congenital heart disease: the glass half empty. Circ. Res. 112, 707-720 10.1161/CIRCRESAHA.112.300853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli B. A., Howard M. J., McDaid J. R., McIlreavey L., Dionne K. M., Centonze V. E., Cserjesi P., Virshup D. M., Firulli A. B. (2003). PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol. Cell 12, 1225-1237 10.1016/S1097-2765(03)00425-8 [DOI] [PubMed] [Google Scholar]
- Firulli B. A., Krawchuk D., Centonze V. E., Vargesson N., Virshup D. M., Conway S. J., Cserjesi P., Laufer E., Firulli A. B. (2005). Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet. 37, 373-381 10.1038/ng1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli B. A., McConville D. P., Byers J. S., III, Vincentz J. W., Barnes R. M., Firulli A. B. (2010). Analysis of a Hand1 hypomorphic allele reveals a critical threshold for embryonic viability. Dev. Dyn. 239, 2748-2760 10.1002/dvdy.22402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., Urasaki A., Kawakami K., McCallion A. S. (2006). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297-1305 10.1038/nprot.2006.230 [DOI] [PubMed] [Google Scholar]
- Garavito-Aguilar Z. V., Riley H. E., Yelon D. (2010). Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function. Development 137, 3215-3220 10.1242/dev.052225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Gemberling M., Karra R., Rosenfeld G. E., Evans T., Poss K. D. (2013). An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 23, 1221-1227 10.1016/j.cub.2013.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y., Shoji W. (2000). Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953-1960. [DOI] [PubMed] [Google Scholar]
- Hinits Y., Pan L., Walker C., Dowd J., Moens C. B., Hughes S. M. (2012). Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 369, 199-210 10.1016/j.ydbio.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606-609 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., MacRae C. A., Stainier D. Y. R., Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601-605 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G., Poss K. D. (2011a). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397-404 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Gupta V., Wang J., Holdway J. E., Wills A. A., Fang Y., Poss K. D. (2011b). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138, 2895-2902 10.1242/dev.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Laflamme M. A., Murry C. E. (2011). Heart regeneration. Nature 473, 326-335 10.1038/nature10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S., Scott I. C. (2011). Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 354, 123-133 10.1016/j.ydbio.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G., Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607-619 10.1016/j.cell.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Li Q., Kannan A., DeMayo F. J., Lydon J. P., Cooke P. S., Yamagishi H., Srivastava D., Bagchi M. K., Bagchi I. C. (2011). The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 331, 912-916 10.1126/science.1197454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Barbosa A. C., Chapman S. L., Bezprozvannaya S., Qi X., Richardson J. A., Yanagisawa H., Olson E. N. (2009). DNA binding-dependent and -independent functions of the Hand2 transcription factor during mouse embryogenesis. Development 136, 933-942 10.1242/dev.034025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably J. D., Burns C. G., Chen J.-N., Fishman M. C., Mohideen M.-A. P. K. (2003). heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138-2147 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- McFadden D. G., McAnally J., Richardson J. A., Charite J., Olson E. N. (2002). Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129, 3077-3088. [DOI] [PubMed] [Google Scholar]
- McFadden D. G., Barbosa A. C., Richardson J. A., Schneider M. D., Srivastava D., Olson E. N. (2005). The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189-201 10.1242/dev.01562 [DOI] [PubMed] [Google Scholar]
- Nam Y.-J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J. A., DiMaio J. M., Baker L. A., Bassel-Duby R., et al. (2013). Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 110, 5588-5593 10.1073/pnas.1301019110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palencia-Desai S., Kohli V., Kang J., Chi N. C., Black B. L., Sumanas S. (2011). Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development 138, 4721-4732 10.1242/dev.064998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G., Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188-2190 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Provost E., Rhee J., Leach S. D. (2007). Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45, 625-629 10.1002/dvg.20338 [DOI] [PubMed] [Google Scholar]
- Raya A., Koth C. M., Buscher D., Kawakami Y., Itoh T., Raya R. M., Sternik G., Tsai H.-J., Rodriguez-Esteban C., Izpisua-Belmonte J. C. (2003). Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 100 Suppl. 1, 11889-11895 10.1073/pnas.1834204100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risebro C. A., Smart N., Dupays L., Breckenridge R., Mohun T. J., Riley P. R. (2006). Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 133, 4595-4606 10.1242/dev.02625 [DOI] [PubMed] [Google Scholar]
- Rychlik J. L., Gerbasi V., Lewis E. J. (2003). The interaction between dHAND and Arix at the dopamine β-hydroxylase promoter region is independent of direct dHAND binding to DNA. J. Biol. Chem. 278, 49652-49660 10.1074/jbc.M308577200 [DOI] [PubMed] [Google Scholar]
- Schoenebeck J. J., Keegan B. R., Yelon D. (2007). Vessel and blood specification override cardiac potential in anterior mesoderm. Dev. Cell 13, 254-267 10.1016/j.devcel.2007.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.-J., Luo X., Qi X., Tan W., Huang G. N., Acharya A., Smith C. L., Tallquist M. D., Neilson E. G., et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599-604 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Thomas T., Lin Q., Kirby M. L., Brown D., Olson E. N. (1997). Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 16, 154-160 10.1038/ng0697-154 [DOI] [PubMed] [Google Scholar]
- Tamura M., Hosoya M., Fujita M., Iida T., Amano T., Maeno A., Kataoka T., Otsuka T., Tanaka S., Tomizawa S., et al. (2013). Overdosage of Hand2 causes limb and heart defects in the human chromosomal disorder partial trisomy distal 4q. Hum. Mol. Genet. 22, 2471-2481 10.1093/hmg/ddt099 [DOI] [PubMed] [Google Scholar]
- Thomas N. A., Koudijs M., van Eeden F. J. M., Joyner A. L., Yelon D. (2008). Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development 135, 3789-3799 10.1242/dev.024083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi T., Maeda J., Shin C. H., Ivey K. N., Black B. L., Olson E. N., Yamagishi H., Srivastava D. (2011). Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Biol. 351, 62-69 10.1016/j.ydbio.2010.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Panakova D., Kikuchi K., Holdway J. E., Gemberling M., Burris J. S., Singh S. P., Dickson A. L., Lin Y.-F., Sabeh M. K., et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421-3430 10.1242/dev.068601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Olson E. N., Bassel-Duby R. (2013). Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14, 529-541 10.1038/nrm3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Firulli A. B., Zhang X., Howard M. J. (2003). HAND2 synergistically enhances transcription of dopamine-β-hydroxylase in the presence of Phox2a. Dev. Biol. 262, 183-193 10.1016/S0012-1606(03)00361-0 [DOI] [PubMed] [Google Scholar]
- Yelon D., Ticho B., Halpern M. E., Ruvinsky I., Ho R. K., Silver L. M., Stainier D. Y. (2000). The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127, 2573-2582. [DOI] [PubMed] [Google Scholar]
- Zeng X.-X. I., Yelon D. (2014). Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 7, 951-960 10.1016/j.celrep.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cashman T. J., Nevis K. R., Obregon P., Carney S. A., Liu Y., Gu A., Mosimann C., Sondalle S., Peterson R. E., et al. (2011). Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645-648 10.1038/nature10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.