Abstract
Alternative splicing of pre-mRNAs is an important means of regulating developmental processes, yet the molecular mechanisms governing alternative splicing in embryonic contexts are just beginning to emerge. Polyglutamine-binding protein 1 (PQBP1) is an RNA-splicing factor that, when mutated, in humans causes Renpenning syndrome, an X-linked intellectual disability disease characterized by severe cognitive impairment, but also by physical defects that suggest PQBP1 has broader functions in embryonic development. Here, we reveal essential roles for PQBP1 and a binding partner, WBP11, in early development of Xenopus embryos. Both genes are expressed in the nascent mesoderm and neurectoderm, and morpholino knockdown of either causes defects in differentiation and morphogenesis of the mesoderm and neural plate. At the molecular level, knockdown of PQBP1 in Xenopus animal cap explants inhibits target gene induction by FGF but not by BMP, Nodal or Wnt ligands, and knockdown of either PQBP1 or WBP11 in embryos inhibits expression of fgf4 and FGF4-responsive cdx4 genes. Furthermore, PQBP1 knockdown changes the alternative splicing of FGF receptor-2 (FGFR2) transcripts, altering the incorporation of cassette exons that generate receptor variants (FGFR2 IIIb or IIIc) with different ligand specificities. Our findings may inform studies into the mechanisms underlying Renpenning syndrome.
Keywords: Alternative splicing, FGF, FGF receptor, Mesoderm, Neural, PQBP1, Renpenning syndrome, WBP11, Xenopus
INTRODUCTION
Polyglutamine binding protein-1 (PQBP1) is a 38 kDa nuclear protein abundantly expressed in the adult mammalian central nervous system (Komuro et al., 1999a; Waragai et al., 1999), and mutations in the human PQBP1 gene cause X-linked intellectual disability (XLID) diseases that include Renpenning, Sutherland–Haan, Hamel cerebropalatocardiac, Golabi–Ito–Hall and Porteous syndromes (Kalscheuer et al., 2003; Lenski et al., 2004; Stevenson et al., 2005; Cossee et al., 2006; Lubs et al., 2006; Martinez-Garay et al., 2007). These are collectively referred to as Renpenning syndrome, and affected patients display unifying clinical features, including intellectual disability, microcephaly and short stature. However, a variety of other physical manifestations can be observed, including midline and cardiac defects, small testes, lean muscle and reduced body mass (Stevenson et al., 2005; Germanaud et al., 2010). These phenotypes suggest that, in addition to brain development and cognition, PQBP1 regulates a broader scope of body patterning events, yet the normal developmental functions of PQBP1 and the pathogenic mechanisms underlying Renpenning syndrome remain largely undeciphered.
At the biochemical level, PQBP1 functions in transcription and pre-mRNA splicing. PQBP1 interacts with the C-terminus of activated RNA polymerase II (Okazawa et al., 2002) and with spliceosome components, including U5-15kD (Dim1p) (Waragai et al., 2000; Zhang et al., 2000) and the U2 snRNP component Sf3b1 (Wang et al., 2013). Most human disease-causing PQBP1 mutations truncate the C-terminal domain of PQBP1 that is required for interaction with the spliceosomal protein U5-15kD (Waragai et al., 2000; Zhang et al., 2000). Knockdown of PQBP1 in mouse embryo primary neurons reduces general splicing efficiency and promotes the use of alternative splice sites in a variety of transcripts (Wang et al., 2013). A missense mutation within the WW domain at the N-terminus of PQBP1 in Golabi–Ito–Hall (GIH) patients (Y65C) causes abnormal pre-mRNA splicing and inhibits PQBP1 binding to spliceosome components and a partner protein, WBP11 (Lubs et al., 2006; Tapia et al., 2010; Sudol et al., 2012). PQBP1 and WBP11 both associate with the BΔU1 or PRP19 spliceosomal complexes (Makarova et al., 2004; Deckert et al., 2006) and colocalize to nuclear speckles enriched in splicing factors (Komuro et al., 1999b; Llorian et al., 2004, 2005; Nicolaescu et al., 2008). A central, polar amino acid-rich domain (PRD) in mammalian PQBP1 can bind to proteins with polyglutamine (polyQ) tracts, such as transcription factors Brn2 and Ataxin-1, and the cytoplasmic transport protein Huntingtin (Htt), which is enhanced in poly(Q)-expanded pathogenic mutants of these human proteins (Waragai et al., 1999; Okazawa et al., 2002; Busch et al., 2003).
Although the molecular effects of PQBP1 have been studied in cultured cells and some model animals, little is known about its physiological function in developing embryos. Partial loss of function of PQBP1 in mice and Drosophila causes neurobehavioral phenotypes with no reported developmental defects (Ito et al., 2009; Tamura et al., 2010). However, a homozygous knockout null mouse line apparently cannot be generated, probably due to lethality, suggesting an essential role for PQBP1 in embryonic development. Overexpression of PQBP1 is also deleterious, promoting neuronal cell death in mice (Okuda et al., 2003; Marubuchi et al., 2005), and impaired long-term memory and behavioral abnormalities in Drosophila (Yoshimura et al., 2006). A PQBP1 gene duplication has been found in a patient with Renpenning syndrome, in which the PQBP1 protein may be overexpressed (Flynn et al., 2011). All of these findings suggest that maintaining PQBP1 within a restricted concentration range is essential for its proper biological actions.
Here, we use the animal model Xenopus laevis to investigate the embryonic expression and functions of PQBP1 and to identify potential molecular targets. We also evaluate one of its physiological partner proteins, WBP11, for which no developmental expression or functional information has been reported. We have found that both genes are expressed in the developing mesoderm and nervous system, and that loss of function (by morpholino knockdown) of either one results in abnormal gastrulation and neurulation. We also find that inhibiting PQBP1 or WBP11 causes similar defects in mesodermal and neural differentiation in Xenopus embryos, particularly in FGF-responsive gene expression. Mechanically, PQBP1 knockdown causes a shift in the alternative splicing of FGF receptor-2 (FGFR2) pre-mRNA, and a loss of MAPK activation in response to FGF4 (eFGF). Our findings may provide clues to the cause of developmental abnormalities associated with human PQBP1 mutations in Renpenning syndrome, with the particular suggestion that PQBP1 mutations may result in aberrant FGF signaling in embryonic and postnatal Renpenning patients.
RESULTS
PQBP1 knockdown causes mesodermal and neural defects in Xenopus embryos
Two X. laevis pqbp1 genes were revealed by BLAST searches of Xenopus expressed sequence tag (EST) and genome databases using mammalian pqbp1 sequences, which are homeologs that result from an ancient interspecies hybridization event that gave rise to the allotetraploid X. laevis lineage (Hughes and Hughes, 1993). The two predicted proteins (PQBP1a and PQBP1b) share 94% sequence identity, and are highly homologous to human PQBP1 (79-80% identity) except for the poly(Q)-binding PRD region, which is less conserved among species (supplementary material Fig. S1A).
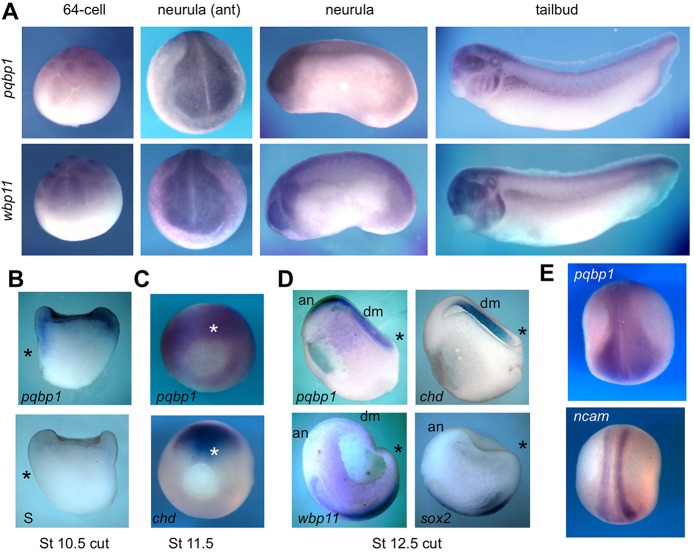
To lay the groundwork for predicting and testing potential developmental functions of PQBP1, we determined the spatiotemporal expression of these genes in developing Xenopus embryos by whole-mount in situ hybridization (WISH) and semi-quantitative RT-PCR (qPCR) (Fig. 1; supplementary material Fig. S2). During early cleavage (∼64 cells), pqbp1 transcripts were present in animal pole blastomeres (Fig. 1A), and in the early gastrula (stage 10.5) pqbp1 was expressed throughout the mesoderm (Fig. 1B,C). In the late gastrula, pqbp1 expression coincided with both chordin (marking the notochord) and sox2 (marking the prospective neural plate) (Fig. 1D). At neurula stages, pqbp1 was expressed in the neural plate and the head primordium (Fig. 1A,E) in a region that overlapped with ncam expression but also encompassed the cranial neural crest (Fig. 1E) and cells in the ventrolateral neural tube (supplementary material Fig. S3). In tailbud tadpole stages, pqbp1 was expressed in head, eyes, spinal cord and branchial arches (Fig. 1A).
Fig. 1.
pqbp1 and wbp11 genes are similarly expressed in mesoderm and neural tissues during Xenopus development. (A) WISH detection of pqbp1 and wbp11 transcripts in 64-cell blastula, neurula and tailbud tadpole; lateral views except anterior (ant) view of neurula. (B-E) WISH on intact (C,E) or longitudinally bisected (B,D) embryos with probes indicated. Asterisks mark the dorsal blastopore lip. (B) pqbp1 transcripts are present in the animal pole ectoderm and marginal zone in early gastrula (stage 10.5), upper panel; sense probe, lower panel. (C) Expression of pqbp1 and chordin in gastrulae, stage 11.5. (D) Expression of pqbp1, wbp11, chordin and sox2 in late gastrulae, dorsal-posterior views. Expression of pqbp1 overlaps with chordin in dorsal/axial mesoderm (arrows) and with sox2 in anterior neurectoderm. (E) Expression of pqbp1 and ncam mRNA in the neural plate of early neurula embryos. Note pqbp1 expression is in a broader region than ncam. an, anterior neurectoderm; chd, chordin; S, pqbp1 sense probe.
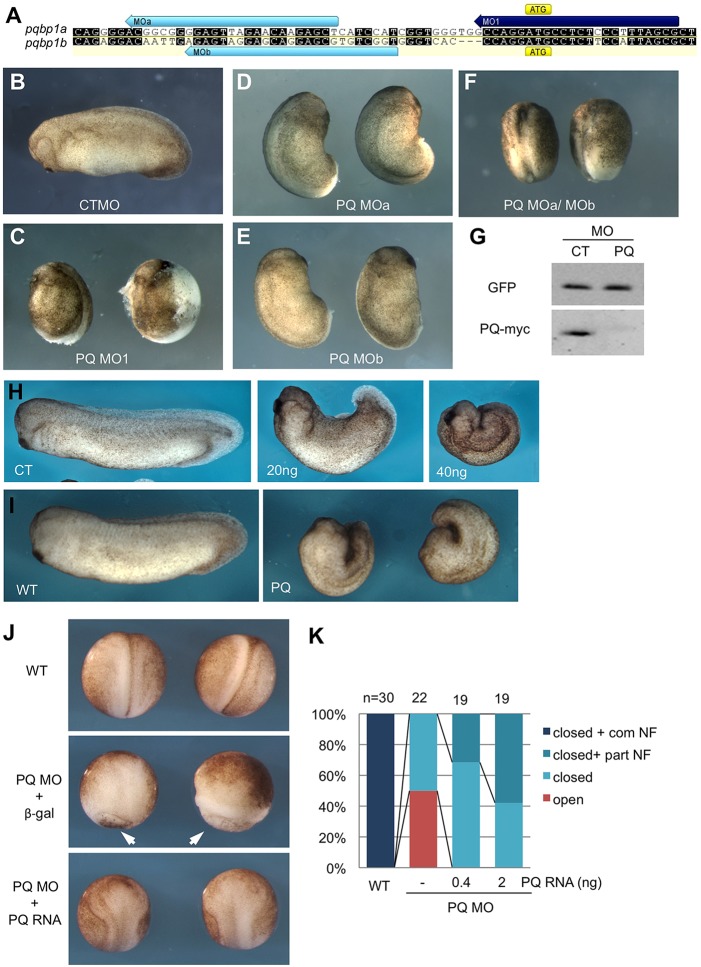
To determine which early developmental processes require PQBP1, we performed gene knockdown in X. laevis embryos with translation-blocking antisense morpholino oligonucleotides (MOs) that target each individual pqbp1 homeolog (MOa and MOb for pqbp1a and pqbp1b, respectively), or another MO predicted to inhibit both homeologs simultaneously (MO1; Fig. 2). The ability of MO1 to block pqbp1 mRNA translation was confirmed by western blot (Fig. 2G). We first examined the effects of PQBP1 knockdown in the mesoderm by bilaterally injecting MOs into the marginal zone of two-cell stage blastulae (Fig. 2). MO-injected embryos are referred to as morphants, and MO1 morphant phenotypes commonly included shortened anteroposterior (AP) body axes, small heads and no tail structures (Fig. 2C). These phenotypes were also accompanied by some cell dissociation at neurula stages. Interestingly, embryos injected singly with either MOa or MOb showed only mild developmental defects (Fig. 2D,E), but when these MOs were combined the resulting morphants lacked head and tail structures that were very similar to, but more severe than, the defects caused by MO1 (Fig. 2F). The congruent effects of knocking down one or both PQBP1 homeologs indicate that the individual PQBP1 morphant phenotypes are specific to the MOs and that both homeologs contribute similarly to normal X. laevis development.
Fig. 2.
Knockdown of PQBP1 causes defective embryo morphogenesis. (A) Design of three pqbp1 antisense morpholino oligonucleotides (MOs). MO1 (purple bar) targets the ATG start codon of pqbp1a and pqbp1b (with three nucleotide mismatches), while MOa and MOb (blue bars) target the 5′ UTRs of pqbp1 a and b, respectively. Identical nucleotides between pqbp1 a and b are indicated by black background. (B-F) Tailbud tadpole stage embryos injected bilaterally at the two-cell stage with 50 ng control (CT) or pqbp1 (PQ) MOs as indicated (embryos in F received 100 ng MOs in total). (G) Translation of C-terminal myc-tagged Xenopus PQBP1 (PQ-myc) was blocked by co-injection of pqbp1 MO1 but not control MO (CT). GFP mRNA co-injected as a negative control for MO targeting and loading control. (H) Phenotypes of tailbud stage embryos dorsally targeted with control (CT) or the indicated amount of pqbp1 MO1. (I) Overexpression of PQBP1 via injected mRNA (PQ) perturbs normal development. Left panel, wild-type embryo (stage 26); right panel, embryos injected dorsally with pqbp1 mRNA (2 ng) at the four-cell stage. (J,K) Gastrulation and neurulation defects are partially rescued by MO-resistant pqbp1 mRNA. PQ MO (30 ng) co-injected dorsally with 2 ng lacZ mRNA encoding β-galactosidase (β-gal), displayed perturbed gastrulation (arrowheads point to open blastopores) substantially rescued by co-injection of morpholino-resistant pqbp1 (2 ng) (J). Embryos injected with PQ MO and either 0.4 or 2 ng of pqbp1 mRNA scored for the following phenotypes (K): closed blastopore with complete neural folds (closed+com NF), closed blastopore with partial neural folds (closed+part NF), closed blastopore without neural folds (closed) or open blastopore (open). WT, wild type.
We next tested the effects of PQBP1 knockdown on development of the dorsal, Spemann–Mangold organizer mesoderm and adjacent neural tissue. MO1 was injected into the dorsal marginal zone (DMZ) of four-cell embryos, and the resulting morphants showed abnormal cell movements associated with gastrulation and neural plate folding, and later stage tadpoles had reduced head and shortened AP axial structures. The severity of the phenotypes was dose-dependent and increased as the amount of MO1 was raised (Fig. 2H). In some cases, open blastopores and abnormally folded neural plates were clearly visible (Fig. 2J,K). Importantly, these morphant defects could be partly rescued by co-injection of synthetic pqbp1 mRNA (0.4 ng or 2.0 ng) resistant to the PQBP1 MOs, indicating that the effects of MO1 are specific (Fig. 2J,K; supplementary material Fig. S5). Consistent with abnormal phenotypes caused by gain of function in other species, overexpression of PQBP1 in Xenopus embryos also caused severe developmental defects in gastrulation and neurulation (Fig. 2I; supplementary material Fig. S4), and those resembled the PQBP1 knockdown phenotypes (Fig. 2H).
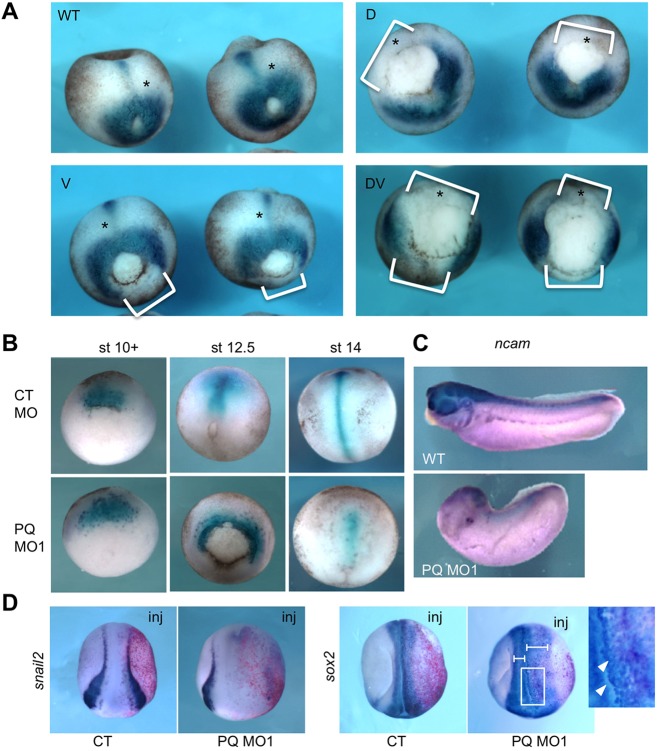
To further characterize the developmental defects in PQBP1 morphants, we examined molecular markers of regional patterning. As a combination of MOa and MOb had the strongest and most consistent effects, this pair was applied together in these and in subsequent PQBP1 knockdown experiments, unless otherwise noted. Examining general mesoderm, injection of PQBP1 MOs into any region of the mesoderm reduced or eliminated brachyury expression in recipient cells (Fig. 3A), which also underwent abnormal gastrulation movements (Fig. 3B). Furthermore, although chordin expression (marking the organizer mesoderm) was not inhibited by PQBP1 knockdown, chordin-expressing cells spread ventrolaterally during gastrulation, indicating abnormal convergence extension movements (Fig. 3B). Blastopore closure was delayed in morphants but eventually finished by the time control embryos reached neurula stage. However, the AP axes of PQBP1 morphants were much shorter than in controls, consistent with insufficient extension movements (Fig. 3B). In the neural domain, expression of the general neural marker ncam and neural crest marker snail2 (slug) were substantially decreased in PQBP1 morphants compared with controls (Fig. 3C,D). Furthermore, unilateral injection of PQBP1 MO1 at the two-cell stage inhibited sox2 expression and perturbed neural folding on the injected side (Fig. 3D). As seen in Fig. 2C, we also observed some cell dissociation at neurula stages, typified by sox2-positive cells detaching from the neural plate in PQBP1 morphants (Fig. 3D, magnified).
Fig. 3.
The effects of PQBP1 knockdown on embryonic mesoderm and neurectoderm. (A) Expression of the pan-mesodermal marker brachyury (bra) in late gastrula (stage 12.5) embryos injected with 20 ng pqbp1 MO1 into dorsal (D), ventral (V) or both (DV) blastomeres at the four-cell stage. Note lack of bra expression wherever the pqbp1 MO was injected (white brackets). (B) Expression of the dorsal (axial) mesoderm marker chordin in embryos injected dorsally with 20 ng of either control MO (CT MO) or pqbp1 MO1 (PQ MO1) at the four-cell stage. (C) Expression of neural marker ncam in uninjected (WT) or pqbp1 MO1-injected embryos targeted dorsally as in B. Lateral views with anterior to left. (D) Expression of the neural crest marker snail2 (slug) and the neural marker sox2 in neurula embryos (stage 19) injected in a single two-cell stage blastomere with 20 ng pqbp1 MO1 or control MO (CT) along with β-galactosidase lineage tracer (red). Dorsal views with anterior down. Perturbed neural folding is shown by differences between width of left and right neural folds (brackets). Loosely adherent sox2-positive cells, marked by arrowheads in the magnified view of the boxed area. WT, wild-type control embryos.
WBP11 is essential for normal development
The spliceosome protein WBP11 is an endogenous partner of mammalian PQBP1 (Nicolaescu et al., 2008; Tapia et al., 2010), but despite its functional importance in pre-mRNA splicing, nothing is known about its developmental expression or function. We identified two homeologs in X. laevis that encode predicted proteins with 95% (604/636) identity to each other and 74% identity to human WBP11 (supplementary material Fig. S1B). In developing Xenopus embryos, wbp11 is expressed maternally, with maternal transcripts persisting through blastula stages. Postzygotically, wbp11 is expressed in ectoderm and mesoderm during gastrulation, in neural plate during neurulation and in spinal cord and brain in tadpole stages (Fig. 1A; supplementary material Fig. S2). Wbp11 expression closely coincides with that of pqbp1, consistent with a physical and functional partnership between these proteins in mammalian systems. We validated the ability of Xenopus PQBP1 and WBP11 proteins to physically associate by co-immunoprecipitation from mammalian cultured cells (supplementary material Fig. S6A), and by observing their ability to colocalize to nuclei (supplementary material Fig. S6B).
Potential embryonic functions for WBP11 have not been evaluated in any species. Therefore, we tested whether WBP11 is required for development by inhibiting expression of the two X. laevis homeologs with translation-blocking MOs (Fig. 4A). We confirmed the specificity of the WBP11 MOs by showing that they block endogenous WBP11 translation in X. laevis embryos (Fig. 4B). Embryos injected bilaterally with WBP11 MOs displayed shortened AP axes, open blastopore remnants, small or absent heads and truncated tails (Fig. 4C). These phenotypes closely resembled those of PQBP1 morphants, so we tested whether combined knockdown of PQBP1 and WBP11 might prove more deleterious than individual gene knockdowns. Embryos were targeted in the marginal zone with low doses of either the PQBP1 or WBP11 MO, which alone allowed normal gastrulation and caused only a slight perturbation of neural folding (Fig. 4D). However, simultaneous low-dose knockdown of PQBP1 and WBP11 blocked gastrulation (note open blastopores) and neural folding (Fig. 4D). These seemingly additive phenotypic effects suggest that these proteins perform similar developmental functions.
Fig. 4.
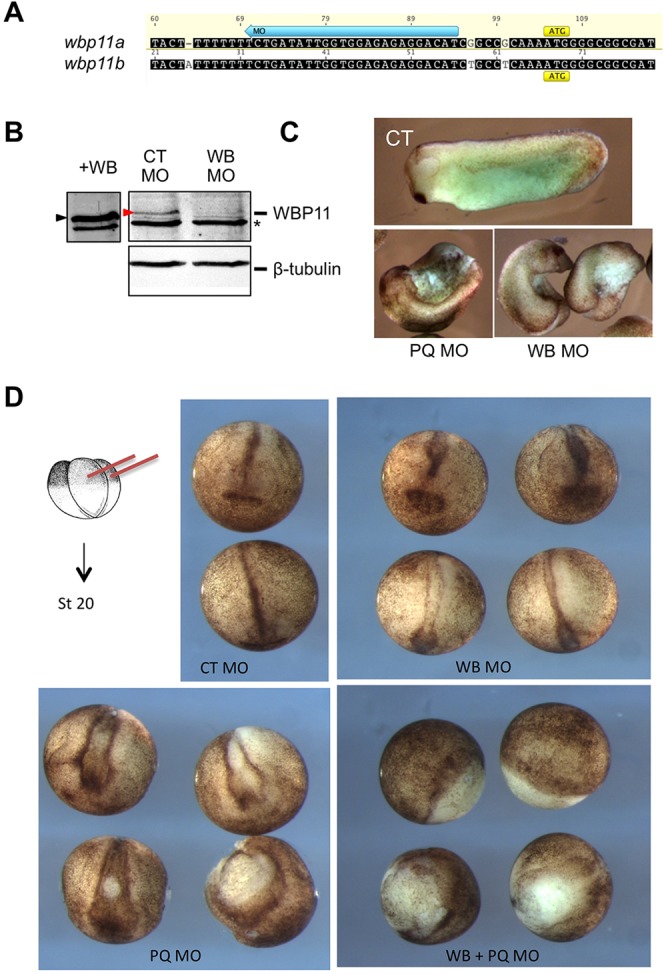
WBP11 knockdown resembles and enhances PQBP1 knockdown phenotypes. (A) WBP11 MO (blue arrow) targets the 5′ UTR of both X. laevis wbp11 homeologs. (B) Expression of endogenous WBP11 was blocked by injection of 40 ng wbp11 MO (WB MO) but not control MO (CT MO). Endogenous (red arrowhead) and overexpressed (black arrowhead) WBP11 were detected by western blot with anti-WBP11 antibodies. An asterisk indicates a non-specific band that, along with β-tubulin staining, controls for sample loading. (C) Tailbud stage embryos injected into two dorsal cells at the four-cell stage with 100 ng control MO (CT), a mix of pqbp1 MOa and MOb (50 ng each; PQ MO) or 50 ng wbp11 MO (WB MO). (D) Neurula (stage 20, anterior view) embryos injected dorsally at the four-cell stage (schematic drawing) with 75 ng control MO (CT), 25 ng wbp11 MO (WB), a mixture of pqbp1 MOa plus MOb (25 ng each; 50 ng total; PQ) or a combination of wbp11 and pqbp1 MOa and MOb (25 ng each; 75 ng total; PQ+WB).
Defective mesodermal and neural marker expression in PQBP1 and WBP11 morphants
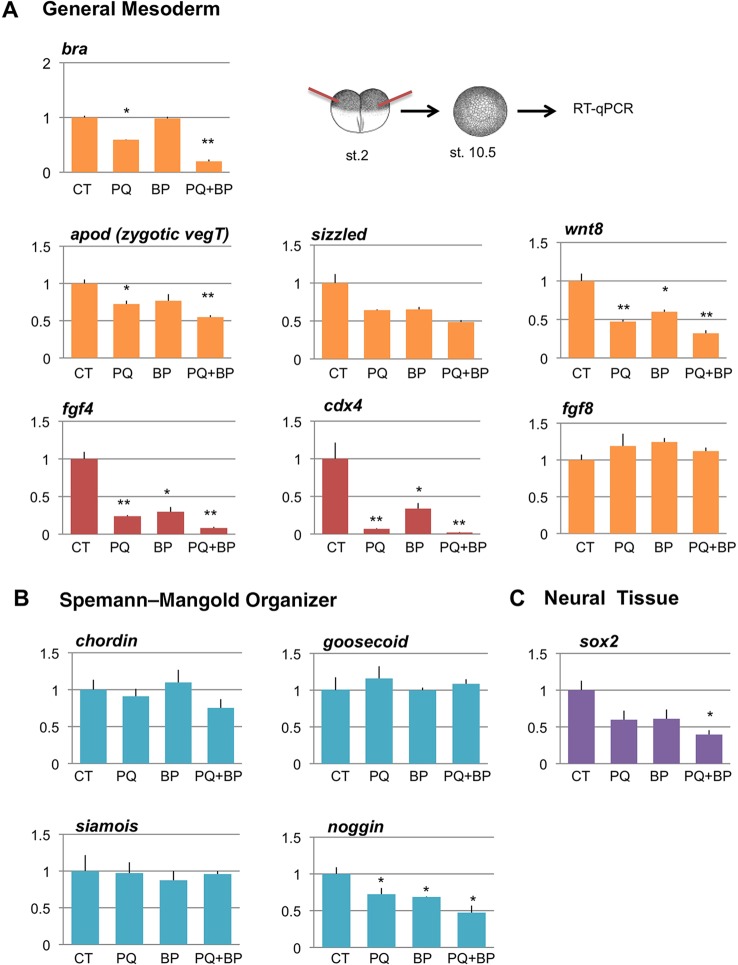
To better understand the PQBP1 and WBP11 morphant phenotypes, we scored regional and tissue-specific marker gene expression by qPCR on embryos injected bilaterally into the marginal zone at the two-cell stage and harvested at early gastrulation (stage 10.5). Results in Fig. 5A show that expression of several general mesodermal markers was reduced in morphants by individual or combined knockdown of PQBP1 or WBP11. To assess general mesoderm specification, we evaluated brachyury and found it was reduced by about 40% of control levels by PQBP1 knockdown, consistent with reduced brachyury expression seen in morphants by WISH (Fig. 3A). Combined knockdown of PQBP1 and WBP11 resulted in an even greater reduction (about 80%) of brachyury expression, despite normal brachyury levels with WBP11 knockdown alone (Fig. 5A). Another general mesodermal marker, zygotic vegT (apod), was reduced by about 50% of control levels by double knockdown of PQBP1 and WBP11.
Fig. 5.
Effects of PQBP1 and WBP11 knockdown on embryonic gene expression. qPCR performed on early gastrula (stage 10.5) morphant embryos injected bilaterally at the two-cell stage with 150 ng control (CT), 100 ng pqbp1 (PQ), 50 ng wbp11 (BP) or a combination of pqbp1 and wbp11 (PQ+BP) MOs (150 ng total). Values were plotted relative to the control cap signal and shown as mean±s.e.m of n=3 with Student's t-test to control embryos (CT) (*P<0.05 or **P<0.01). (A) Expression of general mesodermal markers brachyury (bra), antipodean (apod), sizzled, wnt8, fgf4, cdx4 and fgf8. (B) Expression of Spemann–Mangold organizer-specific markers, chordin, goosecoid, siamois and noggin. (C) Expression of early neural marker sox2.
We also observed distinct differences in the regional mesodermal marker expression in single or double PQBP1 and WBP11 morphants. Expression of the ventrolateral markers sizzled and wnt8 were reduced by about 40 and 60%, respectively, by either single or combined knockdown of PQBP1 and WBP11 (Fig. 5). Several genes that mark the Spemann–Mangold organizer, chordin, goosecoid and siamois, were expressed at near normal levels in single or double PQBP1/WBP11 morphants, but noggin expression was reduced by about 50% in the double morphants (Fig. 5B).
Two other mesodermal markers, fgf4 and cdx4, were the most significantly disrupted in PQBP1 morphants (Fig. 5A). Single knockdown of PQBP1 or WBP11 resulted in 70-90% reduction of fgf4 and cdx4, whereas combined knockdown almost completely eliminated their expression. Expression of another mesodermal FGF gene, fgf8, was normal in all PQBP1 or WBP11 morphants. The specificity of PQBP1 morpholino was further confirmed by the ability of MO-resistant pqbp1 mRNA to rescue fgf4 and cdx4 expression in PQBP1 morphants (supplementary material Fig. S7B), adding support to rescue experiments in whole-embryo morphants (Fig. 2J). Specificity is also supported by results showing that different PQBP1 morpholinos (MO1, MOa or MOb) had similar effects (supplementary material Fig. S7A).
In the neural ectoderm, single or combined knockdown of PQBP1 and WBP11 caused significant reduction (∼60%) in the expression of the early neural marker, sox2 (Fig. 5C), consistent with reduced sox2 and ncam expression in PQBP1 morphants observed using WISH (Fig. 3C,D).
We also tested PQBP1 knockdown in related X. tropicalis embryos, and found phenotypes similar to those of X. laevis PQBP1 morphants, with missing head and tail structures and short body length, as well as reduced fgf4 and cdx4 expression in early gastrulae (supplementary material Fig. S7C). The results of morphant analyses in X. laevis and X. tropicalis are consistent with each other and provide more general evidence that PQBP1 regulates essential developmental functions.
PQBP1 knockdown impairs responses to FGF but not other inductive signals
Multiple signaling pathways regulate mesoderm and neural induction, tissue patterning and morphogenesis in Xenopus embryos, including FGF, Nodal/Vg1, BMP and Wnt. Inhibition of any of these pathways might account for the phenotypes of PQBP1 or WBP11 morphants. To delineate which signaling pathways might be impaired in PQBP1 morphants, we examined the effects of PQBP1 knockdown on the induction of marker genes by FGF, Nodal, BMP or Wnt signals in Xenopus animal caps. Growth factors (as mRNAs) together with morpholinos were injected into animal poles of two-cell embryos, and caps were cut and screened for induction of target genes by qPCR. We found that PQBP1 morphant animal caps responded normally to Wnt8, Nodal2 (Xnr2) and BMP4, but response to FGF4 was significantly inhibited (Fig. 6B). Specifically, induction of cdx4 and bra by FGF4 was diminished in PQBP1 morphants, despite normal induction of cdx4 by Wnt8 or bra by BMP4. Furthermore, induction of fgf4 by FGF4, induced by FGF4/Brachyury-positive feedback loops (Isaacs et al., 1994; Fujii et al., 2008), was also blocked by PQBP1 knockdown.
Fig. 6.
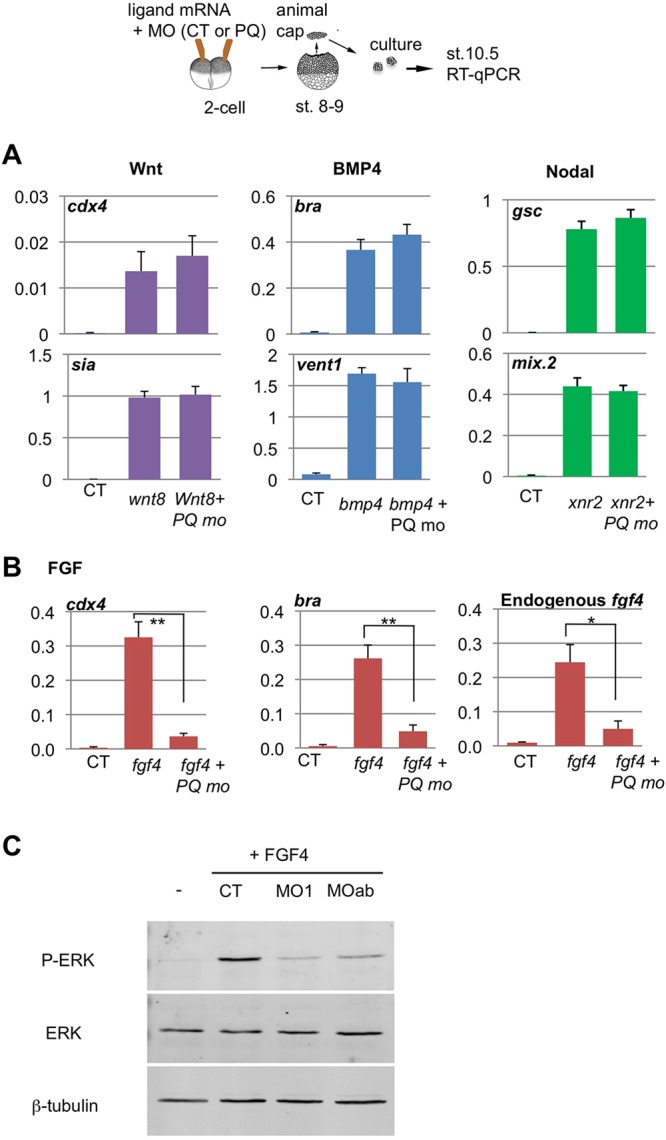
Effects of PQBP1 knockdown on animal cap response to growth factors. (A,B) Two-cell embryos were injected into the animal pole with growth factor mRNAs and MOs, animal caps were cut at mid-blastula and harvested at the equivalent of early gastrula stage, followed by qPCR, as depicted (top). Results were analyzed and plotted as per Fig. 5. (A) Marker gene induction by Wnt8 (50 pg), BMP4 (500 pg) and Nodal2 (Xnr2; 100 pg) was not affected by PQBP1 knockdown (50 ng MO). There was no statistically significant difference between CT and PQBP1 MO-injected caps treated with each ligand (Student's t-test, n=3). (B) Marker gene induction by FGF4 was significantly reduced by PQBP1 knockdown (*P<0.05 or **P<0.01; Student's t-test, triplicate biological replicates). Animal caps were injected with fgf4 mRNA (1 pg) and either control MO (50 ng), pqbp1 MO1 (50 ng) or MOa+MOb (25 ng each). (C) Phosphorylation of MAPK (Erk) was induced in fgf4-injected animal caps, but blocked by co-injection of pqbp1 MO, either MO1 or MOa+MOb. The levels of β-tubulin and total MAPK protein did not change among these samples.
We also tested whether PQBP1 might be required for normal operation of the FGF receptor tyrosine kinase (RTK) signaling cascade by analyzing MAPK (Erk) phosphorylation in PQBP1 morphant animal caps. Injection of 1pg fgf4 mRNA into animal caps induced phosphorylation of MAPK, but co-injection of either PQBP1 MO1 or MOa+MOb blocked MAPK phosphorylation (Fig. 6C). In sum, the results of experiments performed in whole embryos (Fig. 5) and isolated animal caps (Fig. 6) demonstrate that responses to FGF signaling require PQBP1, whereas Nodal, BMP and Wnt pathways do not.
PQBP1 regulates alternative splicing of an FGF receptor
One plausible explanation for the deleterious effects of PQBP1 knockdown on FGF responses is that PQBP1 acts at the level of gene transcription or pre-mRNA splicing for FGF signaling components. At the ligand level, alternative splicing of fgf4 and fgf8 pre-mRNA can generate protein variants with distinct biological activities (Fletcher et al., 2006; Guo and Li, 2007; Mayshar et al., 2008). In particular, two FGF8 isoforms, FGF8a and FGF8b, respectively, induce neural and mesodermal tissues in Xenopus embryos (Fletcher et al., 2006). However, we observed no differences in the splicing patterns of fgf4 or fgf8 transcripts in PQBP1 and/or WBP11 morphants compared with controls (data not shown).
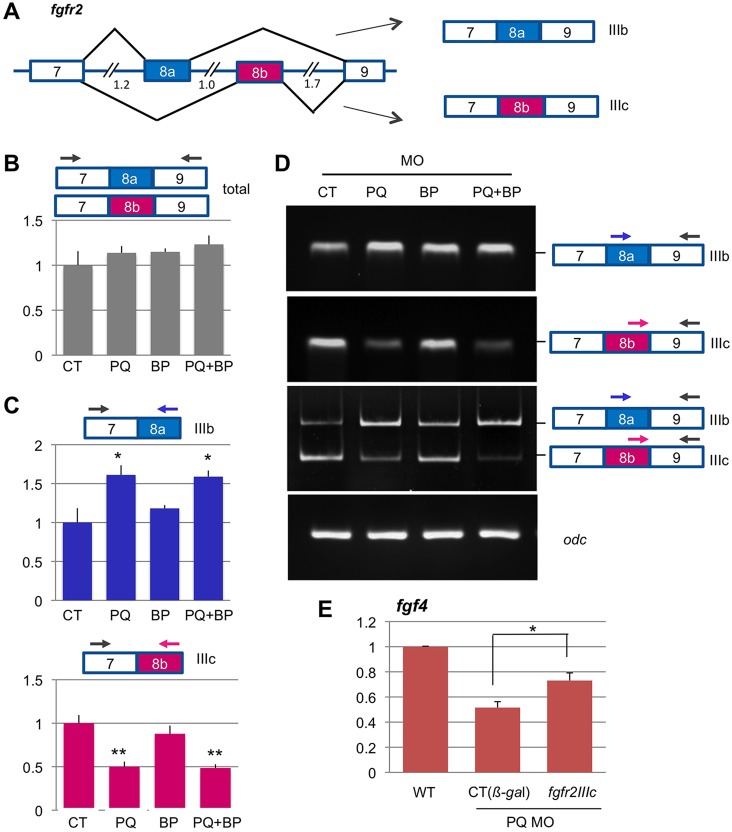
PQBP1 or WBP11 knockdown defects could also be explained by mis-splicing of the FGF receptors. Although only four genes encode FGF receptors, alternative splicing results in numerous isoforms with different ligand specificity (Ornitz et al., 1996; Eswarakumar et al., 2005; Zhang et al., 2006). FGF receptors 1, 2 and 3 incorporate exons 8a or 8b through alternative splicing of their pre-mRNAs, resulting in receptor isoforms IIIb or IIIc, respectively. These isoforms possess distinct ligand specificity resulting from unique residues in the immunoglobulin-like loop III of the extracellular, ligand-binding domain encoded by exon 8. Among mammalian receptors, the IIIb isoforms of FGFR1-3 preferably bind FGF3, FGF7 and FGF10, whereas IIIc isoforms prefer FGF4 and FGF8 (Eswarakumar et al., 2005; Holzmann et al., 2012). FGFR4 does not undergo alternative splicing of exon8, but binds FGF4 and FGF8b (Blunt et al., 1997).
In order to determine whether expression or splicing of FGF receptors was modified, we surveyed the levels of FGFR1-3 IIIb or IIIc isoform transcripts, as well as the total expression levels of all four FGF receptors, in control (control MO-injected), PQBP1 or WBP11 morphant Xenopus embryos at early gastrulation (stage 10.5). To assay IIIb and IIIc isoforms, we used primers that targeted exon8a or exon8b on the upstream side, and thus amplified only spliced transcripts that contained one of these alternative exons in fgfr1-3 (as illustrated for fgfr2 in Fig. 7C,D). In wild-type or control embryos, qPCR indicated the presence of all potential receptor isoforms, but relative isoform levels varied (Fig. 7; supplementary material Fig. S8). We observed a dramatic change in the isoform ratio of fgfr2 IIIb and IIIc transcripts, but no change in the total level of all frfgr2 transcripts were observed (Fig. 7B-D). In control embryos, fgfr2IIIc transcripts (containing exon 8b) were more abundant than those of fgfr2IIIb (containing exon 8a); this ratio was reversed, however, in PQBP1 morphants, as measured by qPCR (Fig. 7C) or by gel-based RT-PCR (Fig. 7D).
Fig. 7.
Alternative splicing pattern of fgfr2 exon 8a/b is altered by PQBP1 knockdown. (A) Alternative splicing incorporates either exon 8a or 8b into fgfr2 transcripts, which generates two isoforms of FGFR2: IIIb and IIIc, respectively (see text). Intron sizes (kb) indicated. (B,C) Sum total of qPCR measurements of fgfr2IIIb/c transcripts (B), and the relative levels of each splicing variant (C), on total RNA extracted from early gastrula (stage 10.5) morphant embryos bilaterally injected at the two-cell stage with MOs: 150 ng control (CT), 100 ng PQBP1 (PQ), 50 ng WBP11 (BP), or combined PQBP1 and WBP11 (PQ+BP) (150 ng total). Samples were prepared and analyzed as per Fig. 5 (*P<0.05 or **P<0.01; Student's t-test, n=3). (D) Semi-quantitative RT-PCR amplification products of alternative spliced exon 8a or 8b from embryos injected with morpholinos (as above). (E) Partial rescue of fgf4 expression in pqbp1 morphants (100 ng pqbp1 MO) by co-injection of 1 ng Xenopus fgfr2IIIc (*P<0.05; Student's t-test, n=3).
The other FGF receptors showed only minor shifts in isoform splicing and total levels in PQBP1 morphants (supplementary material Fig. S8). Knockdown of WBP11 alone had no significant effects on the relative level of these fgfr2 splice isoforms, nor did splicing enhance changes in PQBP1/WBP11 double morphants (Fig. 7; supplementary material Fig. S8). We did not observe an increase in unspliced transcripts resulting from knockdown of either PQBP1 or WBP11 (data not shown), which potentially could have happened if there was a general failure in pre-mRNA splicing, which appears unlikely.
As FGFR2IIIc, but not FGFR2IIIb, can respond to FGF4 or FGF8b ligands, the reduced levels of fgfr2IIIc transcripts in Xenopus PQBP1 morphants might be contributing to lowered FGF signaling and abnormal phenotypes. To address this possibility, we tested whether reduced expression of endogenous FGF response genes in PQBP1 morphants could be rescued by ectopic expression of an FGFR2IIIc isoform (Fig. 7E). To do this, we co-injected a synthetic mRNA encoding the X. tropicalis FGFR2IIIc isoform together with PQBP1 MOs and found that it partially rescued endogenous fgf4 and cdx4 gene expression. This result indicates that altered splicing of fgfr2 transcripts due to PQBP1 knockdown has biological consequences for embryogenesis.
DISCUSSION
PQBP1 and one of its binding proteins, WBP11, are implicated in RNAPII-dependent transcription and pre-mRNA splicing, but neither protein has clearly delineated roles or gene targets in developing embryos. Here, we have shown that during development of Xenopus embryos, PQBP1 and WBP11 are co-expressed in nascent mesodermal and neural tissues, and loss of function of these genes causes defects in mesodermal and neural patterning accompanied by abnormal gastrulation and neurulation, as well as by reduced mesodermal gene expression. At the molecular level, we find that PQBP1 is required for MAPK activation, target gene expression and mesoderm induction by FGF4 ligand, but not for responses to other mesodermal inducers and patterning agents, including Nodal2, BMP4 or Wnt8. Furthermore, we find that PQBP1 regulates alternative splicing of FGF receptor-2 transcripts, influencing the relative abundance of two FGFR2 isoforms (IIIb and IIIc) that have different binding specificities for FGF ligands. Our results reveal important roles for PQBP1 in vertebrate embryonic development, and those may be relevant to the molecular mechanisms of human developmental or neural/cognitive defects caused by pqbp1 mutations. Although WBP11 is not implicated in human disease or birth defects, our study is the first to indicate a normal developmental role for WBP11 in any embryo, raising the possibility that wbp11 mutations could contribute to birth defects.
PQBP1 and WBP11 are required for normal mesodermal and neural development
Despite the recognized importance of pqbp1 mutations in the etiology of human XLID syndromes, information about the embryonic expression and function of PQBP1 or WBP11 is limited to a description of mouse pqbp1 expression (Qi et al., 2005). Our analysis of Xenopus embryos shows that pqbp1 and wbp11 genes are maternally expressed, present throughout cleavage late blastula stages, then zygotically expressed in the mesoderm during gastrulation and the developing neural plate and nervous system of tadpoles.
The expression of pqbp1 in tailbud tadpoles appears homologous to that reported for embryonic and postnatal mice, where pqbp1 mRNA and protein are predominantly expressed in the central nervous system and neuronal stem cells (Waragai et al., 1999; Qi et al., 2005; Wang et al., 2013). The neural expression of pqbp1 in Xenopus and mouse is consistent with the fact that human pqbp1 mutations cause mental retardation and microcephaly. Moreover, the co-expression of pqbp1 and wbp11 in the same tissues of Xenopus embryos is consistent with the known physical and functional links between the two proteins (Komuro et al., 1999b; Llorian et al., 2004, 2005; Nicolaescu et al., 2008). Whether these proteins function together mechanistically to govern the same biochemical or embryonic processes remains to be determined. At neurula stages, PQBP1 or WBP11 morphants display incomplete neural tube closure accompanied by reduced expression of neural marker genes, underscoring their requirement for normal neural development.
The neural expression of pqbp1 in Xenopus embryos might have been anticipated, but our finding that both pqbp1 and wbp11 are expressed and required for normal development of mesoderm is new and reveals a broader role for these genes in early developmental processes, particularly in non-neural tissues. Knockdown of either gene causes abnormal cell migration during gastrulation, notably a failure of dorsal mesodermal convergence-extension movements. These defects in gastrulation are probably linked to reduced brachyury expression in PQBP1 morphants, as this gene is essential for mesodermal cell motility as well as differentiation. Some physical manifestations observed in Renpenning syndrome patients, including midline and cardiac defects, lean muscle and short stature, may be analogous to defects in mesoderm observed in PQBP1 morphants.
PQBP1 is required for FGF signaling and regulates splicing of FGF receptor transcripts
Our work further shows that the molecular and morphological defects observed in PQBP1 morphant embryos appear to be the result of aberrant FGF signaling. Direct tests of mesoderm induction in animal caps showed PQBP1 knockdown is required for signaling triggered by FGF4 but not Nodal, BMP or Wnt ligands. The loss of RTK signal transduction in particular indicates defects upstream of FGF-responsive gene transcription, and not some general defect in transcription or mRNA splicing of FGF target genes. FGF receptors, of which there are four, were foremost among candidates because their transcripts can undergo alternative splicing in various biological systems to generate receptor isoforms with different ligand-binding specificities. Our survey of fgfr expression and splicing shows that among the four receptors, fgfr2 showed drastically altered levels of alternatively spliced transcripts encoding isoforms IIIb and IIIc in PQBP1 morphants (supplementary material Fig. S8). Normally, early gastrula stage embryos express approximately 50% more fgfr2IIIc transcripts than fgfr2IIIb transcripts. We found that PQBP1 knockdown reverses this ratio, lowering fgfr2IIIc transcript levels to about half the level of fgfr2IIIb transcripts. As the FGFR2IIIc, but not IIIb, receptor isoform binds to FGF4 and FGF8, we postulate that the lowered abundance of FGFR2IIIc reduces the ability of the embryo to respond to mesoderm-inducing FGF4/8 ligands.
Furthermore, whereas PQBP1, FGF4 and FGF8 morphants share similar mesodermal, neural and gastrulation defects (Fisher et al., 2002; Fletcher et al., 2006; Isaacs et al., 2007), PQBP1 morphant phenotypes are more severely affected, yet rather similar to those generated by a dominant negative FGF receptor (XFD) or FGFR inhibitor SU5402, which block all FGF signaling, including that stimulated by FGF4 and FGF8b (Amaya et al., 1993; Delaune et al., 2005; Fletcher and Harland, 2008). This greater similarity between PQBP1 knockdown and wholesale loss of FGF signaling is potentially explained by the reduction of the FGFRIIIc isoform in PQBP1 morphants, but we cannot exclude the possibility that PQBP1 inhibition affects expression of other essential embryonic gene targets.
FGFR2 isoform switching via alternative splicing has been observed in multiple contexts, including normal epithelial-mesenchymal transition (Warzecha and Carstens, 2012), embryonic development (Eswarakumar et al., 2002; Rice et al., 2003; Takeuchi et al., 2005; Liu et al., 2011), human birth defects (Hajihosseini et al., 2001; Teebi et al., 2002; Ibrahimi et al., 2005), cancer and various pathologies (Katoh, 2008, 2009; Holzmann et al., 2012; Kelleher et al., 2013). Whether PQBP1 regulates fgfr2 alternative splicing in these normal and disease situations will be worth investigating. Our preliminary results also show changes in relative levels of alternative transcripts for fgfr1-3, but changes in fgfr2 transcript splicing were greater than fgfr1 and fgfr3 upon PQBP1 knockdown (supplementary material Fig. S8). The time and place of expression of all FGF receptors and their splicing variants in Xenopus embryos has not been well delineated, so further investigation is required to understand their precise roles in mesoderm and neural induction by FGFs and how pqbp1 impacts these functions.
In addition to our findings, previous studies have identified other factors regulating fgfr2 pre-mRNA splicing. Epithelial splicing regulatory proteins 1 and 2 (ESRP1 and ESRP2) govern alternative splicing of fgfr2 and other transcripts in a human epithelial cell line, and depletion of ESRP1 and ESRP2 causes splice switching from an epithelial IIIb form to a mesenchymal IIIc form (Warzecha et al., 2009). Fused in sarcoma (FUS) has also been shown to regulate fgfr2 transcript splicing in Xenopus embryos (Dichmann and Harland, 2012), and its knockdown causes a complete loss of fgfr2IIIc transcripts but has no effect on fgfr2IIIb transcript levels. The basis of FUS and PQBP1 target gene specificity is unknown, but FUS can directly bind RNA sequences, whereas direct interaction between PQBP1 and endogenous RNA was not evident by crosslinking immunoprecipitation (Wang et al., 2013). Whether FUS, ESRP1/2, PQBP1 and/or WBP11 interact or function together in the splicing of fgfr2 or other gene transcripts awaits investigation.
Potential implications for XLID
Besides cognitive defects, Renpenning patients have physical abnormalities that suggest underlying defects in the development of mesodermal as well as neural tissues (Stevenson et al., 2005; Germanaud et al., 2010). The details of human tissue pathologies associated with PQBP1 mutations, particularly those of non-brain tissue, are not well described, but the cardiac and skeletal muscular defects and reduced height prevalent in XLID patients may echo the mesodermal defects we observe in Xenopus PQBP1 morphants. FGF signaling is essential not only for development of mesodermal and neural tissues in vertebrate embryos, but also for the ongoing normal functions of adult tissues derived from these germ layers, including brain functions (Reuss and von Bohlen und Halbach, 2003; Iwata and Hevner, 2009; Mudo et al., 2009; Turner et al., 2012). Our findings raise the hypothesis that abnormal FGF signaling may underlie body, brain, and possibly cognitive, defects in Renpenning patients.
One aspect of PQBP1 function suggested by human and animal studies, and reinforced by our work, pertains to the dosage effects of PQBP1 on development and disease. Viable human PQBP1 mutations encode single residue changes, or frame-shifted proteins with C-terminal truncations that are probably partly functional (hypomorphic). No complete deletions of the PQBP1 gene or entire loss of its coding region have been described in PQBP1-linked XLIDs. Mouse gene knockout and overexpression models to investigate PQBP1 developmental functions have not been very successful, which seems to indicate a requirement for PQBP1 protein expression within a critical range: too little or too much protein is apparently lethal (Okuda et al., 2003). Our own studies here support that notion, as we observed dose-dependent embryonic defects in both loss- and gain-of-function tests. The highest doses of pqbp1 morpholinos or injected mRNA inhibit gastrulation and neurulation (Fig. 2; supplementary material Fig. S4), whereas moderate morpholino doses cause mild, dose-dependent phenotypes.
Finally, the PQBP1 partner protein WBP11 has not been implicated in the etiology of Renpenning or other XLIDs, or human diseases in general, but as these proteins appear to have partially overlapping functions in Xenopus development, it will be valuable to evaluate whether altered WBP11 expression or activity might contribute to Renpenning or other human diseases. Further investigation of PQBP1 and WBP11 target genes and molecular mechanisms using Xenopus embryos should contribute to a more complete understanding of how these genes regulate normal vertebrate developmental processes and, when mutated, contribute to human developmental and intellectual disabilities.
MATERIALS AND METHODS
Embryo manipulations
Xenopus laevis embryos were obtained by standard in vitro fertilization, de-jellied in 2% cysteine pH 8.0, microinjected and incubated for several hours in 3% Ficoll+0.5× MMR+10 μg/ml gentamycin and grown in 0.1× MMR+10 μg/ml gentamycin thereafter. All stages are according to Nieuwkoop and Faber (1994). Animal caps were excised at stage 8 and cultured in 0.5× MMR +10 μg/ml gentamycin until the sibling embryos reached stage 10.5.
Microinjection of MO and mRNA
Xenopus embryos were microinjected with MO and/or synthetic mRNA at two-cell or four-cell stages. All MOs were designed and synthesized by Gene Tools. The control MO was the standard scrambled MO, 5′-CCCTTACCT-CAGTTACAATTTATA-3′. pqbp1 MO1 is 5′-CGCTA-AAGGGAGAGGC-ATCCTGGC-3′. pqbp1a MO (MOa) is 5′-AGCTCTT-GTTCTAACTCCC-CGCCGT-3′. pqbp1b MO (MOb) is 5′-ACCGACACG-CTCCTGCTCCTA-CTCT-3′. wbp11 MO is 5′-ATGTCCTCTCTCCACC-AATATCAGA-3′, X. tropicalis pqbp1 MO is 5′-CGGCATCTTGGGCG-GCCAACCACAC-3′. Capped mRNAs were synthesized using AmpliCap SP6 High Yield Message Maker (Epicentre Biotechnologies) or mMESSAGE mMACHINE (Ambion) from the following linearized plasmids: pCS2-pqbp1, pSPYS-chordin, pCS2-bmp4, pCS2-wnt8, pSP64-fgf4, pCS2-wbp11. In some cases, lacZ mRNA encoding β-galactosidase was co-injected as a linage tracer, the activity of which was visualized in fixed embryos using Red-Gal (5-Bromo-6-chloro-3-indolyl β-D-galactopyranoside; Sigma-Aldrich).
X. laevis cDNA cloning and plasmid construction
A TBLASTN search identified two incomplete X. laevis cDNA clones (gi:8320261 and 8319534) homologous to mammalian PQBP1. Based on these EST sequences, the full-length pqbp1 clone was amplified by PCR from Xenopus cDNA (blastula) with the following primers: pqbp1 forward, 5′-GATCGTTGGCGTCATCAACAGG-3′ and pqbp1 reverse, 5′-GCACAA-GCAACTGTACAGCAGT-3′. The sequence of this clone completely matched to another X. laevis EST clone (NM_001091714) except for its shorter 5′ untranslated region (UTR) end. Identical sequences were identified in X. laevis genome scaffolds (Xenbase gbrowse laevis 6.0: Scaffold1707:1728996-1729209). MO-resistant Xenopus pqbp1 cDNA was amplified with the primers forward 5′-CCTCGAGATGCCGTTGCCTT-TAGCGCTTCAGGCT-3′, reverse 5′-CAATCAATAGGGGGCAGCAATG-3′, and subcloned into pCS2+ for in vitro transcription. This resulted in nine nucleotide mismatches at the pqbp1 morpholino recognition site, but normal amino acid coding was retained. C-terminal myc-tagged Xenopus PQBP1 was made by PCR cloning into the myc/pRK5-SK vector. Full-length X. laevis wbp11 was obtained by RT-PCR with the following primers: 5′-CCC-ATCGATATGGGGCGGCGATCCACTTCG-3′ and 5′-CGAATCGATGA-AAAGTTGCTTAGAATTAGCCG-3′ (restriction enzyme sites are underlined) with design based on an EST (BC057737). All cDNA identities were verified by DNA sequencing, and subcloned into pCS2+ unless otherwise noted. Full-length X. tropicalis fgfr2IIIc was a gift from Dr Richard Harland (University of California, Berkeley, USA).
In situ hybridization
In situ hybridization was performed with digoxigenin-labeled probes as previously described (Alexandrova and Thomsen, 2006). BM Purple was used as a chromogenic substrate (Roche). Sagittal or cross sections were performed during MEMPFA fixation. Template plasmids for making probes were pBS-ncam, pBS-sox2, pGEM4Z-chordin, pXT1-brachyury, pMX-snail2, pCS2-pqbp1 and pCS2-wbp11.
Quantitative RT-PCR
Total RNA was extracted from ten animal caps or three embryos and used for cDNA synthesis as previously described by Callery et al. (2005). Quantitative RT-PCR using the LightCycler System and SYBR Green reagent (Roche Applied Science) according to Kalkan et al. (2009) using primers listed in supplementary material Tables S1 and S2. Relative amounts of PCR product were determined based on standard curves derived from 1:1, 1:10 and 1:100 dilutions of the cDNA from control embryos, with target gene expression normalized to the relative levels of ornithine decarboxylase (odc) transcripts. Most primers were designed to have annealing temperature of 60°C, and product sizes between 100-200 bp; most PCR conditions were annealing at 55°C, elongation 12 s at 72°C, denaturation at 94°C for 10 s. cDNA from three or more biological replicate experiments were amplified in most Light Cycler reactions (see text) and statistical analyses were conducted using Student's t-test.
Protein analysis, immunoprecipitation and immunofluorescence
For detection of phosphorylated MAPK, animal caps were lysed in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, together with a proteinase and phosphatase inhibitor cocktail (Roche). Solubilized proteins were separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane (Bio-Rad) and stained with anti-phospho-p44/42 MAPK (Cell Signaling, 4370; 1:2000), anti-p44/42 MAP kinase (Cell Signaling, 4695P; 1:1000) and anti-β-tubulin (Accurate Chemical and Scientific Corporation, BYA6068-1; 1:5000) antibodies (Suga et al., 2006). Signals were visualized by Alexa dye-conjugated goat anti-mouse and anti-rabbit antibody (Molecular Probes), and visualized using the Odyssey Infrared Imager (LI-COR). For co-immunoprecipitation, HA-pqbp1 was co-transfected with myc-wbp11 into COS-1 cells using linear polyethyleneimine transfection reagent. Twenty-four hours after transfection cells were lysed with PBS containing 1% Triton X-100 and complete protease inhibitors (Roche). The lysate was incubated with anti-HA rabbit polyclonal (Y-11, Santa Cruz, sc-805) or anti-myc mouse monoclonal (ATCC, 9E10 hybridoma) antibodies for 2 h at 4°C, then incubated with protein A or G agarose beads for 1 h at 4°C. Agarose beads were spun and washed with cold lysis buffer several times, then boiled in SDS sample buffer before loading onto SDS-PAGE gels. Proteins were immunoblotted and stained with anti-myc (1:1000) or anti-HA (1:500) antibodies. Transfected and endogenous cellular control proteins were visualized on the blots, as above, with mouse anti-β-tubulin (Santa Cruz, sc-58884; 1:500), mouse anti-GFP (Santa Cruz, sc-9996; 1:500) and anti-WBP11 rabbit polyclonal antibodies prepared by our lab using a Xenopus WBP11-GST fusion protein. For immunofluorescence assays, HA-pqbp1 and myc-wbp11 were transfected into COS-1 cells and detected with anti-myc mouse monoclonal 9E10 and anti-HA rabbit polyclonal antibodies, detected with Alexa 488 (Molecular Probes, A-11029, 1:1000) goat anti-mouse and Alexa 594 goat anti-rabbit (Molecular Probes, A-11037, 1:1000) secondary antibodies. COS-1 cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum.
Supplementary Material
Acknowledgements
We thank Dr R. Harland (University of California, Berkeley, USA) for the fgfr2c plasmid and members of the G.H.T. lab for experimental advice and comments on the manuscript. We acknowledge Xenbase for facilitating gene analyses, and the National Xenopus Resource (NXR) and European Xenopus Resource Center (EXRC) for supplying animals and reagents.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
Y.I. and G.H.T. conceived and designed experiments, Y.I. carried out all experiments, and Y.I. and G.H.T. wrote the manuscript.
Funding
This research was supported partially by the U.S. National Institutes of Health [grants R03HD064908 and R01HD032429 to G.H.T.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.106658/-/DC1
References
- Alexandrova, E. M. and Thomsen, G. H. (2006). Smurf1 regulates neural patterning and folding in Xenopus embryos by antagonizing the BMP/Smad1 pathway. Dev. Biol. 299, 398-410 10.1016/j.ydbio.2006.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya, E., Stein, P. A., Musci, T. J. and Kirschner, M. W. (1993). FGF signalling in the early specification of mesoderm in Xenopus. Development 118, 477-487. [DOI] [PubMed] [Google Scholar]
- Blunt, A. G., Lawshé, A., Cunningham, M. L., Seto, M. L., Ornitz, D. M. and MacArthur, C. A. (1997). Overlapping expression and redundant activation of mesenchymal fibroblast growth factor (FGF) receptors by alternatively spliced FGF-8 ligands. J. Biol. Chem. 272, 3733-3738 10.1074/jbc.272.6.3733 [DOI] [PubMed] [Google Scholar]
- Busch, A., Engemann, S., Lurz, R., Okazawa, H., Lehrach, H. and Wanker, E. E. (2003). Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J. Biol. Chem. 278, 41452-41461 10.1074/jbc.M303354200 [DOI] [PubMed] [Google Scholar]
- Callery, E. M., Smith, J. C and Thomsen, G. H. (2005). The ARID domain protein dril1 is necessary for TGF(beta) signaling in Xenopus embryos. Dev. Biol. 278, 542-559 10.1016/j.ydbio.2004.11.017 [DOI] [PubMed] [Google Scholar]
- Cossée, M., Demeer, B., Blanchet, P., Echenne, B., Singh, D., Hagens, O., Antin, M., Finck, S., Vallee, L., Dollfus, H.et al. (2006). Exonic microdeletions in the X-linked PQBP1 gene in mentally retarded patients: a pathogenic mutation and in-frame deletions of uncertain effect. Eur. J. Hum. Genet. 14, 418-425 10.1038/sj.ejhg.5201593 [DOI] [PubMed] [Google Scholar]
- Deckert, J., Hartmuth, K., Boehringer, D., Behzadnia, N., Will, C. L., Kastner, B., Stark, H., Urlaub, H. and Luhrmann, R. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528-5543 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune, E., Lemaire, P. and Kodjabachian, L. (2005). Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 132, 299-310 10.1242/dev.01582 [DOI] [PubMed] [Google Scholar]
- Dichmann, D. S. and Harland, R. M. (2012). fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev. 26, 1351-1363 10.1101/gad.187278.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar, V. P., Monsonego-Ornan, E., Pines, M., Antonopoulou, I., Morriss-Kay, G. M. and Lonai, P. (2002). The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development 129, 3783-3793. [DOI] [PubMed] [Google Scholar]
- Eswarakumar, V. P., Lax, I. and Schlessinger, J. (2005). Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139-149 10.1016/j.cytogfr.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Fisher, M. E., Isaacs, H. V. and Pownall, M. E. (2002). eFGF is required for activation of XmyoD expression in the myogenic cell lineage of Xenopus laevis. Development 129, 1307-1315. [DOI] [PubMed] [Google Scholar]
- Fletcher, R. B. and Harland, R. M. (2008). The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev. Dyn. 237, 1243-1254 10.1002/dvdy.21517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, R. B., Baker, J. C. and Harland, R. M. (2006). FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133, 1703-1714 10.1242/dev.02342 [DOI] [PubMed] [Google Scholar]
- Flynn, M., Zou, Y. S. and Milunsky, A. (2011). Whole gene duplication of the PQBP1 gene in syndrome resembling Renpenning. Am. J. Med. Genet. A 155, 141-144 10.1002/ajmg.a.33756 [DOI] [PubMed] [Google Scholar]
- Fujii, H., Sakai, M., Nishimatsu, S.-I., Nohno, T., Mochii, M., Orii, H. and Watanabe, K. (2008). VegT, eFGF and Xbra cause overall posteriorization while Xwnt8 causes eye-level restricted posteriorization in synergy with chordin in early Xenopus development. Dev. Growth Differ. 50, 169-180 10.1111/j.1440-169X.2008.01014.x [DOI] [PubMed] [Google Scholar]
- Germanaud, D., Rossi, M., Bussy, G., Gerard, D., Hertz-Pannier, L., Blanchet, P., Dollfus, H., Giuliano, F., Bennouna-Greene, V., Sarda, P.et al. (2010). The Renpenning syndrome spectrum: new clinical insights supported by 13 new PQBP1-mutated males. Clin. Genet. 79, 225-235 10.1111/j.1399-0004.2010.01551.x [DOI] [PubMed] [Google Scholar]
- Guo, Q. and Li, J. Y. H. (2007). Distinct functions of the major Fgf8 spliceform, Fgf8b, before and during mouse gastrulation. Development 134, 2251-2260 10.1242/dev.004929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini, M. K., Wilson, S., De Moerlooze, L. and Dickson, C. (2001). A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc. Natl. Acad. Sci. USA 98, 3855-3860 10.1073/pnas.071586898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann, K., Grunt, T., Heinzle, C., Sampl, S., Steinhoff, H., Reichmann, N., Kleiter, M., Hauck, M. and Marian, B. (2012). Alternative splicing of fibroblast growth factor receptor IgIII loops in cancer. J. Nucleic Acids 2012, 950508 10.1155/2012/950508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, M. K. and Hughes, A. L. (1993). Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 10, 1360-1369. [DOI] [PubMed] [Google Scholar]
- Ibrahimi, O. A., Chiu, E. S., McCarthy, J. G. and Mohammadi, M. (2005). Understanding the molecular basis of Apert syndrome. Plast. Reconstr. Surg. 115, 264-270. [PubMed] [Google Scholar]
- Isaacs, H. V., Pownall, M. E. and Slack, J. M. (1994). eFGF regulates Xbra expression during Xenopus gastrulation. EMBO J. 13, 4469-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, H. V., Deconinck, A. E. and Pownall, M. E. (2007). FGF4 regulates blood and muscle specification in Xenopus laevis. Biol. Cell 99, 165-173 10.1042/BC20060103 [DOI] [PubMed] [Google Scholar]
- Ito, H., Yoshimura, N., Kurosawa, M., Ishii, S., Nukina, N. and Okazawa, H. (2009). Knock down of PQBP1 impairs anxiety-related cognition in mouse. Hum. Mol. Genet. 18, 4239-4254 10.1093/hmg/ddp378 [DOI] [PubMed] [Google Scholar]
- Iwata, T. and Hevner, R. F. (2009). Fibroblast growth factor signaling in development of the cerebral cortex. Dev. Growth Differ. 51, 299-323 10.1111/j.1440-169X.2009.01104.x [DOI] [PubMed] [Google Scholar]
- Kalkan, T., Iwasaki, Y., Park, C. Y. and Thomsen, G. H. (2009). Tumor necrosis factor-receptor-associated factor-4 is a positive regulator of transforming growth factor-beta signaling that affects neural crest formation. Mol. Biol. Cell 20, 3436-3450 10.1091/mbc.E08-03-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer, V. M., Freude, K., Musante, L., Jensen, L. R., Yntema, H. G., Gécz, J., Sefiani, A., Hoffmann, K., Moser, B., Haas, S.et al. (2003). Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat. Genet. 35, 313-315 10.1038/ng1264 [DOI] [PubMed] [Google Scholar]
- Katoh, M. (2008). Cancer genomics and genetics of FGFR2 (Review). Int. J. Oncol. 33, 233-237. [PubMed] [Google Scholar]
- Katoh, M. (2009). FGFR2 abnormalities underlie a spectrum of bone, skin, and cancer pathologies. J. Invest. Dermatol. 129, 1861-1867 10.1038/jid.2009.97 [DOI] [PubMed] [Google Scholar]
- Kelleher, F. C., O'Sullivan, H., Smyth, E., McDermott, R. and Viterbo, A. (2013). Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis 34, 2198-2205 10.1093/carcin/bgt254 [DOI] [PubMed] [Google Scholar]
- Komuro, A., Saeki, M. and Kato, S. (1999a). Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res. 27, 1957-1965 10.1093/nar/27.9.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro, A., Saeki, M. and Kato, S. (1999b). Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J. Biol. Chem. 274, 36513-36519 10.1074/jbc.274.51.36513 [DOI] [PubMed] [Google Scholar]
- Lenski, C., Abidi, F., Meindl, A., Gibson, A., Platzer, M., Frank Kooy, R., Lubs, H. A., Stevenson, R. E., Ramser, J. and Schwartz, C. E. (2004). Novel truncating mutations in the polyglutamine tract binding protein 1 gene (PQBP1) cause Renpenning syndrome and X-linked mental retardation in another family with microcephaly. Am. J. Hum. Genet. 74, 777-780 10.1086/383205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D.-W., Hsu, C.-H., Tsai, S.-M., Hsiao, C.-D. and Wang, W.-P. (2011). A variant of fibroblast growth factor receptor 2 (Fgfr2) regulates left-right asymmetry in zebrafish. PLoS ONE 6, e21793 10.1371/journal.pone.0021793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian, M., Beullens, M., Andrés, I., Ortiz, J.-M. and Bollen, M. (2004). SIPP1, a novel pre-mRNA splicing factor and interactor of protein phosphatase-1. Biochem. J. 378, 229-238 10.1042/BJ20030950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian, M., Beullens, M., Lesage, B., Nicolaescu, E., Beke, L., Landuyt, W., Ortiz, J.-M. and Bollen, M. (2005). Nucleocytoplasmic shuttling of the splicing factor SIPP1. J. Biol. Chem. 280, 38862-38869 10.1074/jbc.M509185200 [DOI] [PubMed] [Google Scholar]
- Lubs, H., Abidi, F. E., Echeverri, R., Holloway, L., Meindl, A., Stevenson, R. E. and Schwartz, C. E. (2006). Golabi-Ito-Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J. Med. Genet. 43, e30 10.1136/jmg.2005.037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, O. V., Makarov, E. M., Urlaub, H., Will, C. L., Gentzel, M., Wilm, M. and Lührmann, R. (2004). A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23, 2381-2391 10.1038/sj.emboj.7600241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Garay, I., Tomás, M., Oltra, S., Ramser, J., Moltó, M. D., Prieto, F., Meindl, A., Kutsche, K. and Martínez, F. (2007). A two base pair deletion in the PQBP1 gene is associated with microphthalmia, microcephaly, and mental retardation. Eur. J. Hum. Genet. 15, 29-34 10.1038/sj.ejhg.5201717 [DOI] [PubMed] [Google Scholar]
- Marubuchi, S., Wada, Y.-I., Okuda, T., Hara, Y., Qi, M.-L., Hoshino, M., Nakagawa, M., Kanazawa, I. and Okazawa, H. (2005). Polyglutamine tract-binding protein-1 dysfunction induces cell death of neurons through mitochondrial stress. J. Neurochem. 95, 858-870 10.1111/j.1471-4159.2005.03405.x [DOI] [PubMed] [Google Scholar]
- Mayshar, Y., Rom, E., Chumakov, I., Kronman, A., Yayon, A. and Benvenisty, N. (2008). Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells 26, 767-774 10.1634/stemcells.2007-1037 [DOI] [PubMed] [Google Scholar]
- Mudò, G., Bonomo, A., Di Liberto, V., Frinchi, M., Fuxe, K. and Belluardo, N. (2009). The FGF-2/FGFRs neurotrophic system promotes neurogenesis in the adult brain. J. Neural Transm. 116, 995-1005 10.1007/s00702-009-0207-z [DOI] [PubMed] [Google Scholar]
- Nicolaescu, E., Beullens, M., Lesage, B., Keppens, S., Himpens, B. and Bollen, M. (2008). Nature of the nuclear inclusions formed by PQBP1, a protein linked to neurodegenerative polyglutamine diseases. Eur. J. Cell Biol. 87, 817-829 10.1016/j.ejcb.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop, P. D. and Faber, J. (1994). Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. New York: Garland Publishing. [Google Scholar]
- Okazawa, H., Rich, T., Chang, A., Lin, X., Waragai, M., Kajikawa, M., Enokido, Y., Komuro, A., Kato, S., Shibata, M.et al. (2002). Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 34, 701-713 10.1016/S0896-6273(02)00697-9 [DOI] [PubMed] [Google Scholar]
- Okuda, T., Hattori, H., Takeuchi, S., Shimizu, J., Ueda, H., Palvimo, J. J., Kanazawa, I., Kawano, H., Nakagawa, M. and Okazawa, H. (2003). PQBP-1 transgenic mice show a late-onset motor neuron disease-like phenotype. Hum. Mol. Genet. 12, 711-725 10.1093/hmg/ddg084 [DOI] [PubMed] [Google Scholar]
- Ornitz, D. M., Xu, J., Colvin, J. S., McEwen, D. G., MacArthur, C. A., Coulier, F., Gao, G. and Goldfarb, M. (1996). Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 271, 15292-15297 10.1074/jbc.271.25.15292 [DOI] [PubMed] [Google Scholar]
- Qi, Y., Hoshino, M., Wada, Y.-I., Marubuchi, S., Yoshimura, N., Kanazawa, I., Shinomiya, K.-I. and Okazawa, H. (2005). PQBP-1 is expressed predominantly in the central nervous system during development. Eur. J. Neurosci. 22, 1277-1286 10.1111/j.1460-9568.2005.04339.x [DOI] [PubMed] [Google Scholar]
- Reuss, B. and von Bohlen und Halbach, O. (2003). Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 313, 139-157 10.1007/s00441-003-0756-7 [DOI] [PubMed] [Google Scholar]
- Rice, D. P. C., Rice, R. and Thesleff, I. (2003). Fgfr mRNA isoforms in craniofacial bone development. Bone 33, 14-27 10.1016/S8756-3282(03)00163-7 [DOI] [PubMed] [Google Scholar]
- Stevenson, R. E., Bennett, C. W., Abidi, F., Kleefstra, T., Porteous, M., Simensen, R. J., Lubs, H. A., Hamel, B. C. J. and Schwartz, C. E. (2005). Renpenning syndrome comes into focus. Am. J. Med. Genet. A 134, 415-421 10.1002/ajmg.a.30664 [DOI] [PubMed] [Google Scholar]
- Sudol, M., McDonald, C. B. and Farooq, A. (2012). Molecular insights into the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1. FEBS Lett. 586, 2795-2799 10.1016/j.febslet.2012.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga, A., Hikasa, H. and Taira, M. (2006). Xenopus ADAMTS1 negatively modulates FGF signaling independent of its metalloprotease activity. Dev. Biol. 295, 26-39 10.1016/j.ydbio.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Takeuchi, Y., Molyneaux, K., Runyan, C., Schaible, K. and Wylie, C. (2005). The roles of FGF signaling in germ cell migration in the mouse. Development 132, 5399-5409 10.1242/dev.02080 [DOI] [PubMed] [Google Scholar]
- Tamura, T., Horiuchi, D., Chen, Y.-C., Sone, M., Miyashita, T., Saitoe, M., Yoshimura, N., Chiang, A.-S. and Okazawa, H. (2010). Drosophila PQBP1 regulates learning acquisition at projection neurons in aversive olfactory conditioning. J. Neurosci. 30, 14091-14101 10.1523/JNEUROSCI.1319-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia, V. E., Nicolaescu, E., McDonald, C. B., Musi, V., Oka, T., Inayoshi, Y., Satteson, A. C., Mazack, V., Humbert, J., Gaffney, C. J.et al. (2010). Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J. Biol. Chem. 285, 19391-19401 10.1074/jbc.M109.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teebi, A. S., Kennedy, S., Chun, K. and Ray, P. N. (2002). Severe and mild phenotypes in Pfeiffer syndrome with splice acceptor mutations in exon IIIc of FGFR2. Am. J. Med. Genet. 107, 43-47 10.1002/ajmg.10125 [DOI] [PubMed] [Google Scholar]
- Turner, C. A., Watson, S. J. and Akil, H. (2012). The fibroblast growth factor family: neuromodulation of affective behavior. Neuron 76, 160-174 10.1016/j.neuron.2012.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Moore, M. J., Adelmant, G., Marto, J. A. and Silver, P. A. (2013). PQBP1, a factor linked to intellectual disability, affects alternative splicing associated with neurite outgrowth. Genes Dev. 27, 615-626 10.1101/gad.212308.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waragai, M., Lammers, C.-H., Takeuchi, S., Imafuku, I., Udagawa, Y., Kanazawa, I., Kawabata, M., Mouradian, M. M. and Okazawa, H. (1999). PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum. Mol. Genet. 8, 977-987 10.1093/hmg/8.6.977 [DOI] [PubMed] [Google Scholar]
- Waragai, M., Junn, E., Kajikawa, M., Takeuchi, S., Kanazawa, I., Shibata, M., Mouradian, M. M. and Okazawa, H. (2000). PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5-15kD/dim1p via the carboxyl-terminal domain. Biochem. Biophys. Res. Commun. 273, 592-595 10.1006/bbrc.2000.2992 [DOI] [PubMed] [Google Scholar]
- Warzecha, C. C. and Carstens, R. P. (2012). Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 22, 417-427 10.1016/j.semcancer.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha, C. C., Shen, S., Xing, Y. and Carstens, R. P. (2009). The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6, 546-562 10.4161/rna.6.5.9606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, N., Horiuchi, D., Shibata, M., Saitoe, M., Qi, M.-L. and Okazawa, H. (2006). Expression of human PQBP-1 in Drosophila impairs long-term memory and induces abnormal courtship. FEBS Lett. 580, 2335-2340 10.1016/j.febslet.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Zhang, Y.-Z., Lindblom, T., Chang, A., Sudol, M., Sluder, A. E. and Golemis, E. A. (2000). Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene 257, 33-43 10.1016/S0378-1119(00)00372-3 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Ibrahimi, O. A., Olsen, S. K., Umemori, H., Mohammadi, M. and Ornitz, D. M. (2006). Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 281, 15694-15700 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.