Abstract
We report a remarkably high UV-radiation resistance in the extremely halophilic archaeon Halobacterium NRC-1 withstanding up to 110 J/m2 with no loss of viability. Gene knockout analysis in two putative photolyase-like genes (phr1 and phr2) implicated only phr2 in photoreactivation. The UV-response was further characterized by analyzing simultaneously, along with gene function and protein interactions inferred through comparative genomics approaches, mRNA changes for all 2400 genes during light and dark repair. In addition to photoreactivation, three other putative repair mechanisms were identified including d(CTAG) methylation-directed mismatch repair, four oxidative damage repair enzymes, and two proteases for eliminating damaged proteins. Moreover, a UV-induced down-regulation of many important metabolic functions was observed during light repair and seems to be a phenomenon shared by all three domains of life. The systems analysis has facilitated the assignment of putative functions to 26 of 33 key proteins in the UV response through sequence-based methods and/or similarities of their predicted three-dimensional structures to known structures in the PDB. Finally, the systems analysis has raised, through the integration of experimentally determined and computationally inferred data, many experimentally testable hypotheses that describe the metabolic and regulatory networks of Halobacterium NRC-1.
Biological systems have evolved mechanisms to appropriately respond to environmental stresses that can damage proteins and DNA. Halobacterium NRC-1, for example, has evolved the capacity to withstand up to 4.5M salinity, ultraviolet radiation (UV), and oxidative stress in its natural environment. One such adaptation, the high-density of acidic residues on the surface of almost all halobacterial proteins, is believed to stabilize protein structure and function in high salt (Kennedy et al. 2001). It can also physically relocate to favorable environments using sensors for light, oxygen, and nutrients, and produce energy through respiration, fermentation, or phototrophy (Ng et al. 2000). We have previously reported a response mechanism in Halobacterium NRC-1 that coordinately regulates phototrophy and arginine fermentation, two major sources for anaerobic energy production. The effects of perturbing the function of a regulator in this response mechanism were observed throughout the metabolic network (Baliga et al. 2002).
Systems approaches allow us to raise a whole series of hypotheses regarding the behavior of a system in response to a defined experimental perturbation. These hypotheses then stimulate iterative cycles of perturbations and analyses to verify these hypotheses. Here, we report a systems-level study on the behavior of Halobacterium NRC-1 upon UV-C irradiation. Shortwave ultraviolet light (UV-C) induces two types of mutagenic lesions in DNA—cyclobutane pyrimidine dimers (CPD) formed between the C-4 and C-5 positions of adjacent thymidine or cytosine residues, and pyrimidine (6–4) pyrimidone (6–4) photoproducts formed between the C6 and C4 position of adjacent pyrimidine residues, most often between T-C and C-C residues. Most organisms repair these lesions in two ways—light-dependent photoreactivation catalyzed by a photolyase, and light-independent nucleotide excision repair (NER). Although both mechanisms can repair the two kinds of damage, photore-activation is more efficient in repairing CPDs, whereas NER is more effective in repairing (6–4) photoproducts (Friedberg 1995).
Several archaea, including Halobacterium NRC-1, are capable of photoreactivation (McCready 1996; Wood et al. 1997). Of the two photolyase-homologs, phr1 and phr2, in Halobacterium NRC-1, at least one, phr2, encodes an active CPD photolyase. Residual photoreactivation in the phr2 deletion mutant suggested that phr1 too might encode an active photolyase (McCready and Marcello 2003).
Although different proteins mediate eukaryotic and bacterial NER, in both cases, the DNA damage is recognized by damage-recognition proteins, which recruit nucleases and helicases to excise the damaged nucleotides. The repair process is completed by the action of a DNA polymerase and a ligase (Friedberg 1995). The presence of both eukaryotic (rad1, rad2, rad3, and rad25) and bacterial (uvrA/B/C/D) NER orthologs in Halobacterium NRC-1 suggests the likely occurrence of both types of NER. However, only two NER orthologs, Rad2 and Rad1, have been biochemically characterized in Archaea (Roberts et al. 2003). Moreover, thus far, no damage-recognition proteins have been identified in Archaea, and as a result, the mechanism of NER in these organisms remains a puzzle. Finally, among all Archaea with sequenced genomes, orthologs of bacterial NER (uvrA, uvrB, uvrC, and uvrD) have only been found in Halobacterium NRC-1 and Methanothermobacter thermoautotrophicus (Ogrunc et al. 1998; Ng et al. 2000), strongly suggesting lateral gene transfer from bacteria to these archaea (Diruggiero and Robb 2003).
We have analyzed light and dark repair of UV damage in Halobacterium NRC-1 through a systems approach comprising of genetic analysis of the wild-type and three knockout strains in conjunction with genome-wide analysis of the wild-type strain (Weston et al. 2004). Ideally, in a systems analysis, data from different informational levels, that is, mRNA levels, protein levels, protein modifications, protein–protein interactions, and protein–DNA interactions are integrated and analyzed simultaneously to gain insights into the biology of the whole organism rather than a subset of genes or pathways of interest. We have collected, integrated, and analyzed data at many of the informational levels listed above to provide insights into the behavior of the whole organism following DNA damage. We report extreme UV resistance in Halobacterium NRC-1, evidence for a photolyaselike function for Phr2, and evidence for lack of a photolyase-like function for Phr1. We have also analyzed, at 30 and 60 min during light and dark UV damage repair, mRNA changes for all of the 2400 nonredundant genes in Halobacterium NRC-1. The mRNA changes were analyzed in the context of five types of interactions among halobacterial proteins inferred through analyses of operon-like gene organization, phylogeny of pairs of proteins, and experimentally observed interactions between orthologous pairs in yeast and Helicobacter pylori. The analysis of the integrated genome-wide data has identified several putative repair mechanisms in Halobacterium NRC-1 including photoreactivation, d(CTAG) methylation-directed mismatch repair, glycosylases for repairing photo-oxidatively damaged DNA, and proteases for eliminating damaged proteins. The systems analysis has also provided insights into putative functions for previously unannotated proteins and regulatory mechanisms for various cell processes. Finally, we report a UV-irradiation-specific repression of many global aspects of metabolism within 60 min during light repair.
RESULTS
Survival of Halobacterium NRC-1 to UV-C Irradiation
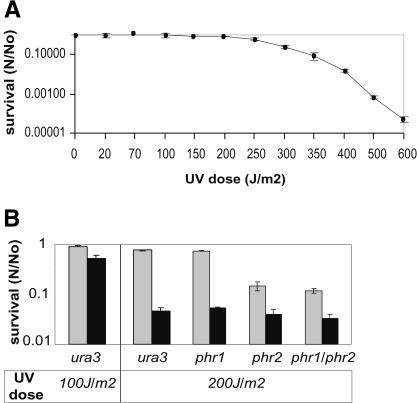
To investigate the UV-C radiation resistance of Halobacterium NRC-1, a monolayer distribution of halobacterial cell suspension in a clear isotonic solution was exposed to increasing doses of UV-C radiation. The survival of irradiated cells was calculated by counting colonies on agar plates post-incubation at 42°C in either light or dark conditions. The survival curve demonstrated high resistance of Halobacterium NRC-1 to UV-C irradiation with no loss of viability up to 110 J/m2, and 37% survival (D37) at 280 J/m2 (Fig. 1A). A 16-fold higher recovery was observed in light relative to dark conditions at a dose of 200 J/m2, suggesting that photo-reactivation was the major pathway for UV lesion repair (Fig. 1B).
Figure 1.
Survival of Halobacterium NRC-1 wild-type and putative photolyase mutants following UV-C irradiation. (A) Wild-type Halobacterium NRC-1 survival (N/No; x-axis) in light following increasing doses of UV irradiation (J/m2; y-axis). (B) Survival of single (phr1 or phr2) and double (phr1/phr2) knockout mutants on recovery in light (gray bars) and dark (black bars) post-UV irradiation at 200 J/m2. Relative difference in effects of UV-C irradiation at 100 and 200 J/m2 on viability of the wild-type organism is shown. (N) Number of viable cells in challenged sample; (No) number of viable cells in control; error bars represent standard error for at least three independent experiments.
Survival of Halobacterium NRC-1 Photolyase Mutants to UV-C Irradiation
The two putative photolyases, Phr1 and Phr2, in Halobacterium NRC-1 share 43% identity with each other and are believed to be CPD photolyases. Classification of photolyases into class I or class II photolyases is not possible through analysis of amino acid sequence alone (Komori et al. 2001). To further investigate the role of each putative photolyase in photoreactivation suggested by the survival experiments, we constructed strains of Halobacterium NRC-1 with single (phr1 or phr2) and double (phr1 and phr2) in frame deletions in the two genes.
Analysis of the three mutants demonstrated that only phr2 was required for UV damage repair during recovery in light. Furthermore, the phr2 and phr1/phr2 mutants had a 3.5-fold higher survival in light, when compared with their survival in dark, suggesting the presence of one or more light-dependent repair (photoreactivation) systems mediated by proteins with no detectable sequence homology to characterized photolyases (Fig. 1B).
We also evaluated possible photoprotection by photoactive pigments such as rhodopsins, carotenoids, and C-50 bacterioruberins in Halobacterium NRC-1 by conducting UV irradiation experiments on two colorless mutants, Pho81 (Spudich and Spudich 1982), which lacks all four rhodopsins (bacteriorhodopsin, halorhodopsin, and the two sensory rhodopsins), and a second pigment-deficient white mutant obtained by ethylmethylsulfonate (EMS) mutagenesis of the wild-type strain. There was no change in UV sensitivity of these mutants relative to the wild-type, ruling out photoprotective roles for the photoactive pigments at this level of irradiation (data not shown).
Global Transcript Analysis in UV-C-Irradiated Cells
The significant residual survival of the Halobacterium NRC-1 photolyase mutant (phr2) upon UV-C irradiation prompted us to undertake a microarray analysis of the global mRNA changes occurring during light and dark repair at 30 and 60 min postirradiation at 200J/m2. The time-point selection was based on the observation that >50% of all UV-induced CPDs are repaired within 30 min during light repair in Halobacterium NRC-1 (McCready 1996). All RNA samples were compared with total RNA from an aliquot of the same culture harvested prior to irradiation (Fig. 2). A likelihood ratio test was performed for each gene to evaluate the statistical significance of the difference in the true intensities over eight replicate spots. The maximum likelihood statistic λ gives a measure of the significance of the observed change in mRNA and is derived from the variation in absolute intensity values within a microarray experiment (Ideker et al. 2000). Comparing different RNA preparations from independently processed, but identically cultured cells consistently yielded λ values <15.0 for >99% of all genes in the array. The results reported below are limited to changes associated with λ >15.0, which correlates to a 99% confidence level.
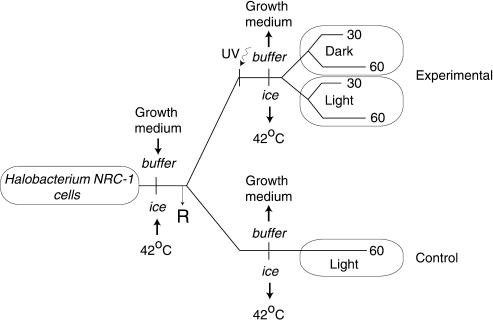
Figure 2.
Experiment design for global analysis of UV-response in Halobacterium NRC-1. Halobacterial cells were harvested from the growth medium, resuspended at a low density in a clear isotonic buffer, and placed on ice. Total RNA prepared from an aliquot of the cells at this point was used as the reference (R) in microarray analyses. Subsequently, one-half of the cell suspension was kept aside on ice for use as control. The remainder was irradiated on ice with UV-C light and post-irradiation the cells were diluted twofold in CM and allowed to recover at 42°C under light or dark conditions. Total RNA was prepared at 30 and 60 min from both the light and dark repair samples. The control was processed in a manner identical to the 60-min light repair sample, except that it was not irradiated.
Recovery in Dark Post-UV Irradiation
30 Min (D30)
At least 11 genes were up-regulated and 31 were down-regulated after 30 min in dark post-irradiation. Fourteen of the 42 translated proteins had no known function, and the remainder was divided into at least seven metabolic categories, including amino acid metabolism, gas vesicle biogenesis, DNA repair, transport, carbohydrate metabolism, metabolism of cofactors and vitamins, and nucleotide metabolism (Supplemental Table 1). At least three previously unannotated proteins (VNG0019H, VNG0765H, VNG1318H) that had increased mRNA at 30 min during dark repair had sequence and/or predicted structural homology to transcription regulators or nucleic acid binding proteins (Supplemental Table 2).
60 Min (D60)
At least 33 genes were differentially regulated in the irradiated cells after 60 min of recovery in dark. Sixteen encoded proteins of unknown function and the remainder were divided among at least seven pathways of metabolism, including amino acid metabolism, transcription, translation, DNA repair, nucleotide metabolism, carbohydrate metabolism, and nucleotide metabolism. Of the 15 up-regulated genes in D60, eight encode proteins involved with nucleic acid metabolism (Supplemental Tables 1 and 2).
Recovery in Light Post-UV Irradiation
30 Min (L30)
Only four genes changed at 30 min during light repair. This was surprising, as >13% of all genes changed subsequently at 60 min during light repair. All four genes that changed were down-regulated [VNG0209H (integrase), VNG2488C (sterol biosynthesis), gvpO (gas vesicle biogenesis), and coxB1 (cytochrome oxidase)] (Supplemental Table 1).
60 Min (L60)
A significantly larger number (315) of mRNAs changed at 60 min during light repair (Supplemental Table 1). Interestingly, only 29 genes were up-regulated, including 12 mRNAs (cspD1, purU, VNG0983C, rpoM, VNG0659H, VNG6332H, VNG6195H, VNG1688C, pyrG, gap, VNG0019H, and VNG6182H) that also increased in the same period during dark repair. The upregulation of these 12 genes is therefore independent of light. Moreover, seven (purU, VNG0983C, rpoM, VNG6195H, VNG1688C, VNG6182H, and gap) of these 12 genes did not change in the control, suggesting that they were induced by UV-irradiation alone, independent of light. Eleven of the 17 genes up-regulated only during light repair were of unknown function.
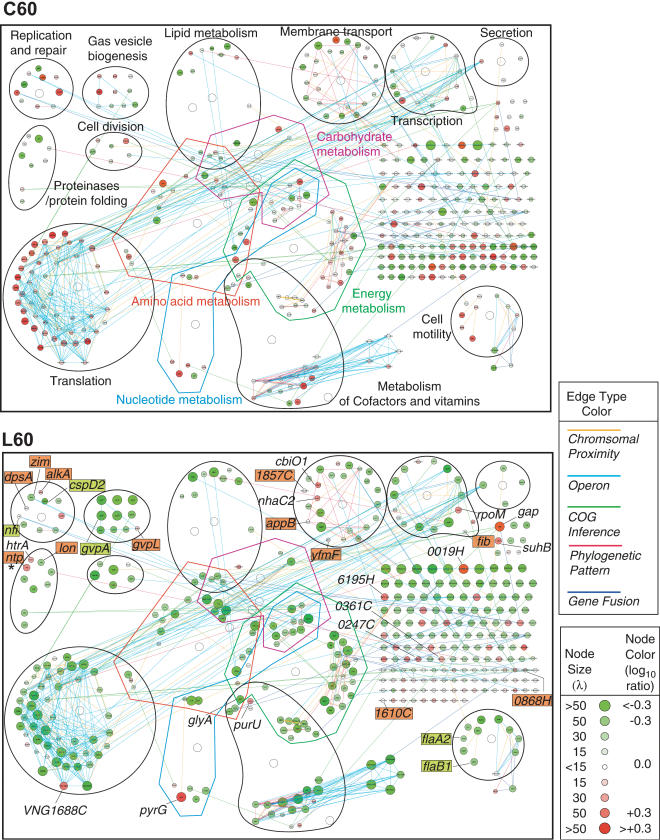
Of the 286 down-regulated genes, 140 were divided into at least 11 categories of metabolism; these included 47 of the 53 ribosomal protein genes and 10 of 12 RNA polymerase genes, suggesting that both transcription and translation were repressed at 60 min during light repair (Fig. 4. below).
Figure 4.
Systems-level visualization of the Halobacterium NRC-1 transcriptome in C60 and L60 with layout organized by general function. The mRNA changes are visualized as a network of genes (nodes) and their interactions (edges). A total of 420 mRNAs that changed during repair or in control are shown with shades of red for increased and shades of green for decreased levels; node size correlates to statistical significance (λ) of change (see inset key). The five types of edges connecting the genes are color coded and described in the text (see inset key for color codes). Representative genes with mRNA changes during repair masked by mRNA changes resulting from the experimental procedure alone are labeled and highlighted with red for a masked up-regulated change during repair and green for a masked down-regulated change during repair. For a complete list of 147 potential masked changes, see Supplemental Table 3. The asterisk indicates the ntp gene, a transposase, which was originally misannotated as a neutral proteinase (see text for details).
Summary of mRNA Changes During Dark and Light Repair
At least 68 mRNAs changed during dark repair. These changes may be mediated by the up-regulation during dark repair of at least four putative transcription regulators (VNG0019H, VNG 0361C, VNG0765H, and VNG1318H). At least three of these putative regulators (VNG0019H, VNG1318H, and VNG0765H) were also up-regulated during light repair. However, VNG0019H mRNA was also increased in the control suggesting that it may play a role in the regulation of genes involved in a generalized stress response (see below).
Most of the changes during light repair occurred after 30 min, the period during which >50% of the UV-induced CPDs are repaired by photoreactivation (McCready 1996). Almost all genes encoding the subunits of RNA polymerase, with the exception of rpoM and VNG0237H, were repressed, correlating with the fact that >90% of all genes that changed were repressed. Likewise, 48 ribosome biogenesis genes were also repressed during light repair, suggesting down-regulation of protein synthesis as well. At least three putative transcription regulators VNG0247C, VNG0019H, and VNG6195H (Supplemental Table 2) that were up-regulated during light repair might mediate the observed mRNA changes.
mRNA Changes at 60 Min in Control (No UV Irradiation): C60
At least 147 genes were differentially regulated in 60 min in light in absence of UV irradiation. Unlike the changes in L60, the differentially regulated genes were evenly distributed among upregulated (67 genes) and down-regulated (80 genes) groups. Twenty-two up-regulated genes encoded proteins of unknown function, and over a third of the remainder (23 genes) were involved with translation. In contrast, none of the RNA polymerase subunits were differentially regulated; however, at least two transcription factors tbpB and tfbB were repressed mRNAs for at least four DNA-repair proteins, dpsA (starvation-induced DNA-binding protein), alka (3-methyl adenine glycosidase), VNG0868H (MutT: 8-oxoguanine glycosylase), and VNG1610C (DNase), were significantly down-regulated (Fig. 4, below).
Summary of All mRNA Changes Observed During Repair and in the Control
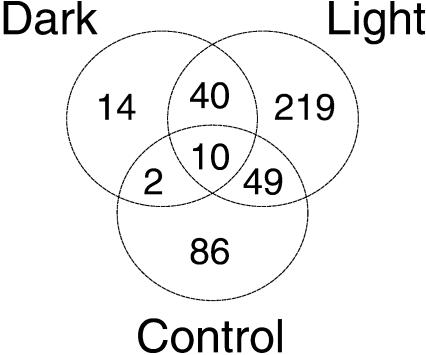
At least 420 mRNAs changed during repair or in the control. Among the 273 mRNAs that changed only during repair, at least 40 changes (30 up-regulated and 10 down-regulated) occurred in both dark and light conditions. Finally, at least 61 mRNAs changed during repair and also in the control; 17 of these changes were anticorrelated between the irradiated cells and the control (Fig. 3; Supplemental Table 1).
Figure 3.
Venn diagram of mRNA changes observed during repair and in the control (see text for details).
These observations indicate that the experimental procedure (Fig. 2) alone results in 147 mRNA changes in the control (Fig. 3) that could be potentially masking or dampening important changes occurring during repair. To verify this, we did a simple linear subtraction of the significant mRNA changes in the control from the corresponding changes occurring at 60 min during light repair. Although the nature of mRNA changes occurring during repair and in the control are complex, the linear subtraction revealed mRNA changes during repair in at least 92 genes that were masked or dampened by changes occurring due to the experimental procedure. The 41 up-regulated mRNA changes included at least five mRNAs with roles in repair of photo-oxidative DNA and protein damage [dpsA, alkA, VNG0868H (mutT), VNG1610C (DNase), and lon (protease)]. Likewise, the 41 down-regulated mRNA changes included 22 ribosomal protein genes (Fig. 4; Supplemental Table 3).
Data Integration and Visualization in Cytoscape
The mRNA changes at 30 and 60 min during dark or light repair and at 60 min in the control were integrated with five types of inferred (or direct) protein–protein interactions to facilitate systems interpretation and the formulation of hypotheses. The putative interactions included associations inferred on the basis of similar phylogeny of pairs of conserved homologs (Pellegrini et al. 1999), associations inferred from gene-fusion events among halobacterial gene orthologs in other organisms (Marcotte et al. 1999), associations inferred from evolutionarily conserved chromosomal proximity of phylogenetically related genes (Overbeek et al. 1999), and associations inferred from operon-like gene organizations (Moreno-Hagelsieb and Collado-Vides 2002). These putative associations were integrated with experimentally determined protein–protein interaction among halobacterial COG (Clusters of Orthologous Groups; Tatusov et al. 2001) counterparts in yeast (Uetz et al. 2000), as well as H. pylori (Rain et al. 2001). Upon integration, these data were displayed in Cytoscape (Shannon et al. 2003), a software tool that displays genes (proteins) and the interactions among them as nodes and edges, respectively. The mRNA changes were then integrated with these interactions and visualized as node colors with shades of red denoting up-regulation, and shades of green denoting down-regulation. The genes in the network were subsequently grouped by function on the basis of their roles in metabolism as described in the Kyoto Encyclopedia of Genes and Genomes KEGG database (Kanehisa 2002). This functional grouping of the nodes provided insights into the physiological relevance of the mRNA changes. For example, the genes involved with translation that are interconnected by at least four types of edges form a function biomodule (a group of genes with related functions), and show coordinated mRNA changes. Almost all of the translation biomodule genes are significantly down-regulated at 60 min during light repair. Surprisingly, VNG1688C, also a putative translation biomodule component, is significantly up-regulated (Fig. 4; see Discussion on regulation of ribosome biogenesis).
Functional Annotation of Previously Unannotated Proteins
We are using sequence homologies as well as protein-fold predictions to assign functions to previously unannotated proteins. The structural homology approach is applied only in cases in which there is no detectable PSI-BLAST match or protein-family signature (Pfam; Bateman et al. 2000). Rosetta is a method for predicting three-dimensional structures of proteins in the absence of global sequence homologies to characterized proteins (Bonneau et al. 2002). Consensus fold recognition, on the other hand, searches a query sequence for a match to known structures using a number of methods, including threading and fold recognition; the results from several methods are then collected and compiled into a single consensus structure. The Meta server (PCONS/3D-jury) is one such Web-based consensus fold recognition server (Bourne 2003). Both Rosetta and PCONS/3D-jury have performed well in blind tests (Bonneau et al. 2001; Bourne 2003). Below, we provide examples in which these approaches provided insights into functions of two proteins that were upregulated during repair.
Example 1
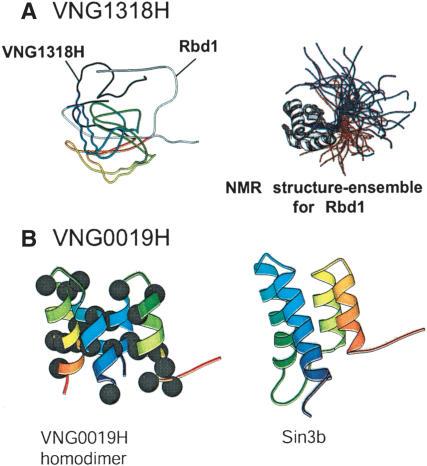
The Rosetta-predicted three-dimensional structure for VNG1318H matched to that of Rbd1 (PDB: 1fj7), a nucleolin from the golden hamster. The lack of structure homologies between the corresponding N- and C-termini of the two proteins is due to the lack of a defined NMR structure for the two terminii of Rbd1 (Fig. 5A). On the basis of this observation, VNG1318H is functionally annotated as a putative DNA-binding protein with likely transcription regulatory function.
Figure 5.
Functional annotation of proteins through ab inito structure prediction by Rosetta. (A) Alignment of the Rosetta-predicted structure for VNG1318H and Rbd1 (left). The NMR structure for Rbd1 is shown on the right. (B) Rosetta-predicted structure for the VNG0019H homodimer (left) and its closest match, the NMR structure for Sin3b. The black dots indicate hydrophobic residues located primarily at the predicted protein–protein interface.
Example 2
The VNG0019H structure matched the first and the last 32 residues of Sin3b (a transcriptional repressor in Mus musculus twice the size of VNG0019H). To verify the possibility that VNG0019H is a homodimer with structure similarity to Sin3b (PDB: 1e91), we duplicated its sequence and resubmitted it to the Meta server. The duplicated structure had a significantly better match to Sin3B, suggesting that VNG0019H might function as a homodimeric transcription regulator (Fig. 5B).
Altogether, 26 proteins were annotated by sequence and/or structure-based methods as follows: 18 proteins were annotated by sequence-based methods, four proteins were annotated by structure-prediction approaches, and at least four proteins were annotated by a combination of both approaches (Supplemental Table 2).
DISCUSSION
We have demonstrated a high-UV resistance in Halobacterium NRC-1, and elucidated, through classical genetics and a systems approach, the putative mechanisms underlying this extreme resistance. The systems analysis reported here integrates information at many different levels including gene expression, gene localization, gene function, evolutionarily conserved functional associations among genes, and physical interactions among corresponding orthologs in other organisms. The simultaneous analysis and integration of the different data types has provided insights into Halobacterium NRC-1 biology that would otherwise have not been possible by analyzing mRNA levels alone. The systems approach has allowed us to formulate many hypotheses that can now be addressed through iterative cycles of additional perturbations. Below, we discuss the various aspects of the regulatory and metabolic networks in Halobacterium NRC-1 that together constitute the UV response.
Photoreactivation by the Photolyase Phr2 Is the Major Mechanism of UV-C Damage Repair in Halobacterium NRC-1
The increased UV-sensitivity of the Halobacterium NRC-1 phr2 mutant confirms that Phr2 is a photolyase that plays a major role in photoreactivation (Fig. 1B). However, it was surprising that phr2 mRNA did not increase during light or dark repair, because Halobacterium NRC-1 can repair 50% of CPDs in <30 min in light (McCready 1996). This is similar to the Arabidopsis thaliana 6-4PP photolyase, which is constitutively expressed and not regulated by either white or UV-B light (Waterworth et al. 2002). However, further experiments are necessary to rule out post-transcriptional regulation of phr2.
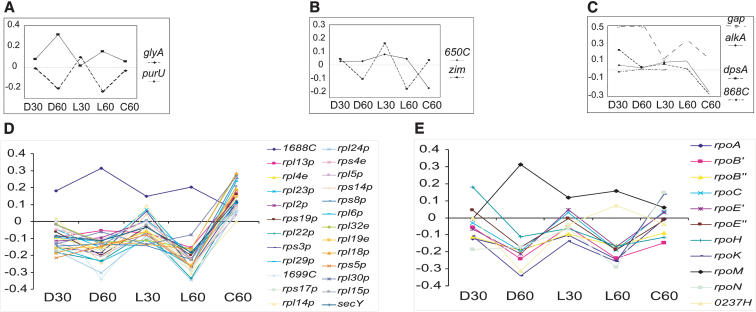
Repair of CPDs by class I photolyases requires 5, 10-methenyl tetrahydrofolate (5, 10-CH=THF) as a cofactor (Jorns 1990). We observed inversely correlated changes in the mRNAs for at least two genes, purU and glyA (Fig. 6A), each with a role in one of two opposing branches in the biosynthesis pathway of this cofactor. The differential regulation of purU and glyA is perhaps a mechanism to increase the synthesis of 5, 10-CH=THF for photolyase function.
Figure 6.
Transcript levels for select genes involved with DNA repair, transcription, and translation. (A) The glyA and purU genes are involved with 5,10-CH=THF biosynthesis pathway. (B) The two genes, zim (d(CTAG) methylase) and VNG0650C (RecJ exonuclease), are involved with a putative d(CTAG) methylation-directed mismatch repair genes. (C) Four genes involved with photo-oxidative DNA damage repair. (D) The large ribosomal operon (cluster 1). (E) RNA polymerase synthesis. The y-axis indicates log10 ratios of mRNA changes with respect to the reference RNA, and the x-axis indicates 30 and 60 min during dark and light repair, as well as the control. The lines connecting the dots are for the benefit of the reader to correlate mRNA changes for a given gene in the five experiments, and do not indicate a temporal ordinate except from D30 to D60 and L30 to L60.
phr1 Does Not Encode a Photolyase and the Photoactive Pigments Do Not Provide Photoprotection in Halobacterium NRC-1
The residual survival upon UV irradiation of the phr2 mutant in both light and dark conditions suggests additional repair mechanisms. However, the lack of change in UV resistance of the rhodopsin and pigment deficient mutants (data not shown), as well as the phr1 mutant (Fig. 1B), rules out their role in photoprotection and repair. Instead, phr1 might encode a cryptochrome, a photolyase paralog that can control circadian rhythms in response to blue light, as suggested for Synechocystis sp. (Hitomi et al. 2000).
Several Mechanisms Including a Putative Methyl-Directed Mismatch Repair System and Photo-Oxidative Damage Repair Systems Play a Role in UV Repair
In addition to photoreactivation, our studies have indicated the likely roles in the UV response of a putative mismatch repair system, absence of an Escherichia coli-like SOS response, and six enzymes for repair of photo-oxidative damage.
Mismatch Repair (MMR)
In most bacteria, mismatches in DNA are repaired by the MutS/L/H, UvrD, and RecJ proteins through a dam [d(GATC)-specific methylase] methylation-directed mismatch repair (MMR) pathway. MMR is initiated by the mismatch-stimulated d(GATC) endonuclease activity of MutH, which takes advantage of transient undermethylation of the DNA during repair, and specifically nicks the newly synthesized and unmethylated daughter strand. Subsequently, the RecJ exonuclease selectively degrades the nicked strand, and the mutations are corrected by the action of a DNA polymerase, a helicase, and a ligase (Marti et al. 2002).
Halobacterium NRC-1 does not have a dam methylase or a MutH endonuclease. Instead, it has a d(CTAG) methylase (Zim), MutS/L, four RecJ-like exonucleases (VNG0650C, VNG2333C, VNG2519H, and VNG6183C) and the UvrD repair helicase (Ng et al. 2000). However, we could not detect a d(CTAG)-specific endonuclease using either sequence- or structure-based annotation methods. Unlike MutS and MutL, which are absolutely necessary for MMR, and therefore evolutionarily highly conserved, MutH is significantly less conserved. In fact, no MutH homolog has been identified in either the Eukaryotes or the Archaea. Moreover, phylogenetic studies have suggested that most species might have evolved separate restriction modification and restriction systems for strand recognition and cleavage during MMR (Eisen and Hanawalt 1999).
In our studies, zim mRNA decreased significantly at 60 min during repair and at least one of the four RecJ homolog (VNG0650C) mRNAs was moderately up-regulated during repair and significantly down-regulated in the control (Fig. 6B). This is strongly suggestive of a transient undermethylation upon UV irradiation of the Halobacterium NRC-1 DNA that might trigger a novel MMR mechanism.
Absence of an SOS-Like Response
In E. coli, UV exposure transiently inhibits replication and transcription, and up-regulates recA within 5 min of irradiation. RecA elicits, for at least 60 min, a strong SOS response involving >20 genes, including recA, uvrABD, sulA, umuCD, lexA, and ruvAb (Courcelle et al. 2001). In our experiments, there was a moderate increase in radA1 mRNA at 60 min during dark repair. However, we did not find a coordinated change in other repair genes, suggesting that UV irradiation does not activate an SOS response in Halobacterium NRC-1.
Repair of Photo-Oxidative Damage
In addition to pyrimidine dimers, UV irradiation can also cause photoxidative damage in both DNA and proteins (Friedberg et al. 1995). At least six genes that were up-regulated upon irradiation may play a role in preventing and/or repairing such photooxidative damage in Halobacterium NRC-1. DpsA might bind the DNA, protecting it from further photo-oxidation, whereas AlkA (3-methyladenine and 5-formyluracil DNA glycosylase; Masaoka et al. 1999) and VNG0868H (MutT: 8-oxoguanine glycosylase) might play a role in base-excision repair of photo-oxidized bases. The glycolytic enzyme Gap, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), in eukaryotes, also has a uracil DNA glycosylase activity (Meyer-Siegler et al. 1991). Therefore, the upregulation of GAPDH upon irradiation suggests that it too might play a role in repair of oxidative-DNA damage in Halobacterium NRC-1.
We were surprised that there was no up-regulation of the proteasome subunit mRNAs (psmA/B) during repair. mRNAs for three putative proteases Lon, Ntp, and HtrA did, however, increase at 60 min during repair (Figs. 4 and 6C; Supplemental Table 3). However, the Ntp sequence matched a family of transposases (PF01385), suggesting that this protein was misannotated as a protease. The two putative serine-proteases, Lon and HtrA, on the other hand, are hypothesized to play a role in degrading photo-oxidized proteins.
A Global Down-Regulation of Physiology Occurs During UV-Damage Repair in Halobacterium NRC-1
Altogether, nearly 12% of all Halobacterium NRC-1 genes were down-regulated at 60 min during light repair. These genes were divided into at least 10 major aspects of metabolism (Fig. 4). Global repression of metabolism during DNA repair has been reported in E. coli, Deinococcus radiodurans, and Saccharomyces cerevisiae, suggesting that this might be a generalized stress-response mechanism shared by all three domains of life (Causton et al. 2001; Courcelle et al. 2001; Liu et al. 2003).
Global down-regulation of genes results from the necessity of the cells to maintain internal homeostasis in response to abrupt changes in the environment. In yeast, for instance, during the environmental stress response (ESR), expression levels for >900 genes (∼14% of the yeast genes) change. Among these 14% genes that change, similar to our observations, the protein synthesis and transcription genes are down-regulated and the protein folding and degradation, the oxidative stress response, and the DNA damage-repair genes are all up-regulated (Gasch and Werner-Washburne 2002). This global down-regulation might also help conserve energy required for costly DNA repair processes, as previously proposed for down-regulation of ribosomal protein genes (Warner 1999).
Down-regulation of metabolic processes can also serve specific purposes. For example, the repression of gas vesicle biogenesis during repair in Halobacterium NRC-1 might prevent the cells from floating toward the source of the damage, that is, UV radiation. Likewise, the down-regulation of cell cycle genes, for example, the ftsZ2 and ftsZ3 (ring formation during cell division), the sojA and sojB (partitioning of replicated DNA to daughter cells), and the cdc48c (cell division control) genes, like the down-regulation of minCDE (septum formation) genes during repair in E. coli (Courcelle et al. 2001), suggests cell-division arrest, perhaps to prevent the inheritance of mutations by daughter cells.
The up-regulation during repair of only a fraction of all genes involved with the repair of DNA and protein damage implies that perhaps the other repair proteins are either posttranscriptionally regulated or their endogenous levels are sufficient to provide protection against sublethal doses of damaging treatment. Because many of the lesions such as oxidative damage by reactive oxygen species, stalled replication forks, and double-stranded DNA breaks occur naturally as by-products during general metabolism, the constitutive presence of the DNA-repair proteins may be necessary to repair these damages (Birrell et al. 2002). We believe the post-transcriptional control mechanisms will be critical in many aspects of Halobacterium NRC-1 physiology (Baliga et al. 2002).
The Global mRNA Changes May Be Mediated by at Least Three General-Transcription Factors and at Least Eight Regulators
The global changes observed in our studies are likely to be mediated by the differential regulation of three general transcription factors, tbpB, tfbB, and tfbG, and at least seven regulators (VNG0019H, VNG0247C, VNG0361C, VNG1318H, VNG1564H, VNG0207H, and VNG0482H) that were all differentially regulated during this study. The up-regulation of three putative transcriptional regulators, VNG0247C, VNG0361C, and VNG1318H, was exclusively in response to UV irradiation, as their transcript levels were either unchanged or repressed in the control.
RpoM Might Modulate RNA Polymerase Function During Repair
The halobacterial RNA polymerase (Rpo) is similar to the eukaryotic RNA polymerase II, and is comprised of at least 10 subunits encoded by genes organized as three clusters (cluster1: rpoA, rpoB', rpoB”, rpoC, and rpoH; cluster 2: rpoE' and rpoE”; cluster 3: rpoN, rpoK, and rpb3). At least two additional putative RNA polymerase subunits are encoded elsewhere in the genome as singletons [rpoM and VNG0237H (DNA-directed RNA polymerase subunit P)] (Ng et al. 2000).
All 10 Rpo genes in the three clusters were down-regulated at 60 min during repair. In contrast, rpoM was up-regulated during both light and dark repair (Fig. 6E). RpoM contains a TFIIS signature domain that, in eukaryotes, is involved in reinitiating transcription by RNA polymerase past template-encoded pause sites (Szentirmay and Sawadogo 1993). These observations lead us to the hypothesis that subsequent to repair in Halobacterium NRC-1, RpoM facilitates reinitiation of transcription from Rpo complexes stalled at DNA lesions.
Ribosome Biogenesis and/or Function in Halobacterium NRC-1 May Be Regulated by an Evolutionarily Conserved Nucleic Acid Methylase
The halobacterial ribosome is comprised of at least 53 protein subunits encoded by two gene clusters and several other genes located elsewhere in the genome (Ng et al. 2000). VNG1688C is encoded immediately upstream to the larger 24-gene ribosomal protein operon. However, in contrast to up-regulation of VNG1688C at 60 min during repair, all 24 genes in this operon including VNG1699C [P29_RnaseP] and secY (protein secretion) were down-regulated. Moreover, at least 24 mRNAs involved with protein synthesis, but not a part of this operon, also decreased. Altogether, transcript levels for 47 of the 53 ribosomal protein genes changed in our studies; 46 of these changes were positively correlated to each other; in contrast, VNG1688C mRNA changed in an anticorrelated manner (Figs. 4 and 6D).
The three-dimensional structure for the VNG1688C ortholog MTH1 in Methanothermobacter thermoautotrophicus (PDB: 1k3r) matches two superfamilies in SCOP (Hubbard et al. 1997), b.40.4 (a nucleic acid binding domain) and c.116.1 (a SAM-dependent methytransferases). Like VNG1688C, MTH1 is also encoded immediately upstream to the ribosomal protein operon (Smith et al. 1997; Ng et al. 2000). This gene organization is also conserved in at least eight other archaeal genomes (Archaeoglobus fulgidus, Aeropyrum pernix, Pyrococcus abyssi, Pyrococcus furiosus, Pyrococcus horikoshii, Haloarcula marismortuii, Sulfolobus solfatarius, Sulfolobus tokodaii). These observations taken together suggest that VNG1688C might play an evolutionarily conserved regulatory role in ribosome biogenesis and/or function.
Systems Analysis Provides Insights Into the Global Regulatory Networks and Putative Functions for Previously Unannotated Proteins
The grouping of genes by function together with the mRNA changes and predicted interactions among proteins has facilitated the functional annotation, by sequence- and/or structure-based methods of previously unannotated proteins. Moreover, the systems analysis has provided insights into the regulation of various biochemical pathways. The example below illustrates one of many discoveries made possible by data integration and visualization in Cytoscape.
Cobalamin Biosynthesis
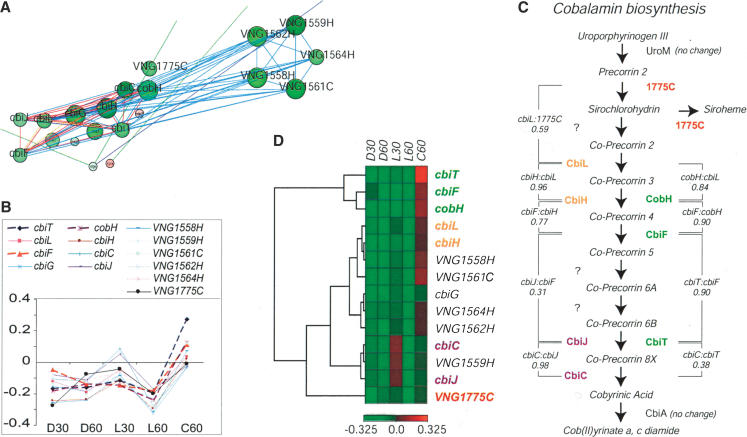
Eight of the nine cobalamin biosynthesis genes in Halobacterium NRC-1 have evolutionarily conserved phylogenetic associations (red edges in the Cytoscape network) and are organized in the genome as three operons (blue edges), members of which are five genes of unknown function (Fig. 7A). The nine genes encode a contiguous segment of the cobalamin biosynthesis pathway in Halobacterium NRC-1, which converts Precorrin 2 to Cobyrinic acid. This is the only segment of the pathway that is differentially regulated and is flanked by biochemical steps catalyzed by the enzymes UroM and CbiA that do not change during repair.
Figure 7.
Cobalamin biosynthesis pathway in Halobacterium NRC-1. (A) Cofactor and vitamin biosynthesis biomodules are linked to five genes of unknown function visualized in Cytoscape; (see Fig. 4 for key to network display). Genes implicated in cobalamin biosynthesis are shown in a larger font size. (B) mRNA changes for the cobalamin biosynthesis genes (the x- and y-axes are defined in Fig. 7). (C) Cobyrininc acid biosynthesis pathway. The proteins catalyzing the various steps are shown along with the standard correlation between mRNA changes in adjacent biochemical steps. The question mark (?) indicates enzymes yet to be identified. (D) Hierarchical clustering of mRNA changes using standard correlation. Red denotes up-regulation and green denotes down-regulation. The dendogram on the left represents closeness between two genes on the basis of the similarity in their expression level changes in the five perturbations.
Hierarchical clustering of the mRNA changes across the various perturbations (Fig. 7B) divided the nine cobalamin biosynthesis genes and the five genes of unknown function into four groups and one orphan (Fig. 7D). These coregulated groups are remarkably well correlated with the order of the biochemical steps in the conversion of precorrin-2 into cobyrinic acid (Fig. 7C). For example, cbiL and cbiH have highly correlated mRNA changes (Fig. 7D); CbiL catalyzes the conversion of Co-precorrin 2 into Co-precorrin 3, which, in turn, is converted into Coprecorrin 4 through the action of CbiH (Fig. 7C). Three genes, cbiT, cbiF, and cobH, form a distinct group; two of the three genes, cobH and cbiT, according to KEGG, catalyze biochemical steps similar to those of cbiH and cbiJ, respectively, and the third gene, cbiF, catalyzes an intermediate reaction (Fig. 7C). These observations suggest that perhaps at least two parallel pathways of cobalamin biosynthesis may exist in Halobacterium NRC-1 and each pathway is regulated differently. Finally, VNG1775C is an orphan unrelated to any of the four clustered groups, perhaps because it catalyzes two reactions in tandem at a branch point in the pathway (Fig. 7C), and therefore, might require an independent regulatory mechanism. Consistent with this interpretation, VNG1775C is not a part of the cobalamin biosynthesis operons (Fig. 7A).
The mRNA changes for the five previously unannotated genes place them into three of the four coregulated groups, implicating them as the potential enzymes in, or regulators of the cobalamin biosynthesis pathway. At least three of the five genes could be annotated as follows. The revised sequence of VNG1561C in NCBI (VNG1561Cm) matched in PDB a protoheme ferrolyase (PDB: 1ak1), a terminal enzyme of heme biosynthesis. The Rosetta-predicted structure for VNG1562C matched a cytochrome c protein from Rhodospirllum molischianum (PDB: 2ccy). Finally, the Rosetta-predicted structure for VNG1564H matched the three-dimensional structure of the heme biosynthesis regulator Tup1 (PDB: 1erj; Amillet et al. 1995). Given the similarity of the molecular structure of precorrin to porphyrin, the predicted structural homology of VNG1562H to 2ccy suggests VNG1562H binds a porphyrin or a precorrin-like molecule. A second hypothesis is that these genes are involved in the conversion of precorrin-2 to protoporphyrinogen III, an intermediate of the heme biosynthesis pathway (Ishida et al. 1998). The putative function for VNG1564C, taken together with its cotranscription with the cobalamin biosynthesis genes, implicates it as the putative regulator of this pathway. In summary, systems approaches can facilitate the discovery of regulatory mechanisms as well as the functional annotation of proteins.
Conclusion
The systems analysis has identified several putative DNA repair mechanisms in Halobacterium NRC-1 and provided insights into regulatory networks modulated by DNA damage. The study has confirmed the role of Phr2 in photoreactivation, ruled out a photolyase function for Phr1, ruled out photoprotective roles for the photoactive pigments, and has raised the potential for several mechanisms in repair of photo-oxidative damage and mismatches in the DNA duplex. The experimental procedure alone caused changes in ∼6% of all mRNAs; 61 changes observed during repair also occurred in the control. Therefore, the control was key in identifying changes that were exclusively a result of UV irradiation. With this report, the phenomenon of global down-regulation of many aspects of metabolism, hypothesized to be necessary to maintain internal homeostasis, seems to be a general response to stress shared by all three domains of life. We also observed that light not only provided energy for photolyase function, but also facilitated global changes in regulation during DNA repair. This is not surprising, given that Halobacterium NRC-1 has evolved mechanisms to use light as a source of energy through the action of photoactive ion pumps, as well as information through sensory rhodopsins and transducers.
We are now testing many of these hypotheses experimentally by systematic genetic and/or genome-wide analyses as reported for the wild-type and the three knockout mutant strains of Halobacterium NRC-1 in this work. These iterative cycles of experimental perturbations, genome-wide analysis, and formulation of hypotheses will continue until the deciphered models are in agreement with the experimental observations, and are capable of predicting the behavior of the system in response to new environmental perturbations.
Finally, the function annotation of 26 proteins through sequence and structure-based approaches was key in inferring many hypotheses. Moreover, we have demonstrated the power of comparative genomics in driving systems approaches through providing evolutionary, and therefore, functional relationships among proteins. Such approaches to function annotation in Halobacterium NRC-1 will be further facilitated upon completion of the ongoing genome-sequencing projects on two related halophiles, Haloferax volcanii and Haloarcula marismortui. Therefore, the future for systems analysis in Halobacterium NRC-1 holds great potential, especially with regard to understanding cellular responses to environmental changes.
METHODS
Strains, Culturing, and Mutant Construction
Halobacterium NRC-1 is the wild-type strain (Ng et al. 2000). Gene knockout strains were constructed in an ura3 mutant of Halobacterium NRC-1 as described previously (Peck et al. 2000). The rhodopsin-deficient strain of Halobacterium NRC-1, Pho81, has been characterized previously (Spudich and Spudich 1982); the white, colorless mutant of Halobacterium NRC-1 was obtained in this study through EMS mutagenesis. Culturing of all strains was done at 42°C in liquid nutrient-rich Complete Medium (CM) as previously described (DasSarma and Fleischmann 1995).
UV Irradiation
For UV irradiation, cell pellets from early stationary phase cultures (OD600nm = 1.0, ∼5 × 109 colony-forming units (cfu/mL) were resuspended in an isotonic salt buffer at ∼5 × 108 cfu/mL. A total of 1 mL of cell suspension was irradiated in the dark (254 nm, 1.90 J/s/m2) on ice in a sterile rectangular dish (24 mm × 67 mm), placed 29 cm below a UVP Pen-Ray lamp (228.6 mm × 9.5 mm, UVP Inc.). Post-irradiation, the cell suspension was diluted twofold in CM, and incubated at 42°C either in light or dark; the nonirradiated control was processed identically (Fig. 2). Cell viability was determined by counting colonies on agar plates. At least three independent measurements were made for determining radiation resistance profiles.
Fabrication of Microarrays and Microarray Analysis
Unique 70-mer oligonucleotides spanning the 2400 nonredundant genes in the Halobacterium NRC-1 genome were purchased from QIAGEN. Microarray fabrication, RNA preparation, labeling with Alexa 594, and Alexa 660 dyes (Molecular Probes), hybridization, and washing have been described previously (Baliga et al. 2002). Each comparison was performed twice with total RNA from two independently processed cultures. A reversal in the labeling dyes (dye-flip) was included to rule out bias in dye incorporation. Raw data was processed, log10 ratios were estimated, and the significance of change was determined by the maximum likelihood method (Ideker et al. 2000).
Annotation of Proteins of Unknown Function
Proteins were first divided into domains by sequence-based methods (PSI-BLAST, Pfam, TMHMM). Functions were also assigned by comparing with known structures in the PDB the predicted three-dimensional structures for proteins or domains that resisted initial sequence analysis. The hierarchy for function annotation was as follows. (1) BLAST match to the PDB, (2) HMMER match to Pfam, (3) BLAST match to characterized sequence in nr, and (4) Rosetta or PCONS match to known structure.
Protein Structure Prediction Using Rosetta
Three-dimensional structures for proteins or domains (<150 amino acids) were predicted using the Rosetta method as described previously (Bonneau et al. 2002). Twenty top-ranked Rosetta-predicted structures for each query were searched against a nonredundant subset of the PDB, and matches were reported as CATH or SCOP structural superfamily classifications (Lo Conte et al. 2002; Orengo et al. 2002).
Consensus Fold Recognition
Proteins or domains that escaped Rosetta structure prediction were analyzed by PCONS and 3D-jury using the Meta server (http://bioinfo.pl/meta/, http://www.sbc.su.se/∼arne/pcons/), a consensus fold recognition sever (Bourne 2003).
Acknowledgments
We acknowledge support from HFSP (#RG522002 to J.D.R.), NASA (#NCC9147 to J.D.R.), and NSF (#0220153 to L.H. and N.B.). We thank John L. Spudich for the rhodopsin-deficient strain Pho81, Paul Shannon, Michael Johnson, Kerry Deutsch, and Eric Deutsch for assistance with databases and Cytoscape, and Marc Facciotti for helpful comments. The authors would like to acknowledge that the technology for RNA labeling using Alexa fluorophores was developed by Kreatech Biotechnologies, Amsterdam, NL.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1993504. Article published online before print in May 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GEO under accession no. GSE1040.]
References
- Amillet, J.M., Buisson, N., and Labbe-Bois, R. 1995. Positive and negative elements involved in the differential regulation by heme and oxygen of the HEM13 gene (coproporphyrinogen oxidase) in Saccharomyces cerevisiae. Curr. Genet. 28: 503–511. [DOI] [PubMed] [Google Scholar]
- Baliga, N.S., Pan, M., Goo, Y.A., Yi, E.C., Goodlett, D.R., Dimitrov, K., Shannon, P., Aebersold, R., Ng, W.V., and Hood, L. 2002. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. 99: 14913–14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L. 2000. The Pfam protein families database. Nucleic Acids Res. 28: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell, G.W., Brown, J.A., Wu, H.I., Giaever, G., Chu, A.M., Davis, R.W., and Brown, J.M. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. 99: 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau, R., Tsai, J., Ruczinski, I., Chivian, D., Rohl, C., Strauss, C.E., and Baker, D. 2001. Rosetta in CASP4: Progress in ab initio protein structure prediction. Proteins Suppl: 119–126. [DOI] [PubMed] [Google Scholar]
- Bonneau, R., Strauss, C.E., Rohl, C.A., Chivian, D., Bradley, P., Malmstrom, L., Robertson, T., and Baker, D. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322: 65–78. [DOI] [PubMed] [Google Scholar]
- Bourne, P.E. 2003. CASP and CAFASP experiments and their findings. Meth. Biochem. Anal. 44: 501–507. [PubMed] [Google Scholar]
- Causton, H.C., Ren, B., Koh, S.S., Harbison, C.T., Kanin, E., Jennings, E.G., Lee, T.I., True, H.L., Lander, E.S., and Young, R.A. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle, J., Khodursky, A., Peter, B., Brown, P.O., and Hanawalt, P.C. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158: 41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma, S. and Fleischmann, E.M. 1995. Halophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Diruggiero, J. and Robb, F.T. 2003. Evolution of DNA repair mechanisms. In The genetic code and the origin of life (ed. L. Ribas de Pouplana). Landes Bioscience, Georgetown, TX.
- Eisen, J.A. and Hanawalt, P.C. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435: 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg, E.C. 1995. Out of the shadows and into the light: The emergence of DNA repair. Trends Biochem. Sci. 20: 381. [DOI] [PubMed] [Google Scholar]
- Friedberg, E., Walker, G.C., and Siede, W. 1995. DNA repair and mutagenesis, ASM Press, Washington, D.C.
- Gasch, A.P. and Werner-Washburne, M. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2: 181–192. [DOI] [PubMed] [Google Scholar]
- Hitomi, K., Okamoto, K., Daiyasu, H., Miyashita, H., Iwai, S., Toh, H., Ishiura, M., and Todo, T. 2000. Bacterial cryptochrome and photolyase: Characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res. 28: 2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, T.J., Murzin, A.G., Brenner, S.E., and Chothia, C. 1997. SCOP: A structural classification of proteins database. Nucleic Acids Res. 25: 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Siegel, A.F., and Hood, L.E. 2000. Testing for differentially-expressed genes by maximum-likelihood analysis of microarray data. J. Comput. Biol. 7: 805–817. [DOI] [PubMed] [Google Scholar]
- Ishida, T., Yu, L., Akutsu, H., Ozawa, K., Kawanishi, S., Seto, A., Inubushi, T., and Sano, S. 1998. A primitive pathway of porphyrin biosynthesis and enzymology in Desulfovibrio vulgaris. Proc. Natl. Acad. Sci. 95: 4853–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorns, M.S. 1990. DNA photorepair: Chromophore composition and function in two classes of DNA photolyases. Biofactors 2: 207–211. [PubMed] [Google Scholar]
- Kanehisa, M. 2002. The KEGG database. Novartis Found Symp. 247: 91–101; discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- Kennedy, S.P., Ng, W.V., Salzberg, S.L., Hood, L., and DasSarma, S. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori, H., Masui, R., Kuramitsu, S., Yokoyama, S., Shibata, T., Inoue, Y., and Miki, K. 2001. Crystal structure of thermostable DNA photolyase: Pyrimidine-dimer recognition mechanism. Proc. Natl. Acad. Sci. 98: 13560–13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Zhou, J., Omelchenko, M.V., Beliaev, A.S., Venkateswaran, A., Stair, J., Wu, L., Thompson, D.K., Xu, D., Rogozin, I.B., et al. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. 100: 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte, L., Brenner, S.E., Hubbard, T.J., Chothia, C., and Murzin, A.G. 2002. SCOP database in 2002: Refinements accommodate structural genomics. Nucleic Acids Res. 30: 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., and Eisenberg, D. 1999. Detecting protein function and protein–protein interactions from genome sequences. Science 285: 751–753. [DOI] [PubMed] [Google Scholar]
- Marti, T.M., Kunz, C., and Fleck, O. 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191: 28–41. [DOI] [PubMed] [Google Scholar]
- Masaoka, A., Terato, H., Kobayashi, M., Honsho, A., Ohyama, Y., and Ide, H. 1999. Enzymatic repair of 5-formyluracil. I. Excision of 5-formyluracil site-specifically incorporated into oligonucleotide substrates by alka protein (Escherichia coli 3-methyladenine DNA glycosylase II). J. Biol. Chem. 274: 25136–25143. [DOI] [PubMed] [Google Scholar]
- McCready, S. 1996. The repair of ultraviolet light-induced DNA damage in the halophilic archaebacteria, Halobacterium cutirubrum, Halobacterium halobium and Haloferax volcanii. Mutat. Res. 364: 25–32. [DOI] [PubMed] [Google Scholar]
- McCready, S. and Marcello, L. 2003. Repair of UV damage in Halobacterium salinarum. Biochem. Soc. Trans. 31: 694–698. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler, K., Mauro, D.J., Seal, G., Wurzer, J., deRiel, J.K., and Sirover, M.A. 1991. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. 88: 8460–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G. and Collado-Vides, J. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18: S329–S336. [DOI] [PubMed] [Google Scholar]
- Ng, W.V., Kennedy, S.P., Mahairas, G.G., Berquist, B., Pan, M., Shukla, H.D., Lasky, S.R., Baliga, N.S., Thorsson, V., Sbrogna, J., et al. 2000. From the cover: Genome sequence of halobacterium species NRC-1. Proc. Natl. Acad. Sci. 97: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrunc, M., Becker, D.F., Ragsdale, S.W., and Sancar, A. 1998. Nucleotide excision repair in the third kingdom. J. Bacteriol. 180: 5796–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo, C.A., Bray, J.E., Buchan, D.W., Harrison, A., Lee, D., Pearl, F.M., Sillitoe, I., Todd, A.E., and Thornton, J.M. 2002. The CATH protein family database: A resource for structural and functional annotation of genomes. Proteomics 2: 11–21. [PubMed] [Google Scholar]
- Overbeek, R., Fonstein, M., D'Souza, M., Pusch, G.D., and Maltsev, N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. 96: 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, R.F., Dassarma, S., and Krebs, M.P. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35: 667–676. [DOI] [PubMed] [Google Scholar]
- Pellegrini, M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96: 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain, J.C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. 2001. The protein–protein interaction map of Helicobacter pylori. Nature 409: 211–215. [DOI] [PubMed] [Google Scholar]
- Roberts, J.A., Bell, S.D., and White, M.F. 2003. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 48: 361–371. [DOI] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.R., Doucette-Stamm, L.A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J. Bacteriol. 179: 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, E.N. and Spudich, J.L. 1982. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc. Natl. Acad. Sci. 79: 4308–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentirmay, M.N. and Sawadogo, M. 1993. Synthesis of reinitiated transcripts by mammalian RNA polymerase II is controlled by elongation factor SII. EMBO J. 12: 4677–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Waterworth, W.M., Jiang, Q., West, C.E., Nikaido, M., and Bray, C.M. 2002. Characterization of Arabidopsis photolyase enzymes and analysis of their role in protection from ultraviolet-B radiation. J. Exp. Bot. 53: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Weston, A.D., Baliga, N.S., Bonneau, R., and Hood, L. 2004. Systems approaches applied to the study of Saccharomyces cerevisiae and Halobacterium sp. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Wood, E.R., Ghane, F., and Grogan, D.W. 1997. Genetic responses of the thermophilic archaeon Sulfolobus acidocaldarius to short-wavelength UV light. J. Bacteriol. 179: 5693–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bioinfo.pl/meta/; A Meta server provides access to various fold recognition and local structure prediction methods. This server also provides translation of the standard formats like PDB, CASP, or PIR, and is coupled to several consensus servers such as pcons.
- http://www.sbc.su.se/∼arne/pcons/; A consensus fold recognition predictor.