Abstract
Alu elements are primate-specific members of the SINE (short interspersed element) retroposon family, which comprise ∼10% of the human genome. Here we report the first chromosomal-level comparison examining the Alu retroposition dynamics following the divergence of humans and chimpanzees. We find a twofold increase in Alu insertions in humans in comparison to the common chimpanzee (Pan troglodytes). The genomic diversity (polymorphism for presence or absence of the Alu insertion) associated with these inserts indicates that, analogous to recent nucleotide diversity studies, the level of chimpanzee Alu diversity is ∼1.7 times higher than that of humans. Evolutionarily recent Alu subfamily structure differs markedly between the human and chimpanzee lineages, with the major human subfamilies remaining largely inactive in the chimpanzee lineage. We propose a population-based model to account for the observed fluctuation in Alu retroposition rates across primate taxa.
Alu elements are primate-specific members of the SINE (short interspersed element) family of retroposons. They have enjoyed enormous success over the course of primate evolution and, by conservative estimates, comprise some 10% of the human genome (Schmid 1996; Lander et al. 2001). Largely as a result of the human genome project, a wealth of knowledge has been accumulated concerning the underlying biology, retroposition activity, and associated population genetics of Alu repeats (Schmid 1998; Batzer and Deininger 2002). The ubiquitous presence of Alu sequences within primate genomes has been the cumulative result of a “copy and paste” mechanism, in which an RNA polymerase III–generated transcript is reverse-transcribed and integrated into the genome (Burke et al. 1999). In addition to being wholly dependent upon host cellular processes for their transmission through the germline, Alu elements also lack the ability to generate the endonuclease and reverse transcriptase necessary for their own retroposition. Instead, they must appropriate the necessary enzymatic machinery from L1, a member of the LINE (long interspersed element) retroposon family (Jurka 1997; Kajikawa and Okada 2002). As a result of this obligatory relationship with their genomic host and other transposable elements, the Alu family has been characterized as a “parasite's parasite” (Schmid 2003). Despite the family's various designations as “junk,” “parasites,” and “selfish DNA,” researchers have been reluctant to dismiss them as entirely self-serving genomic entities. A number of investigators have suggested a potential role for Alu elements within their host genomes, and recent implications of Alu element involvement in alternative splicing, segmental duplications, and DNA repair serve to further fuel these arguments (Morrish et al. 2002; Bailey et al. 2003; Lev-Maor et al. 2003; Salem et al. 2003a). Whether these observations constitute adaptations, exaptations (i.e., they have been commandeered for their current roles, despite not having been evolved for them; Brosius 1999), or are simply coincidental by-products of their presence in the genome remains a subject of debate. To address these and other questions will require a better understanding of the manner in which Alu elements have propagated and adapted themselves within nonhuman primate lineages. As the fate of the Alu retroposon is necessarily linked to that of its genomic host, major events in primate evolutionary history will likely have left their mark within the Alu “fossil record” that is present in the genomes of all living primates.
Given the relatively recent divergence time (5 to 6 Mya) of the human and chimpanzee lineages (Wildman et al. 2003), it would be reasonable to expect Alu transpositional activity and the underlying molecular biology associated with retrotransposition in the chimpanzee might closely parallel that of humans. However, initial examination of ∼10.6 Mb of sequence from multiple primate genomes by Liu et al. (2003) revealed a significant deficit in chimpanzee Alu insertions compared with humans and baboons. Their results suggest that substantial variation in transposition and/or fixation rates may exist among primate lineages. Whether these differences are attributable to underlying differences in biology, stochastic fluctuations in Alu proliferation, and/or broader population–genetic processes remains to be determined.
Here we present the first chromosomal-level comparison of Alu retroposition dynamics and associated polymorphism between chimpanzees and humans. We have surveyed common chimpanzee chromosome 22, and its human homolog, chromosome 21, for lineage-specific Alu sequences and determined the insertion polymorphism associated with each of these insertions. We also examined the nucleotide composition of the observed inserts to better understand evolutionarily recent Alu activity. Finally, we propose a population-based model to account for fluctuations in Alu activity within and between primate lineages. In contrast to prior studies of Alu diversity, which have largely relied upon inferred “young” Alu sequence characteristics to identify loci for investigation, the present comparative approach allows for a more unfiltered appraisal of Alu retroposition activity since we last parted ways with our chimpanzee relatives.
RESULTS
Alu Insertion Levels
For the purpose of our comparison, all available sequence from human chromosome 21 and chimpanzee chromosome 22 was first aligned by using a local installation of BLAT (Kent 2002), resulting in ∼32 Mb of aligned sequence that was subsequently screened for evidence of lineage specific Alu insertions (see Methods). To reduce the likelihood of misidentifying deletion events in one lineage as insertions in the other, the identification of Alu insertions was restricted to loci exhibiting distinct, individually inserted Alu elements (see Methods). As a consequence, several questionable insertion/deletions from both the human and chimpanzee were excluded as probable lineage-specific deletion events. Of the remaining putative insertions, the possibility of deletion events masquerading as Alu insertion events was further excluded by using the gorilla as an outgroup to determine the ancestral state of the locus. In all, 46 lineage-specific Alu insertions were identified in chimpanzee chromosome 22, whereas 101 lineage-specific elements were identified in human chromosome 21, demonstrating a 2.2× increase in the number of detectable human insertions (Table 1). These results are in excellent agreement with those of Liu et al. (2003), who found 11 chimpanzee and 23 human insertions (2.1×) in their ∼10.6-Mb human–chimp comparison; as their sequence data was derived from multiple genomic locations, this correspondence suggests that our data are reflective of the genome as a whole and not endemic to the particular chromosomes surveyed.
Table 1.
Lineage-specific Alu Insertions
| Human | Human/Chimp ratio | Chimpanzee | |
|---|---|---|---|
| Observed inserted total | 101 | 2.20 | 46 |
| PCR tested | 78 | — | 43 |
| Fixed present | 63 | — | 26 |
| Observed polymorphic | 16 | — | 18 |
| Observed polymorphic fraction | 0.21 | 0.50 | 0.41 |
| Adjusted polymorphica | 31-33 | — | 35-37 |
| Adjusted polymorphic fraction | 0.33-0.34 | 0.56-0.60 | 0.57-0.59 |
| Adjusted inserted total | 116-118 | 1.84-1.93 | 61-63 |
Adjusted polymorphic fraction was calculated based upon simulation of the frequency of polymorphic Alu elements observed in a given genome by sampling alleles from a uniform frequency distribution (see Methods). Ranges indicated were generated based on 95% confidence intervals derived by simulation.
Although the cross-species comparison allowed us to classify loci as putatively specific to either the human or chimpanzee lineage, there remained the possibility that (1) some of the insertions were shared polymorphisms in which only one lineage's sequenced individual possessed the insertion, and (2) there were “fixed present” insertions in one species that remained polymorphic in the other. Extensive surveys of hundreds of human AluYa5, AluYb8, and AluYc1 insertions in which representative common chimpanzee and bonobo (Pan paniscus) samples were analyzed in nonhuman primate controls have demonstrated that the sharing of Alu polymorphism between species for these young Alu subfamilies would be negligible (Carroll et al. 2001; Roy-Engel et al. 2001, 2002a). In addition, theoretical estimates of the rate of decay of shared polymorphism (Clark 1997), as well as empirical nucleotide data from human, chimpanzee, and gorilla sequences (Hacia et al. 1999), indicate that the number of shared polymorphisms expected given the number of loci involved in our study would be at most one, and therefore, this effect would not appreciably alter our results. However, to address the possibility that some unknown property of Alu insertions might cause them to deviate substantially from these expectations, we evaluated all non-Ya5/Yb8/Yc1 human insertions (most likely to be shared) and 25 chimpanzee-specific insertions in population panels (80 humans and 12 common chimpanzees) from the opposite species and found no instances of shared Alu polymorphism. In addition, these results also give no indication that an appreciable number of elements fixed in human populations remain polymorphic in the chimpanzee. This is further evidenced by the fact that surveys of human Alu elements found that shared insertion in chimpanzee was extremely rare (Carroll et al. 2001; Roy-Engel et al. 2001). Were there a significant number of fixed human elements remaining polymorphic in the chimpanzee, insertion status of the chimpanzee reference samples in these large surveys would have occurred with higher frequency.
To aid in distinguishing whether the observed Alu insertion disparity represents a decrease in the chimpanzee Alu retroposition rate or an increase in the human retroposition rate within a local phylogenetic context (human, chimpanzee, gorilla), we examined a 1.5-Mb segment of homologous 7q31 sequence available in all three species for Alu insertions specific to a given species. The results of this comparison indicate a gorilla Alu transposition/fixation level that is near that of P. troglodytes, with four Alu inserts in Gorilla gorilla compared with three in P. troglodytes and eight in humans. The small amount of gorilla sequence available for comparison resulted in too few Alu insertions to yield significant results (P ∼ 0.25). However, the trend exhibited between humans and chimpanzees in this region (8:3) echoes that of our larger chromosome 21 survey, leading us to believe that the gorilla insertion numbers are also representative of its genome. Although more extensive sequence comparisons using gorillas and orangutans will be required before definitive conclusions can be drawn, our data favor a human-specific increase in Alu retroposition activity within the local phylogenetic context. Examination of the subfamily composition of human and chimpanzee elements (see below) lends further support to this interpretation.
Distribution of Insertions
Qualitatively, the evolutionarily recent Alu insertions were found distributed relatively evenly throughout the chimpanzee and human chromosomes, with expected lower densities near telomeric and centromeric regions primarily due to unsequenced heterochromatic regions. Alu density has previously been established to be strongly correlated with both GC-content and gene density (Schmid 1996; Lander et al. 2001). Chromosome 21 exhibits a 42% GC content, compared with 48% on chromosome 22 and 49% on chromosome 19, which contains both the highest GC content and highest gene density (Lander et al. 2001). Correspondingly, overall Alu density is highest on chromosome 19, followed by chromosome 22 (Chen et al. 2002). Chromosome 21 is relatively gene poor, with an average density of approximately seven genes per megabase compared with the 11.1 per megabase genomic average (Hattori et al. 2000). However, recent genomic surveys of young AluYb8 and AluYa5 subfamilies demonstrate no significant deficit of young subfamily insertions on chromosome 21 (Carter et al. 2004; data not shown). This may partially be attributable to the fact that the Alu GC and genic distribution bias appears to be more pronounced for evolutionarily older insertions (Lander et al. 2001; Jurka et al. 2004). As a result of the relatively small numbers of recently inserted Alu elements in our survey, larger genome-wide comparisons of young Alu inserts will be necessary for adequately detecting any changes in distribution between species. However, we do note here that, in agreement with previous studies of total Alu content (Lander et al. 2001; Chen et al. 2002), human- and chimpanzee-specific insertions on chromosomes 21/22 had a tendency to insert in GC-rich genic regions, with >20% of the insertions in our survey being located within the introns of known genes, and an even higher frequency (>50%) when predicted genes are considered. Based on estimates of known and predicted gene number and average chromosome 21 gene sizes, we estimate that these gene categories span ∼20% and 8% of the sequenced region of the chromosome, respectively. In addition, DSCAM, an alternatively spliced gene involved in neural development (Yamakawa et al. 1998), demonstrated a total of five human-specific insertions. This may not in itself be remarkable, as DSCAM spans 840 kb, making it a rather large target for insertion. However, all five inserts are in the antisense orientation relative to gene transcription, a feature that has been linked to alternative splicing (Lev-Maor et al. 2003). Given intronic Alu orientation frequencies of 0.47 (sense) and 0.53 (antisense) calculated from a survey of 179 AluYb8 and AluYa5 gene insertions, this configuration of antisense Alu elements deviates significantly from expectation (P < 0.05).
Anomalous Loci
In addition to the lineage-specific insertions found in our study, one element, designated CS12, was determined to be exclusive to gorilla and chimpanzee genomes and not present in human, implying a relationship contrary to the orthodox phylogeny of ([HC],[G]). Such discrepancies have been reported elsewhere (Salem et al. 2003b) and most likely represent lineage sorting of an ancestral polymorphism present in the common ancestor of humans, chimpanzee, and gorilla. The existence of such sorting events serves to highlight the relatively short period of time, evolutionarily speaking, during which these three lineages emerged. For the purposes of this study, however, putative lineage sorting events were excluded from further analysis, as they could not be classified as lineage specific for either humans or chimpanzee.
Another locus, HS6, exhibited phylogenetic inconsistencies that were less readily explained. PCR analysis of the locus showed insertions in orangutan, gorilla, and human to the exclusion of chimpanzee. The maintenance of a polymorphism over this period of time—approximately 6 Myr from the branching of orangutan to the divergence of humans and chimpanzees—would be unlikely, prompting us to consider the possibility of an Alu excision at the chimpanzee locus. For further examination, we sequenced the orthologous loci in G. gorilla, P. paniscus, and Pongo pygmaeus (Fig. 1). The HS6 insertions in human, gorilla, and orangutan contained direct repeats that were identical in both sequence and length, strongly indicating identical by descent insertions. Unexpectedly, the chimpanzee locus was a perfect preintegration site, consisting of only one copy of the direct repeat (Fig. 1). In the only previously reported instance in which an Alu element appeared to be excised from a genome, remnants of the Alu insertion remained in the sequence (Edwards and Gibbs 1992). As the precise excision of an Alu insertion appeared to be a remote possibility, we began to explore other potential explanations for our observations. One such possibility is that a segmental duplication in a great ape common ancestor produced a pair of paralogous loci, only one of which received an Alu insertion. This paralogous locus, which would itself be polymorphic and subject to lineage sorting, could have resolved itself into the observed phylogenetic situation. Our inability to detect evidence through PCR for more than one uninserted locus among the tested species indicates that this long-term maintenance of a duplication polymorphism is no more probable than that of a long-lived Alu insertion polymorphism. However, when considered together, these alternative pathways to the same observed state makes the observed insertion states somewhat more likely. On further examination of the HS6 locus, we discovered two immune-related genes, CXADR and CHODL, within 1 Mb of HS6. It is conceivable that balancing selection acting at these nearby loci served to maintain the HS6 polymorphism, ultimately resulting in the unusual phylogenetic distribution of this Alu insertion. Additional investigation of the genes at this locus will be required to verify this hypothesis.
Figure 1.
Reconstructed Alu HS6 insertion sites in human and nonhuman primates. Shaded area indicates direct repeat region. Chimpanzee site demonstrates no evidence for an extracted insertion.
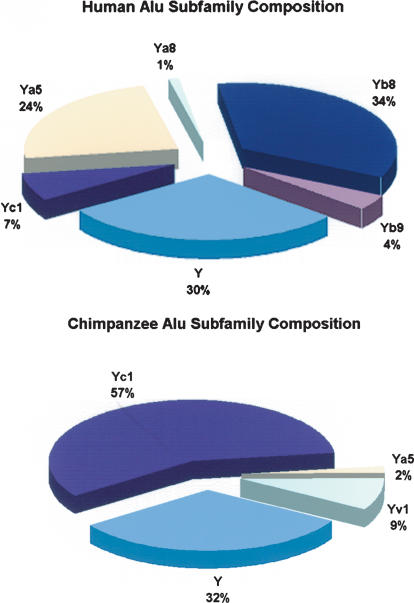
Subfamily Composition
Human Alu elements inserted on chromosome 21 were classified according to subfamily structure as previously reported (Fig. 2; Batzer et al. 1996). All human-specific insertions were members of the AluY subfamily or one of its derivatives. Of these, the AluYa5 and AluYb8 subfamily constituted the largest percentage, comprising 25% and 38% of the loci, respectively. For those elements categorized as members of AluY, their sequences were screened against the human genome database to determine if they belonged to previously uncharacterized subfamilies. Several of these elements appeared to be members of small (10- to 100-member) Alu subfamilies that had previously remained unidentified. Comparative analysis of additional chromosomes will likely reveal additional small subfamily structure that remained undetected by previous molecular and computational methods.
Figure 2.
Subfamily composition of lineage-specific Alu insertions in humans and common chimpanzee.
At present, very little is known about the subfamily structure of Alu elements within the chimpanzee genome. Multiple alignments of all observed P. troglodytes chromosome 22 lineage-specific inserts uncovered two candidates for active subfamilies. The first group, consisting of 27 elements, has a consensus sequence identical to that of AluYc1 in humans. Whether this subfamily is identical by descent or state to its human counterpart is unclear, as AluYc1 differs from the canonical AluY sequence by a single G→A nucleotide substitution. Human AluYc1 insertions exhibit a relatively young (1 to 3 Myr) average age (Garber et al. 2004). Our estimates of the chimpanzee AluYc1 family place it between 1.2 and 2.6 Myr old. Although this is suggestive of an independent parallel mutation, the human AluYc1 elements may have remained relatively dormant in the human genome until some time subsequent to Pan-Homo split. To better localize the chimpanzee AluYc1 activity in time, we examined the insertion status of 18 P. troglodytes–specific AluYc1-like elements in a representative bonobo (P. paniscus), estimated to have diverged from Pan troglodytes ∼1.8 Mya (Yu et al. 2003). Eleven elements were present in the P. troglodytes population but absent from our P. paniscus individual and seven elements were present in both species, indicating that the chimpanzee AluYc1-like subfamily had began amplifying prior to the P. troglodytes–P. paniscus divergence. This places a lower bound on the chimpanzee AluYc1 family age of ∼2 Mya, not ruling out the possibility that these subfamilies are of common descent.
The second group of four elements (designated YV1) were distinguished by five diagnostic mutations from the AluY consensus. Screening of the human genome database revealed several matches within humans, indicating that this subfamily was not restricted to the chimpanzee lineage and has been amplifying, albeit slowly, since before the human–chimpanzee split. Here, there is little possibility of a parallel forward mutation event, as YV1 is distinguished by five mutations.
Alu Insertion Polymorphism
To assess the diversity of individual lineage-specific Alu insertions on human chromosome 21, 78 Alu elements that were amenable to PCR were amplified on a panel of 80 human individuals from four geographically diverse populations (African American, Asian, German Caucasian, and South American). Among the four represented populations, 16 of 78 (20.51%) elements demonstrated polymorphism in our panel. Allele frequencies of all polymorphisms, as well as primers used in this study, are available at our Web site (http://batzerlab.lsu.edu). Forty-three chimpanzee-specific insertions were evaluated on our chimpanzee panel of 12 unrelated P. troglodytes. Because of the small size of our P. troglodytes sample, we assessed its adequacy in evaluating loci for polymorphism (see Methods). Assuming a uniform distribution of Alu allele frequencies, we estimated that our 12 individual (24-chromosome) sample would capture ∼88% to 93% of the polymorphism present at the examined loci. In all, 18 of 43 (41.86%) elements exhibited polymorphism in our chimpanzee panel. The 2.0 ratio of human-to-chimpanzee polymorphism fraction is somewhat higher than the 1.5 ratio of a recent nucleotide heterozygosity study (Yu et al. 2003). If adjustments for unequal polymorphism levels are made, however, the values become closer (see Discussion).
DISCUSSION
Alu Transposition Levels and Subfamily Structure
Our results suggest that an elevation in human Alu retroposition activity, largely mediated by two human Alu subfamilies (AluYa5 and AluYb8), occurred some time subsequent to the divergence of the human and chimpanzee lineages. The most current estimates for the ages of these subfamilies place them amplifying between 2.5 and 3.5 Mya (Carroll et al. 2001). A survey of a 4-Mb X-Y translocation event (Schwartz et al. 1998), which has previously been dated to ∼3.5 to 4 Mya (Sargent et al. 2001), suggests no appreciable retroposition activity of AluYa5 and AluYb8 families prior to that time period. This is indicated by the absence of AluYb8 and AluYa5 elements duplicated at the time of the translocation event. These observations place the onset of significant AluYa5 and AluYb8 mobilization subsequent to the divergence of the human and chimpanzee lineages, indicating that a contraction in population size during or immediately following speciation does not account for the chimpanzee–human Alu disparity.
The question arises as to whether or not the AluYa5 and AluYb8 subfamily expansions were simultaneous or distinct events. Although current age estimates date them to roughly the same period, polymorphism levels of AluYb8 (20%) and AluYa5 (25%) suggest a somewhat younger overall age for the AluYa5 subfamily, as more of its members remain unfixed in the population (Carroll et al. 2001). However, the polymorphism fraction may only serve to indicate that the bulk of AluYa5 insertions are distributed closer to the present than that of AluYb8, and is not necessarily reflective of the initial appearance date of the subfamily.
An additional factor with the potential to influence the estimated ratio of Alu insertion numbers in species is the existence of unequal diversity levels within humans and chimpanzees for Alu insertions. By using the observed Alu diversity in chimpanzee and human, we estimated the extent to which this effect may have skewed our results (see Methods). Our estimates suggest that in 95% of cases, 42% to 58% of the polymorphic Alu insertion loci would be missed by sequencing a single representative human genome or chimpanzee genome. When we adjust insertion numbers within both lineages for these missed Alu loci, our estimate of the human/chimpanzee insertion ratio is 1.84 to 1.93 (Table 1).
The paucity of evolutionarily recent Alu insertions observed on the P. troglodytes chromosome 22 restricts our ability to completely capture the chimpanzee Alu substructure. However, assuming that young Alu subfamily dispersal in humans is distributed proportional to chromosome size, the chance of missing a major young Alu family (>300 elements) in our chimpanzee chromosome 22 survey would be remote (<5%). Our data indicate that the major lineages that constitute the bulk of recent human activity, AluYa5 and AluYb8, are only present at negligible levels in P. troglodytes. A solitary AluYa5 element was found on chimpanzee chromosome 22, and although GenBank database queries indicate that a small number of authentic AluYb8 chimpanzee insertions are present in the P. troglodytes genome, quantitative PCR results suggest that their copy number is negligible compared with that of humans (Walker et al. 2003). The AluYc1-type subfamily appears to dominate the Pan lineage (Fig. 2), but we can not conclusively say if it is identical by descent to the subfamily that is found in humans. If it is indeed the same family, it would be curious that, given their estimated ages (1 to 3 Myr), the source sequence would have remained relatively dormant in both lineages only to become active, independently, at a later time. Alternatively, the independent parallel success of these source mutations may suggest a selective advantage for the G→A consensus substitution, or it could simply be a base position where such change is tolerated in the Alu source or “master” genes.
Although several of the Alu polymorphic loci in chimpanzee contained sequence characteristics that were present in only a single copy on chromosome 22, these insertions will serve as excellent starting points to search for further chimpanzee Alu family substructure, as they likely represent chromosome 22 representatives of smaller, active Alu subfamilies analogous to those recovered in the human sequence.
The presence of AluYb8 and AluYa5 members in small copy numbers within the chimpanzee and gorilla genomes (Leeflang et al. 1993) demonstrates that the sequence evolution of successful subfamilies begins well before their peak activity. These subfamilies appear to undergo a lengthy period during which low baseline mobilization occurs. A chance insertion within a suitable genomic context, however, could initiate a burst of activity from the locus within a given host lineage. In conjunction with L1 enzyme availability and population genetic factors (see below), such fortuitous insertions would initiate the expansion phase of the Alu subfamily.
Alu Insertion Polymorphism
Our Alu insertion diversity data demonstrate two times higher Alu polymorphism in chimpanzee compared with humans. If we adjust the estimates of polymorphic Alu loci by accounting for the insertion polymorphisms that were predicted to be missed in chimpanzee and human sequences (see Methods), our ratio of chimpanzee-to-human Alu polymorphism decreases to 1.67 to 1.78. A number of previous studies, making use of multiple genetic systems, have attempted to assess the level of genetic diversity of chimpanzees relative to that of humans. Mitochondrial and nuclear genome surveys have generated seemingly conflicting depictions of chimpanzee diversity. Mitochondrial diversity has been estimated to be as much as 10 times higher among chimpanzees than humans (Rogers and Jorde 1995). Nuclear nucleotide diversity estimates, in contrast, have yielded chimpanzee heterozygosities that are lower than human levels for protein-coding loci (King and Wilson 1975; Satta 2001). Surveys of additional coding and noncoding loci have reported nucleotide heterozygosity estimates three to four times higher in chimpanzee than humans (Deinard and Kidd 1999; Kaessmann et al. 1999). Our range of 1.67 to 1.78 times higher common chimpanzee diversity best corresponds to that of Yu et al. (2003), who estimated nucleotide diversity in common chimpanzee at 1.5 times higher than that of human, with a lower value for bonobo.
The previously reported disparity of heterozygosity values exhibited by different genetic systems (mitochondrial, microsatellite, nuclear SNPs) can potentially be explained by a population bottleneck in humans that had a more severe effect on mitochondrial diversity due to its smaller (1/4 autosomal) effective population size (Yu et al. 2003). The existence of a bottleneck in human evolutionary history has been suggested by many studies (Harpending et al. 1998; Chen and Li 2001; Lonjou et al. 2003). Although our chromosome 21/22 data are consistent with this scenario, we can not exclude other possibilities, such as selective sweeps reducing mitochondrial diversity.
If the correspondence between Alu insertion polymorphism ratios and the nucleotide diversity ratios between humans and chimpanzees is not simply coincidental, it would appear that the effective population size is the dominant influence determining the fraction of Alu insertion polymorphisms in these genomes. That is, despite markedly different subfamily composition and retroposition histories between the two lineages, Alu insertion polymorphism generally parallels nucleotide polymorphism in behavior. This is a somewhat surprising result, given that fluctuations in Alu activity over time could result in one lineage having an excess or deficit of younger polymorphic Alu insertions relative to the other lineage, largely independent of effective population size. However, this situation could conceivably be explained if the more dramatic changes in Alu insertion rates occurred in more distant evolutionary history and have had little influence on current polymorphism levels. In this scenario, relatively uniform insertion rates within individual lineages over recent evolution history have resulted in effective population size being the dominant determinant of polymorphism levels. Further resolution of the insertion dates of human and chimpanzee Alu elements will be necessary to clarify this issue.
A Population-Based Model for Fluctuations in Alu Mobilization
Under standard neutral or “nearly neutral” population genetics theory, three scenarios could conceivably account for the relative increase in fixed Alu insertions within humans compared with chimpanzees. First, a smaller long-term effective population size in the human lineage could have resulted in the fixation of otherwise slightly deleterious Alu insertions at a higher rate in humans. Under this scenario, the roughly twofold increase in observed human insertions would need to be accounted for by deleterious elements. Although this possibility can not presently be excluded, the fixation of hundreds of deleterious Alu insertion loci would no doubt represent a considerable burden to a population. An explanation that avoided such a genetic calamity would appear to be more parsimonious. A second scenario would be that the existing Alu polymorphism that was present at the time of human–chimpanzee speciation was funneled through a Homo lineage bottleneck, resulting in an increased fixation of Alu elements within humans. In this situation, the differences in Alu insertion number would be attributable to many more of these ancestral polymorphisms fixing in the human lineage than the chimpanzee. This scenario is unlikely as well, however, as the sequence structure of Alu insertions of humans, comprised largely of two young subfamilies, differs considerably from that of chimpanzee (Fig. 2). This suggests that they were not derived from a common pool of Alu insertions that were polymorphic at the time of speciation. In addition, the major retroposition activity within the AluYa5 and AluYb8 subfamilies can be reasonably dated by independent lines of evidence to a period subsequent to the human–chimpanzee speciation (see Results). The third possibility, which we favor, is an increase in the Alu retroposition rate itself. This would be analogous to an increased nucleotide mutation rate within a given lineage. However, in the case of retroposition, there is an added layer of complexity in the interaction among insertion rates, fixation rates, and population size that must be addressed.
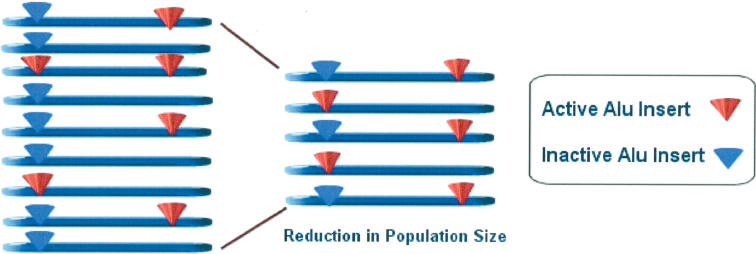
The population dynamics of Alu elements within their hosts can account for much of the insertion variance observed within and between primate lineages. The basic components of our model are as follows: (1) variation in source Alu-producing loci exist in the population, (2) stochastic sampling of these source variants either at speciation or during bottleneck events alters the population-level Alu transposition activity (insertions per birth), and (3) although the previous two conditions are sufficient to produce variation within and between lineages, smaller effective population sizes will both increase the sampling variance of Alu sources and reduce a given population's ability to select against deleterious source loci. This may result in a substantially increased population-level Alu activity (insertions per birth) brought about by environmental insults, speciation events, etc.
Aside from their observed GC-rich distribution bias, there has been no evidence indicating that Alu insertions behave appreciably different than nucleotide polymorphisms as genetic markers once inserted in the genome (Perna et al. 1992; Stoneking et al. 1997; Watkins et al. 2001, 2003; Bamshad et al. 2003). As such, the behavior of Alu elements should be consistent with other neutral or “nearly neutral” characters. The probability of a given Alu insertion reaching fixation in a population is therefore contingent upon its initial frequency in the population, 1/2N, where N is the population size (Kimura 1983). In the context of Alu retrotransposition, however, not all of the further assumptions of neutral theory hold. Although the number of novel nucleotide mutations arising each generation in a population is dictated by the size of the population (i.e., total number of mutable sites) and the frequency of mutations arising each generation, the number of novel Alu insertions has a more complex relationship with population size. As the majority of new Alu copies are known to arise from a select number of “master” or source loci, these loci themselves will be subject to allelic variation in both transpositional competency and/or insertion status. Evidence for such allelic variation in retrotransposition capability has been observed in members of the L1 subfamily (Lutz et al. 2003) and within Alu may be attributable to variation at PolIII promoter efficiency, variation in target-primed reverse transcription, oligo dA tail instability (Roy-Engel et al. 2002b), and insertion status polymorphism for the source locus itself. Additional evidence from L1 sequence transduction events demonstrate that retroposon source sequences can produce “offspring” that proceed to fixation, whereas the parent sequences are ultimately lost (Boissinot et al. 2001). As a consequence of this source allele variation, a reduction in overall human population size may occur, whereas the number of novel Alu insertions per individual birth actually increases due to the stochastic effects of sampling the active source variants (Fig. 3). In effect, unlike nucleotide substitution rates, the equivalent of the Alu substitution rate will itself fluctuate along with population size. The intensity of these fluctuations will increase as the population size becomes smaller. Simultaneously, a reduced effective population size is less capable of selecting against detrimental source variants as the population size grows smaller. This effect is exacerbated because the Alu source is effectively “screened” by its indirect relationship to the deleterious insertion loci it generates. As a consequence, transposition may run rampant when the population size is no longer large enough to effectively select against Alu “hyperactivity.” Within a window of selective pressure, deleterious insertions would still be effectively removed from the genome, but the source or sources generating the deleterious insertions become(s) essentially neutral (i.e., having a selective coefficient ≪1/2N).
Figure 3.
Variation in the insertion status and retroposition capability of Alu elements at two loci. Reduction in population size leads to variation in the number of active elements.
An attractive feature of this explanation is that it does not necessitate the presence of a large number of fixed deleterious loci to account for differential lineage Alu insertion counts. Furthermore, it does not require the invocation of any novel biology to account for changes in the relative number of insertions per generation. One prediction of the model is that the onset of increased Alu transposition activity would tend to be coincident with population size decreases, and as a consequence, Alu transposition rates may change rapidly within and between lineages. By developing better analytical tools to estimate the ages of individual Alu insertions, it may be possible to localize transposition events in time and estimate the rate at which Alu transposition activity fluctuates. A further prediction is that isolated inbred populations would be at an increased risk for Alu “hyperactivity,” as they would experience a decreased capacity to select against active source loci. Genomic display, ATLAS, and similar methodologies that have the potential to exhaustively examine retroposon insertions within individual genomes will allow testing in extant populations for evidence of this effect.
METHODS
DNA Samples
Cell lines used to isolate DNA samples were as follows: A chimpanzee diversity panel of 12 P. troglodytes of unknown geographic origin was obtained from the SouthWest foundation for Biomedical Research, gorilla (G. gorilla), lowland gorilla Coriell AG05253A, owl monkey (A. trivirgatus), ATCCCRL1556, and pygmy chimpanzee (P. paniscus), Coriell AG05253A. Human DNA from South American populations was purchased as part of the Human Variation Panel available from the Coriell Institute for Medical Research. DNA samples from the European, African American, and Asian population groups were isolated from peripheral blood lymphocytes available from previous studies.
Human–Chimpanzee Comparison
DNA sequences for chromosome 22 (∼43 Mb, including overlapping sequence) were obtained from The Chimpanzee Chromosome 22 Sequencing Consortium (http://chimp22pub.gsc.riken.go.jp). Sequence for human chromosome 21 was obtained from the University of California, Santa Cruz (UCSC) June 2003 assembly data. Human chromosome 21 and chimpanzee chromosome 22 alignments were generated by using a local installation of BLAT (Blast-like Alignment Search Tool; Kent 2002), resulting in ∼32 Mb of aligned sequence out of an estimated 33.8 Mb total chromosome 21 sequence (Hattori et al. 2000). BLAT results were subsequently screened by using a Perl script for all insertions/deletions of sizes 100 to 1000 bp. These sequences, along with 200 bp of flanking sequence, were extracted for further examination. In addition, a separate manual BLAT screen of the human genome database (using UCSC Web interface) using the chimpanzee chromosome 22 sequence was conducted to assess the accuracy of our script-generated results. Indel sequences were screened by using a local installation of RepeatMasker (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker) to determine their repetitive element content. Subsequent sequence alignments were done with MEGALIGN program, part of the DNASTAR package. Redundant repeat insertions resulting from overlapping BLAT fragments were excluded by verifying unique flanking sequence. An additional ∼1.5Mb of human, chimpanzee, and gorilla homologous sequence from chromosome 7 was obtained from the National Institutes of Health (NIH) Intramural Sequencing Center (www.nisc.nih.gov). Sequences were aligned with BLAT and/or MEGALIGN to identify species-specific indels, and RepeatMasker was used to determine their repetitive element content.
All putative Alu insertions were manually verified as authentic by determining if the insertions met established criteria for evolutionarily recent Alu insertions. Authentic Alu insertions were required to have only 5′ truncations, as 3′ truncations have not been observed to occur upon insertion. Any “partial” Alu indels in which a fragment of the Alu is already present at the locus prior to the indel event were excluded, as these are more characteristic of partial deletions of elements. Alus that were contained within larger insertion/deletion events were also excluded, as these did not represent authentic Alu transposition events. To further resolve ambiguities, all putative insertions were amplified from the gorilla genome to determine the ancestral state of the insertion.
Statistical Methods
Estimating the Number of Detected Polymorphic Alu Insertions
Estimations of the number of polymorphic insertions that would be detected in a single sequenced genome were conducted by generating 1000 samples of a genome (set of detectable alleles) from a uniform distribution of Alu insertion frequencies. This choice of distribution was based on observations of the allele frequencies of human Alu inserts (Carroll et al. 2001; Roy-Engel et al. 2001), and reasoning that the higher long-term effective population size of chimpanzee would result in an even more uniform (flat) distribution of Alu insertion frequencies due to the lack of recent bottlenecks and/or expansions (Harpending et al. 1998). In our simulation, the probability of discovering a given allele was proportional to its frequency in the population. The mean fraction of detections was 0.5, with a variance inversely proportional to the number of actual polymorphic loci. Our 1000 replicates using 100 loci yielded a standard deviation of 4%, which was used to calculate a 95% confidence interval for unsampled polymorphisms of 42% to 58%.
Detection of Polymorphism
The probability of detecting an Alu insertion polymorphism at a given locus is contingent upon its minor allele frequency 1 – [(1 – q)^N], where q is the minor allele frequency and N is the number of sampled chromosomes. Consequently, the number of detectable Alu variants will be subject to the distribution of allele frequencies in the population. If we assume this is roughly uniform, then summing over i minor allele frequencies Σ[1 – [(1 – qi)^N]] yields the fraction of polymorphic sequences detected. By simulating 1000 trial detections of uniformly distributed minor alleles, we estimate that 95% of the time our human panel of 80 individuals (160 chromosomes) would detect 97.3% to 99.7%, and our chimpanzee panel of 12 individuals (24 chromosomes) would detect 89% to 93% of the polymorphism at PCR evaluated loci. Within the observed polymorphism, there should be a skew toward higher frequency alleles, as these are more likely to appear in a given sequenced genome. Because we restricted our analysis to polymorphic/fixed status, this bias should not affect our conclusions.
PCR Analysis
Oligonucleotide primers for the PCR amplification of each Alu element were designed using the 700- to 1200-bp flanking unique sequence fragments and Primer3 software (Whitehead Institute for Biomedical Research; http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The sequences of the oligonucleotide primers, annealing temperatures, PCR product sizes and chromosomal locations for all Alu elements in this study can be found on our Web site (http://batzerlab.lsu.edu). PCR amplification was performed in 25 μL reactions using 10 to 50 ng target DNA, 200 nM each oligonucleotide primer, 200 μM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.4), and 1 U Taq DNA polymerase. Each sample was subjected to an initial denaturation step of 150 sec at 94°C, followed by 32 cycles of PCR at 1 min of denaturation at 94°C, 1 min at the annealing temperature, 1 min of extension at 72°C, followed by a final extension step of 10 min at 72°C. The resulting products were then evaluated for polymorphism on Ethidium Bromide (EtBr)-stained 2% agarose gels and visualized with UV lighting.
DNA Sequencing
DNA sequencing was performed on gel-purified PCR products that had been cloned by using the TOPO TA cloning vector (Invitrogen) using chain termination sequencing on an Applied Biosystems 3100 automated DNA sequencer. All sequences generated in this study are available in the GenBank database (accession nos. AY569161–AY569170).
Acknowledgments
We thank Dr. Yoshiyuki Sakaki and the rest of the chimpanzee chromosome 22 sequencing consortium for providing us with access to their sequence data. We would also like to thank Daniel Ortiz and the anonymous referees for helpful comments on the manuscript. This research was supported by National Science Foundation BCS-0218338 (M.A.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2530404.
Footnotes
[The sequence data from this study have been submitted to GenBank under accession nos. AY569161–AY569170.]
References
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad, M.J., Wooding, S., Watkins, W.S., Ostler, C.T., Batzer, M.A., and Jorde, L.B. 2003. Human population genetic structure and inference of group membership. Am. J. Hum. Genet. 72: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. [DOI] [PubMed] [Google Scholar]
- Batzer, M.A., Deininger, P.L., Hellmann-Blumberg, U., Jurka, J., Labuda, D., Rubin, C.M., Schmid, C.W., Zietkiewicz, E., and Zuckerkandl, E. 1996. Standardized nomenclature for Alu repeats. J. Mol. Evol. 42: 3–6. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., Entezam, A., and Furano, A.V. 2001. Selection against deleterious LINE-1–containing loci in the human lineage. Mol. Biol. Evol. 18: 926–935. [DOI] [PubMed] [Google Scholar]
- Brosius, J. 1999. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238: 115–134. [DOI] [PubMed] [Google Scholar]
- Burke, W.D., Malik, H.S., Jones, J.P., and Eickbush, T.H. 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16: 502–511. [DOI] [PubMed] [Google Scholar]
- Carroll, M.L., Roy-Engel, A.M., Nguyen, S.V., Salem, A.H., Vogel, E., Vincent, B., Myers, J., Ahmad, Z., Nguyen, L., Sammarco, M., et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311: 17–40. [DOI] [PubMed] [Google Scholar]
- Carter, A.B., Salem, A.-H., Hedges, D.J., Nguyen Keegan, C., Kimball, B., Walker, J.A., Watkins, W.S., Jorder, L.B., and Batzer, M.A. 2004. Genome wide analysis of the human Yb lineage. Hum. Genomics 1: 167–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Gentles, A.J., Jurka, J., and Karlin, S. 2002. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 99: 2930–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.G. 1997. Neutral behavior of shared polymorphism. Proc. Natl. Acad. Sci. 94: 7730–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinard, A. and Kidd, K. 1999. Evolution of a HOXB6 intergenic region within the great apes and humans. J. Hum. Evo.l 36: 687–703. [DOI] [PubMed] [Google Scholar]
- Edwards, M.C. and Gibbs, R.A. 1992. A human dimorphism resulting from loss of an Alu. Genomics 14: 590–597. [DOI] [PubMed] [Google Scholar]
- Garber, R.K., Hedges, D.J., Herke, S.W., Hazard, N.W., and Batzer, M.A. 2004. The Alu Yc1 subfamily: Sorting the wheat from the chaff. Cytogenet. Genome Res. (in press). [DOI] [PubMed]
- Hacia, J.G., Fan, J.B., Ryder, O., Jin, L., Edgemon, K., Ghandour, G., Mayer, R.A., Sun, B., Hsie, L., Robbins, C.M., et al. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22: 164–167. [DOI] [PubMed] [Google Scholar]
- Harpending, H.C., Batzer, M.A., Gurven, M., Jorde, L.B., Rogers, A.R., and Sherry, S.T. 1998. Genetic traces of ancient demography. Proc. Natl. Acad. Sci. 95: 1961–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, Y., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21. Nature 405: 311–319. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. 94: 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J., Kohany, O., Pavlicek, A., Kapitonov, V.V., and Jurka, M.V. 2004. Duplication, coclustering, and selection of human Alu retrotransposons. Proc. Natl. Acad. Sci. 101: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann, H., Wiebe, V., and Paabo, S. 1999. Extensive nuclear DNA sequence diversity among chimpanzees. Science 286: 1159–1162. [DOI] [PubMed] [Google Scholar]
- Kajikawa, M. and Okada, N. 2002. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111: 433–444. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT: The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge.
- King, M.C. and Wilson, A.C. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Leeflang, E.P., Liu, W.M., Chesnokov, I.N., and Schmid, C.W. 1993. Phylogenetic isolation of a human Alu founder gene: drift to new subfamily identity [corrected]. J. Mol. Evol. 37: 559–565. [DOI] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., and Ast, G. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300: 1288–1291. [DOI] [PubMed] [Google Scholar]
- Liu, G., Zhao, S., Bailey, J.A., Sahinalp, S.C., Alkan, C., Tuzun, E., Green, E.D., and Eichler, E.E. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonjou, C., Zhang, W., Collins, A., Tapper, W.J., Elahi, E., Maniatis, N., and Morton, N.E. 2003. Linkage disequilibrium in human populations. Proc. Natl. Acad. Sci. 100: 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, S.M., Vincent, B.J., Kazazian Jr., H.H., Batzer, M.A., and Moran, J.V. 2003. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 73: 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish, T.A., Gilbert, N., Myers, J.S., Vincent, B.J., Stamato, T.D., Taccioli, G.E., Batzer, M.A., and Moran, J.V. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31: 159–165. [DOI] [PubMed] [Google Scholar]
- Perna, N.T., Batzer, M.A., Deininger, P.L., and Stoneking, M. 1992. Alu insertion polymorphism: A new type of marker for human population studies. Hum. Biol. 64: 641–648. [PubMed] [Google Scholar]
- Rogers, A.R. and Jorde, L.B. 1995. Genetic evidence on modern human origins. Hum. Biol. 67: 1–36. [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., Vogel, E., Garber, R.K., Nguyen, S.V., Salem, A.H., Batzer, M.A., and Deininger, P.L. 2001. Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Carroll, M.L., El-Sawy, M., Salem, A.H., Garber, R.K., Nguyen, S.V., Deininger, P.L., and Batzer, M.A. 2002a. Non-traditional Alu evolution and primate genomic diversity. J. Mol. Biol. 316: 1033–1040. [DOI] [PubMed] [Google Scholar]
- Roy-Engel, A.M., Salem, A.H., Oyeniran, O.O., Deininger, L., Hedges, D.J., Kilroy, G.E., Batzer, M.A., and Deininger, P.L. 2002b. Active Alu element “A-tails”: Size does matter. Genome Res. 12: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, A.H., Kilroy, G.E., Watkins, W.S., Jorde, L.B., and Batzer, M.A. 2003a. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20: 1349–1361. [DOI] [PubMed] [Google Scholar]
- Salem, A.H., Ray, D.A., Xing, J., Callinan, P.A., Myers, J.S., Hedges, D.J., Garber, R.K., Witherspoon, D.J., Jorde, L.B., and Batzer, M.A. 2003b. Alu elements and hominid phylogenetics. Proc. Natl. Acad. Sci. 100: 12787–12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent, C.A., Boucher, C.A., Blanco, P., Chalmers, I.J., Highet, L., Hall, N., Ross, N., Crow, T., and Affara, N.A. 2001. Characterization of the human Xq21.3/Yp11 homology block and conservation of organization in primates. Genomics 73: 77–85. [DOI] [PubMed] [Google Scholar]
- Satta, Y. 2001. Comparison of DNA and protein polymorphisms between humans and chimpanzees. Genes Genet. Syst. 76: 159–168. [DOI] [PubMed] [Google Scholar]
- Schmid, C.W. 1996. Alu: Structure, origin, evolution, significance and function of one-tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol. 53: 283–319. [DOI] [PubMed] [Google Scholar]
- Schmid, C.W. 1998. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26: 4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, C.W. 2003. Alu: A parasite's parasite? Nat. Genet. 35: 15–16. [DOI] [PubMed] [Google Scholar]
- Schwartz, A., Chan, D.C., Brown, L.G., Alagappan, R., Pettay, D., Disteche, C., McGillivray, B., de la Chapelle, A., and Page, D.C. 1998. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum. Mol. Genet. 7: 1–11. [DOI] [PubMed] [Google Scholar]
- Stoneking, M., Fontius, J.J., Clifford, S.L., Soodyall, H., Arcot, S.S., Saha, N., Jenkins, T., Tahir, M.A., Deininger, P.L., and Batzer, M.A. 1997. Alu insertion polymorphisms and human evolution: Evidence for a larger population size in Africa. Genome Res. 7: 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.A., Kilroy, G.E., Xing, J., Shewale, J., Sinha, S., and Batzer, M.A. 2003. Human DNA quantitation using Alu element based PCR. Anal. Biochem. 315: 122–128. [DOI] [PubMed] [Google Scholar]
- Watkins, W.S., Ricker, C.E., Bamshad, M.J., Carroll, M.L., Nguyen, S.V., Batzer, M.A., Harpending, H.C., Rogers, A.R., and Jorde, L.B. 2001. Patterns of ancestral human diversity: An analysis of Alu-insertion and restriction-site polymorphisms. Am. J. Hum. Genet. 68: 738–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, W.S., Rogers, A.R., Ostler, C.T., Wooding, S., Bamshad, M.J., Brassington, A.M., Carroll, M.L., Nguyen, S.V., Walker, J.A., Prasad, B.V., et al. 2003. Genetic variation among world populations: Inferences from 100 Alu insertion polymorphisms. Genome Res. 13: 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman, D.E., Uddin, M., Liu, G., Grossman, L.I., and Goodman, M. 2003. Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: Enlarging genus Homo. Proc. Natl. Acad. Sci. 100: 7181–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, K., Huot, Y.K., Haendelt, M.A., Hubert, R., Chen, X.N., Lyons, G.E., and Korenberg, J.R. 1998. DSCAM: A novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum. Mol. Genet. 7: 227–237. [DOI] [PubMed] [Google Scholar]
- Yu, N., Jensen-Seaman, M.I., Chemnick, L., Kidd, J.R., Deinard, A.S., Ryder, O., Kidd, K.K., and Li, W.H. 2003. Low nucleotide diversity in chimpanzees and bonobos. Genetics 164: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://batzerlab.lsu.edu; allele frequencies of all polymorphisms, as well as primers used in this study.
- http://chimp22pub.gsc.riken.go.jp; The Chimpanzee Chromosome 22 Sequencing Consortium.
- http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker.
- http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi; Primer3 software, Whitehead Institute for Biomedical Research.