Abstract
Light microscopic analysis of cell morphology provides a high-content readout of cell function and protein localization. Cell arrays and microwell transfection assays on cultured cells have made cell phenotype analysis accessible to high-throughput experiments. Both the localization of each protein in the proteome and the effect of RNAi knock-down of individual genes on cell morphology can be assayed by manual inspection of microscopic images. However, the use of morphological readouts for functional genomics requires fast and automatic identification of complex cellular phenotypes. Here, we present a fully automated platform for high-throughput cell phenotype screening combining human live cell arrays, screening microscopy, and machine-learning-based classification methods. Efficiency of this platform is demonstrated by classification of eleven subcellular patterns marked by GFP-tagged proteins. Our classification method can be adapted to virtually any microscopic assay based on cell morphology, opening a wide range of applications including large-scale RNAi screening in human cells.
Genomewide cDNA overexpression and gene knock-down by RNA interference enable gain and loss of function screens in many systems of cultured cells that were traditionally not accessible to genetic screens (e.g., Drosophila Schneider cells, human cell cultures). In addition to gain and loss of function, the subcellular localization of the whole proteome can be determined by expressing tagged proteins (Pepperkok et al. 2001; Huh et al. 2003; Simpson and Pepperkok 2003) providing essential indications for protein function. Similar to classical genetics, the phenotype of cultured cells is typically determined by morphological analysis and a more detailed phenotypic characterization can be obtained by single-cell fluorescence microscopy of appropriate marker proteins (Rolls et al. 1999). Cell arrays (Ziauddin and Sabatini 2001) and microwell transfection assays (Liebel et al. 2003) on cultured cells have made cell phenotype analysis accessible to high-throughput. While transfection and gene tagging procedures for such studies have been adapted to automation (Simpson et al. 2000), phenotypes are currently typically determined by manual microscopy (Kiger et al. 2003) even when thousands of full-length GFP-tagged proteins are localized (Huh et al. 2003).
Such a manual approach is a source for bias in data analysis and causes a bottleneck for large-scale experiments. Automated systems for the interpretation of cell images from cell arrays would provide three important advantages over manual practice: (1) high-throughput performance; (2) quantitative and reproducible identification of cellular phenotypes; and therefore (3) consistent and unbiased phenotypic information in protein databases.
Automatic classification of microscopic images typically requires the extraction of quantitative parameters (features) from the digital image (Egmont-Petersen et al. 2002). Such features are based on morphology (e.g., number of objects), texture (e.g., granularity), or other gray-level-based measures. Selecting subsets of the many possible features is then necessary to reduce the complexity of the classifier. Different feature selection algorithms for elimination of noninformative features have been suggested in the literature (Leray and Gallinari 1999). After feature extraction, a statistical model needs to be learned from data that accurately associates image features with predefined phenotype classes. In the field of machine-learning, this is usually referred to as supervised training of a classifier. The performance of a classifier is measured by the accuracy to correctly predict unseen test images using the same set of input features (Jain et al. 2000). Stepwise discriminant analysis and genetic algorithms so far produced the highest accuracies (∼87%; Huang et al. 2003). For automated prediction of the localization of 46 randomly GFP-tagged proteins (Jarvik et al. 1996), hierarchical clustering was proposed (Murphy et al. 2002). However, even though 3D image features were used to identify twelve classes derived from hierarchical clustering, recognition was less successful, with only 70% accuracy (Chen et al. 2003).
Importantly, none of the existing studies on automatic classification of subcellular localization has used images that were captured automatically such as in high-throughput microscopy screens that yield images of inherently lower quality. Manual selection, centering, focusing, and control of the illumination and detection parameters in the imaging process evidently leads to better classification results than in an automated imaging system. In this study, we present a fully automated system for the production and imaging of human cell arrays, and most importantly, an automated classification of phenotypes. As a case study, we set up an overexpression screen, in which we marked eleven different subcellular patterns by GFP-tagged proteins, each of them characterizing a distinct phenotype.
RESULTS
Workflow Concept
The focus of this study was to set up a framework for high-throughput cell phenotyping. To make this screen scalable to the high number of human genes, we based our screen on human cell arrays (Fig. 1). We generated a variety of phenotypes in a proof-of-principle study by an overexpression screen, in which we marked eleven different subcellular patterns by GFP-tagged proteins, each of them characterizing a distinct phenotype. To allow automation of cell array production, we developed an improved protocol for solid phase transfection by mixing all required transfection components prior to robotic spotting. Thus, transfection-ready DNA arrays on chambered coverglasses compatible with direct live cell observation can be printed, dried, and stored, and cells are simply seeded on the array one day prior to phenotype analysis.
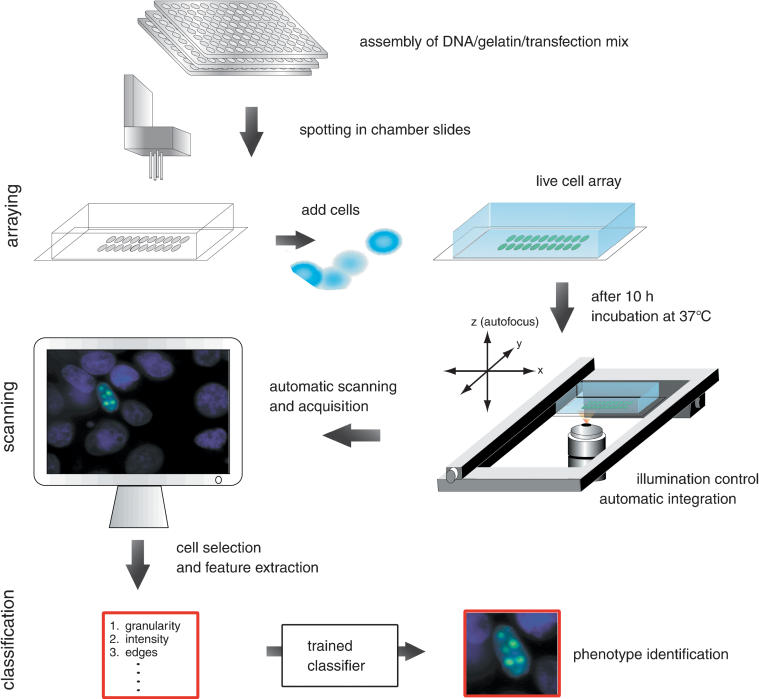
Figure 1.
Workflow of phenotype classification system. The mix of targeted DNA (e.g., subcellular clones–GFP fusion), gelatin, and transfection reagents are prepared in microwells and spotted in arrays into chamber slides by a DNA-spotting robot. Cultured cells are added to the chamber slides and transfected with the DNA on the arrayed spots. After 10 h incubation at 37°C the fluorescence signal of the expressed GFP signal can be visualized. On the automatic scanning platform a motorized stage and motorized z-stepper perform the scanning of the live cells. The control of the illumination and the automatic calculation of the integration time allow an automatic acquisition of cell images. Cells with GFP-signals are captured and image features are extracted. These features serve as input for training of the automated classification system.
For fully automated phenotype analysis, we designed an imaging strategy to automatically capture single-cell fluorescence images from entire live cell arrays with high resolution. We implemented our imaging strategy by adapting a commercial widefield fluorescence scanning microscope (Mehes et al. 2000), which automatically located each spot in the array, auto-focused the cell layer, and then captured high resolution images from each GFP-expressing cell.
For enhanced detection of cells an auto-focus algorithm is applied first to cell nuclei counterstained by Hoechst, then the GFP or YFP signal is automatically integrated and the auto-focus algorithm is executed again on the protein signal. Auto-focusing works reliably in cells growing as monolayers, making this method applicable to a wide variety of cultured mammalian cells. For this study we chose the MCF7 breast cancer cell line but we have obtained similar results with HeLa cells (data not shown). A series of images is then acquired with variable integration times of the CCD camera, thereby allowing selection of cells with good signal-to-noise ratio from a field with cells at different expression levels. In the next step, objects potentially representing cell nuclei are segmented based on the counterstained signal. Finally, the system selects `valid' cell nuclei out of the set of candidate objects based on morphological parameters (for details see Methods).
Subcellular Class Design With Additional Artifact Class
The above-described strategy allowed us to generate large amounts of images, which we then used as input for our automated phenotype classification system. The described scanning system captured 2182 images automatically. This set of images was used to build an automated classification system that could identify the 11 different classes of subcellular localizations. For the phenotypes and number of analyzed cells per protein class refer to Figure 2. Upon manual inspection all images in each class that could not be properly assigned were labeled as artifacts. By a random selection every fifth image was set aside as test set for independent evaluation of the classifier constituting the test set. Thus, 1755 (80%) of the images were used for the training of the classifier (training set), and 427 (20%) of all images were assigned to the test set. All classification results given below refer to the performance of the classifier on this test set. Training of an accurate classification system was greatly corrupted by the high number (∼50%) of all images representing cells with obvious artifacts such as dead or extremely overexpressed cells (Fig. 2B). The frequency of false-positive artifact capture could not be further reduced because more stringent selection criteria during acquisition would have lead to rejection also of certain true-positive subcellular localizations (Supplemental Fig. 1). Artifacts are unavoidable in a fully automated screen, since the quality control for cell selection cannot be as stringent as with manually acquired images, where typically only evenly shaped cells are chosen in a biased way by a microscopist. Thus, we designed our classification method to deal with artifacts by including an artifact class into the training procedure and by explicitly defining discriminating features of artifact images.
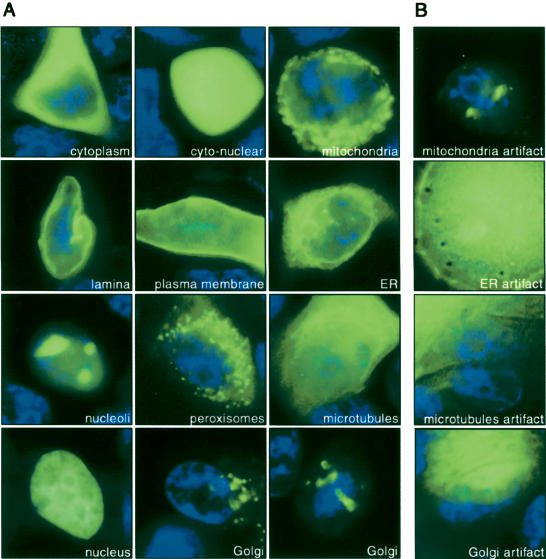
Figure 2.
Automatically captured images of live MCF7 cells representing localization classes. (A) Cells were counterstained by Hoechst (blue) and different GFP-tagged cDNAs were expressed in cells transfected on cell arrays (green). Image acquisition is designed in such a way that each image contains only one cell. From top left to bottom right: GFP-tagged NES→cytoplasm (96 images), EGFP→cyto-nuclear (170 images), GFP-tagged cDNA #488→mitochondria (108 images), YFP-tagged LB1→nuclear lamina (108 images), GFP-tagged ErbB1→plasma membrane (70 images), YFP-tagged SRb→endoplasmatic reticulum (84 images), GFP-tagged cDNA #351→nucleoli (116 images), YFP-tagged cDNA #447→peroxisomes (119 images), YFP-tagged cDNA #22f21→microtubules (78 images), YFP-tagged H2B→nuclear (111 images), YFP-tagged GalT→Golgi (93 images) imaged from two different spatial directions. For cDNA reference see Supplemental Table 1 (see also http://www.dkfz.de/LIFEdb/ and http://harvester.embl.de/; Wiemann et al. 2001). (B) Artifacts of the corresponding subcellular localization class are shown in the right column: mitochondria artifacts (116 images), endoplasmatic reticulum artifacts (99 images), microtubules artifact (67 images), Golgi artifact (77 images). In total 1035 images were labeled as artifacts and assigned to this class. Note that artifact cells show expression levels both below and above the level of expression typically observed for valid cells (left). Hence, the level of expression is not sufficient to discriminate artifact cells from valid cells (see also Supplemental Fig. 1).
Designing Optimal Classification Schemes
Image classification is the critical next step in the analysis stream. To find the optimal method for classification of automatically captured images, we compared two well-known methods in machine learning, namely, Artificial Neural Networks (ANN; MacKay 1992; Bishop 2000) and Support Vector Machines (SVMs; Vapnik 1995; Smola and Schölkopf 1998). SVM is a well-regularized method (Vapnik 1995; Smola and Schölkopf 1998)—that is, it performs well on images that were not used for training of the classifier. In contrast, ANN tend to overfit the training set during learning, therefore it was suggested to combine it with Bayesian learning to achieve regularization (MacKay 1992). This approach is referred to as BayesANN. Alternatively, evolutionary search using genetic algorithms (GA) can be used for global optimization of parameters in the ANN. This approach is referred to as ANN/GA. For all suggested methods, different classification models corresponding to different parameter settings were trained and validated (model selection by cross-validation) on the training set. The optimal models were then used to predict phenotype classes for the test images (for details see Methods). The classification methods were combined with three feature selection methods: Mutual Information (MI; Ragg 2002), Significance Analysis of Microarrays (SAM; Tusher et al. 2001), and stepwise discriminant analysis (STEPWISE; Leray and Gallinari 1999). Note, that SAM is a statistical method that has been developed for identification of differentially expressed genes in microarray gene expression studies. As the method is based on commonly used statistical tests like t-test and permutation test, we readily applied it for image feature selection (for details on feature selection see Methods).
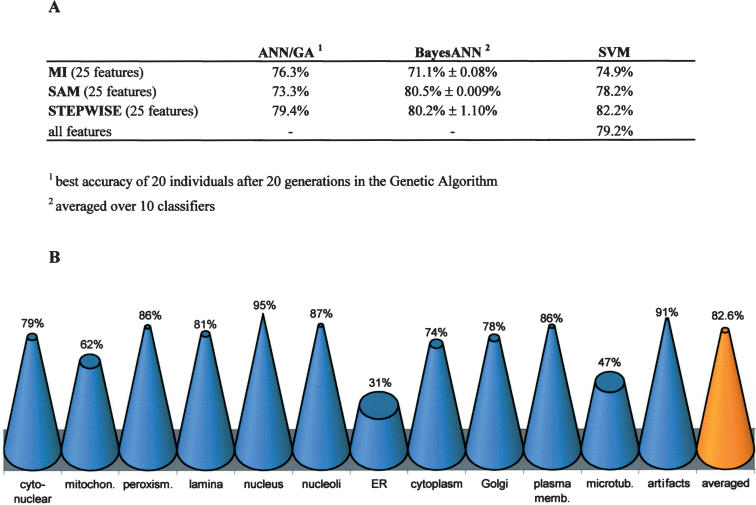
Since the optimal number of image features was typically between 20 and 25 features for the BayesANN approach (data not shown), we based the training of all classification methods on only 25 features for better comparison. The 25 best-ranked features of the STEPWISE or SAM selections contained predominately texture-related features such as granularities and co-occurrence (Supplemental Table 2). Texture-related features thus appear to be at least as efficient as morphological object features for automatic classification of subcellular patterns. Further, they are more robust with respect to biological variation. Figure 3 summarizes the results from these classification algorithms. The ANN/GA performed worse than the other two methods (Fig. 3A) and was therefore not further considered in our study. BayesANN and SVM classification algorithms performed well in combination with the feature selection methods SAM and STEPWISE. The two best combinations of algorithms were generated through SVM/STEPWISE and BayesANN/SAM with accuracies of 80.5% and 82.2%, respectively (Fig. 3B; Supplemental Table 3). In contrast to ANNs, the training of SVM does not strongly depend on feature preselection (Ramaswamy et al. 2001). Accordingly, the recognition rate of the SVM classifier based on the full set of 323 features instead of 25 selected features is only reduced by 3% despite the much higher dimensionality of the classification task (Fig. 3A). Thus, considering a comparable prediction accuracy at a much lower sensitivity to feature selection SVMs appear to be more suited than ANNs when analyzing a set of very variable images. Through the combination of different imaging strategies, automated object detection and feature subset selection we were able to set up a robust and fully automated framework for phenotype classification that achieves high accuracy even for applications with large biological variability and a considerably high number of artifacts.
Figure 3.
Accuracy of phenotype classification. (A) Classification accuracy determined on the test set for three different classification algorithms using the first 25 ranked features obtained by three different feature selection methods. In contrast to the BayesANN approach, the ANN/GA runs with nonconvergent fivefold cross-validation as regularization. As the result of the BayesANN classifier is dependent on random initialization of network weights, the averaged classification accuracies and the corresponding standard deviations of 10 classification runs of the same test set are shown, while the ANN/GA generates many similar networks during the evolutionary search process. Only the SVM was trained with the entire set of 323 features. (B) While BayesANN performed best on average in combination with SAM for feature selection, the overall best classifier was obtained by combination of BayesANN with STEPWISE (82.6% accuracy). The cone diagram shows single class accuracies obtained by this overall best classifier for all subcellular classes and artifact class.
DISCUSSION
In the present study, we developed a fully automated workflow from cell array production to phenotype analysis. As a case study we chose to phenotype cells by identification of the subcellular localization of marker proteins that can be used as indicators for the cellular state. We achieved a very high overall accuracy of more than 80% prediction for eleven localization classes with our fully automated system. An even better accuracy was affected mostly by three problematic localizations that were difficult to distinguish at the resolution of our imaging system. The localization class in our study that could be classified most inefficiently was endoplasmatic reticulum (accuracy 31%), which was frequently incorrectly classified as microtubules (accuracy 47%) or mitochondria (accuracy 62%). These misclassifications are in agreement with the visual similarities of the corresponding images (Fig. 2). All other automatically imaged subcellular patterns were recognized with accuracy between 74% and 95%.
A key for achieving this high degree of accuracy was to include all artifacts into one additional localization class. Despite its heterogeneity, the artifact class (Fig. 2) could be accurately distinguished from all other subcellular classes (accuracy 91%), but receives false positives from other classes. Importantly, the prediction within all other classes of subcellular localizations achieved a specificity per class of more than 97% (Supplemental Table 3). Thus, only a very small proportion of all images is assigned incorrectly to an individual class.
Biological variation of the same phenotype in different cells remained the most challenging aspect of our fully automated workflow from cell array production to phenotype analysis. By choosing transient overexpression of GFP-tagged cDNAs as a proof-of-principle application this problem was probably more severe than in immunofluorescence screens based on endogenous proteins (Kiger et al. 2003), where one would expect variations in protein levels between different cells to be less pronounced. However, changes of cellular morphology could be much more subtle and heterogeneous in RNAi screens, where each class lacks a different gene. To address such subtle variations, colocalization of several domains (e.g., microtubules and plasma membrane) labeled by other spectral mutants of GFP would help to extract appropriate morphological image features. A two-level hierarchal classification could first predict the cellular compartment of interest in the image of the first label. Subsequently, the distinct morphology within this compartment would be predicted based on the image of the second label. In addition, we are confident that classification rates could be further improved with a larger training data set, which was beyond the scope of this study.
To assess the classification ability of our system for different cDNA clones not being used in the training step, we automatically captured 20 images of microtubule-related Ensconsin (Bulinski et al. 2001) and mitochondrial cDNA clone 1692 or DKFZp434D0421 (Accession no. AL136804). The overall best BayesANN classifier achieved accuracies of 55% and 45%, respectively, for these clones, which was in the same range (47% and 62%, respectively) as for the cDNA clones originally used as markers for the respective phenotype classes. This assessment further supports the generalization ability of our classification system.
The automated captured images of endoplasmatic reticulum and microtubles are partly very similar. The lack of sharp images with a low depth of image field in this study appeared to be problematic for the separation of the fine tubular structure of microtubules and the membrane network of endoplasmatic reticulum. Using a cell line that has a flatter morphology than the MCF7 cells employed in our study, where a significant overlap between ER and microtubules in the rounded up cytoplasm is observed, can most likely solve this problem. For subtle phenotypic differences we anticipate that a confocal screening microscope will improve accuracy in the future and we have started work on such a system. The essential advance of this study is the integration of automated classification with automated microscopy and production of cell arrays, which can potentially be applied to large, genomewide cell arrays. Our system can also be easily extended to extract dynamic features from time-lapse screening studies (Gonczy et al. 2000). Unbiased and robust analysis of the subcellular localizations of proteins and their behavior over time on a large-scale will be essential to efficiently assess the dynamic topology of protein networks. In summary, the framework presented here provides a platform for a number of functional genomics applications, like large-scale automated morphological cell assays for gain and loss of function, as well as chemical screening in human cells.
METHODS
Live Cell Arrays
The plasmid-gelatin-transfection solution was prepared in 384-well plates (Nunc) as follows: 500 ng of GFP or YFP tagged plasmid, 7.5 μL EC buffer and 0.75 μL Enhancer were incubated for 10 min at room temperature and then mixed with 2.5μL Effectene (Effectene Transfection Kit, Qiagen) and again incubated for 10 min at room temperature in 7.25 μL of 0.08% Gelatin (G-9391, Sigma). The plasmid-gelatin-transfection solution was arrayed onto 1-well Labtek (Nunc) slides using the ChipWriter Compact robot (Biorad). The spot diameter was 400 μm for all experiments. After printing, 6.5 × 105 MCF7 cells were plated on the Labtek slide and cultured for 10 h in growth medium containing 10% inactivated fetal calf serum, 1% glutamine, and 1% penicillin-streptomycin. Thereafter, Hoechst stain (33342, Sigma, 1 μg/mL final concentration) was added for 10 min to stain cell nuclei. For live cell data acquisition the growth medium was replaced by imaging medium (DMEM without carbonate but supplemented with 30 mM Hepes, pH 7.4, obtained from Sigma). The transfection efficiency varied between 1% and 30% depending strongly on the cDNA to be transfected.
Automatic Image Acquisition
For image acquisition the Metafer4 system (MetaSystems) based on an Axiovert 200M microscope (Zeiss) equipped with 40×/0.95 air Plan-Apochromat objective (Zeiss), a motorized scanning stage (Märzhäuser), and a CCD camera (Jai M1, 1280 × 1024 pixels, 6.7 μm × 6.7 μm square pixels, 60 sec maximum integration) was used. Its integrated automated functions include a motorized stage (1 μm resolution, maximum speed of 10 cm/sec, 0.025 μm focus step size) and a motorized reflector revolver (Hoechst filter: excitation 390/22, dichroic beam splitter 420, emission filter 460/50; GFP/YFP filter: excitation 500/20, dichroic beam splitter 515, emission filter 535/30).
For auto-focusing, the stage is first moved down and then up to a number of focus planes to minimize the effects of mechanical focus drive backlash. At each position an image is captured. The number of planes and distance between consecutive focus planes is defined in the parameter set of the imaging microscope. For each of the captured images a focus criterion is computed. The stage is then moved to the Z-position corresponding to the plane optimizing the focus criterion.
After global or local background correction, the gray-level histogram of the counterstain channel image is computed and analyzed. Based on the maximum and minimum gray levels in the image and a threshold factor defined in the parameter set, the system calculates a global segmentation threshold. A fast-contour-following algorithm is used to isolate the objects defined by this thresholding operation. Finally, the system accepts a candidate as a `valid' cell nucleus if the object area ranges from 50–500 μm2, concavity depth <0.8, and aspect ratio <3.0.
Only those valid cells are selected for imaging of subcellular localization. Multiple image acquisition with three integration times (1×, 2×, and 3× the calculated integration time, respectively, with maximum integration of 2 sec) was applied to the GFP/YFP signal. A selection of the brightest unsaturated (threshold for saturation: 4 saturated pixels) GFP/YFP cell image (ROI of 148×148 pixels, centered by center of nucleus) from the three integrated GFP/YFP images was determined. Tiles are considered positive if total intensity >400 and relative center intensity >20. The relative center intensity is the ratio between minimum gray level within and maximum gray level outside of half of the radius of the cell. The scanning time for 150 spots imaged in this study was in the range of ∼12 hours (∼300 sec for three times three images per spot). Even though the time required for scanning can become a bottleneck in more extended screens, it should be noted that the scanning time could be dramatically decreased if the number of focal sections and the maximum integration time are reduced.
Feature Generation and Classification
Four hundred and forty-eight different image features comprising the following feature groups with different parameterization were extracted: granularity (position-depending differentiation), object-related features, intensity distribution, main momentum axis, Karhunen-Loève transform (Bishop 2000), tree-structured wavelets (Chang and Kuo 1993), gray scale invariants (Burhardt and Siggelow 2001), Haralick texture features (Haralick 1979), Zernike moments (Zernike 1934), and Sobel edge features. All extracted features with a standard deviation of zero (calculated on images in the training set) were discarded, resulting in 323 remaining features. The feature selection of stepwise discriminant analysis (STEPWISE; Leray and Gallinari 1999) is a Fisher test with Wilk's λ as a measure, while the Mutual Information (MI) measures the reduction of uncertainty of a feature due to a given class (Ragg 2002). Significance Analysis Microarray (SAM) is derived from a permutation test on modified t-test statistic (Tusher et al. 2001). As input for SAM, we used the class-labeled image feature values rather than gene expression values. This results in a score for every image feature on the bases of the relative differences of feature means between phenotype classes. Thus, the SAM approach provides feature selection by ranking the calculated feature scores and by selecting the best features.
The Support Vector Machines (http://www.csie.ntu.edu.tw/∼cjlin/libsvm/) and Bayesian learning ANN (http://www.ncrg.aston.ac.uk/netlab/) are integrated in our propriety data mining software `mine-it.' As kernel function for the SVM, we utilized the Radial Base Function. In a threefold cross-validation on the training set (model selection) the kernel parameter γ for the Radial Base Function and the misclassification parameter C were varied in the ranges of 20–2–30 and 20–230, respectively. BayesANN consists of a multilayer backpropagation neural network with one hidden layer, sigmoid transfer function, cross-entropy as error measure, and scaled conjugate gradient error minimization. The initial weight decay parameter was 0.01, reestimated automatically by the Bayesian learning after every 20 epochs until the maximal number of 400 epochs was reached. For model selection, the network topology was varied over the range of 10–20 hidden neurons and 10–50 input features. The second version of ANN in this study uses genetic algorithms (ANN/GA), based on the NevProp3 package by Philip Goodman for parameter optimization (http://brain.unr.edu). In contrast to BayesANN, the ANN/GA was used with Quickprop error minimization for computational reasons. ANN/GA software was further developed for parallelization with Message Passing Interface (MPI) for an optimized runtime on a 20-node Debian/Linux cluster. The optimization procedure aims at identifying the optimal values of number of input features, number of hidden neurons, learning rate, momentum rate, and epochs. The genetic algorithm was mutated by 80% crossover, 3% mutation rate, and reproduced through binary tournament selection. The fitness criterion was the ANN fivefold cross-validation error on the training set.
Acknowledgments
We thank Benedikt Brors and Daniel Gerlich for suggestions on the manuscript. We thank Carl Zeiss Inc. (Göttingen, Germany) for microscope support to the ALMF at EMBL. Stefan Wiemann and Annemarie Poustka have kindly provided some of the cDNA clones. This work was supported by grants from Federal Ministry of Education and Research (DHGP:01KW9937, NGFN:01GR0101, BioFuture: 0311880A) and Human Frontiers Science Program (RGP0031/2001-M).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2383804.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S. Wiemann and A. Poustka.]
References
- Bishop, C.M. 2000. Neural network for pattern recognition. Oxford University Press, New York.
- Bulinski, J.C., Odde, D.J., Howell, B.J., Salmon, T.D., and Waterman-Storer, C.M. 2001. Rapid dynamics of the microtubule binding of ensconsin in vivo. J. Cell. Sci. 114: 3885–3897. [DOI] [PubMed] [Google Scholar]
- Burhardt, H. and Siggelow, S. 2001. Invariant features in pattern recognition—fundamentals and application. In Nonlinear model-based image/video processing and analysis (eds. C. Kotropoulos and I. Pitas), pp. 269–307. John Wiley & Sons, New York.
- Chang, T. and Kuo, C.C. 1993. Texture analysis and classification with tree-structured wavelet transform. IEEE Transactions on Image Processing 2: 429–441. [DOI] [PubMed] [Google Scholar]
- Chen, X., Velliste, M., Weinstein, S., Jarvik, J.W., and Murphy, R.F. 2003. Location proteomics—Building subcellular location tree from high resolution 3D fluorescence microcope images of randomly-tagged proteins. Manipulation and Analysis of Biomolecules, Cells, and Tissues, Proceedings of SPIE 4962: 298–306. [Google Scholar]
- Egmont-Petersen, M., de Ridder, D., and Handels, H. 2002. Image processing with neural networks—A review. Pattern Recognition 35: 2279–2301. [Google Scholar]
- Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Haralick, R.M. 1979. Statistical and structural approaches to texture. Proceedings of the IEEE 67: 768–804. [Google Scholar]
- Huang, K., Velliste, M., and Murphy, R.F. 2003. Feature reduction for improved recognition of subcellular location pattern in fluorescence microscope images. Manipulation and Analysis of Biomolecules, Cells, and Tissues, Proceedings of SPIE 4962: 298–306. [Google Scholar]
- Huh, W.-K., Falvo, J.V., Gerke, L.C., Caroll, A.S., Howson, R.W., Weissmann, J.S., and O'Shea, E.K. 2003. Global analysis of the protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jain, A.K., Duin, R.P.W., and Mao, J. 2000. Statistical pattern recognition: A review. IEEE Transactions on Pattern Analysis and Machine Intelligence 22: 4–37. [Google Scholar]
- Jarvik, J.W., Adler, S.A., Telmer, C.A., Subramaniam, V., and Lopez, A.J. 1996. CD-tagging: A new approach to gene and protein discovery and analysis. Biotechniques 20: 896–904. [DOI] [PubMed] [Google Scholar]
- Kiger, A., Baum B., Jones, S., Jones, M., Coulson, A., Echeverri, C., and Perrimon, N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leray, P. and Gallinari, P. 1999. Feature selection with neural networks. Behaviormetrika 26: 1–27. [Google Scholar]
- Liebel, U., Starkuviene, V., Erfle, H., Simpson, J.C., Poustka, A., Wiemann, S., and Pepperkok, R. 2003. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 554: 394–398. [DOI] [PubMed] [Google Scholar]
- MacKay, D.J.C. 1992. A practical Bayesian framework for backpropagation networks. Neural Comput. 4: 448–472. [Google Scholar]
- Mehes, G., Lorch, T., and Ambros, P.F. 2000. Quantitative analysis of disseminated tumor cells in the bone marrow by automated fluorescence image analysis. Cytometry 42: 357–362. [DOI] [PubMed] [Google Scholar]
- Murphy, R.F., Velliste, M., and Porreca, G. 2002. Robust classification of subcellular location patterns in fluorescence microscope images. In Proceedings of the 2002 IEEE International Workshop on Neural Networks Signal Processing (NNSP 12), pp. 67–76.
- Pepperkok, R., Simpson, J.C., and Wiemann, S. 2001. Being in the right location at the right time. Genome Biol. 2: REVIEWS1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragg, T. 2002. Bayesian learning and evolutionary parameter optimization. AI Communications 15: 61–74. [Google Scholar]
- Ramaswamy, S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C.H., Angelo, M., Ladd, C., Reich, M., Latulippe, E., Mesirov, J.P., et al. 2001. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. 98: 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, M.M., Stein, P.A., Taylor, S.S., Ha, E., McKeon, F., and Rapoport, T.A. 1999. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell. Biol. 146: 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J. and Pepperkok, R. 2003. Localizing the proteome. Genome Biol. 4: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J.C., Wellenreuther, R., Poustka, A., Pepperkok, R., and Wiemann, S. 2000. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 1: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola, A.J. and Schölkopf, B. 1998. On a kernel-based method for pattern recognition, regression, approximation and operator inversion. Algorithmica 22: 211–231. [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik, V.N. 1995. The nature of statistical learning theory. Springer-Verlag, New York.
- Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Böcher, M., Blöcker, H., Bauersachs, S., Blum, H., et al. 2001. Towards a catalog of human genes and proteins: Sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 11: 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernike, F. 1934. Beugungstheorie des schneidenverfahrens und seiner verbesserten form, der phasenkontrastmethode. Physika 1: 689–704. [Google Scholar]
- Ziauddin, J. and Sabatini, D.M. 2001. Microarrays of cells expressing defined cDNAs. Nature 411: 107–110. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.dkfz.de/LIFEdb/; cDNA database.
- http://harvester.embl.de/; Database cross linker.
- http://www.csie.ntu.edu.tw/∼cjlin/libsvm/; Support Vector Machine implementation.
- http://www.ncrg.aston.ac.uk/netlab/; Neural Network toolbox using Matlab.
- http://brain.unr.edu; Source code of ANN (NevProp3) by Philip Goodman.