Abstract
Several transcription factors (TFs) have been implicated in neuroectoderm (NE) development, and recently, the TF PAX6 was shown to be critical for human NE specification. However, microRNA networks regulating human NE development have been poorly documented. We hypothesized that microRNAs activated by PAX6 should promote NE development. Using a genomics approach, we identified PAX6 binding sites and active enhancers genome-wide in an in vitro model of human NE development that was based on neural differentiation of human embryonic stem cells (hESC). PAX6 binding to active enhancers was found in the proximity of several microRNAs, including hsa-miR-135b. MiR-135b was activated during NE development, and ectopic expression of miR-135b in hESC promoted differentiation toward NE. MiR-135b promotes neural conversion by targeting components of the TGF-β and BMP signaling pathways, thereby inhibiting differentiation into alternate developmental lineages. Our results demonstrate a novel TF-miRNA module that is activated during human neuroectoderm development and promotes the irreversible fate specification of human pluripotent cells toward the neural lineage.
Keywords: BMP, hsa-miR-135b, human embryonic stem cells, microRNA, neuroectoderm, PAX6, TGF-β, transcription factor
Introduction
An early step during vertebrate neural development is the specialization of a group of ectodermal cells as precursors of the entire nervous system (Hemmati-Brivanlou & Melton, 1997). This process, termed neuroectoderm (NE) development, is one of the best studied processes occurring during gastrulation, but a clear understanding of the molecular details governing this transition is still lacking. Several transcription factors (TFs) have been implicated in promoting NE specification including SOX family proteins specifically SOX2, OTX family members, and zinc finger proteins (Acampora et al, 2000; Kishi et al, 2000; Sheng et al, 2003).
Recently, by employing neural differentiation of human embryonic stem cells (hESC) as an in vitro model of NE specification, PAX6 was uncovered as a major determinant of human NE development (Zhang et al, 2010). NE differentiation from hESC was severely affected by RNAi-mediated knockdown of PAX6, and overexpression of a specific isoform of PAX6, PAX6a, in hESC was sufficient to induce NE tissue (Zhang et al, 2010). Importantly, PAX6 was found to be dispensable for mouse NE development as RNAi-mediated knockdown of PAX6 in mouse ES cells gave rise to functional neuroectodermal cells (Zhang et al, 2010). However, PAX6−/− mice die immediately after birth and display a malformed cerebral cortex along with aberrant cortical neuron projections (Jones et al, 2002; Pratt et al, 2002). Thus, PAX6 is crucial for both human and mouse neural development.
MicroRNAs (miRNAs), a class of small non-coding RNAs, regulate gene expression post-transcriptionally and have been found to regulate key steps in development with several miRNAs being stem cell- and lineage-specific (Espinoza-Lewis & Wang, 2012; Sittka & Schmeck, 2013; Sun et al, 2013; Wilson & Doudna, 2013). Interestingly, miRNAs and TFs reciprocally regulate each other, and such TF-miRNA regulatory modules have been shown to modulate many aspects of neuronal identity (Zhao et al, 2009; Chen et al, 2011; Otaegi et al, 2011). MicroRNAs have been shown to regulate PAX6 during neural development (de Chevigny et al, 2012) but whether PAX6 regulates microRNAs and whether PAX6-regulated microRNAs play a role in neural development has not been elucidated. We hypothesized that microRNAs activated by PAX6 would promote NE specification in humans. By employing an unbiased genomics approach based on ChIP followed by high-throughput sequencing of ChIP-enriched DNA (ChIP-Seq), we identified genome-wide PAX6 binding sites in human neuroectodermal cells (NEC). We found that PAX6 activates several TFs implicated in neural development as well as microRNAs by direct binding to proximal enhancers. One of the identified microRNAs, miR-135b, is activated by PAX6 during human NE development. Activation of miR-135b during NE specification inhibits the BMP and TGF-β pathway, thereby inhibiting differentiation into non-neural lineages. Thus, we have identified a regulatory network whereby PAX6 activates miR-135b to inhibit TGF-β and BMP signaling thereby differentiating hESC toward a neural phenotype.
Results
Derivation of neuroectodermal cells from human embryonic stem cells
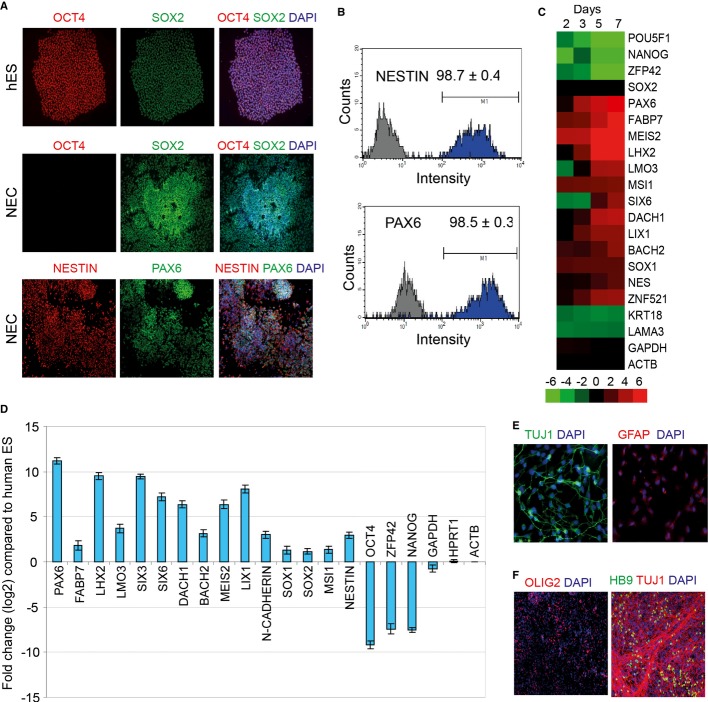
A robust source of NEC was produced from hESC by dual SMAD inhibition (Chambers et al, 2009). To improve yields and reduce costs, we implemented some modifications based on published protocols. First, we plated human ES cells as colonies in feeder-free media onto matrigel. Second, we replaced biologic SMAD inhibitor noggin with the small-molecule inhibitor dorsomorphin, which targets SMAD5 phosphorylation and inhibits BMP signaling (Zhou et al, 2010). Treatment of human ES cells with SB431542 and dorsomorphin efficiently converted OCT4+, SOX2+ pluripotent H1 hES cells into OCT4−, SOX2+, PAX6+, and NESTIN+ NEC (Fig 1A). Flow cytometry analysis showed that > 90% of the cells were PAX6+ and NESTIN+ NEC (Fig 1B). To assay pluripotent and neural markers during the time course of differentiation, the cells were fixed for immunostaining at various time points. PAX6 protein expression was readily apparent as early as day 3 of differentiation while peak levels were obtained at day 6, after which the expression remained steady (Supplementary Fig S1A). OCT4 levels started decreasing by day 3 and disappeared by day 6 (Supplementary Fig S1A). A previous study showed high efficiency conversion of human pluripotent stem cells to NEC by using the small molecule dorsomorphin alone (Zhou et al, 2010). We compared the efficacy of using the combination of dorsomorphin and SB431542 with dorsomorphin alone in neural differentiation of human ES cells. Cultures treated with dorsomorphin alone still displayed OCT4+ cells by day 6 of differentiation, whereas the combination treatment suppressed OCT4 expression completely (Supplementary Fig S1B). Hence, we decided to use the combination to obtain NEC.
Figure 1. Derivation of human neuroectodermal cells (NEC) from embryonic stem cells.
- Dual SMAD inhibition using SB431542 and dorsomorphin efficiently differentiates H1 human embryonic stem cells into SOX2+, PAX6+, and NESTIN+ human NEC in 8 days.
- Quantitation of PAX6- and NESTIN-expressing cells by flow cytometry indicates that > 90% of the cells are PAX6+ and NESTIN+ NECs.
- Gene expression analysis of the neural differentiation time course by microarrays.
- Confirmation of downregulation of pluripotent markers and upregulation of neural markers by quantitative RT-PCR. Error bars shown indicate s.e.m. (n = 3 independent experiments).
- Differentiation of NECs into TUJ1+ neurons and GFAP+ astrocytes.
- NECs can be patterned into OLIG2+ spinal motor neuron progenitors and further differentiated into Hb9+ motor neurons.
We harvested the cells at various time points in the course of differentiation and profiled gene expression using microarrays. Microarray analysis showed upregulation of several neuroectodermal markers as early as day 2. PAX6 transcript levels started increasing by day 2 and steadily increased during the time course of differentiation. Markers of pluripotency such as OCT4, NANOG, and ZFP42 started decreasing from day 2 and were strongly downregulated by day 7 of differentiation (Fig 1C). To assess the robustness of the protocol, we generated NEC from hESC in an independent set of biological replicates and profiled expression of several genes using quantitative reverse transcription PCR. As shown in Fig 1D, neuroectodermal markers such as PAX6, FABP7, LHX2, LMO3, MEIS2, DACH1, LIX1, SOX1, and SIX6 were strongly upregulated, while pluripotency markers (OCT4, NANOG, ZFP42) were strongly downregulated as observed in our previous microarray analysis. A hallmark of NEC is their ability to differentiate into neurons and glial cells. We were able to differentiate the NECs obtained by our dual SMAD inhibition method into TUJ1+ neurons and GFAP+ astrocytes (Fig 1E). Additionally, we were able to pattern the NECs into OLIG2+ progenitors of spinal motor neurons by treatment with retinoic acid and purmorphamine and further differentiate these progenitors into HB9+ motor neurons (Fig 1F). These results establish that our protocol generates functional NEC with high efficiency.
Identifying PAX6 binding sites genome-wide in human NECs
We identified PAX6 binding sites across the genome by performing PAX6 chromatin immunoprecipitation (ChIP) in ES-derived NECs. Deep sequencing of the ChIP-enriched DNA (ChIP-Seq) identified approximately 16,000 PAX6 binding sites using stringent criteria (FDR < 0.1%). We counted PAX6 binding sites in different genomic features such as promoters, exons, introns, UTRs, and intergenic regions. More than 80% of the binding sites were found to reside within introns and intergenic regions while < 2% of the total sites mapped close to the transcriptional start site (TSS) of protein-coding genes (Fig 2A). Interestingly, about 7% of PAX6 binding sites were found proximal to non-coding RNAs including microRNAs (Fig 2A).
Figure 2. Identification of PAX6 binding sites in neuroectodermal cells genome-wide using ChIP-Seq.
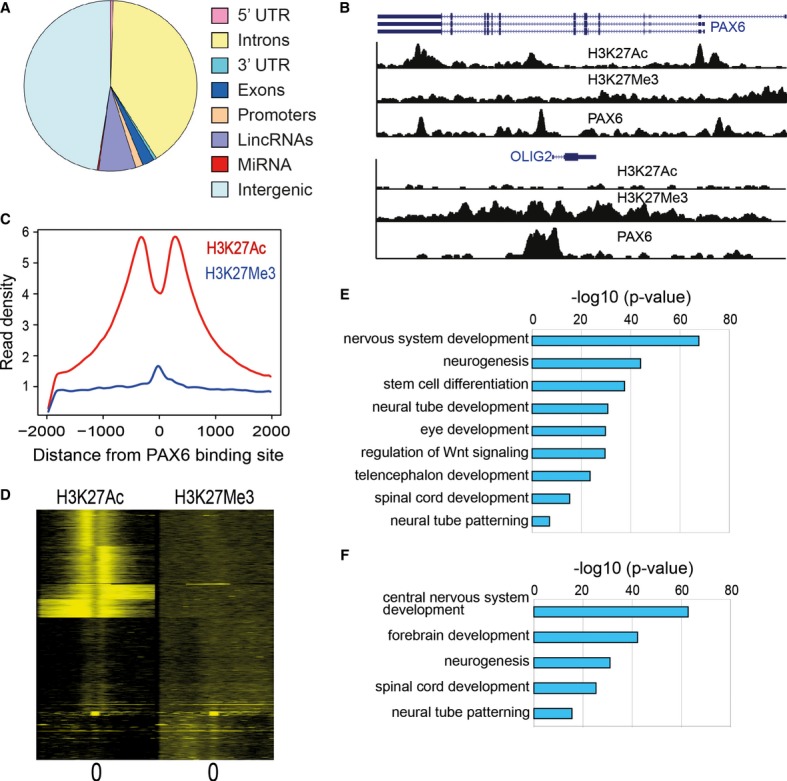
- Distribution of PAX6 binding sites in different genomic regions. Promoters were defined as ±2 kb from the transcriptional start site (TSS). For miRNAs, we mapped binding sites to within 20 kb from the mature sequence. Intergenic regions were defined as non-genic regions that did not map to any promoters or gene bodies or non-coding RNAs.
- Genome browser snapshot of PAX6 binding sites overlapping with H3K27Ac and H3K27Me3 signal proximal to PAX6 and OLIG2 TSS.
- Average distribution of H3K27Ac and H3K27Me3 signal relative to the center of PAX6 binding sites. On the x-axis, 0 indicates the PAX6 peak center as identified from ChIP-Seq data. Read densities were normalized for sequencing depth.
- Heatmap of H3K27Ac and H3K27Me3 signals relative to the center of PAX6 binding sites. A window of 4,000 bp centered on the PAX6 peaks and binned into 50 bp bins has been displayed. Individual profiles were clustered by k-means clustering.
- Enriched functional categories derived by GREAT analysis for all PAX6 targets (threshold P-value = 1e−10).
- Enriched functional categories derived by GREAT analysis for PAX6 transcriptional targets (threshold P-value = 1e−10).
Binding alone does not indicate whether the TF acts as an activator or repressor at a particular site. Genomic loci that function as activators or repressors are marked by distinct histone marks. Acetyl-ation of the histone H3 at Lysine 27 (H3K27Ac) has been shown to be a reliable indicator of active regulatory regions namely enhancers and promoters, while tri-methylation on H3 at Lysine 27 (H3K27Me3) has been shown to associate with repressed regions (Creyghton et al, 2010; Rada-Iglesias et al, 2011; Gifford et al, 2013). To distinguish sites where PAX6 mediates activation or repression, we also mapped locations of H3K27Ac and H3K27Me3 by ChIP-Seq in NEC. Previous ChIP-chip (ChIP followed by hybridization of the enriched DNA on microarrays) studies have identified PAX6 binding to its own promoter (Sansom et al, 2009). We identified PAX6 binding sites not only near the PAX6 TSS, but also within the gene body, overlapping with distinct H3K27Ac peaks, indicating that PAX6 activates its own transcription (Fig 2B upper panel). PAX6 has been shown to inhibit OLIG2 transcription though this inhibition was suggested to be via indirect means (Sansom et al, 2009). We observed PAX6 binding sites proximal to the OLIG2 TSS overlapping with H3K27Me3 signal, indicating that PAX6 inhibits OLIG2 gene by directly binding to the OLIG2 promoter (Fig 2B lower panel). We plotted the average distribution of H3K27Ac and H3K27Me3 ChIP signal in a 4-kb window centered on PAX6 binding sites (Fig 2C). H3K27Ac signal showed the characteristic double peak pattern around the center of the PAX6 site indicating that nucleosomes adjacent to PAX6 occupancy on the genome were acetylated. On the other hand, we identified a peak for H3K27Me3 occupancy directly overlapping PAX6 binding though the signal was much weaker as compared to H3K27Ac, indicating that PAX6 acts primarily as an activator. In order to analyze PAX6 occupancy of enhancers and repressor in detail, we analyzed H3K27Ac and H3K27Me3 at individual PAX6 peaks and clustered these peaks in distinct groups (Fig 2D). In concordance to our previous observation, the majority of PAX6 binding sites overlapped with enhancers (i.e. H3K27Ac signal), while only a minor fraction overlapped with H3K27Me3 signal. To understand the functional significance of genome-wide PAX6 occupancy, we analyzed PAX6 binding sites using the gene ontology tool GREAT (McLean et al, 2010). GREAT analysis showed significant enrichments for functional categories such as nervous system development, neural tube patterning, and spinal cord, forebrain and eye development recapitulating previously known functions of PAX6 (Fig 2E). To identify direct PAX6 target genes, we mapped PAX6 sites proximal to protein-coding genes. A gene was deemed a PAX6 target if a PAX6 binding site was located in the gene body or within 50 kb upstream from the TSS. Strikingly, we observed that a significant fraction of PAX6 target genes encoded for TFs. Out of approximately 4,000 PAX6 targets identified by the above criteria, almost 700 genes were annotated as proteins that regulate transcription (P-value = 5.3e−5, hypergeometric test). Gene ontology analysis using GREAT on PAX6 peaks that mapped to TFs revealed enrichment of functional categories pertaining to neural development such as forebrain development, neural tube development, and spinal cord patterning (Fig 2F). Indeed, PAX6 binding sites were found upstream of several TFs implicated in neural development such as SOX2, LHX2, LMO3, MEIS2, DACH1, and LIX1 (Supplementary Fig S2). Additionally, all of these binding sites overlapped with H3K27Ac enhancer marks, indicating that PAX6 activates these neural TFs. Indeed, PAX6 has been shown to activate SOX2 in neural progenitors (Wen et al, 2008). SELEX experiments have identified two different DNA motifs that can bind PAX6 in vitro (Epstein et al, 1994; Czerny & Busslinger, 1995). However, de novo motif analysis of PAX6-bound sites in NEC only uncovered the motif identified by Epstein et al, although at a much lower significance (P-value = 1e−46; Supplementary Fig S3). Additionally, this motif was present in < 1% of the ChIP-Seq-identified binding sites, suggesting that the in vitro identified motif may not be the predominant binding motif in vivo. We also identified motifs for other neural factors, including SOX2, LHX2, TCF12, and RFX, in close proximity to PAX6 binding sites, suggesting that PAX6 cooperates with these neural TFs in regulating target genes (Supplementary Fig S3). Interestingly, the top motif that was identified as bound by LHX2/LHX3 TFs was very similar to another PAX6 motif recorded in the motif database JASPAR (Mathelier et al, 2013). It is possible that PAX6 binding profiles are influenced by its interacting partners, which is reflected in the absence of a clear canonical motif that can be identified for PAX6. Indeed, cooperation between PAX6 and SOX2 has been shown in lens development and eye morphogenesis (Smith et al, 2009).
PAX6 activates miR-135b
We hypothesized that microRNAs activated by PAX6 during neural differentiation would be involved in promoting hESC to differentiate into neural cells. We identified PAX6-activated miRs by: (i) identifying enhancers specifically activated in NECs; (ii) overlapping NEC-specific enhancers with PAX6 binding sites; and (iii) identifying miRs proximal to PAX6-bound NEC-specific enhancers (Fig 3A). We identified several miRs that displayed overlapping PAX6 and H3K27Ac occupancy within 20 kb of the mature miR sequence (Supplementary Table S2). One of these miRs, hsa-miR-135b, showed multiple PAX6 binding sites in its immediate proximity, and all binding sites overlapped with H3K27Ac signal (Fig 3B). MiR-135b has been found to be overexpressed in several cancers (Bandres et al, 2006; Matsuyama et al, 2011; Munding et al, 2012; Lin et al, 2013) and shows brain-specific expression in the developing mouse embryo (Sempere et al, 2004). However, the role of this microRNA in human neural development has not yet been documented.
Figure 3. PAX6 activates miR-135b during neural induction.
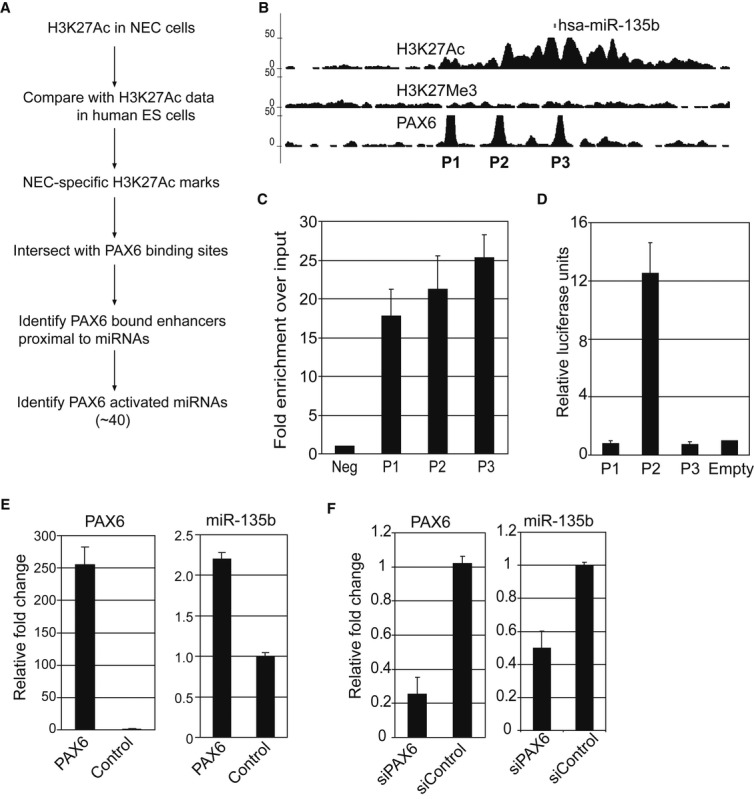
- Strategy to identify PAX6-activated microRNAs.
- Genome browser snapshot of PAX6 sites proximal to miR-135b mature sequence. All three PAX6 sites overlap with strong H3K27Ac signal.
- ChIP-PCR for confirmation of PAX6 binding to sites identified by ChIP-Seq in neuroectodermal cells (NEC) in the proximity of miR-135b. P1, P2, and P3 indicates loci displayed in (B).
- Enhancer luciferase assays conducted in NEC indicate that P2 is a bona fide enhancer. Relative luciferase values were normalized to data from empty vector transfections.
- Overexpression of PAX6 in differentiating H1 ESC activates miR-135b. Left panel shows qRT-PCR data for PAX6 expression. Right panel shows qRT-PCR for miR-135b expression. Values obtained were normalized to control GFP expression.
- Knockdown of PAX6 in differentiating H1 ESC by siRNAs inhibits miR-135b activation. Left panel shows qRT-PCR data for PAX6 expression. Right panel shows qRT-PCR for miR-135b expression. siPAX6: siRNA specific to PAX6, siControl: control siRNA. Values obtained were normalized to control expression.
Data information: All error bars shown indicate s.e.m. (n = 3 independent experiments).
We confirmed that PAX6 binds to the ChIP-Seq-identified sites by performing ChIP-PCR analysis in a set of independent biological replicates. The sites identified by ChIP-Seq showed significantly higher PAX6 occupancy, as compared to the negative control (Fig 3C). To confirm whether the PAX6-bound sites are functional enhancers, we performed luciferase enhancer assays by cloning the genomic sequence of these regulatory regions upstream of a Firefly luciferase gene and transfecting these constructs into NEC. One of the three regions we tested showed significantly higher activation as compared to the control, indicating that PAX6 regulates miR-135b expression by direct binding to a proximal enhancer (Fig 3D). To examine PAX6-mediated regulation of miR-135b, we overexpressed PAX6 in differentiating H1 hESC using lentiviruses and assayed expression of miR-135b by quantitative RT–PCR (qRT–PCR). Cells overexpressing PAX6 showed higher expression of miR-135b as compared to a control GFP expression (Fig 3E). In contrast, knockdown of PAX6 using siRNAs in differentiating H1 hESC suppressed miR-135b activation (Fig 3F). Similar results for PAX6 overexpression and knockdown were obtained using the H9 ESC line (Supplementary Fig S4). These data indicate that PAX6 activates miR-135b by direct binding to enhancers proximal to the miRNA.
miR-135b promotes neural conversion of human ES cells
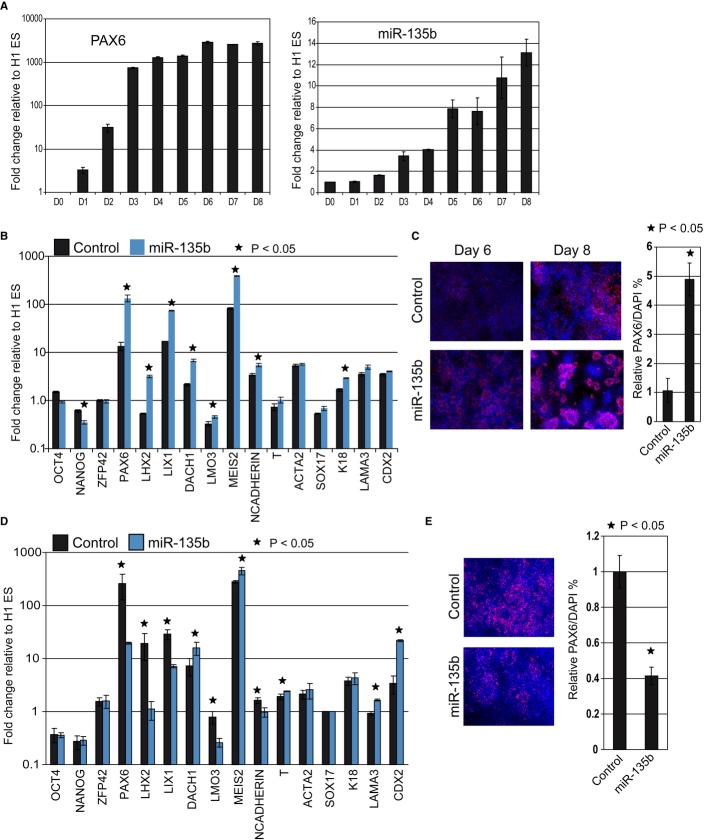
We first confirmed that miR-135b expression is activated during neural differentiation. Human ES cells were induced to differentiate into NEC as described above (2i method), and cells were harvested at various time points for assaying miR-135b expression. Mir-135b was activated as early as day 2, and its expression steadily increased during the time course of differentiation correlating with the activation profile of PAX6 (Fig 4A). We also differentiated the H9 hESC line using the 2i method and confirmed that miR-135b activation correlates well with PAX6 expression (Supplementary Fig S5A). To confirm that activation of miR-135b was not restricted to the method of neural induction, we differentiated hESC (both the H1 and H9 ES lines) using a recently developed protocol based on simultaneously inhibiting the TGF-β and notch pathways in parallel to activating the Wnt pathway using small-molecule inhibitors (3i method; Li et al, 2011). We observed activation of miR-135b using this alternate 3i method for both ESC lines (Supplementary Fig S5B). This showed that miR-135b was activated upon neural differentiation of hESC irrespective of the ESC line or neural induction method used.
Figure 4. MiR-135b promotes neuroectodermal fate in human pluripotent cells.
- Time course of miR-135b and PAX6 expression during neural differentiation of H1 ESC. MiR-135b levels were quantified by Taqman assays.
- Quantitative RT-PCR data to measure gene expression changes after miR-135b transfection. Fold changes were compared to untransfected H1 ESC.
- Immunostaining for PAX6 in miR-135b-transfected H1 ES cells at day 6 and day 8 after induction of differentiation.
- Quantitative RT-PCR data to assay gene expression after miR-135b knockdown in H1 ESC. Fold changes were compared to untransfected H1 ESC.
- Immunostaining for PAX6 after miR-135b knockdown in differentiated H1 ESC at day 8.
Data information: All error bars shown indicate s.e.m. (n = 3 independent experiments). Images were quantified as described in Materials and Methods. P-values indicated were calculated using the Student's one-tailed t-test.
We examined whether miR-135b can promote neural differentiation by transfecting hESC with miR-135b mimics and allowing cells to differentiate by withdrawal of growth factors that maintain pluripotency. To evaluate the effects of miR-135b expression in hESC, we assayed gene expression of the transfected cells by qRT–PCR. Transfection of hESC with miR-135b mimics significantly induced neural genes such as PAX6, LHX2, LIX1, DACH1, MEIS2, and N-CADHERIN as compared to the control transfection. Although OCT4 levels did not change significantly, miR-135b overexpression downregulated the pluripotency marker NANOG, indicating that miR-135b can inhibit pluripotency signals. OCT4 downregulation has been shown to precede PAX6 activation during neural differentiation of human ESC (Rosa & Brivanlou, 2011). It may be possible that, in the time frame of our experiment, PAX6 activation was not yet accompanied by a detectable downregulation of OCT4 transcripts. This raises the possibility that OCT4 may be regulated at a post-transcriptional level to de-repress the PAX6 promoter. In addition, markers for ectoderm (LAMA3), mesoderm (BRACHYURY and ACTA2), endoderm (SOX17), and trophectoderm (CDX2) were not induced relative to control transfections. Though the ectodermal marker K18 was activated significantly more than control, its activation was modest as compared to the neural markers (Fig 4B). This indicated that miR-135b did not induce a pan-lineage differentiation of human ESC but specifically promoted neural differentiation. Immunostaining of transfected cells for PAX6 revealed that miR-135b expression induced strong PAX6 expression as compared to control (Fig 4C left panel). Remarkably, when allowed to differentiate further, miR-135b-transfected cells organized into characteristic neural rosettes, while no such rosettes were seen in control transfected cells (Fig 4C right panel). These results demonstrate that miR-135b promotes neuro-ectodermal differentiation of hESC. Similar results were obtained for miR-135b overexpression in the H9 ESC line, where miR-135b overexpression specifically increased neural lineage markers but non-neural markers were either not changed or showed only modest changes (Supplementary Fig S6). Next, we wanted to ascertain whether miR-135b is required for NE development. We transfected H1 ESC with miR-135b antagomirs and allowed cells to differentiate as described above. After 8 days of differentiation, we assayed for markers of pluripotency, neural, and non-neural lineages as described for the overexpression experiment. MiR-135b knockdown significantly inhibited expression of PAX6 as well as other neural markers compared to control (Fig 4D and E). Additionally, LAMA3 (ectoderm marker) and CDX2 (trophectoderm marker) expression were significantly induced upon miR-135b knockdown compared to the control indicating differentiation of the transfected cells toward non-neural lineages (Fig 4D). Surprisingly, DACH1 as well as MEIS2, which are commonly used as neuroectodermal markers, were also induced upon miR-135b knockdown although their activation relative to control was modest as compared to other neural markers such as PAX6 and LHX2. In addition to the nervous system, DACH1 is also expressed in the ectoderm (Backman et al, 2003), which correlates with upregulation of the ectodermal marker LAMA3 upon miR-135b knockdown. MEIS2 expression has been identified in other non-neural tissues such as the somitic mesoderm, dorsal ectoderm, and otic epithelium (Cecconi et al, 1997; Sanchez-Guardado et al, 2011). Hence, the modest upregulation of DACH1 and MEIS2 could reflect differentiation of transfected cells toward non-neural phenotypes.
miR-135b targets the TGF-β and BMP signaling pathways
To uncover the mechanism by which miR-135b mediates neural differentiation, we used the web-based computational tool DIANA-mirPath (Papadopoulos et al, 2009) to predict miR-135b targets. DIANA-mirPath first predicts miRNA targets using established prediction softwares such as TargetScan, DIANA-microT, and PicTar. The predicted genes are then assigned to signaling pathways based on published data. Predictions from any single software tend to be noisy; however, candidate genes or pathways predicted by all three softwares would be expected to be high-confidence targets. When applied to predict miR-135b targets, one of the pathways identified from this analysis was the TGF-β/BMP signaling pathway. MiR-135b was predicted to target BMPR1, BMPR2, TGF-βR1, TGF-βR2, SMAD5, and ACVR1b. The TGF-β/BMP signaling pathway is known to be essential in maintaining pluripotency and self-renewal of human pluripotent stem cells (James et al, 2005). On the other hand, suppression of these pathways by small-molecule inhibitors such SB431542, noggin, dorsomorphin, and LDN193189 has been used to differentiate human ES and iPS cells into neural progenitors (Chambers et al, 2009; Zhou et al, 2010; Amoroso et al, 2013). We cloned the 3′UTRs of the predicted target genes downstream of the firefly luciferase enzyme and assayed the ability of miR-135b to inhibit luciferase translation by performing a dual luciferase assay in 293T cells. A preliminary screen using the 3′UTR luciferase reporter assay revealed that BMPR2, TGF-βR1, SMAD5, and ACVR1b were direct targets (Supplementary Fig S7). To confirm that translational suppression of these targets was dependent upon miR-135b binding to its cognate site in the 3′ UTR, we introduced mutations in the 3′UTR region overlapping with the miRNA seed sequence (nucleotides 2–8; Fig 5A). Introducing mutations in the seed region relieved miR-135b-mediated translational inhibition of the target 3′UTRs, thereby increasing relative luciferase enzyme activity. This confirmed that miR-135b targets BMPR2, TGF-βR1, SMAD5, and ACVR1b by directly binding to recognition sites in their 3′UTR (Fig 5B). Next, we assayed whether miR-135b is able to suppress endogenous transcript and protein levels of the identified candidates by overexpression of miR-135b in hESC. Though transcript levels of all four candidates remained unchanged, miR-135b was able to suppress BMPR2, SMAD5, and ACVR1b at the protein level (Fig 5C and D). Overall, our results indicated that miR-135b targets the TGF-β and BMP pathways by directly binding cognate sites in the 3′UTR of the signaling receptors and signaling intermediates.
Figure 5. MiR-135b inhibits TGF-β and BMP signaling.
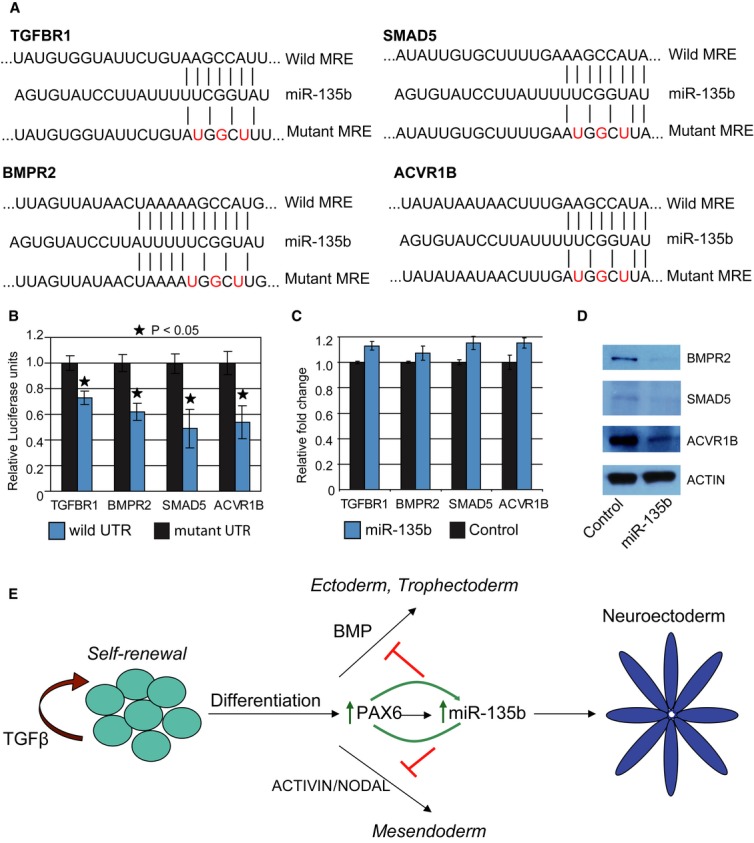
- Predicted miR-135b regulatory elements within TGF-βR1, BMPR2, SMAD5, and ACVR1b 3′UTRs. Nucleotides shown in red were mutated for the luciferase assays.
- Relative luciferase expression of wild-type and mutant UTR-bearing luciferase vectors co-transfected with miR-135b expression vectors in 293T cells. All error bars shown indicate s.e.m. (n = 3 independent experiments). P-values indicated in the graph were calculated using the Student's one-tailed t-test.
- Quantitative RT-PCR data for analyzing expression of miR-135b target transcript levels upon miR-35b transfection in H1 ESC. Cells were harvested 96 h post-transfection.
- Western blot showing proteins levels of miR-135b targets after transfection of H1 ESC with miR-135b mimics. Cells were harvested 96 h post-transfection.
- Model illustrating the role of miR-135b in neuroectoderm specification. Human ESC can self-renew indefinitely in the presence of cytokines such as TGF-β or differentiate into specific lineages in response to specific factors. Treatment with BMP promotes extraembryonic trophectoderm or ectoderm formation while activation of the ACTIVIN/NODAL axis drives differentiation toward mesoendoderm. Activation of PAX6 strongly promotes neural conversion of human embryonic stem cells (hESC), in part by activating miR-135b, which positively regulates PAX6 levels. MiR-135b inhibits BMP and activin signaling thereby inhibiting differentiation toward trophectoderm, ectoderm, and mesoendoderm, which allows irreversible neural fate specification of hESC.
Discussion
Extensive work has been carried out to understand the role of signaling pathways such as TGF-β and BMP in NE development. The ‘default model’ of neural induction proposes that ectodermal cells have an intrinsic tendency to form neural tissue that is inhibited by BMPs, especially BMP4 and inhibition of BMP signaling is sufficient to induce neural conversion (Streit & Stern, 1999). In line with this model, many neural differentiation protocols rely on small-molecule inhibition of the BMP pathway to generate neural cells in vitro from human ES cells. However, the default model has been deemed to be a simplistic explanation of neural induction in the embryo. The involvement of other signaling molecules, such as FGFs and WNTs in promoting neural conversion independently of BMP inhibition in parallel with studies that showed inhibition of BMP signaling alone is not sufficient to induce neural conversion, has highlighted that NE development might be a highly involved and coordinated process (Linker & Stern, 2004; Wawersik et al, 2005; Greber et al, 2011; Mulligan & Cheyette, 2012). However, the regulatory networks mediating human NE development at the transcriptional and post-transcriptional level are just beginning to be unraveled. The importance of post-transcriptional regulation mediated by microRNAs in development, including neural development, has been firmly established. For example, the microRNA miR-125 was shown to potentiate early neural specification of human ES cells (Boissart et al, 2012). Interestingly, microRNAs can target TFs while reciprocally being regulated by TFs, and a few examples of such TF-miRNA regulatory modules have been shown to regulate ES self-renewal and neural differentiation (Rosa & Brivanlou, 2011; Sun et al, 2013). For example, OCT4 in concert with miR-302 was shown to inhibit NR2F2 at the transcriptional as well as post-transcriptional level, respectively, to prevent precocious neural differentiation (Rosa & Brivanlou, 2011). In this study, we have demonstrated that the master transcriptional regulator PAX6 activates the microRNA miR-135b which in turn inhibits the TGF-β and BMP signaling pathways, thereby promoting NE development.
We implemented an in vitro model of human NE development by differentiating human ES cells into neuroectodermal progenitors in a cost-effective and efficient manner, thereby generating cells in large amounts for use in genomic assays such as ChIP-Seq. Genome-wide identification of PAX6 binding sites in these progenitors revealed that PAX6 regulates several genes implicated in mediating neuroectodermal differentiation by direct binding to enhancers proximal to the TSS or within the introns of these genes. Gene ontology analysis of PAX6-activated target genes implicated a functional role of PAX6 in central nervous system as well as spinal development. Strikingly, a significant proportion of these genes are TFs. Thus, PAX6 may represent a ‘hub’ node that connects to several other genes including TFs involved in neural development. Indeed, motif analysis of PAX6 binding sites identified motifs for other TFs implicated in neural development. Some of these TFs, such as SOX2 and LHX2, have been known to cooperate with PAX6 to regulate downstream genes in other developmental contexts (Smith et al, 2009; Tetreault et al, 2009). Our data suggest that such cooperation may also be functional in neural development. Importantly, we observed PAX6 binding sites overlapping with enhancers proximal to the TSS of all these TFs. This strongly indicates that PAX6 activates its own binding partners to facilitate target recognition.
We also identified enhancer-associated PAX6 binding sites proximal to several microRNAs, including hsa-miR-135b. We confirmed that PAX6 activated miR-135b during differentiation of human ES cells into NE, and miR-135b expression closely paralleled PAX6 expression. Since PAX6 can act as a repressor, it is possible that the activation of miR-135b may be via an indirect mechanism where PAX6 represses a repressor of miR-135b. Or, either mechanisms may play a role in miR-135b activation, that is, PAX6 directly activates miR-135b as well as inhibits a suppressor of miR-135b. However, our data show that activation of PAX6 upregulates miR-135b while inhibition of PAX6 downregulates miR-135b levels. Additionally, PAX6 binding sites were found to overlap a functional enhancer in the proximity of miR-135b. Taken together, our data support the case that PAX6 acts as an activator, at least in part, to upregulate miR-135b expression. Human ESC pluripotency has been postulated to be maintained by a fine balance of several signaling pathways including TGF-β, WNT, and FGF2 (Dalton, 2013). Activation of the TGF-β pathway via the Activin/Nodal branch maintains self-renewal and pluripotency of hESC, while inhibition of this pathway by the small molecule SB431542 leads to rapid differentiation of hESC toward the trophoblast lineage (James et al, 2005). On the other hand, treatment of hESC in low-serum conditions with activin A, a TGF-β protein agonist, promotes rapid differentiation into mesoderm or endoderm in a dose-dependent fashion, while treatment with BMP4 promotes generation of extraembryonic tissue (D'Amour et al, 2005; Amita et al, 2013). Suppression of these two pathways is the basis for the dual SMAD inhibition protocol (Chambers et al, 2009) that results in efficient neural conversion of human ESC by suppressing differentiation into trophectoderm, mesoderm, and endoderm. We found that miR-135b promotes neural conversion in a similar fashion by targeting the TGF-β and BMP pathways. Our results provide evidence for a model where PAX6-mediated neural conversion of human ES cells is brought about not only by activation of neural-promoting TFs, but also microRNAs that suppress differentiation into non-neural lineages (Fig 5E). It must be noted that we did not observe an explicit downregulation of non-neural lineage transcripts after miR-135b overexpression. It's possible that in the short time frame of the transfection experiments, the non-neural markers are not strongly induced in the control transfections. Secondly, transfection efficiencies of hESC are usually < 30%. Such low transfection efficiencies combined with minimal induction of the non-neural markers might be the reason why we could not detect a significant downregulation of the non-neural markers. This is consistent with findings from a previous study on the role of PAX6 in NE development, where overexpression of PAX6a-GFP in human ESCs strongly upregulated neural markers without significant changes in the non-neural markers (Zhang et al, 2010). Our data suggest that PAX6 and miR-135b may be acting in a feed-forward loop positively enforcing neural conversion of human ES cells and that inhibition of any one component of this loop severely impairs neural development. However, miR-135b failed to downregulate pluripotency markers or upregulate the neural program when overexpressed in hESC cultured under conditions optimized to maintain pluripotency, that is, cells that were maintained in mTeSR1 (Supplementary Fig S8), indicating that the microRNA alone is not sufficient to override pluripotency signals. Neural differentiation of hESC is specific to the PAX6a isoform while overexpression of the PAX6b isoform results in loss of pluripotency without a specific neural conversion (Zhang et al, 2010). Since the antibody for ChIP used in this study cannot distinguish between the two isoforms, we cannot ascertain which isoform binds proximal to miR-135b. However, overexpression of the PAX6a isoform upregulated miR-135b. Since miR-135b specifically promotes neural development, it is likely that the feed-forward loop involves PAX6a and not PAX6b. Pax6 levels have been shown to regulate the timing of NEC differentiation into neurons and astrocytes as well as specification of NEC into specific regional progenitors such as spinal motorneuron and interneuron progenitors (Hochstim et al, 2008; Sansom et al, 2009; Balaskas et al, 2012). Hence, it is possible that miR-135b-mediated regulation of Pax6 could also be involved in the differentiation of NEC into specific neuronal subtypes as well as glial cells.
These data provide a new angle to the PAX6-mediated regulatory network and open up the possibility that PAX6 regulates other non-coding RNAs as well, such as lincRNAs. Further, our study underscores the importance of non-coding RNAs, especially the interplay of TFs and microRNAs, in transducing extracellular signaling into distinct developmental cellular transitions.
Materials and Methods
Maintenance and differentiation of human embryonic stem cells
Human ES cell lines H1 and H9 were maintained in mTeSR1 media on matrigel-coated plates and routinely split in 1:6 ratios. For neural differentiation, ES colonies were split in a 1:8 ratio onto matrigel-coated plates and allowed to grow in mTeSR1 for 2 days. Neural differentiation was initiated by changing media to neural induction medium (NIM) comprising of DMEM/F12 and Neurobasal media in a 1:1 ratio (Invitrogen), N2 supplement 1% v/v (Invitrogen), B27 supplement without A 2% v/v (Invitrogen), 2 mM l-glutamine (Invitrogen), SB43542 10 μM (Tocris), and dorsomorphin 1 μM (Tocris). Colonies were allowed to differentiate for 8 days with media changes every other day. Neuroectodermal colonies were then dissociated using Accutase into single cells and plated onto Matrigel in neural precursor media comprising of DMEM/F12, 2 mM l-glutamine, N2 supplement 1%, B27 supplement without A 2%, insulin 20 μg/ml, bFGF 20 ng/ml, EGF 20 ng/ml. Rock inhibitor (10 μM) was used in the initial 2 passages to enhance cell survival.
Immunostaining and flow cytometry
For immunostaining, cells were fixed with 4% paraformaldehyde for 25 min at room temperature and permeabilized with PBS containing 0.1% Triton X-100 and 10% serum. Cells were incubated with primary antibodies against OCT4, SOX2 (SCBT), PAX6 (Covance), NESTIN (Millipore) diluted into blocking buffer containing 10% serum and incubated overnight at 4°C. Next day, after washing, cells were incubated with Alexa-fluor-conjugated secondary antibodies (Molecular probes) for 45 min at room temperature and nuclei were stained with DAPI. Imaging was performed on a Nikon fluorescent microscope. To quantify the immunostaining experiments, at least eight random fields (˜1,000 cells/field) were captured for each experiment. DAPI-stained nuclei and PAX6-positive cells were detected and quantified using Cellprofiler (Carpenter et al, 2006) using standard pipelines available with the software. For flow cytometry, cells were incubated with primary antibodies for PAX6 and NESTIN for 30 min at room temperature, washed and then incubated with Alexa-fluor-conjugated secondary antibodies for 30 min at room temperature. Cells incubated with secondary antibodies only were used as control. Flow cytometry analysis was done using a FACSCalibur flow cytometer, and at least 1,000 events above threshold levels were counted for each sample. Data analysis was done using CellQuest Pro.
Terminal differentiation of neuroectodermal cells
For undirected neuronal differentiation, NECs were plated onto laminin-coated culture wells and media were changed to neuronal differentiation medium (N2B27+ BDNF 10 ng/ml + GDNF 10 ng/ml). Cells were fixed 14 days post-induction. Motor neuron differentiation was carried out based on a protocol developed by Hu and Zhang (2009). NECs colonies were caudalized with retinoic acid (1 μM) and ventralized by purmorphamine (1 μM) for 2 weeks to generate OLIG2-positive motor neuron progenitors (MNPs). MNPs were then dissociated into single cells using Accutase and plated onto laminin for neuronal differentiation. For astrocyte differentiation, NEC colonies were grown in suspension to form spheres and treated with N2B27 media containing 10% FBS for 3 weeks after which spheres were dissociated into single cells and plated onto matrigel. Dissociated cells were allowed to grow for 3–4 days in the same media used for astrocyte induction and then fixed for staining.
PAX6 ChIP sequencing
Chromatin immunoprecipitation was performed as previously described in Johnson et al (2012). Briefly, 3 million cells were cross-linked using 1% formaldehyde, lysed, and chromatin was sonicated to < 500 bp using a bioruptor (Diagennode). Immunoprecipitation was carried out using antibodies specific for PAX6 (SCBT), H3K27Ac (Active Motif), and H3K27Me3 (Millipore). Libraries for Illumina sequencing were prepared according to manufacturer's protocol and sequenced on the GAII system. Raw reads were quality filtered and mapped to human genome hg19 using BWA (Li & Durbin, 2009). Bioinformatic analysis including peak finding, heatmaps, and density profiles was performed using HOMER (http://homer.salk.edu/homer/ngs/index.html) and custom PERL scripts.
PAX6 overexpression and knockdown
The PAX6a isoform cDNA was cloned into an EF1-alpha promoter-driven lentiviral expression plasmid and transfected into 293T cells along with the viral packaging plasmids (psPAX2 and VSVg, obtained from http://www.addgene.org). GFP-expressing lentiviral plasmids were used as control. Human ES cells were transduced with the respective viruses in the presence of 6 μg/ml polybrene and differentiated by culturing them in ES cell media without any growth factors, that is, DMEM:F12, 20% knock-out serum, 2 mM l-glutamine, β-mercaptoethanol, and 10 μM ROCK inhibitor. PAX6 knockdown was performed using 100 nM of the Dharmacon SmartPool PAX6 siRNAs. Cells were harvested 96 h after initiating differentiation.
MicroRNA overexpression and knockdown
ES cells were plated as single cells at a density of 50,000 cells/cm2, transfected with the microRNA mimics or antagomirs with the corresponding controls (Dharmacon) and differentiated by culturing them in ES cell media without any growth factors, that is, DMEM:F12, 20% knock-out serum, 2 mM l-glutamine, β-mercaptoethanol, and 10 μM ROCK inhibitor. For the miRNA overexpression experiments, cells were harvested at day 5 for gene expression analysis and day 6 or 8 for immunostaining. For the miRNA knockdown experiments, cells were harvested at day 7 or 8 for gene expression and immunostaining.
Luciferase assays
Enhancer luciferase vectors were constructed by cloning approximately 350 bp of genomic loci into the pGL4.23 vectors from Promega and co-transfected with pRL-CMV (Promega) plasmid into NEC. For 3′UTR luciferase assays, at least 500 bp of the 3′UTR of the target gene surrounding the miR target site was cloned downstream of the Firefly luciferase gene into the Psicheck2 vector from Ambion. MicroRNA seed site mutants were made by mutating three base pairs in the 6-mer seed sequence by amplifying the entire wild-type plasmid with primers harboring the necessary mutations. MiR-135b expression vectors were constructed by PCR amplifying 150 bp flanking the mature miR-135b sequence from human genomic DNA and cloning the PCR product downstream of an EF1-alpha promoter. Psicheck vectors were co-transfected with the miR-135b expression vector into 293T cells in a 96-well format. All luciferase assay readouts were performed in a dual injector system from Promega.
Quantitative RT-PCR
For miRNA assays, RNA was extracted using the miRNeasy kit from Qiagen. Quantitative real-time PCR for miR-135b was performed using Applied Biosystems TaqMan MicroRNA Assays according to the manufacturer's protocol. For assaying cDNA expression, RNA was extracted with the TRIzol reagent (Invitrogen) and reverse transcribed using random hexamers and the high-capacity reverse transcription system from Applied Biosystems. PCR was performed using the SYBR GREEN PCR Master Mix from Applied Biosystems. The target gene mRNA expression was normalized to the expression of Hprt1, and relative mRNA fold changes were calculated by the ΔΔCt method. The primer sequences are shown in Supplementary Table S1.
Western blots
Cell lysates were separated on 10% SDS–PAGE gels, and proteins were transferred onto PVDF membranes. Membranes were blocked with 5% milk in TBST (25 mM Tris pH 8.0, 150 mM NaCl, 0.05% Tween-20) and probed with corresponding primary antibodies against specific proteins (SMAD5, ACVR1b, BMPR2; Cell Signaling Technology). HRP-conjugated secondary antibodies (SCBT) were used to detect primary antibodies, and proteins were visualized by chemiluminescence.
Acknowledgments
The authors wish to thank Ang Sida Rene (GIS imaging facility) for help with imaging, Sathiyanathan Padmapriya (GIS) for help with the flow cytometry, and Lavanya Adusumilli (GIS) for help with luciferase assays. This study was supported by intramural funding from A*STAR (Agency for Science, Technology and Research, Singapore), Biomedical Research Council.
Author contributions
AB and LWS designed the research and wrote the paper. AB performed research and analyzed data. JP, SCN, XT, SJHL, and AT performed the research. SP contributed significant reagents, material, and analytical help.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Acampora D, Postiglione MP, Avantaggiato V, Di Bonito M, Simeone A. The role of Otx and Otp genes in brain development. Int J Dev Biol. 2000;44:669–677. [PubMed] [Google Scholar]
- Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110:E1212–E1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso MW, Croft GF, Williams DJ, O'Keeffe S, Carrasco MA, Davis AR, Roybon L, Oakley DH, Maniatis T, Henderson CE, Wichterle H. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman M, Machon O, Van Den Bout CJ, Krauss S. Targeted disruption of mouse Dach1 results in postnatal lethality. Dev Dyn. 2003;226:139–144. doi: 10.1002/dvdy.10210. [DOI] [PubMed] [Google Scholar]
- Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Proetzel G, Alvarez-Bolado G, Jay D, Gruss P. Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somitic mesoderm. Dev Dyn. 1997;210:184–190. doi: 10.1002/(SICI)1097-0177(199710)210:2<184::AID-AJA10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69:721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chevigny A, Core N, Follert P, Gaudin M, Barbry P, Beclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S. Signaling networks in human pluripotent stem cells. Curr Opin Cell Biol. 2013;25:241–246. doi: 10.1016/j.ceb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol. 2012;100:279–317. doi: 10.1016/B978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Coulon P, Zhang M, Moritz S, Frank S, Muller-Molina AJ, Arauzo-Bravo MJ, Han DW, Pape HC, Scholer HR. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30:4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton D. Vertebrate neural induction. Annu Rev Neurosci. 1997;20:43–60. doi: 10.1146/annurev.neuro.20.1.43. [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Johnson R, Richter N, Bogu GK, Bhinge A, Teng SW, Choo SH, Andrieux LO, de Benedictis C, Jauch R, Stanton LW. A genome-wide screen for genetic variants that modify the recruitment of REST to its target genes. PLoS Genet. 2012;8:e1002624. doi: 10.1371/journal.pgen.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Lopez-Bendito G, Gruss P, Stoykova A, Molnar Z. Pax6 is required for the normal development of the forebrain axonal connections. Development. 2002;129:5041–5052. doi: 10.1242/dev.129.21.5041. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, Kim WR, Lipton SA, Zhang K, Ding S. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci USA. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, Yang PC. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131:5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2013;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, Mano H, Sugimoto K, Miyazono K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood. 2011;118:6881–6892. doi: 10.1182/blood-2011-05-354654. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan KA, Cheyette BN. Wnt signaling in vertebrate neural development and function. J Neuroimmune Pharmacol. 2012;7:774–787. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding JB, Adai AT, Maghnouj A, Urbanik A, Zollner H, Liffers ST, Chromik AM, Uhl W, Szafranska-Schwarzbach AE, Tannapfel A, Hahn SA. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer. 2012;131:E86–E95. doi: 10.1002/ijc.26466. [DOI] [PubMed] [Google Scholar]
- Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- Pratt T, Quinn JC, Simpson TI, West JD, Mason JO, Price DJ. Disruption of early events in thalamocortical tract formation in mice lacking the transcription factors Pax6 or Foxg1. J Neurosci. 2002;22:8523–8531. doi: 10.1523/JNEUROSCI.22-19-08523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Guardado LO, Ferran JL, Rodriguez-Gallardo L, Puelles L, Hidalgo-Sanchez M. Meis gene expression patterns in the developing chicken inner ear. J Comp Neurol. 2011;519:125–147. doi: 10.1002/cne.22508. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Sittka A, Schmeck B. MicroRNAs in the lung. Adv Exp Med Biol. 2013;774:121–134. doi: 10.1007/978-94-007-5590-1_7. [DOI] [PubMed] [Google Scholar]
- Smith AN, Miller LA, Radice G, Ashery-Padan R, Lang RA. Stage-dependent modes of Pax6-Sox2 epistasis regulate lens development and eye morphogenesis. Development. 2009;136:2977–2985. doi: 10.1242/dev.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Stern CD. Neural induction. A bird's eye view. Trends Genet. 1999;15:20–24. doi: 10.1016/s0168-9525(98)01620-5. [DOI] [PubMed] [Google Scholar]
- Sun AX, Crabtree GR, Yoo AS. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 2013;25:215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault N, Champagne MP, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol. 2009;327:541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Evola C, Whitman M. Conditional BMP inhibition in Xenopus reveals stage-specific roles for BMPs in neural and neural crest induction. Dev Biol. 2005;277:425–442. doi: 10.1016/j.ydbio.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Wen J, Hu Q, Li M, Wang S, Zhang L, Chen Y, Li L. Pax6 directly modulate Sox2 expression in the neural progenitor cells. NeuroReport. 2008;19:413–417. doi: 10.1097/WNR.0b013e3282f64377. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M, Bondarenko GI, Du ZW, Jin Y, Golos TG, Zhang SC. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Su P, Li D, Tsang S, Duan E, Wang F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.