Abstract
A method for conjugation of ligands to the surface of exosomes was developed using click chemistry. Copper-catalyzed azide alkyne cycloaddition (click chemistry) is ideal for biocojugation of small molecules and macromolecules to the surface of exosomes, due to fast reaction times, high specificity, and compatibility in aqueous buffers. Exosomes cross-linked with alkyne groups using carbodiimide chemistry were conjugated to a model azide, azide-fluor 545. Conjugation had no effect on the size of exosomes, nor was there any change in the extent of exosome adherence/internalization with recipient cells, suggesting the reaction conditions were mild on exosome structure and function. We further investigated the extent of exosomal protein modification with alkyne groups. Using liposomes with surface alkyne groups of a similar size and concentration to exosomes, we estimated that approximately 1.5 alkyne groups were present for every 150 kDa of exosomal protein.
Introduction
Exosomes are membrane vesicles 30–100 nm in diameter that are released by cells and present in most, if not all, biological fluids including blood, urine, saliva, and so forth. Exosomes are believed to facilitate cell-to-cell communication, as there is increasing evidence that exosomes can deliver functional cargo in the form of proteins, mRNA, and miRNA, to recipient cells in vitro and in vivo.(1−7) The innate ability of exosomes to affect the phenotypes of recipient cells by delivery of naturally incorporated cargo has excited the drug delivery field. While there has been some success in delivering drugs using exosomes,8,9 there is growing evidence that a surface targeting moiety must be present for efficient delivery. A number of groups have recently reported that intravenously injected exosomes quickly localize in the liver and spleen.10−12 Conjugation of targeting ligands, such as antibodies and peptides, to the surface of exosomes may enable specific interactions of exosomes with target cells. To this end, we propose the use of click chemistry to functionalize the surface of exosomes. In addition to being used to affect exosome biodistribution, we foresee click chemistry as an efficient tool to label exosomes with fluorescent, radioactive, and MRI agents for precise in vivo tracking of injected exosomes.
Copper-catalyzed azide alkyne cycloaddition (click chemistry) is a highly efficient reaction between an alkyne and an azide that forms a triazole linkage. Click chemistry reactions are significantly accelerated in the presence of a Cu(I) catalyst, and proceed in a variety of solvents including water, alcohols, and DMSO.13 Use of click chemistry has grown exponentially since its introduction in 1999 due to mild reaction conditions, easily available reagents,14 and most importantly, high efficiency compared to traditional cross-linking chemistries (e.g., carbodiimide).15,16 Use of click chemistry with biomacromolecules is especially advantageous as there are rarely any nonaromatic double bonds for undesirable side reactions to take place.13,17
In this study we report the successful conjugation of azide-fluor 545 to exosomes chemically modified with alkyne groups. Conjugation chemistry had no impact on the size of exosomes, nor was there any change in the extent of exosome association with recipient cells. A goal of this study was to determine the number of alkyne groups cross-linked, with our methods, to exosomal proteins. Overmodification of antibodies with small molecules has been shown to cause a decrease in antibody binding affinities, while minimal modifications (1–5) have resulted in little to no loss in function.18−22 Similarly, we believe overmodification of exosomal proteins with alkyne groups, which are necessary for click chemistry reactions, might inhibit exosomal protein function. Because we could not directly measure alkyne modification on exosomes, liposomes of similar size and concentration to exosome samples were prepared with varying concentration of terminal alkyne groups extending out from the liposome surface. Using a standard curve of the extent of azide-fluor 545 conjugation to liposomes, we were able to estimate the extent of alkyne modification on exosomal proteins. Due to its specificity and efficiency, we believe that this robust technique is superior to other approaches to exosome labeling, and could benefit the burgeoning field of exosome research.
Results
Conjugation of Azide-Fluor 545 to Exosomes
Exosomes used for conjugation were prepared as previously described.23 To ensure we were working with isolated exosomes, 4T1 exosome samples were analyzed using mass spectrometry (Supporting Information Table 1). Examination of the results revealed the presence of 54 unique proteins in 4T1 exosome samples including many of the canonical markers of exosomes: Heat Shock Proteins 70 and 90; Ras-related protein 10 (RAB-10 and others); Annexins 1, 2, 5, and 6; Pyruvate kinase (PKM); Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (YWHAG); and actin.24 A Vesiclepedia (http://microvesicles.org/) search in June 2014 revealed that each protein found, using LC/MS, in 4T1 exosomes samples had been previously linked with exosomes. Additionally, nearly all the proteins identified also are classified in the UniProtKB Gene Ontology Cellular Component sections as members of “extracellular vesicles exosomes”, adding more support to the notion that the proteins identified were derived from exosomes (http://www.uniprot.org/).
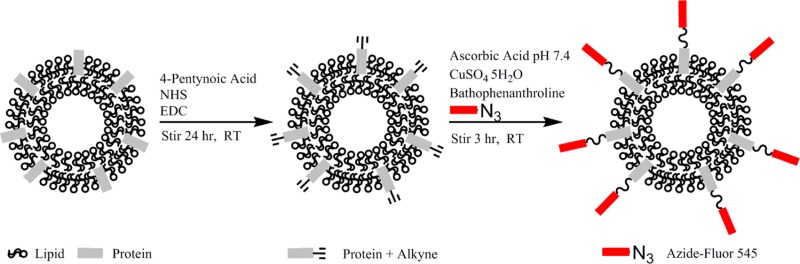
In order to conjugate exosomes with azide-fluor 545, exosomes were functionalized with a terminal alkyne group. Amine groups found on exosomal proteins or, likely to a lesser degree, amines on the head groups on the exosomal membrane lipid phosphatidylethanolamine, were cross-linked with the carboxyl group of 4-pentynoic acid using carbodiimide activation. Following the functionalization of the surface of exosomes with a terminal alkyne, exosomes were conjugated with azide-fluor 545 using click chemistry (Scheme 1).
Scheme 1. Modification of Exosomal Amine Groups with a Terminal Alkyne.
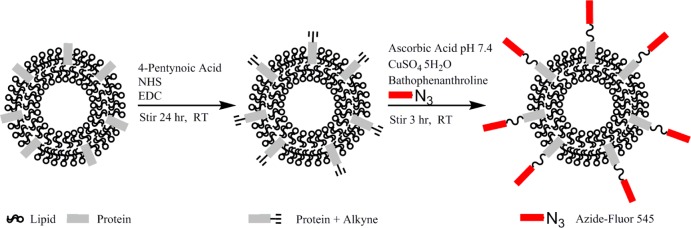
Followed by conjugation of azide-fluor 545 to exosomes using click chemistry.
Exosome Functionality Post Conjugation
To elucidate whether cross-linking of exosomal amines to 4-pentynoic acid and subsequent conjugation of azide-fluor 545 caused 4T1 exosomes to aggregate, the mean size and distribution of 4T1 exosomes was measured using a NanoSight NS300 prior to chemical modification, and after conjugation of azide-fluor 545 (Figure 1). No change in the mean diameter or size distribution was observed prior to modification (120 ± 54 nm) or after conjugation (128 ± 60 nm), suggesting the conjugation chemistry did not perturb exosome structure.
Figure 1.
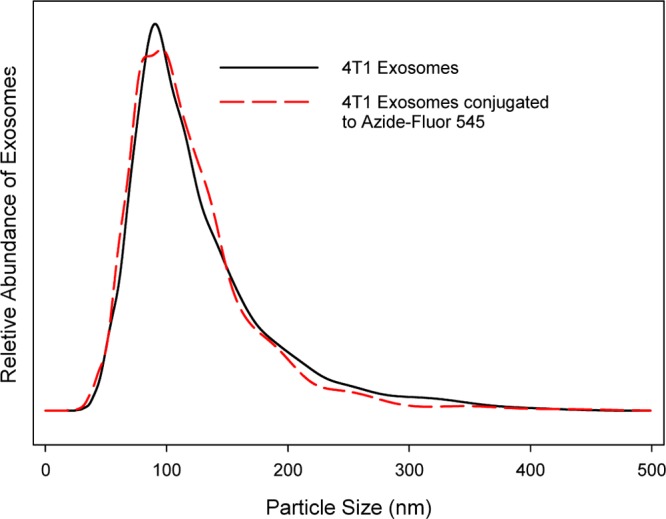
Mean size and distribution of 4T1 exosomes were analyzed using a Nanosight NS300. 4T1 exosomes had a mean size of 120 ± 54 nm, and exosomes after azide-fluor 545 conjugation had a mean size of 128 ± 60 nm.
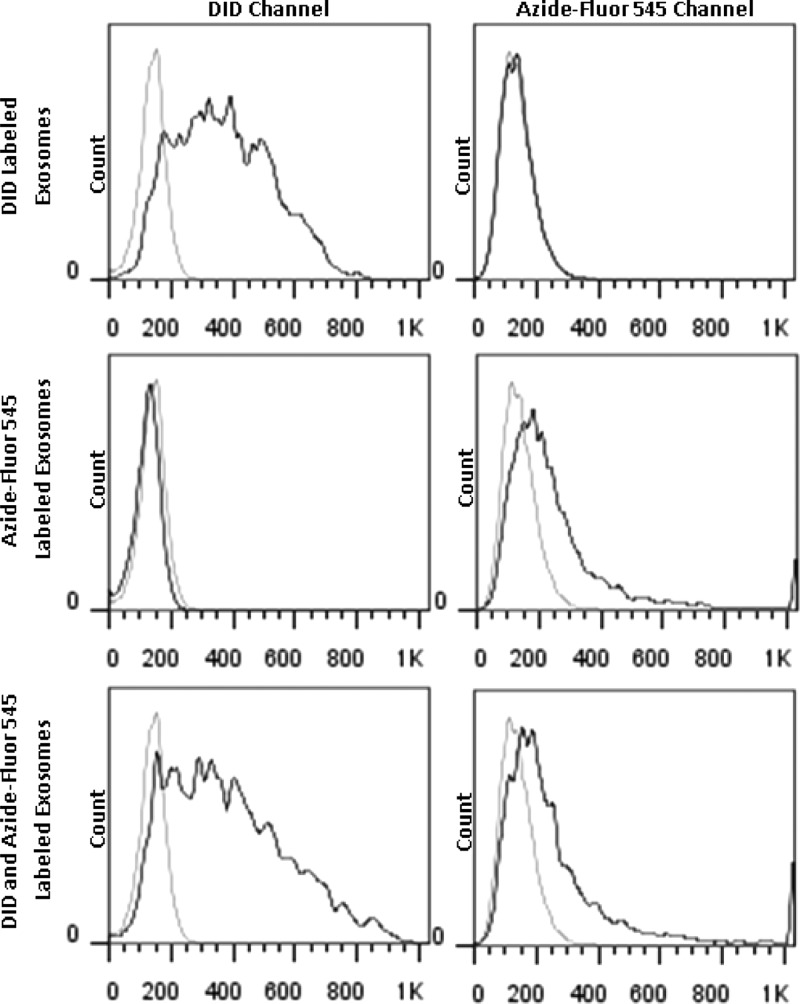
We next set out to investigate any changes in the ability of 4T1 exosomes to adhere/internalize with their parent cell line in vitro before and after conjugation. To compare association of unmodified exosomes and azide-fluor 545 conjugated exosomes with 4T1 cells, 4T1 exosomes from a single preparation were split into three sample fractions. Fraction one was labeled with the lipophilic fluorescent marker DID and served as the unmodified control. The second sample fraction was conjugated with azide-fluor 545, and the third fraction was labeled with DID and conjugated with azide-fluor 545. In order to ensure the concentration of exosomes remained equivalent in all three sample fractions, each fraction was diluted and processed identically, treating all three fractions as if they had been labeled with both fluorescent probes. Exosomes from each fraction were added to 4T1 cells cultured in a 96 well plate at a concentration of 50 μg/mL. After 4 h incubation, cells were trypsinized and placed on ice before being analyzed using flow cytometry. The extent of adherence/internalization for each exosome fraction with their parent cell line can be seen in Figure 2. Analysis of the data suggests that association of exosomes with their parent cell line is unaffected by azide-fluor 545 conjugation. The adherence/internalization profile of unmodified exosomes labeled with DID closely resembles that of exosomes labeled with DID and conjugated to azide-fluor 545. Similarly, incorporation of DID into exosomes does not alter the extent of azide-fluor 545 conjugation to exosomes. The extent of adherence/internalization of exosomes labeled with azide-fluor 545 is remarkably similar to exosomes labeled with DID and azide-fluor 545.
Figure 2.
Exosomes fluorescently labeled with DID, azide-fluor 545, or both were incubated with 4T1 cells for 4 h. Cells were analyzed using flow cytometry for increases in fluorescence caused by adherence/internalization of DID or azide-fluor 545 labeled exosomes. The light gray profile in each panel represents background autofluorecence of 4T1 cells.
To further visualize the conjugation of azide-fluor 545 to 4T1 exosomes, conjugated exosomes were incubated with 4T1 cells for 4 h. After incubation, cells were rinsed with PBS, the nucleus of cells were fluorescently stained, and the cells were fixed before being imaged using an OperettaTM High Content Imaging Systems instrument. Uptake of 4T1 exosomes can be seen in Figure 3.
Figure 3.
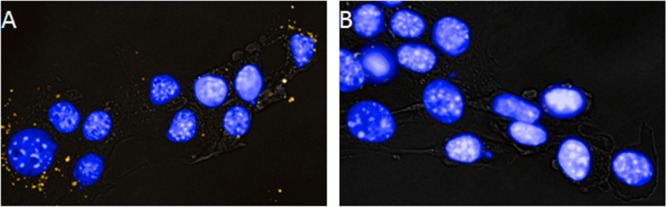
4T1 cells were incubated for 4 h with 50 μg/mL azide-fluor 545 conjugated exosomes (yellow) in panel A or with unmodified control exosomes seen in panel B. The nuclei of cells were stained with Hoeschst 33342 (Blue). Bright field images are overlaid to visualize cell borders.
Estimating the Extent of Exosomal Alkyne Modification
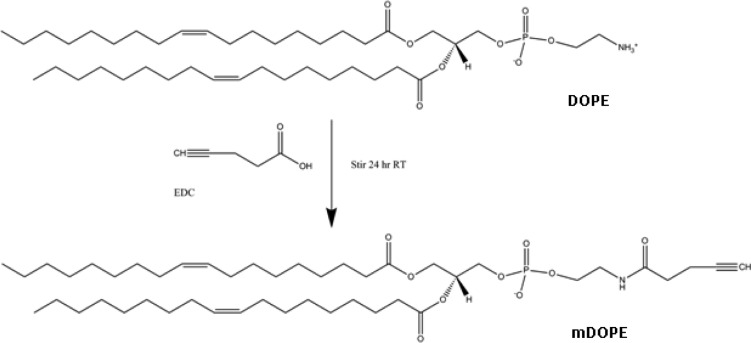
To estimate the extent of alkyne modification to exosomal proteins, liposomes with varying concentrations of terminal alkynes extending from the lipid bilayer were formulated and conjugated with azide-fluor 545. A standard curve of azide-fluor 545-conjugated liposomes was used to extrapolate the number of alkyne groups in exosome samples. In order to make liposomes with a terminal alkyne group, the terminal amine on the headgroup of DOPE was cross-linked with the carboxyl group of 4-pentynoic acid using carbodiimide coupling chemistry (Scheme 2). An HPLC chromatogram of modified DOPE (mDOPE) samples revealed the presence of a new peak relative to controls (unmodified DOPE), suggesting that DOPE modification with an alkyne group was successfully accomplished (Figure 4). Twenty-one percent of DOPE molecules were modified with an alkyne as measured with HPLC (See Materials and Methods section).
Scheme 2. Modification of DOPE.
Amine group of DOPE was reacted with 4-pentynoic acid using carbodiimide activation chemistry.
Figure 4.
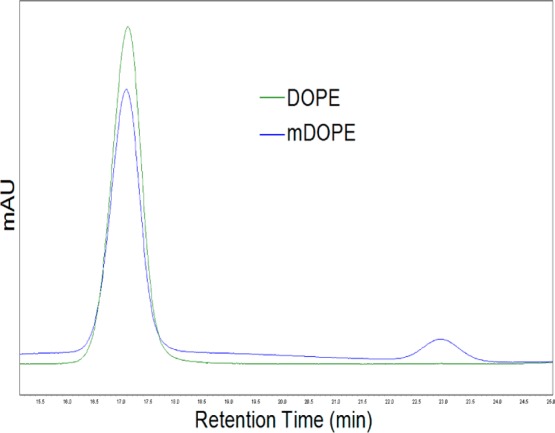
HPLC chromatogram of DOPE control and reacted sample containing mDOPE. Retention time of DOPE was 15.5 min. A new peak (retention time 22.5 min) appeared for samples cross-linked with 4-pentynoic acid.
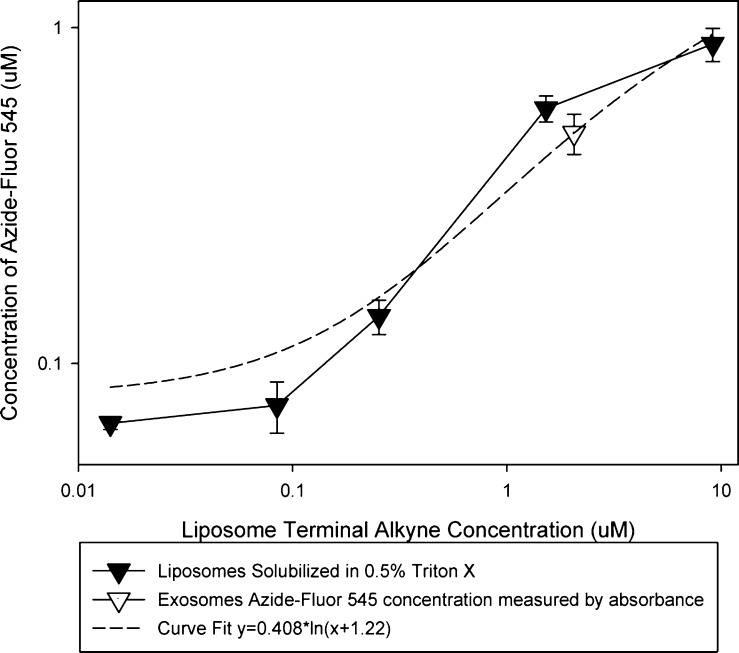
Liposomes were formulated with PC:Cholesterol:DOPE/mDOPE at a mole ratio of 1:1:1 with the DOPE/mDOPE fraction containing varying percentages of mDOPE. Liposomes were extruded to a similar size (125 ± 20 nm) to exosome samples (120 ± 54 nm) and have equivalent particle concentrations as measured by a NanoSight NS300 (data not shown). Liposomes and exosomes were conjugated to azide-fluor 545 using the same reaction conditions (Schemes 1 and 3). For liposome samples, only mDOPE molecules on the outer leaflet of the liposomal bilayer were available for conjugation to azide-fluor 545, cutting the effective concentration of mDOPE roughly in half. The extent of azide-fluor 545 conjugation to liposomes was quantified using fluorescence. Liposomes were solubilized using Triton X-100 to ensure azide-fluor 545 was not being quenched when conjugated to liposomes. From a fluorescent standard curve of free azide-fluor 545, the concentration of azide-fluor 545 conjugated to liposomes, containing varying concentration of mDOPE, could be calculated. Considering the potential for the fluorescence of azide-fluor 545 to be quenched, at least partially, after conjugation to exosomal proteins, absorbance was used to calculate the concentration of azide-fluor 545 in exosome samples. Using a logarithmic regression line fit to the data, we calculated that a 200 μL sample containing 40 μg exosomes conjugated with azide-fluor 545 had an alkyne group concentration of 2.06 μM (Figure 5). To estimate the number of alkyne modifications per exosomal protein we assumed an average protein molecular weight of 150 kDa. Our results imply that for every 150 kDa of protein, 1.5 alkyne groups modification were made.
Scheme 3. Conjugation of Azide-Fluor 545 to Liposomes (125 ± 20 nm) Using Click Chemistry.
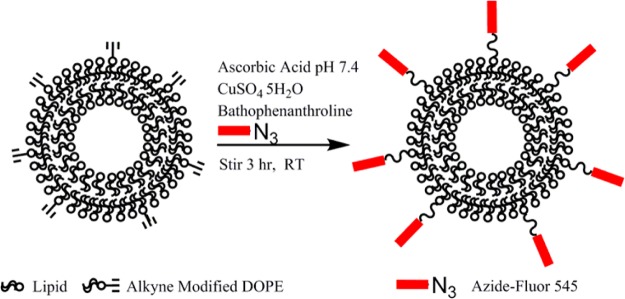
Figure 5.
Use of liposomes to estimate the extent of exosome alkyne modification. The extent of azide-fluor 545 conjugation to liposomes (0.2 mg/mL) with varying concentrations of terminal alkyne groups was measured using a fluorometer. Liposomes were solubilized with Triton X-100 to ensure fluorescence of azide-fluor 545 was not being quenched. The concentration of azide-fluor 545 conjugated to exosomes was measured using absorbance. A logarithmic regression line fitting the data was used to estimate the concentration of alkynes in an exosome sample.
Discussion
Click chemistry is rapidly becoming a popular tool to functionalize the surface of biomacromolecules, including viruses, DNA, peptides, antibodies, liposomes, micelles, and nanoparticles with a wide variety of conjugates.16,25−30 In this study, we have presented a novel technique that can be used to functionalize the surface of exosomes with small molecules, large biomacromolecules, and polymers. Functionalization of the surface of exosomes, using click chemistry, can be foreseen to affect exosome biodistribution through conjugation of targeting moieties, and potentially lead to improved methods for tracking exosomes in vivo through conjugation of fluorescent, radioactive, and MRI contrast agents. Reaction conditions, using our methods, did not alter exosome size and function. Furthermore, using liposomes with known alkyne concentrations, we estimated approximately 1.5 alkyne modifications were made for every 150 kDa of exosomal protein. In order to gauge the impact of alkyne modification on exosomal function, a comparison can be drawn to the chemical modification of antibodies. Small molecule modification of antibodies has been shown to have little to no effect on antibody binding affinities when approximately five or fewer modifications are made.18−22 This bolsters our conclusion that exosome modification using our methods has little to no impact on exosome functionality.
Materials and Methods
Materials
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE: 18:1 (Δ9-Cis) was purchased from Avanti Polar Lipids (Alabaster, AL). N-(3-(Dimethylamino)propyl)-N′-ethylcarbodiimide (EDC), N-hydroxysuccinimide (NHS), 4-pentynoic acid, copper(II) sulfate pentahydrate, l-ascorbic acid, bathophenanthroline, Triton X-100, and sodium bicarbonate were obtained from Sigma–Aldrich Chemical Co. (St. Louis, MO). BCA Protein Assay Reagent and HPLC grade methanol and water were purchased from Fisher Scientific (Pittsburgh, PA). Mouse mammary carcinoma (4T1) cells were acquired from ATCC (Manassas, VA). Roswell Park Memorial Institute (RPMI) medium 1640, 1× phosphate buffered saline (PBS), fetal bovine serum (FBS), trypsin, and penicillin–streptomycin were all purchased from Mediatech, Inc. (Manassas, VA). Plasmocin was obtained from InvivoGen (San Diego, CA). Hoechst 33342 nucleic acid stain, and DID (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) was obtained from Life Technologies (Carlsbad, CA). Azide-Fluor 545 was purchased from KeraFAST, Inc. (Boston, MA). Formalin was purchased from JT Baker (Center Valley, PA). Sepharose CL-4B was purchased from GE Healthcare (Uppsala, Sweeden). Sequencing grade trypsin was obtained from Promega Corporation (Madison, WI).
Exosome Isolation
Exosomes were isolated from the supernatant of 4T1 cells as previously described.23 Briefly, 4T1 cells lines were subcultured 3 days prior to collecting the cell culture supernatant, allowing the cells to become 75% confluent. 4T1 cells were cultured in 5-layer BD tissue culture-treated flasks (875 cm2). Exosomes were isolated from the supernatant by a series of centrifugation steps: 10 min at 300g, 20 min at 20 000g, and 2 h at 120 000g. Concentrated exosomes were then washed in PBS and centrifuged at 200 000g for 2 h on a sucrose density cushion. The sucrose cushion consisted of three distinct layers, 12% sucrose, 30% sucrose, and 50% sucrose. Exosomes have been previously reported to have a density ranging from 1.1 to 1.2 g/cm3.1,31,32 Subsequently, the 30% sucrose fraction and the top of the 50% sucrose fraction were collected, washed with PBS, and centrifuged at 120 000g for 2 h. The pelleted exosomes were resuspended in 200 μL PBS. Exosome protein content was quantified using the BCA protein assay. All exosomes used in this study were derived from 4T1 cells.
Mass Spectrometry Analysis
100 μg of isolated exosomes from 4T1 cells were digested using performic acid. A performic acid solution was prepared in a 1:19 ratio of 30% hydrogen peroxide:formic acid. Perfromic acid solution was warmed to 55 °C for 3 min immediately prior to use. Three volumes of performic acid were added to the exosome sample and incubated on ice for 3 h. The reaction was quenched using five volumes of ice-cold double-distilled water. Samples were then dried to completion using a speed vacuum and subsequently resuspended in 50 μL of 50 mM ammonium bicarbonate. To the sample, sequencing grade trypsin was added in a 1:50 trypsin:protein ratio and incubated overnight at 37 °C. The following morning the samples were thoroughly dried using a speed vacuum and the pellet resuspended in 30 μL of 0.1% formic acid in water. The tryptic digests was separated using a 5–50% ACN gradient over 120 min on a C18 column (Michrocom, Agilent). MS/MS spectra were collected using the Amazon Speed ion ETD trap equipped with CaptiveSpray nanoBooster ionization source (Bruker Daltonics) at the University of Colorado School of Pharmacy Mass Spectrometry Core. Acetonitrile enriched nitrogen gas was used as a sheath gas to increase the charge state of peptide ions and enhance identifications. Data was processed using ProteinScape 3.1. Database searches were performed against all mouse entries in the Swiss Prot database using the Mascot Server using 0.6 Da peptide mass tolerance and 0.5 Da MS/MS tolerance allowing for 1 missed cleavage and modifications for dioxidation of methionine and trioxidation of cysteine. Identification of proteins was considered significant if at least 2 unique peptides were used for identification.
Alkyne Modification of Exosomal Proteins
To 1 mL PBS (10 mM PO43–, 137 mM NaCl, and 2.7 mM KCl) were added n-hydroxysuccinimide (35 mg, 0.3 mmol, 1 equiv) and 4-pentynoic acid (29 mg, 0.3 mmol, 1 equiv), the pH buffered to 7.4 with sodium bicarbonate, and the resulting solution stirred on ice for 1 h. To the reaction mixture 1-ethyl-3-(3-carbodiimide dimethylaminopropyl) (46 mg, 0.3 mmol, 1 equiv) was added and the solution was stirred on ice for 1 h. Four microliters of the reaction mixture were added to 160 μg exosomes in 150 μL PBS and stirred for 24 h (Scheme 1). Exosomes were purified from excess reaction material using a Sepharose CL-4B column conditioned with PBS, pH 7.4.
Alkyne Modification of DOPE
To a 100 μL solution of chloroform containing DOPE (2.0 mg, 2.7 μmol, 1 equiv), 4-pentynoic acid (290 μg, 3.0 μmol, 1.1 equiv) was added, followed by 1-ethyl-3-(3-carbodiimide dimethylaminopropyl) (620 μg, 3.2 μmol, 1.2 equiv). To facilitate the coupling of 4-pentynoic acid to DOPE, 200 μL of methanol was added, followed by 80 μL of ddH2O. The reaction mixture (all one phase) was mixed using a magnetic stir bar at room temperature for 3 h. A set of control samples were prepared without 4-pentyonic acid in parallel to reactant samples. Reactant and control samples were subsequently dried using a nitrogen stream and stored at −20 °C.
HPLC-UV Analysis
To measure the extent of DOPE modification with an alkyne group, a Shimadzu analytical HPLC system (LC-20AB, DGU-20A, CTO-20A, Sil-20A HT) equipped with SPD-20A UV–vis detector was used (Shimadzu Scientific Instruments, Inc.; Columbia, MD). A guard column and an Aligent Zorbax extended-C18 50 × 4.6 mm (5 μm) column (Santa Clara, CA) were installed. The column temperature was set to 40 °C with a flow rate of 0.6 mL/min. An isocratic method (20 min run time; 19:1 methanol/water mobile phase) was used, and UV detection was monitored at 205 nm. A 10 μL volume was injected for reactant samples, control samples, and DOPE standards of known concentrations. A standard curve was used to quantify the concentration of DOPE in reactant and control samples. The extent of DOPE modification was quantified by subtracting the concentration of DOPE in reactant samples from control samples. We concluded that a decrease in DOPE in reactant samples was due to modification with an alkyne group. Consistent with this conclusion, we observed a new peak, not present in the DOPE control samples, in the reactant samples (Figure 4). Modified DOPE within this text will be referred to as mDOPE.
Liposome Preparation
Liposomes were formulated by mixing egg PC with cholesterol and DOPE/mDOPE at a 1:1:1 molar ratio in chloroform in glass vials. Liposomes were formulated with varying molar percentages of mDOPE (7.3%, 1.21%, 0.202%, 0.033%, 0.006%). Lipid mixtures in glass vials were dried under nitrogen gas and placed under vacuum to remove residual chloroform. To the glass vials containing the dried lipids, 550 μL PBS was added and subsequently sonicated to remove lipids from the glass vial walls. The lipid/PBS mixture was removed and extruded through 100 nm pore size polycarbonate membranes (Avestin, Ottawa, ON). Liposome formulations had a mean diameter of 125 ± 20 nm as measured using a Nanosight NS300 (Amesbury, Wiltshire, UK).
Click Chemistry
Click chemistry methods were adapted from the works of Hassane et al., Zhang et al., and Kumar et al.29,33,34 To 80 μg of liposome or 80 μg of exosomes (weight based on protein content) in 150 μL PBS, 7 μL of 0.32 M copper(II) sulfate pentahydrate, 35.5 μL of 1.44 M l-ascorbic acid (pH buffered to 7.4 with sodium bicarbonate), 16.4 μL of 0.27 M bathophenanthrolinedisulfonic acid disodium salt trihydrate, and 4 μL of 8.5 mM azide-fluor 545 were added stepwise at room temperature in a 1.5 mL glass vial. All reactants were dissolved in PBS immediately before use. Total sample volume was 212.9 μL. The head space of each sample vial was purged with nitrogen before mixing with a magnetic stir bar at room temperature for 3 h. To separate liposomes and exosomes from the reaction mixture, samples were run through a Sepharose CL-4B column conditioned with PBS (pH 7.4) or RMPI supplemented with 10% FBS, diluting the sample to 400 μL.
Sizing
Exosomes and liposomes were sized using Nanoparticle Tracking Analysis. Nanoparticle Tracking Analysis was performed on a NanoSight NS300. Nanoparticle Tracking Analysis visualizes particles by imaging the light scattered when exposed to the light of a laser, and relates the rate of particle movement in liquid, due to Brownian motion, to particle size. Exosomes were analyzed prior to and after addition of azide-fluor 545. Briefly, 1 μg of exosomes (based on protein weight) or liposomes was diluted in 1000 μL PBS. Samples were run and analyzed with Nanosight NTA 2.3 software. For analysis, samples were infused into the NanoSight NS300 at rate of 10 (arbitrary units) using a syringe pump. Data was collected for 60 s and analyzed using NanoSight NTA 2.3 software.
Fluorescent Labeling of Exosomes
DID was used to fluorescently label the bilayer of exosomes. Five microliters of DID at a concentration of 64 μg/mL in ethanol was mixed with 160 μg exosomes in 100 μL PBS for 1 h. Any unincorporated DID was removed using a Sepharose CL-4B column conditioned with PBS, pH 7.4.
Flow Cytometry
All flow cytometry experiments were performed on a Becton Dickinson FACScan. Briefly, 10 μg exosomes in 200 μL cell culture media (RPMI supplemented with 10% FBS) labeled with the membrane stain DID at 0.2% by exosome weight, azide-fluor 545, or both, were added to cells cultured in a Costar 96 well cell culture flat bottom tissue culture plate. After 4 h incubation, cells were rinsed with PBS, removed by trypsinization, suspended in PBS, and put on ice. Cells were immediately analyzed by flow cytometry. A minimum of ≥5000 events were acquired per sample. The y-axis of flow cytometry data is presented as a relative count of cells, where each data set is fit to have equivalent maximum peak heights in order to facilitate visual comparison between samples. Using this type of analysis, the area under the curve does not indicate the absolute number of cells analyzed. Manual compensation was performed to account for fluorescent spillover between detectors.
Fluorescent Imaging of Exosome Uptake
Fluorecent images were taken using an Operetta High Content Imaging System instrument (PerkinElmer; Waltham, MA). 4T1 cells were grown on a 96-well black walled plate made by Greiner Bio-One (Monroe, NC). Five micrograms of exosomes in 100 μL RMPI supplemented with 10% FBS, labeled with azide-fluor 545, were incubated with cells for 4 h. Cell media was subsequently removed and cells were rinsed with PBS twice. Cells were then stained with 20 μL Hoechst 33342 nucleic acid stain at a concentration of 10 μg/mL in PBS for 15 min. Hoeschst 33342 stain was then removed and cells washed with PBS. Cells were fixed for 10 min with 3.7% formalin followed by washing with PBS. Images were processed on Harmony 3.5.1 software manufactured by PerkinElmer.
Exosome Absorbance
To measure the extent of azide-fluor 545 conjugation to exosomes, a UV spectrophotometer (Agilent 8453 UV–visible Spectroscopy System) was used. Absorbance was used to measure azide-fluor 545 conjugation to exosomes, because we were concerned that the fluorescence of azide-fluor 545 might be quenched, at least partially, after conjugation to exosomal proteins. The molar extinction coefficient of azide-fluor 545 (92 000 cm–1 M–1 at 545 nm) was used to calculate the molar concentration of azide-fluor 545 in exosome samples.
Liposome Fluorescence
Fluorescence was used to measure the extent of azide-fluor 545 conjugation to liposomes containing varying levels of modified DOPE. Using a Fluoromax, (Photon Technology International, Birmingham, New Jersey) with 2 nm slit widths, samples were excited at 540 nm and emission analyzed at 560 nm. To ensure azide-fluor 545 was not being quenched, liposomes were mixed with Triton X at a final concentration of 0.5% and vortexed for 1 min prior to analysis. A standard curve of free azide-fluor 545 was used to calculate the molar concentration of azide-flour 545 in each liposome sample.
Acknowledgments
The authors would like to thank John Carpenter for use of the Nanosight NS300. We would also like to thank Mike Wempe for use of the Shimadzu analytical HPLC system. Additionally, we would like to thank Brian Reid and the High Throughput and High Content Screening Core at the University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences for use of the Operetta High Content Imaging System instrument. This work was supported by NIH/NIBIB grant #1RO1 EB016378 to T.J.A. and M.W.G.
Supporting Information Available
Proteomic analysis of 4T1 exosomes. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Valadi H.; Ekstrom K.; Bossios A.; Sjostrand M.; Lee J. J.; Lotvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–9. [DOI] [PubMed] [Google Scholar]
- Hergenreider E.; Heydt S.; Treguer K.; Boettger T.; Horrevoets A. J.; Zeiher A. M.; Scheffer M. P.; Frangakis A. S.; Yin X.; Mayr M.; Braun T.; Urbich C.; Boon R. A.; Dimmeler S. (2012) Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–56. [DOI] [PubMed] [Google Scholar]
- Umezu T.; Ohyashiki K.; Kuroda M.; Ohyashiki J. H. (2013) Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 32, 2747–55. [DOI] [PubMed] [Google Scholar]
- Aliotta J. M.; Pereira M.; Johnson K. W.; de Paz N.; Dooner M. S.; Puente N.; Ayala C.; Brilliant K.; Berz D.; Lee D.; Ramratnam B.; McMillan P. N.; Hixson D. C.; Josic D.; Quesenberry P. J. (2010) Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp. Hematol. 38, 233–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon H.; Heikamp E.; Turley H.; Dragovic R.; Thomas P.; Oon C. E.; Leek R.; Edelmann M.; Kessler B.; Sainson R. C.; Sargent I.; Li J. L.; Harris A. L. (2010) New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood 116, 2385–94. [DOI] [PubMed] [Google Scholar]
- Atay S.; Gercel-Taylor C.; Suttles J.; Mor G.; Taylor D. D. (2011) Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 65, 65–77. [DOI] [PubMed] [Google Scholar]
- Webber J.; Steadman R.; Mason M. D.; Tabi Z.; Clayton A. (2010) Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 70, 9621–30. [DOI] [PubMed] [Google Scholar]
- Sun D.; Zhuang X.; Xiang X.; Liu Y.; Zhang S.; Liu C.; Barnes S.; Grizzle W.; Miller D.; Zhang H. G. (2010) A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 18, 1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L.; Seow Y.; Yin H.; Betts C.; Lakhal S.; Wood M. J. (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–5. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Li S.; Song J.; Ji T.; Zhu M.; Anderson G. J.; Wei J.; Nie G. (2014) A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35, 2383–90. [DOI] [PubMed] [Google Scholar]
- Lai C. P.; Mardini O.; Ericsson M.; Prabhakar S.; Maguire C. A.; Chen J. W.; Tannous B. A.; Breakefield X. O. (2014) Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 8, 483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y.; Nishikawa M.; Shinotsuka H.; Matsui Y.; Ohara S.; Imai T.; Takakura Y. (2013) Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J. Biotechnol. 165, 77–84. [DOI] [PubMed] [Google Scholar]
- Algar W. R.; Prasuhn D. E.; Stewart M. H.; Jennings T. L.; Blanco-Canosa J. B.; Dawson P. E.; Medintz I. L. (2011) The controlled display of biomolecules on nanoparticles: a challenge suited to bioorthogonal chemistry. Bioconjugate Chem. 22, 825–58. [DOI] [PubMed] [Google Scholar]
- Hein C. D.; Liu X. M.; Wang D. (2008) Click chemistry, a powerful tool for pharmaceutical sciences. Pharm. Res. 25, 2216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolley J.; Guenin E.; Lievre N.; Lecouvey M.; Soussan M.; Lalatonne Y.; Motte L. (2013) Carbodiimide versus click chemistry for nanoparticle surface functionalization: a comparative study for the elaboration of multimodal superparamagnetic nanoparticles targeting alphavbeta3 integrins. Langmuir 29, 14639–47. [DOI] [PubMed] [Google Scholar]
- Thorek D. L.; Elias D. R.; Tsourkas A. (2009) Comparative analysis of nanoparticle-antibody conjugations: carbodiimide versus click chemistry. Mol. Imaging 8, 221–9. [PubMed] [Google Scholar]
- Best M. D. (2009) Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 48, 6571–84. [DOI] [PubMed] [Google Scholar]
- Kulkarni P. N.; Blair A. H.; Ghose T. I. (1981) Covalent binding of methotrexate to immunoglobulins and the effect of antibody-linked drug on tumor growth in vivo. Cancer Res. 41, 2700–6. [PubMed] [Google Scholar]
- Guillemard V.; Saragovi H. U. (2001) Taxane-antibody conjugates afford potent cytotoxicity, enhanced solubility, and tumor target selectivity. Cancer Res. 61, 694–9. [PubMed] [Google Scholar]
- Boghaert E. R.; Sridharan L.; Armellino D. C.; Khandke K. M.; DiJoseph J. F.; Kunz A.; Dougher M. M.; Jiang F.; Kalyandrug L. B.; Hamann P. R.; Frost P.; Damle N. K. (2004) Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-acetyl gamma calicheamicin dimethyl hydrazide targets Lewisy and eliminates Lewisy-positive human carcinoma cells and xenografts. Clin. Cancer Res. 10, 4538–49. [DOI] [PubMed] [Google Scholar]
- McDonagh C. F.; Turcott E.; Westendorf L.; Webster J. B.; Alley S. C.; Kim K.; Andreyka J.; Stone I.; Hamblett K. J.; Francisco J. A.; Carter P. (2006) Engineered antibody-drug conjugates with defined sites and stoichiometries of drug attachment. Protein Eng., Des. Sel. 19, 299–307. [DOI] [PubMed] [Google Scholar]
- Junttila T. T.; Li G.; Parsons K.; Phillips G. L.; Sliwkowski M. X. (2011) Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib insensitive breast cancer. Breast Cancer Res. Treat. 128, 347–56. [DOI] [PubMed] [Google Scholar]
- Smyth T. J.; Redzic J. S.; Graner M. W.; Anchordoquy T. J. (2014) Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim. Biophys. Acta 1838, 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S.; Simpson R. J. (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000. [DOI] [PubMed] [Google Scholar]
- Prasuhn D. E. Jr.; Yeh R. M.; Obenaus A.; Manchester M.; Finn M. G. (2007) Viral MRI contrast agents: coordination of Gd by native virions and attachment of Gd complexes by azide-alkyne cycloaddition. Chem. Commun. (Cambridge, U.K.) 1269–71. [DOI] [PubMed] [Google Scholar]
- Gierlich J.; Burley G. A.; Gramlich P. M.; Hammond D. M.; Carell T. (2006) Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 8, 3639–42. [DOI] [PubMed] [Google Scholar]
- Li Z. B.; Wu Z.; Chen K.; Chin F. T.; Chen X. (2007) Click chemistry for (18)F-labeling of RGD peptides and microPET imaging of tumor integrin alphavbeta3 expression. Bioconjugate Chem. 18, 1987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman M. A.; Kaur G.; Lee L. A.; Xie F.; Sepulveda J.; Breitenkamp R.; Zhang X.; Joralemon M.; Russell T. P.; Emrick T.; Wang Q. (2008) Surface modification of tobacco mosaic virus with ″click″ chemistry. ChemBioChem 9, 519–23. [DOI] [PubMed] [Google Scholar]
- Said Hassane F.; Frisch B.; Schuber F. (2006) Targeted liposomes: convenient coupling of ligands to preformed vesicles using ″click chemistry″. Bioconjugate Chem. 17, 849–54. [DOI] [PubMed] [Google Scholar]
- O’Reilly R.; Joralemon M.; Wooley K. L.; Hawker C. J. (2005) Functionalization of micelles and shell cross-linked nanoparticles using click chemistry. Chem. Mater. 17, 5976–5988. [Google Scholar]
- Fevrier B.; Vilette D.; Archer F.; Loew D.; Faigle W.; Vidal M.; Laude H.; Raposo G. (2004) Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U.S.A. 101, 9683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C.; Regnault A.; Garin J.; Wolfers J.; Zitvogel L.; Ricciardi-Castagnoli P.; Raposo G.; Amigorena S. (1999) Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Chan K. H.; Prud’homme R. K.; Link A. J. (2012) Synthesis and evaluation of clickable block copolymers for targeted nanoparticle drug delivery. Mol. Pharmaceutics 9, 2228–36. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Erasquin U. J.; Qin G.; Li K.; Cai C. (2010) ″Clickable″, polymerized liposomes as a versatile and stable platform for rapid optimization of their peripheral compositions. Chem. Commun. 46, 5746–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.